Abstract
The antibody-based immune system (AIS) is one of many means by which organisms protect themselves against pathogens and parasites. The AIS is present in jawed vertebrates (gnathostomes) but absent in all other taxa, including jawless vertebrates (agnathans). We argue that the AIS has been assembled from elements that have primarily evolved to serve other functions and incorporated existing molecular cascades, resulting in the appearance of new organs and new types of cells. Some molecules serving other functions have been appropriated by the AIS, whereas others have been modified to serve new functions, either after the duplication of their encoding genes or through the acquisition of an additional function without gene duplication. A few molecules may have been created de novo. The deployment and integration of the ready-made elements gives the impression of a sudden origin of the AIS. In reality, however, the AIS is an example of an organ system that has evolved gradually through a series of small steps over an extended period.
Keywords: gnathostomes, agnathans, immunoglobulin, lymphocyte, thymus
Immunologists classify immune responses into two categories: adaptive (acquired, anticipatory) immune system and innate immune system (1). Of the two, however, only the former is a single system in the sense that it encompasses specialized organs such as the thymus, cells such as the T lymphocytes, B lymphocytes, and antigen-presenting cells, and molecules such as the MHC glycoproteins, T cell receptors (TCRs), and B cell receptors (BCRs), together with their soluble forms, the Igs or antibodies, and the recombination-activating gene (RAG) proteins. The innate immune system, on the other hand, is a collective term lumping together numerous independent systems, which often have only one thing in common, their distinctiveness from the adaptive immune system. Placing the adaptive immune system and innate immune system on equal footing is, therefore, misleading: the adaptive immune system is not one of two systems but one of many immune systems. Moreover, recent studies suggest that there might be more than one “adaptive” or “acquired” immune system, operating on the same principle (somatic diversification of genes) but based on different molecules (2). Therefore, we use the acronym AIS to mean “antibody-based immune system.”
Originally, immunologists believed that the AIS was widely distributed among animal phyla (3), but now the view that the system is restricted to vertebrates (4) is generally accepted. Recent evidence indicates that the system is present in all gnathostomes (jawed vertebrates) but absent in agnathans (jawless vertebrates represented by lampreys and hagfishes). Some 25,000 expressed sequence tags obtained from lamprey and hagfish lymphocyte-like cells (LLCs) have failed to provide any evidence for the presence of MHC, TCR, BCR, or RAG genes, which define the AIS (5–8). It appears, therefore, that the AIS arose in the gnathostome lineage after its divergence from the agnathan lineage (9). This seemingly sudden appearance of a complex body system (10, 11) presents a challenge to evolutionary biology. Here, we sketch out some of the changes that the emergence of the AIS entailed and speculate how they may have come about. We argue that the origin of the AIS only appears to be sudden, whereas in reality it represents a culmination of a long preparatory phase characterized by gradual accumulation of small changes over an extended period. In what follows, we provide examples of such changes for each of the three levels constituting the system: organs, cells, and molecules.
The Thymus
An essential feature of an immune system is its potential to deploy its effector functions at any site in the body (12). The consequences of this feature are the system's disseminate nature, reliance on mobile cells and soluble molecules, absence of genuine effector organs, and close ties to the circulatory system. One of the essential features of the AIS is the ability to distinguish nonself from self. This feature calls for a sequestered environment in a specialized organ, the thymus, in which the discrimination can take place (13). The thymus is the only AIS organ that all of the gnathostomes possess and that the agnathans lack. The gnathostome thymus has three essential components (epithelial tissue, capsular elements, and lymphocytes), each of which is of a different ontogenetic origin (14, 15). The epithelial component is derived from the endoderm of the pharyngeal region, the capsules from the neural crest cells in the dorsal part of the hindbrain, and the lymphocytic tissue from the hemopoietic mesoderm. These three sources are present in all vertebrates, jawed and jawless. The first steps toward the specialization of the pharyngeal region were undertaken by ancestral protochordates. In modern protochordates, lateral paired outpocketings of the endoderm produce a series of pharyngeal pouches, ultimately opening to the surface as slits, separated by pharyngeal arches (16). Neural crest, a group of migratory cells arising from the ectoderm of the neural tube and possessing high regulatory potential, is a landmark innovation of the vertebrates (17). The origin of the hemopoietic tissue can be traced back to protochordates (18). The forces driving the evolution of the pharyngeal pouches were at first the shift from arm to filter feeding, then a switch to oral feeding, and finally, the assumption of a respiratory function by the pharyngeal region (16). These changes were accompanied by modifications of the different pharyngeal arches, including the development of jaws from the first arch and of the thymus from some of the more posterior arches in the gnathostomes. Most of these modifications resulted from the interaction between the incoming neural crest cells and the pharyngeal endoderm.
In the case of the mammalian thymus, the neural crest cells are derived from rhombomere 6 of the hindbrain and the region of the neural tube posterior to it (14, 15). The outpocketing of the pharyngeal pouches is regulated by the paired box genes 1 and 9 (PAX1 and PAX9) and the fibroblast growth factor gene 8 (FGF8). The specification of the pouches involved in the development of the thymus requires also the expression of the homeobox A3 (HOXA3) gene, which initiates a gene interaction cascade HOX–PAX–EYA–SIX in the endodermal cells, and the expression of the HOXA3, EYA1, and SIX1 genes in the neural crest cells (EYA1 = eyes absent 1 homolog; SIX1 = sine oculis-related homeobox 1 homolog). The thymic rudiment formed under the influence of these genes then differentiates into two domains distinguished by the expression of glial cells missing 2 (GCM2) homolog in one and of forkhead box N1 (FOXN1) gene in the other. The former domain develops into the parathyroid and the latter into the thymus. Further development of the thymic rudiment depends on the arrival of lymphocyte progenitor cells. Ultimately, the endodermal cells develop into the thymic epithelium and the neural crest cells condense into the capsular elements.
The HOX, PAX, EYA, SIX, GCM2, and FOXN1 are all old genes, present in both vertebrates and invertebrates. Similarly, the PAX–EYA–SIX is an ancient cascade deployed by both vertebrates and invertebrates in the formation of different organs, for example in the development of the fruit-fly eye (19). It seems likely, therefore, that in agnathans, both the histological and molecular requirements for the development of the thymus are fulfilled. All that might be missing is an adjustment in the regulatory network. The situation might be similar to that described recently for the molecular mechanism underlying the development of the jaws in gnathostome embryos (20). The essential step in jaw development during the agnathan–gnathostome transition might have been the topographical restriction of existing molecular cascades for oral patterning. Some such steps might also be necessary for the development of the thymus. Furthermore, agnathans might also lack a chemoattractant capable of guiding lymphocyte progenitors into the rudiment, a factor responsible for the progression of the lymphocyte progenitors to cells committed to become thymocytes, and MHC molecules, whose presence on the surface of the thymic epithelial cells is required for self/nonself discrimination (13).
The Lymphocyte
Self/nonself discrimination is one characteristic of the AIS; another is the possession of cells capable of clonal expansion (12). In the gnathostomes, this characteristic is the hallmark of the lymphocytes, which are generated together with other blood cells during the process of hemopoiesis. In both the phylogeny and the ontogeny of the gnathostomes, the site of hemopoiesis has moved from tissue to tissue and from organ to organ (21). Apparently, an environment supporting hemopoiesis can arise at different locations in the body in association with the circulatory system. The presence of blood cells in agnathans (22) indicates that the environment and hemopoiesis have evolved before the emergence of jawed vertebrates. But did this evolution include also the appearance of lymphocytes? Comparative morphologists have known for some time that, among the blood cells of lamprey and hagfish, there also are cells that morphologically resemble mammalian lymphocytes (23). This resemblance has now been demonstrated to extend to the physicochemical properties (5–8) and gene expression profiles of the cells (6, 8). Moreover, among the genes expressed in agnathan LLCs are also homologs of those that, in gnathostomes, guide the differentiation of lymphocytes from their progenitors. In gnathostome hemopoiesis, all blood cells derive from a common stem cell by progressive differentiation, each step in the progression being controlled by a set of regulatory molecules (24). The differentiation into lymphocytes is regulated by a set that includes genes such as the spleen focus-forming virus integration B (SPI-B), globin transcription factor homolog 3 (GATA3), and genes of the IKAROS family (8, 25–27). Expression of the homologs of these genes in agnathan LLCs suggests that, in jawless vertebrates, the hemopoietic differentiation pathway progresses at least to the stage of lymphocyte progenitors. Finally, studies on purified lamprey LLCs grown in culture have demonstrated the ability of these cells to respond to mitogenic stimuli by proliferation (2). Nevertheless, the agnathan cells are not fully equivalent to gnathostome lymphocytes, for they do not express the receptor molecules idiosyncratic of the latter, the TCR and BCR. Here, as in the case of the thymus, the evolution of an essential component of the AIS, the lymphocyte, has come close to the gnathostome stage but has not quite reached it. And here, too, one can imagine that a limited number of steps could convert the agnathan cell into a primitive lymphocyte capable for integration into the AIS.
One point cannot be overemphasized, however. Notwithstanding their ontogenic, morphological, gene-profile, and functional similarities with gnathostome lymphocytes, the agnathan cells are not lymphocytes. The agnathan immune system is not a primitive form of the AIS. The agnathan hemopoietic progression does not yield T and B cells. And the agnathan SPI, IKAROS, and GATA genes are homologous but not orthologous to their gnathostome counterparts and very likely do not have the same function (8, 25–27). Both the cells and the molecules have their own functions, which are not components of AIS. Furthermore, we wish to distinguish two phases in the evolution of lymphocytes: one from no-lymphocyte to LLC and the other from LLC to the lymphocyte. The evolution of the complex internal structure, intricate network of molecular interactions, and the wide range of functional expressions was a long process, unachievable in the time interval from the separation of the agnathan and gnathostome lineages to the divergence of the various gnathostome lineages. The evolution must have started long before this interval, and the majority of innovations, which would ultimately make the cell uniquely suited for becoming a key player in the AIS, must have been introduced before this period. Compared with these innovations, the changes that occurred during the much shorter second phase and that integrated the cell into the emerging AIS might have been relatively minor.
Three Categories of Molecules
In humans, the genes encoding molecules deployed by the AIS constitute ≈5% of the transcribed and translated genome. In terms of their origin, the molecules fall into three categories. The first category consists of molecules that evolved long before the emergence of the AIS for functions in other systems and that were then recruited to work also for the immune system. An example is provided by molecules assisting in the synthesis of MHC class I proteins: heat-shock protein A5, calnexin, calreticulin, glucose-regulated protein of 78 kDa, and others (28). All these molecules are widely distributed among eukaryotes and function as molecular chaperons for a variety of different proteins.
The second category contains already established molecules that were modified after the duplication of their encoding genes. The modification adapted the molecules to the specific needs of the AIS. The category is exemplified by the three pairs of proteasome subunits of β type (29): PSMB5–PSMB8, PSMB6–PSMB9, and PSMB7–PSMB10, and the activation-induced deaminase (AID) (30). The members of each pair originated from a common ancestor through duplications of their encoding genes in the gnathostome lineage (31, 32). In gnathostomes, in the absence of immune stimulation, proteasomes containing subunits PSMB5, PSMB6, and PSMB7 degrade unused or defective proteins into peptides, which are reduced further by cytoplasmic enzymes (29). During an immune response, a new population of proteasomes appears in which these three subunits have been replaced by their corresponding partners (31). These “immunoproteasomes” then produce peptides that, when delivered into the endoplasmic reticulum, fit into the groove of the MHC class I molecules. Agnathans lack the ability to produce immunoproteasomes; their proteasomes contain subunits encoded in the preduplication genes of each of the three pairs. The evolution from the housekeeping to the immunoproteasomal subunits embodied three relatively inextensive changes: (i) gene duplication, which is anything but rare in a gene-family cluster; (ii) a modification of the enzymatically active site to produce a new type of peptides; and (iii) the subjugation of the encoding genes to the control of a signaling pathway that involves IFN-γ.
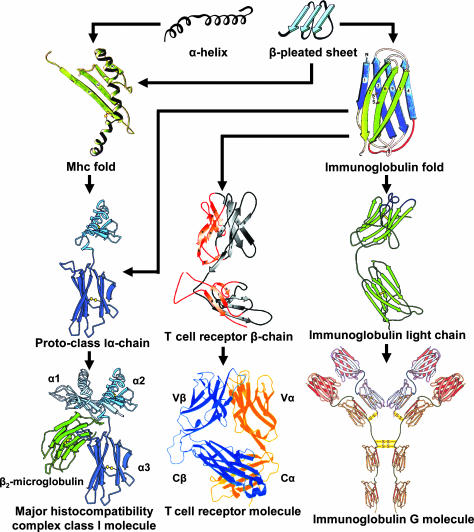
The third category contains molecules that did not exist before the emergence of the AIS. The novelties consist of new combinations of protein domains, of new domain designs, or of both. The category includes the key molecular players of the AIS (Fig. 1): the MHC molecules, TCRs, BCRs, and RAGs. An MHC molecule is an example of a novelty generated by bringing together different domains, presumably by shuffling gene segments at the genomic level, modifying preexisting domains, and possibly generating a new design of a domain, the peptide-binding domain (PBD).
Fig. 1.
Hypothetical scenario for the emergence of the MHC, TCR, and BCR molecules by gradual evolution, which encompassed modification of preexisting domains, joining together of different domains, and possibly generation of new domain designs.
The PBD of MHC Molecules
The 90 or so N-terminal amino acid residues of a class II monomer are arranged into a sheet of four antiparallel β-strands, rimmed on one side by an α-helix, the PBD (12). In a class II dimer, the two closely apposed PBDs form a peptide-binding module (PBM) in which the two antiparallel α-helices are separated by the peptide-binding groove. In a class I molecule, a similar module is formed by a single polypeptide chain. The origin of the PBD is not known, and it remains unresolved whether in evolution the class II arrangement preceded that of the class I molecule or the other way around (33). Folds of the polypeptide chain resembling the PBM have been found in three other molecules: the coat protein of the RNA bacteriophage MS2 (34), the mammalian interleukin-8 (IL-8) molecule (35), and the endothelial cell protein receptor of the mammalian blood clotting pathway (36). Because in each case conversion of these structures into a PBM would require major rearrangement of the polypeptide chain, the three molecules are not considered related to MHC molecules. Several other mammalian molecules possess somewhat modified PBMs (including some consisting of PBM only; see ref. 37), but because they resemble class I molecules also in other domains, they are thought to be derivatives of MHC molecules, rather than being their precursors (12, 38). In short, no candidate aspiring for the title of a PBD ancestor has been found. Assuming that it does not exist, two possibilities remain to explain the origin of the PBD: creation from a noncoding sequence and genesis by interdomain exchange. Of the two, the latter is the more likely possibility. One of the two domains involved in the putative recombination could have been an Ig-like domain (ILD), which could have contributed a part of its β-sheet, whereas the α-helix could derive from another domain (e.g., from the phage MS2 coat protein by a transposon-mediated transfer) (Fig. 1). Not only do the β-sheets of the ILDs resemble the sheet of the PBD, but also ILDs constitute the remainder of the extracellular part in the MHC polypeptide, and genes encoding other ILD-bearing molecules are abundantly represented in the MHC region.
The ILDs
ILDs are found in a number of molecules participating in the AIS: MHC molecules, TCR, BCR, CD4 and CD8 coreceptors, and others (12). These domains, which can be distinguished into seven main types (V, C1, C2, C3, C4, I, and FnIII) (39), all share the basic Ig-fold structure consisting of two β-pleated sheets of a sandwich rolled into a cylinder (Fig. 1). ILDs differ in the arrangement and number of the β-strands and the length of the interconnecting loops. These domains all belong to a single Ig superfamily, which is widely distributed among both eukaryotes and prokaryotes (40). It is commonly believed that all of the members of the Ig superfamily derive from a single common ancestor. However, many Ig superfamily members do not show significant sequence similarity to one another and are assigned to the superfamily only on the basis of a few shared, conserved amino acid residues and similarity in predicted or determined tertiary structure. Hence, independent origin by convergent evolution of some of the members cannot be excluded. This fact, combined with cases in which different authors assign different types to the same ILD (41), makes the resolution of phylogenetic relationships among the types difficult. A particularly contentious group is the set of V-type ILDs. The shared rearrangement mechanism suggests that the V-domains of the TCRs and BCRs derive from a common ancestral domain, which must have existed before the divergence of the extant gnathostome classes (42). The databases contain numerous invertebrate sequences annotated as V-type ILDs, but upon closer scrutiny, most if not all of these have been reassigned to the I type (41). The gnathostome V-type domains present in molecules other than TCRs and BCRs (40) are probably genuine, but their appearance might have postdated the divergence of gnathostomes. Nevertheless, genuine V-domains of the subtype found in the TCRs and BCRs have been found in both agnathans (7) and cephalochordates (43, 44), albeit in these they are not rearranging. It seems, therefore, that genuine, but nonrearranging V-type domains arose before the advent of the AIS.
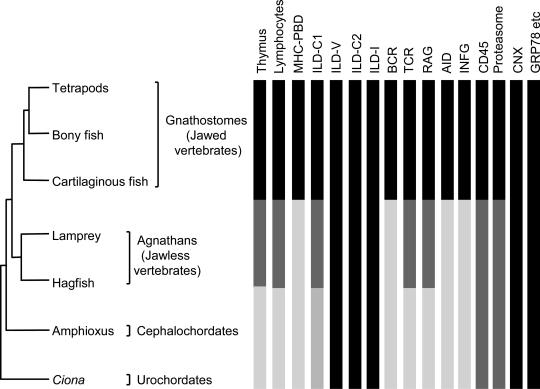
The C1-type ILDs have been documented in MHC, TCR, BCR, and a few other gnathostome molecules, some of which do not participate directly in the AIS (40). A C1-like domain may be present in the urochordate Ciona (45), so the evolution of this ILD type might have also preceded the appearance of the AIS (Fig. 2).
Fig. 2.
Emergence of the organs, cells, and molecules of the AIS during the evolution of chordates. Black bars indicate the presence of a fully developed trait. Darkly shaded bars indicate the ancestral form of a trait. Lightly shaded bars indicate that an ancestral form of a trait has not been identified.
The C2-type ILD is present in the CD4 and CD8 coreceptors of the TCR, in an assortment of other gnathostome proteins, particularly adhesion molecules, and in a variety of invertebrate molecules (40). In agnathans, it is linked to a V-domain of a molecule resembling the TCR and occurs also in a CD4-like molecule (7). The phylogenetic relationship among the seven ILD types is contentious, except that the FnIII, C3 and C4 types are generally agreed to be in an outgroup position relative to the remaining four types. In the V, C1, C2, and I group, the V and the C1 types appear to be derived from the C2 type, either directly or through the I type (46). The three types deployed by the AIS (V, C1, and C2) probably evolved before the divergence of agnathans and gnathostomes.
Molecules Encoded in Recombination-Activating Genes
In gnathostomes, somatic V(D)J recombination assembles the V segments of the TCR and BCR genes from two (V, J) or three (V, D, J) pieces selected randomly from the corresponding arrays, and in the process diversifies the segments. Key players in V(D)J recombination are two enzymes encoded in the RAG1 and RAG2 genes (47). The RAG1 protein is a large multifunctional recombinase, which binds to specific recombination signal sequences flanking the V, D, and J gene segments, cleaves the DNA between these sequences and the coding sequence, opens hairpins formed by the broken ends, joins broken DNA ends, and acts as a transposase, at least in vitro. The RAG2 protein seems to act primarily as a stabilizing cofactor of the RAG1 protein. No RAG transcripts have been found in agnathan LLCs (6, 8), no RAG genes are present in the urochordate (Ciona) genome (45), and the RAG proteins are not closely related to any other eukaryotic recombinase (48). The DNA-binding region of RAG1 does, however, contain stretches of high sequence similarity to the DNA-binding regions of the Caenorhabditis elegans Tc3 transposase and to the bacterial Hin DNA recombinase (49, 50). The Tc3 transposase belongs to the Tc1/mariner family of transposable elements widely distributed from fungi to humans. The Hin recombinase is a member of a family of bacterial DNA invertases, which catalyze site-specific recombination reactions. These similarities and other features of the RAGs, together with the ability of the RAG1 protein to function as a transposase in vitro, have led to the suggestion that the RAG genes were transferred horizontally into the eukaryotic genome by a prokaryote transposon (49, 50); however, the suggestion remains controversial (51).
AID
In mammalian B lymphocytes, V(D)J recombination is followed by further diversification of the BCR genes by two or three mechanisms, depending on the species: untemplated somatic hypermutation, pseudogene-templated gene conversion, and switch recombination, which places the rearranged V segment next to one of several C segments (12). All three mechanisms rely on the participation of the AID, an enzyme that deaminates cytosine residues and thus converts them into uracil (52). Whether the deamination takes place at the DNA or the RNA levels is still uncertain, as is the actual mechanism of the diversification process. AID is a member of a vertebrate family of related enzymes, which perform a variety of functions (53). Thus, the apolipoprotein B-editing catalytic 1 (APOBEC1) enzyme deaminates cytosine residues at a specific site of the RNA transcribed from the apolipoprotein B gene, thereby truncating the protein through a premature stop codon. Certain APOBEC3 enzymes deaminate cytosine residues of the RNA that the HIV uses for reverse transcription into the proviral DNA. Other members of the APOBEC family carry out other functions. The vertebrate APOBEC proteins are related to RNA-editing enzymes of nonmetazoan eukaryotes (e.g., yeast) (54). Hence, the ability to convert cytosine to uracil by deamination was established early in eukaryote evolution and was then deployed repeatedly to serve specific needs as they arose in the various emerging taxa. Neither AID nor APOBEC homologs have been found in the agnathan LLC transcriptome or the Ciona genome, so these subfamilies of the deaminating enzymes might represent a gnathostome innovation. In the case of the AID, the innovation had not been anything more than an application of an existing catalytic activity in yet another circumstance, which arose with the emergence of the AIS. Because presumably the catalytic site of the deaminases has not been changed, all that might have been needed for the integration of the enzyme into the AIS would have been a few mutations in the regulatory region of the encoding gene enabling the expression of the deaminase in the right cell at the right time. Similar mutations are presumably occurring in genes all of the time, but only those favored by selection have an increased probability of fixation (55). The fact that some gnathostome species possess only one of the three AID functions suggests that the functions may have been acquired sequentially in the evolution of the jawed vertebrates. An early form of the AIS might have functioned without the three additional diversification mechanisms, but the integration of the deaminases into the system improved the system's efficiency.
Signaling Molecules
The emergence of the AIS must have involved deployment of a number of existing signaling cascades, which evolved earlier to serve other functions. The integration of these cascades into the AIS was presumably accomplished by the insertion of a few regulatory molecules into the pathways. These molecules themselves may have served other functions before being drafted by the AIS. Here, we provide two examples of such molecules, IFN-γ and CD45. IFN-γ, a molecule secreted by activated T lymphocytes, binds either to IFN-γ receptors linked to the Janus kinase/signal transducer and activator of transcription signaling pathway or to other signal-transduction proteins linked to several other pathways (56). Through these pathways, IFN-γ regulates the expression of a wide spectrum of genes containing the appropriate sequence motifs in their regulatory regions. IFN-γ is evolutionarily related to the cytokines interleukin 4 and interleukin 10 (57), whereas its relationship to the group of proteins with which it shares its name (interferons α, β, and others) is unclear. There is no significant sequence similarity between IFN-γ and the interferons of the α/β group and the similarities in function (all interferons induce resistance of cells to virus infection and all act as cytokines regulating cellular activities) and in tertiary structure (the presence of a bundle of α-helices) could be the result of either divergence or convergence. Although orthologs of IFN-γ exist in bony fishes (58), no corresponding sequence could be found in either the agnathan transcriptome (6, 8) or the urochordate genome (45). The gene might have therefore arisen during the agnathan–gnathostome transition as an evolutionary novelty. On the other hand, if IFN-γ is truly related to the other interferons, the great depth of the divergence could be indicative of its origin before the rise of the gnathostomes. The signaling pathways that it regulates are all much older than the AIS. The Janus kinase/signal transducer and activator of transcription pathway, for example, not only arose before the divergence of protostomes and deuterostomes but is known to regulate other immune systems in insects (59).
CD45 or protein-tyrosine phosphatase receptor type C is a member of a family of receptor-type protein tyrosine phosphatases found in both protostomes and deuterostomes (60). The individual members of the family have specialized in dephosphorylating different tyrosine kinases and thus regulating signaling cascades in different cells. The CD45 expressed in lymphocytes has two functions (61). First, by dephosphorylating the src-family tyrosine kinases, such as lck or fyn, associated with TCRs and BCRs, it regulates antigen-induced activation of T and B cells in the AIS. And second, by dephosphorylating the Janus kinases, it regulates signaling pathways via type I and type II cytokine receptors in both the AIS and non-AIS immune responses. Both the lamprey (6) and the hagfish (62) LLCs express an ortholog of the gnathostome CD45. However, because these cells have cytokine receptors (8, 63) but lack TCRs and BCRs, the enzyme presumably performs only one of the two functions. Nevertheless, it might have the potential of signaling through TCRs or BCRs, because it seems to possess all of the motifs presumably needed to perform this function (6). Even if it does not, however, the acquisition of the ability to dephosphorylate src-family protein kinases probably would require a few substitutions, a supposition that can be tested experimentally. Because both agnathans and gnathostomes possess only one CD45 gene in their genomes (6, 8, 62), the acquisition of the second function has apparently, in this case, not involved gene duplication.
Conclusion and Prospects
According to a currently popular view, the “big bang” hypothesis (10, 11), the AIS arose suddenly, within a relatively short time interval, in association with the postulated two rounds of genome-wide duplications (64). That a number of genes have duplicated during the transition period from agnathans to gnathostomes is undeniable. However, how extensive this genomic expansion was remains controversial (65). If the duplication rate did indeed increase in the agnathan–gnathostome transition interval, it might have contributed to the appearance of AIS by providing the necessary “raw material” for the final integration of the emerging AIS into the other immune systems. The evolution of the AIS itself, however, must have begun long before the divergence of agnathans and gnathostomes from their common ancestor. The evolution consisted initially of changes unrelated to immune response that were selected to serve other functions. The different functions may have been unrelated to one another, but ultimately, a combination of these functions arose by chance, which presented the potential for the development, in a not too large number of small steps, of a qualitatively new system. The actualization of the potential required integration of the different functions into one whole. The necessary steps for this integration were undertaken in the gnathostome lineage, whereas the agnathans evolved in a different direction (66). Once the critical steps were accomplished in the gnathostome lineage, the integration created the illusion of a sudden, explosive change, a big bang. However, in reality, the entire process was gradual, consisting of accumulation of small changes over an extended period (Fig. 2).
The agnathan genomes should bear witness to how close they have come to acquiring the AIS. The closeness can be determined experimentally in two ways: by introducing agnathan genes into the gnathostome genome or the other way around. It should be possible to determine which changes are necessary for agnathan genes, such as the PSMB, ATP-binding cassette transporter, or CD45, to replace the functions of their gnathostome counterparts. In the opposite direction, introducing gnathostome genes such MHC, BCR, or RAG, into agnathan cells or animals should reveal to what extent the cells' or animals' physiology is “ready” for some of the functions associated with the AIS. Extant agnathans are, of course, evolutionarily far away from the agnathan–gnathostome ancestors, but this kind of experimental molecular evolution should nevertheless shed light on events that would otherwise remain hopelessly in the realm of mere speculations.
Acknowledgments
We thank Dimitra Chalkia for discussion of the manuscript. This work was partially supported by National Institutes of Health Grant GM20293 (to Masatoshi Nei).
Author contributions: J.K. designed research; and J.K. and N.N. performed research, analyzed data, and wrote the paper.
Abbreviations: AID, activation-induced deaminase; AIS, antibody-based immune system; APOBEC, apolipoprotein B-editing catalytic; BCR, B cell receptor; ILD, Ig-like domain; LLC, lymphocyte-like cell; PBD, peptide-binding domain; PBM, peptide-binding module; RAG, recombination-activating gene; TCR, T cell receptor.
References
- 1.Janeway, C. A., Jr., & Travers, P. (1994) Immunobiology: The Immune System in Health and Disease (Garland, New York).
- 2.Pancer, Z., Amemiya, C. T., Ehrhardt, G. R. A., Ceitlin, J., Gartland, G. L. & Cooper, M. D. (2004) Nature 430, 174-180. [DOI] [PubMed] [Google Scholar]
- 3.Cooper, E. L. (1976) Comparative Immunology (Prentice–Hall, Englewood Cliffs, NJ).
- 4.Klein, J. (1989) Scand. J. Immunol. 29, 499-505. [DOI] [PubMed] [Google Scholar]
- 5.Mayer, W. E., Uinuk-ool, T., Tichy, H., Gartland, G. L., Klein, J. & Cooper, M. D. (2002) Proc. Natl. Acad. Sci. USA 99, 14350-14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Uinuk-ool, T., Mayer, W. E., Sato, A., Dongak, R., Cooper, M. D. & Klein, J. (2002) Proc. Natl. Acad. Sci. USA 99, 14356-14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pancer, Z., Mayer, W. E., Klein, J. & Cooper, M. D. (2004) Proc. Natl. Acad. Sci. USA 101, 13273-13278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki, T., Shin-i, T., Kohara, Y. & Kasahara, M. (2004) Dev. Comp. Immunol. 28, 993-1003. [DOI] [PubMed] [Google Scholar]
- 9.Takezaki, N., Figueroa, F., Zaleska-Rutczynska, Z. & Klein, J. (2003) Mol. Biol. Evol. 20, 287-292. [DOI] [PubMed] [Google Scholar]
- 10.Abi Rached, L., McDermott, M. F. & Pontarotti, P. (1999) Immunol. Rev. 167, 33-44. [DOI] [PubMed] [Google Scholar]
- 11.Kasahara, M., Suzuki, T. & Pasquier, L. D. (2004) Trends Immunol. 25, 105-111. [DOI] [PubMed] [Google Scholar]
- 12.Klein, J. & Horejší, V. (1997) Immunology (Blackwell, Oxford).
- 13.Miller, J. (2004) C. R. Biol. 327, 399-408. [DOI] [PubMed] [Google Scholar]
- 14.Blackburn, C. C. & Manley, N. R. (2004) Nat. Rev. Immunol. 4, 278-289. [DOI] [PubMed] [Google Scholar]
- 15.Manley, N. R. (2000) Semin. Immunol. 12, 421-428. [DOI] [PubMed] [Google Scholar]
- 16.Kent, G. C. & Miller, L. (1997) Comparative Anatomy of the Vertebrates (Brown, Dubuque, IA).
- 17.Mark, M., Rijli, F. M. & Chambon, P. (1995) Semin. Dev. Biol. 6, 275-284. [Google Scholar]
- 18.Kampmeier, O. F. (1969) Evolution and Comparative Morphology of the Lymphatic System (Thomas, Springfield, IL).
- 19.Pignoni, F., Hu, B., Zavitz, K. H., Xiao, J., Garrity, P. A. & Zipursky, S. L. (1997) Cell 91, 881-891. [DOI] [PubMed] [Google Scholar]
- 20.Shigetani, Y., Sugahara, F., Kawakami, Y., Murakami, Y., Hirano, S. & Kuratani, S. (2002) Science 296, 1316-1319. [DOI] [PubMed] [Google Scholar]
- 21.Orkin, S. H. (1996) Curr. Opin. Genet. Dev. 6, 597-602. [DOI] [PubMed] [Google Scholar]
- 22.Good, R. A., Finstad, J. & Litman, G. W. (1972) in The Biology of Lampreys, eds. Hardisty, M. W. & Potter, I. C. (Academic, London), Vol. 2, pp. 405-432. [Google Scholar]
- 23.Fichtelius, K. E. (1970) Lymphology 3, 50-59. [PubMed] [Google Scholar]
- 24.Georgopoulos, K. (1997) Curr. Opin. Immunol. 9, 222-227. [DOI] [PubMed] [Google Scholar]
- 25.Shintani, S., Terzic, J., Sato, A., Saraga-Babic, M., O'hUigin, C., Tichy, H. & Klein, J. (2000) Proc. Natl. Acad. Sci. USA 97, 7417-7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haire, R. N., Miracle, A. L., Rast, J. P. & Litman, G. W. (2000) J. Immunol. 165, 306-312. [DOI] [PubMed] [Google Scholar]
- 27.Mayer, W. E., O'Huigin, C., Tichy, H., Terzic, J. & Saraga-Babic, M. (2002) Scand. J. Immunol. 55, 162-170. [DOI] [PubMed] [Google Scholar]
- 28.Watts, C. & Powis, S. (1999) Rev. Immunogenet. 1, 60-74. [PubMed] [Google Scholar]
- 29.Kloetzel, P.-M. & Ossendorp, F. (2004) Curr. Opin. Immunol. 16, 76-81. [DOI] [PubMed] [Google Scholar]
- 30.Conticello, S. G., Thomas, C. J. F., Petersen-Mahrt, S. & Neuberger, M. S. (2005) Mol. Biol. Evol., 22, 367-377. [DOI] [PubMed] [Google Scholar]
- 31.Kasahara, M., Hayashi, M., Tanaka, K., Inoko, H., Sugaya, K., Ikemura, T. & Ishibashi, T. (1996) Proc. Natl. Acad. Sci. USA 93, 9096-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takezaki, N., Zaleska-Rutczynska, Z. & Figueroa, F. (2002) Gene 282, 179-187. [DOI] [PubMed] [Google Scholar]
- 33.Klein, J. & O'hUigin, C. (1993) Curr. Opin. Genet. Dev. 3, 923-930. [DOI] [PubMed] [Google Scholar]
- 34.Valegard, K., Liljas, L., Fridborg, K. & Unge, T. (1990) Nature 345, 36-41. [DOI] [PubMed] [Google Scholar]
- 35.Baldwin, E., Weber, I., Charles, R., Xuan, J., Appella, E., Yamada, M., Matsushima, K., Edwards, B., Clore, G., Gronenborn, A. & Wlodawer, A. (1991) Proc. Natl. Acad. Sci. USA 88, 502-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oganesyan, V., Oganesyan, N., Terzyan, S., Qu, D., Dauter, Z., Esmon, N. L. & Esmon, C. T. (2002) J. Biol. Chem. 277, 24851-24854. [DOI] [PubMed] [Google Scholar]
- 37.Cosman, D., Müllberg, J., Sutherland, C. L., Chin, W., Armitage, R., Fanslow, W., Kubin, M. & Chalupny, N. J. (2001) Immunity 14, 123-133. [DOI] [PubMed] [Google Scholar]
- 38.Loconto, J., Papes, F., Chang, E., Stowers, L., Jones, E. P., Takada, T., Kumanovics, A., Fischer-Lindahl, K. & Dulac, C. (2003) Cell 112, 607-618. [DOI] [PubMed] [Google Scholar]
- 39.Halaby, D. M., Poupon, A. & Mornon, J.-P. (1999) Protein Eng. 12, 563-571. [DOI] [PubMed] [Google Scholar]
- 40.Halaby, D. M. & Mornon, J. P. (1998) J. Mol. Evol. 46, 389-400. [DOI] [PubMed] [Google Scholar]
- 41.Teichmann, S. A. & Chothia, C. (2000) J. Mol. Biol. 296, 1367-1383. [DOI] [PubMed] [Google Scholar]
- 42.Hood, L., Kronenberg, M. & Hunkapiller, T. (1985) Cell 40, 225-229. [DOI] [PubMed] [Google Scholar]
- 43.Cannon, J. P., Haire, R. N. & Litman, G. W. (2002) Nat. Immunol. 3, 1200-1207. [DOI] [PubMed] [Google Scholar]
- 44.Sato, A., Mayer, W. E. & Klein, J. (2003) Immunogenetics 55, 423-427. [DOI] [PubMed] [Google Scholar]
- 45.Azumi, K., De Santis, R., De Tomaso, A., Rigoutsos, I., Yoshizaki, F., Pinto, M., Marino, R., Shida, K., Ikeda, M., Ikeda, M., et al. (2003) Immunogenetics 55, 570-581. [DOI] [PubMed] [Google Scholar]
- 46.Smith, D. K. & Xue, H. (1997) J. Mol. Biol. 274, 530-545. [DOI] [PubMed] [Google Scholar]
- 47.De, P. & Rodgers, K. K. (2004) Immunol. Rev. 200, 70-82. [DOI] [PubMed] [Google Scholar]
- 48.Agrawal, A., Eastman, Q. M. & Schatz, D. G. (1998) Nature 394, 744-751. [DOI] [PubMed] [Google Scholar]
- 49.Banerjee-Basu, S. & Baxevanis, A. (2002) Genome Biol. 3, interactions1004.1-1004.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bernstein, R. M., Schluter, S. F., Bernstein, H. & Marchalonis, J. J. (1996) Proc. Natl. Acad. Sci. USA 93, 9454-9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hughes, A. L. (1999) Arch. Immunol. Ther. Exp. (Warsz) 47, 347-353. [PubMed] [Google Scholar]
- 52.Honjo, T., Muramatsu, M. & Fagarasan, S. (2004) Immunity 20, 659-668. [DOI] [PubMed] [Google Scholar]
- 53.Beale, R. C. L., Petersen-Mahrt, S. K., Watt, I. N., Harris, R. S., Rada, C. & Neuberger, M. S. (2004) J. Mol. Biol. 337, 585-596. [DOI] [PubMed] [Google Scholar]
- 54.Xie, K., Sowden, M. P., Dance, G. S. C., Torelli, A. T., Smith, H. C. & Wedekind, J. E. (2004) Proc. Natl. Acad. Sci. USA 101, 8114-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takahata, N. (1990) Proc. Natl. Acad. Sci. USA 87, 2419-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramana, C. V., Gil, M. P., Schreiber, R. D. & Stark, G. R. (2002) Trends Immunol. 23, 96-101. [DOI] [PubMed] [Google Scholar]
- 57.Nagem, R. A. P., Colau, D., Dumoutier, L., Renauld, J.-C., Ogata, C. & Polikarpov, I. (2002) Structure 10, 1051-1062. [DOI] [PubMed] [Google Scholar]
- 58.Zou, J., Yoshiura, Y., Dijkstra, J. M., Sakai, M., Ototake, M. & Secombes, C. (2004) Fish Shellfish Immunol. 17, 403-409. [DOI] [PubMed] [Google Scholar]
- 59.Agaisse, H. & Perrimon, N. (2004) Immunol. Rev. 198, 72-82. [DOI] [PubMed] [Google Scholar]
- 60.Glover, N. R. & Tracey, A. S. (2000) Biochem. Cell Biol. 78, 39-50. [PubMed] [Google Scholar]
- 61.Hermiston, M. L., Xu, Z. & Weiss, A. (2003) Annu. Rev. Immunol. 21, 107-137. [DOI] [PubMed] [Google Scholar]
- 62.Nagata, T., Suzuki, T., Ohta, Y., Flajnik, M. F. & Kasahara, M. (2002) Immunogenetics 54, 286-291. [DOI] [PubMed] [Google Scholar]
- 63.Kuroda, N., Uinuk-ool, T. S., Sato, A., Samonte, I. E., Figueroa, F., Mayer, W. E. & Klein, J. (2003) Immunogenetics 54, 884-895. [DOI] [PubMed] [Google Scholar]
- 64.Kasahara, M. (1998) Immunol. Rev. 166, 159-175. [DOI] [PubMed] [Google Scholar]
- 65.Hughes, A. (1998) Mol. Biol. Evol. 15, 854-870. [DOI] [PubMed] [Google Scholar]
- 66.Klein, J., Sato, A. & Mayer, W. E. (2000) in Major Histocompatibility Complex: Evolution, Structure, and Function, ed. Kasahara, M. (Springer, Tokyo), pp. 3-26.