ABSTRACT
Engineering Saccharomyces cerevisiae for the utilization of pentose sugars is an important goal for the production of second-generation bioethanol and biochemicals. However, S. cerevisiae lacks specific pentose transporters, and in the presence of glucose, pentoses enter the cell inefficiently via endogenous hexose transporters (HXTs). By means of in vivo engineering, we have developed a quadruple hexokinase deletion mutant of S. cerevisiae that evolved into a strain that efficiently utilizes d-xylose in the presence of high d-glucose concentrations. A genome sequence analysis revealed a mutation (Y353C) in the general corepressor CYC8, or SSN6, which was found to be responsible for the phenotype when introduced individually in the nonevolved strain. A transcriptome analysis revealed altered expression of 95 genes in total, including genes involved in (i) hexose transport, (ii) maltose metabolism, (iii) cell wall function (mannoprotein family), and (iv) unknown functions (seripauperin multigene family). Of the 18 known HXTs, genes for 9 were upregulated, especially the low or nonexpressed HXT10, HXT13, HXT15, and HXT16. Mutant cells showed increased uptake rates of d-xylose in the presence of d-glucose, as well as elevated maximum rates of metabolism (Vmax) for both d-glucose and d-xylose transport. The data suggest that the increased expression of multiple hexose transporters renders d-xylose metabolism less sensitive to d-glucose inhibition due to an elevated transport rate of d-xylose into the cell.
IMPORTANCE The yeast Saccharomyces cerevisiae is used for second-generation bioethanol formation. However, growth on xylose is limited by pentose transport through the endogenous hexose transporters (HXTs), as uptake is outcompeted by the preferred substrate, glucose. Mutant strains were obtained with improved growth characteristics on xylose in the presence of glucose, and the mutations mapped to the regulator Cyc8. The inactivation of Cyc8 caused increased expression of HXTs, thereby providing more capacity for the transport of xylose, presenting a further step toward a more robust process of industrial fermentation of lignocellulosic biomass using yeast.
KEYWORDS: sugar transporter, xylose transport, evolutionary engineering, transcriptome, yeast
INTRODUCTION
An increasing energy demand and concerns of obtaining this energy from fossil fuels have stimulated the development of liquid fuels from renewable feedstock. Bioethanol, mostly used as a fuel additive, produced from readily fermentable agricultural feedstocks, such as sugar cane and corn, is less desired because the production of these feedstocks requires large amounts of arable land and competes with the food supply (1). A more sustainable source of feedstock is lignocellulosic biomass from hardwood, softwood, and agricultural residues (2). However, a major drawback of lignocellulosic feedstocks is the inability of the most commonly used yeast in industry, Saccharomyces cerevisiae, to ferment the substantial fraction of pentose sugars, such as d-xylose, released upon conversion of lignocellulose besides the hexose sugar fraction (3). In recent years, two strategies have been developed to equip S. cerevisiae with the ability to convert d-xylose into bioethanol: (i) the xylose reductase (XR)-xylitol dehydrogenase (XDH) pathway, a two-step redox pathway in which XR first catalyzes the reduction of xylose to xylitol, which is subsequently oxidized via XDH to form xylulose (4, 5); and (ii) the xylose isomerase (XI) pathway, a one-step conversion from xylose into xylulose using either a bacterial or a fungal xylose isomerase (6–8). The XI pathway overexpressing the fungal xylose isomerase of Piromyces sp. E2 was used in this study. To further optimize the flux of xylose fermentation toward ethanol, the endogenous genes of the nonoxidative pentose phosphate pathway were overexpressed (8).
Although overexpression of xylose isomerase results in the desired d-xylose fermentation, the consumption of d-xylose in the presence of a high glucose concentration remains difficult (9). All the xylose-fermenting S. cerevisiae strains currently used first consume d-glucose, before d-xylose is metabolized. To generate an economically feasible process in an industrial setting, it is preferred that both sugars are fermented simultaneously and at high consumption rates (10). Pentose transport and the quest for the coconsumption of d-xylose and d-glucose are important topics in xylose-fermenting strains. Various approaches have been used, including the introduction of specific xylose transporters derived from other organisms (11, 12), but these support only low rates of xylose transport (13–16). Another approach, reported recently, is the mutagenesis of the hexose transporter genes HXT7 and GAL2, yielding mutants that were found to be defective in glucose uptake while still retaining substantial xylose transport activity (17). In another study, it was shown via saturated mutagenesis of conserved amino acid sequence motifs that HXT7 could be converted into a d-xylose transporter. However, this rewired transporter remained sensitive to glucose inhibition (18). The extensively studied hexose transporter (HXT) family of sugar transporters mediate glucose transport in S. cerevisiae (19, 20). In a strain lacking the main hexose transporters, HXT1 to HXT7 and GAL2, uptake of d-xylose could be restored by the reintroduction of HXT1, HXT2, HXT4, and HXT7 (21). In another study, HXT3 and a chimeric HXT36, in which HXT3 and HXT6 are fused, were shown to complement growth on d-xylose (22). Furthermore, in a d-xylose-fermenting S. cerevisiae strain in which all hexose transporters were deleted, it was shown that HXT5 also transports d-xylose (9). However, what is common among all of these expressed HXTs is that the preferred substrate is d-glucose and not d-xylose. In the quest for a specific d-xylose transporter, we recently showed via a combinatorial approach of evolutionary engineering and directed evolution that the HXT36 chimeric could be converted into a specific d-xylose transporter by a single amino acid change, allowing cometabolism of d-glucose and d-xylose (22). However, the best mutant showed a rather low maximum rate of metabolism (Vmax) on d-xylose, which limited the growth rate on this sugar.
To further optimize d-xylose transport in the presence of d-glucose, we have used an in vivo evolutionary engineering method using a xylose-fermenting S. cerevisiae strain that lacks the four hexokinase genes (17, 22). This strain is therefore unable to grow on d-glucose, but still ferments d-xylose. By growing this strain on repeated batches of d-xylose in the presence of increasing concentrations of d-glucose, an evolved strain was obtained in which the transport and metabolism of d-xylose is highly resistant to d-glucose. Genome sequencing and expression analysis indicated that the growth phenotype can be explained by a mutation in CYC8, or SSN6, which leads to increased expression levels of the HXTs causing a higher transport flux of d-xylose into the cell in the presence of d-glucose.
RESULTS
Evolutionary engineering of an S. cerevisiae quadruple hexokinase deletion strain on d-xylose in the presence of increasing d-glucose concentrations.
The S. cerevisiae quadruple hexokinase (GLK1, HXK1, HXK2, and GAL1) deletion strain, DS69473 (22, 23), was grown in a fermentor to select for improved growth on d-xylose in the presence of d-glucose. This strain contains an engineered d-xylose metabolic pathway, based on a fungal xylose isomerase, and is thus capable of growing on d-xylose (6–8), but it does not grow on d-glucose. The experiment was designed to isolate an evolved d-xylose-fermenting strain that is less sensitive to d-glucose inhibition. The DS69473 strain was grown aerobically in batch culture on 1% d-xylose in the presence of 3% d-glucose and, in time, increasing concentrations of d-glucose up to 8% (see Fig. S1 in the supplemental material). The growth was assayed via measuring CO2 production and optical density during the fermentation. Because of the experimental batch culture setup, the strain consumes the d-xylose whereas the d-glucose concentration remains unaltered. This leads, in time, to increasing d-glucose to d-xylose ratios and therefore decreased growth rates as observed by the CO2 measurements. If needed, d-xylose was added to maintain growth. To further challenge the strain, the d-glucose concentration was increased slowly over a 40-day time frame, during which the wild-type DS69473 adapted to the increasing ratio of d-glucose to d-xylose. This led to the evolved DS69473Evo strain, which was able to grow on d-xylose (1%) in the presence of an 8-fold excess of d-glucose (8%). The growth of the DS69473Evo strain in shake flasks with 2% d-xylose in the presence of various concentrations of d-glucose was analyzed and compared with that of the progenitor DS69473. Both strains showed comparable growth profiles on 2% xylose without glucose. The growth of the original DS69473 strain and the evolved DS69473Evo strain were inhibited at 6% d-glucose, although the DS69473Evo strain grew to higher optical density at 600 nm (OD600) levels compared with that of the parental strain (Fig. 1). The growth of the DS69473 strain on 2% d-xylose in the presence of 12% d-glucose was completely inhibited, while the DS69473Evo strain was still able to grow. As expected, control experiments demonstrated that these strains were unable to consume or grow on d-glucose (data not shown).
FIG 1.
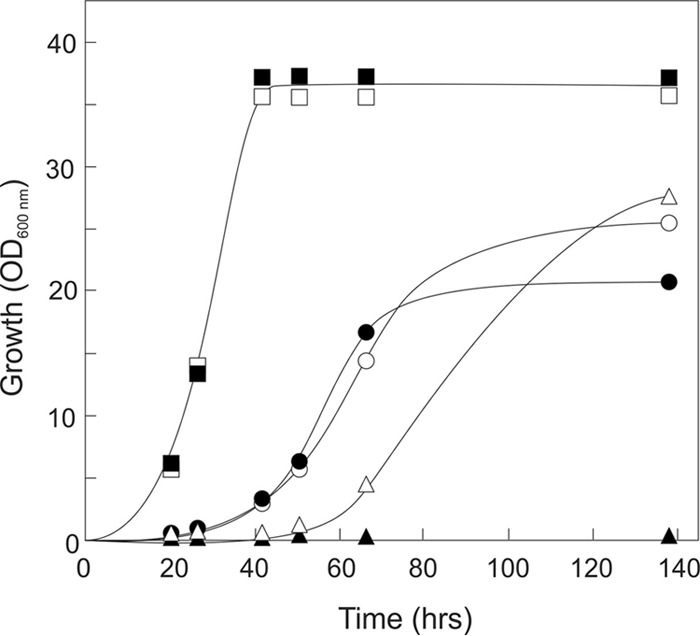
Growth of the original DS69473 strain (closed symbols) and the DS69473Evo strain (open symbols) on 2% d-xylose and 0% (■ and □), 6% (● and ○), and 12% (▲ and △) d-glucose.
d-Xylose uptake in the presence of d-glucose.
To investigate the molecular basis that allows growth of the DS69473Evo strain on d-xylose in the presence of competing concentrations of d-glucose, d-[14C]xylose uptake experiments were carried out. With 100 mM (∼1.5%) xylose and without glucose in the buffer, the DS69473Evo strain already showed an increased rate of d-xylose uptake (49.8 ± 3.7 nmol/mg [dry weight] · h) compared with that of the original DS69473 strain (32.2 ± 1.3 nmol/mg [dry weight] · h) (Fig. 2). With both strains, d-xylose transport was inhibited by d-glucose, but a substantial residual d-xylose uptake rate remained with the DS69473Evo strain at the highest d-glucose concentrations tested. If the uptake rates were compared relative to the d-xylose uptake in the absence of d-glucose, the difference between both stains was negligible (Fig. 2, inset). This suggests that the evolution experiment resulted in an increased d-xylose transport activity but that the d-glucose sensitivity remained unchanged.
FIG 2.
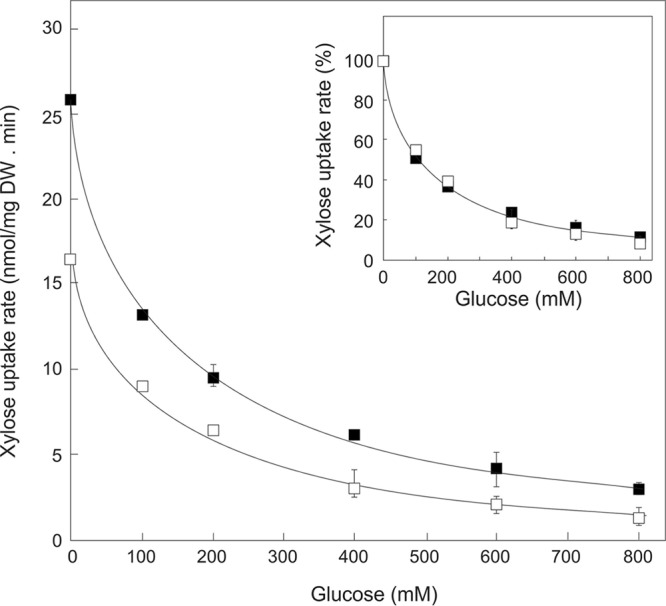
d-Xylose uptake by the in vivo engineered S. cerevisiae strain. Uptake of 100 mM d-[14C]xylose by the DS69473 (□) and DS69473Evo (■) strains in the presence of competing concentrations of d-glucose ranging from 0 to 800 mM. (inset) Xylose uptake normalized to the rate observed in the absence of competing glucose.
Genome sequencing.
Genome sequencing of DS69473 and DS69473Evo was conducted to find mutations that enabled the evolved strain to grow on 1% d-xylose in the presence of 8% d-glucose. Sequencing data were mapped to the CEN.PK113-7D strain (24). Unique variants (insertion, deletion, multinucleotide variant [MNV], and single nucleotide polymorphisms [SNP]) were detected by comparing the DS69473 and DS69473Evo genomes. When selecting variants that were found only in DS69473Evo, one interesting mutation was obtained in CYC8, or SSN6, (see Table S1 for all variants of DS69473Evo). This point mutation (A1,058G) in CYC8 resulted in an amino acid change, Y353C, and was found in 335 of 336 reads that mapped at that position, meaning it was present with high coverage and at a high frequency. The presence of the mutation was checked in the predecessors' strains but found to be absent in the lineage (data not shown). Since the glucose/xylose uptake was changed in the DS69473Evo strain, we focused especially on all the HXTs in the genome, and the comparison between the DS69473Evo strain and DS69473 yielded no differences. However, both strains showed stop codons in HXT13 at position 842 (W281stop), which is also present in the CEN.PK113-7D strain (24). Furthermore, both strains have synonymous substitutions in the HXT10 gene (A1,587G), which is not present in the genome of CEN.PK113-7D.
Reverse engineering of CYC8 in the original DS69473 strain.
To validate the significance of the Y353C mutation from CYC8, the same mutation was reverse-engineered in strain DS69473. A similar strain containing the kanMX marker but not the CYC8 mutation was constructed as a control (DS69473-Y353). The obtained strains were tested for aerobic growth on 2% d-xylose and 12% d-glucose in shake flasks (Fig. 3) in a manner analogous to the evolution experiment. The growth pattern of the DS69473-Y353C strain was highly similar to that of the DS69473Evo strain (Fig. 1). By contrast, the DS69473-Y353 control strain was unable to grow on 2% d-xylose or 12% d-glucose, and showed normal growth on 2% d-xylose only (data not shown). This demonstrates that the Y353C mutation from CYC8 is the main element responsible for the obtained phenotype in the DS69473Evo strain.
FIG 3.
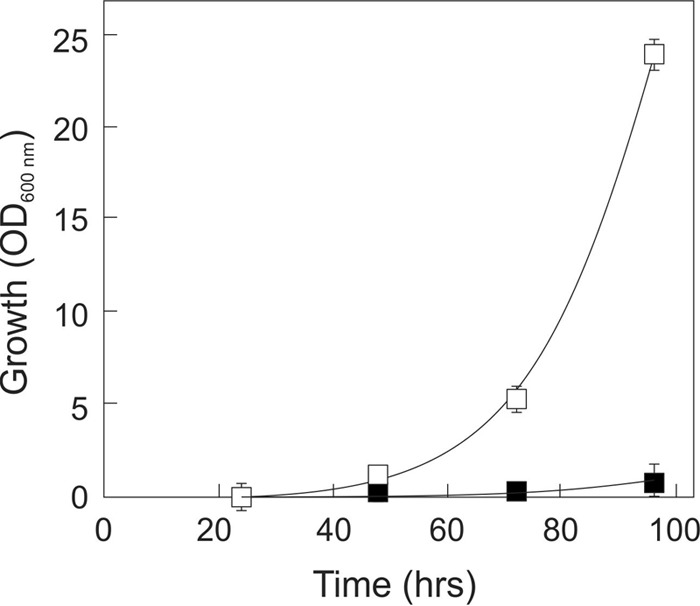
Growth of the DS69473-Y353 strain (■) and the DS69473-Y353C strain (□) on 2% d-xylose and 12% d-glucose.
Saturation mutagenesis of residue 353 of Cyc8.
To explore the sequence space of position Y353 in the protein encoded by CYC8, all further amino acid substitutions were individually introduced into CYC8 using clustered regularly interspaced short palindromic repeat(s) (CRISPR)/Cas9 technology. The CYC8(Y353) mutants were transformed to the original DS69473 hexokinase deletion strain and tested for growth on 2% d-xylose and 12% d-glucose. Improved growth (within 48 h) occurred with 10 additional mutants relative to that with the wild-type CYC8 (see Fig. S2). However, none of these mutants showed a significantly improved growth rate compared with that of the original CYC8(Y353C) mutant. Therefore, in all further experiments, the CYC8(Y353C) mutant was used.
Transcriptome analysis of the CYC8 mutant strain.
To further examine the transcriptional impact of the CYC8(Y353C) mutation, transcriptome sequencing (RNA-seq) analysis was carried out with the DS69473-Y353C and DS69473-Y353 strains, grown for 29 h on 2% d-xylose in the presence of 6% d-glucose. In the comparison, 93 genes were upregulated and 2 genes were downregulated in strain DS69473-Y353C (see Table S2). The up- and downregulated genes are distributed over almost all chromosomes, and four main functional clusters of genes could be identified that included: (i) the downregulation of two maltose permeases (MAL11 and MAL31) and 2 maltases (MAL12 and MAL32), (ii) the upregulation of members of the seripauperin multigene family (10 PAU genes upregulated), (iii) the upregulation of the cell wall mannoprotein family (8 genes upregulated, e.g., TIR1 and FIT2), and, most notably, (iv) the upregulation of members of the hexose transporter family, including highly (HXT1, HXT2, HXT36, HXT5, and HXT7) and silent or lowly expressed (HXT10, HXT13, HXT15, and HXT16) genes (Table 1). Combining the absolute expression levels of all HXTs, the overall expression was nearly 2-fold higher in the DS69473-Y353C mutant compared with that in the DS69473-Y353 control strain (Table 1). RNA-seq data of all the HXT genes were confirmed by quantitative PCR (qPCR) (data not shown). Of the 93 upregulated genes, 34 are located in the 30 kbp of the telomeric regions of the various chromosomes, which is consistent with data published before (25).
TABLE 1.
Transcriptome data of all HXTs expressed in DS69473 strains carrying CYC8(Y353) and CYC8(Y353C)a
| Probe | Gene | Chromosome |
CYC8(Y353) |
CYC8(Y353C) |
FC | ||
|---|---|---|---|---|---|---|---|
| mRNA level | % of total | mRNA level | % of total | ||||
| YMR011W | HXT2 | XIII | 13,238 | 53.7 | 24,749 | 51.5 | 1.9 |
| YDR343C | HXT36 | IV | 2,947 | 12.0 | 6,643 | 13.8 | 2.3 |
| YDR342C | HXT7 | IV | 1,307 | 5.3 | 6,234 | 13.0 | 4.8 |
| YHR092C | HXT4 | VIII | 5,093 | 20.7 | 4,750 | 9.9 | 0.9 |
| YHR096C | HXT5 | VIII | 358 | 1.5 | 1,903 | 4.0 | 5.3 |
| YHR094C | HXT1 | VIII | 528 | 2.1 | 1,629 | 3.4 | 3.1 |
| YJL214W | HXT8 | X | 917 | 3.7 | 797 | 1.7 | 0.9 |
| YDL245C | HXT15 | IV | 54 | 0.2 | 485 | 1.0 | 9.0 |
| YJR158W | HXT16 | X | 38 | 0.2 | 316 | 0.7 | 8.3 |
| YEL069C | HXT13 | V | 11 | <0.1 | 367 | 0.8 | 34.3 |
| YFL011W | HXT10 | VI | 16 | 0.1 | 127 | 0.3 | 8.1 |
| YJL219W | HXT9 | X | 53 | 0.2 | 46 | 0.1 | 0.9 |
| YOL156W | HXT11 | XV | 53 | 0.2 | 37 | 0.1 | 0.7 |
| YLR081W | GAL2 | XII | 25 | 0.1 | 14 | <0.1 | 0.6 |
| YNL318C | HXT14 | XIV | 9 | <0.1 | 4 | <0.1 | 0.4 |
| Sum of all HXTs | 24,647 | 48,102 | 1.95 | ||||
| Sum of all genes | 2,706,604 | 2,655,949 | 0.98 | ||||
HXTs are ranked based on total expression level as measured by RNA-seq. The percentages of transcripts for all genes combined are indicated, showing the relative abundance of the transcripts, as well as the ratio (fold change [FC]) of the expression in the DS69473 strain carrying CYC8(Y353C) versus DS69473 carrying CYC8(Y353). Also indicated are the sums of HXT gene transcripts and of all genes, showing no major global increase in mRNA.
Next, uptake experiments with d-[14C]xylose were performed to study the effect of the increased expression of HXTs in the DS69473-Y353C mutant strain. The collective kinetic parameters for d-xylose uptake were improved in the DS69473-Y353C strain compared with that in the DS69473-Y353 strain, showing an increased Vmax (287.7 ± 14.5 nmol/mg [dry weight] · h versus 244.1 ± 24.1 nmol/mg [dry weight] · h, respectively) and an improved apparent Km (368.5 ± 48.0 mM versus 486.8 ± 60.9 mM) (Fig. 4). The uptake of d-glucose was also increased with a Vmax of 147.5 ± 7.2 nmol/mg (dry weight) · h in the DS69473-Y353C mutant versus 99.3 ± 4.5 nmol/mg (dry weight) · h in the DS69473-Y353 control strain, with apparent Km values of 48.2 ± 2.9 mM and 29.9 ± 2.3 mM, respectively. It should be stressed that the above kinetic parameters are an approximation as they reflect the overall transport activity of the yeast strains. Summarizing, these data demonstrate that the evolved strain exhibits enhanced rates of d-glucose and d-xylose transport.
FIG 4.
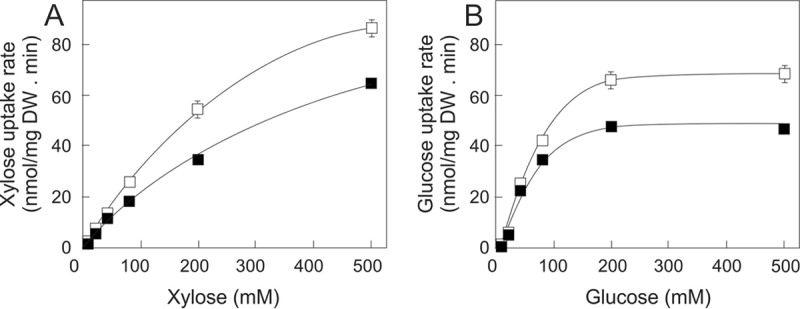
Kinetic parameters for d-xylose (A) and d-glucose (B) uptake. Uptake was measured in nmol/mg (dry weight) · min in the DS69473-Y353 (□) and DS69473-Y353C (■) strains. The uptake levels of both sugars for the DS68625 strain, in which HXT1 to HXT7 and GAL2 were deleted, were subtracted from those of the DS69473 and DS69473-Y353C strains to correct for background sugar uptake and cellular binding.
Characterization of a CYC8 deletion strain.
To investigate if the Y353C mutation for the CYC8 gene is a functional change or inactivates the protein, a CYC8 deletion strain was studied. Although various attempts were made with the HIS3 marker as well as the kanMX marker, and also with different flanking regions, no inactivation mutant of the CYC8 gene could be obtained in the original DS69473 hexokinase deletion strain. When the same constructs were transformed to the DS68616 strain, a xylose-fermenting strain with intact hexokinase genes, the appropriate deletion was obtained in which CYC8 was replaced with the kanMX resistance marker as confirmed by PCR (data not shown). Additionally, the CYC8(Y353C) mutation was introduced into DS68616, yielding strain DS68616-Cyc8-Y353C and the control DS68616-Cyc8-Y353, both harboring the kanMX resistance marker. All three strains were grown on 2% d-xylose and 6% d-glucose to the mid-exponential growth phase and total RNA was isolated. Using qPCR, ADH1, FIT2, and HXT13 were found to be 0.3- (P = 0.028), 8.7- (P = 0.034), and 36.2-fold (P = 0.000) upregulated in the DS69473-Y353C strain, respectively. The DS68616-Cyc8-Y353C mutant strain showed results similar to those from the DS69473-Y353C mutant strain (fold expression: ADH1, 0.25 ± 0.02; FIT2, 2.9 ± 0.10; and HXT13, 16.62 ± 1.06). Likewise, the DS68616 Δcyc8 strain showed a similar upregulation of HXT13 (14.9 ± 1.5) and a downregulation of ADH1 (0.42 ± 0.08). In this strain, FIT2 (0.10 ± 0.01) was downregulated. HXT10 and HXT15-HXT16, both characteristic for the HXT upregulated phenotype, were 5.82 (±0.64)- and 13.36 (±1.04)-fold upregulated, respectively, in the DS68616 Δcyc8 strain (see Fig. S3). It is of interest to note that the silent and inactivated HXT13 gene was also upregulated in the DS69473Evo and DS69473-Y353C strains (data not shown), suggesting a common mechanism.
Characterization of a xylose-fermenting industrial strain expressing CYC8(Y353C).
The impact of the Y353C mutation from CYC8 on d-xylose consumption was further studied in an industry-relevant strain that carries the four hexose kinases (GLK1, HXK1, HXK2, and GAL1) and thus is capable of glucose consumption. DS68616-Cyc8-Y353, DS68616-Cyc8-Y353C, and DS68616 ΔCyc8 strains were inoculated at an OD600 of 10 and grown for 8 h on 2% d-glucose and 2% d-xylose. Since the DS68616 Δcyc8 strain flocculated and showed a severely reduced growth rate (data not shown), it was not further analyzed. Strain DS68616-Cyc8-Y353C showed a reduced growth rate on the glucose-xylose mixture compared with that of the DS68616-Cyc8-Y353 wild-type strain (see Fig. S4C), but the initial d-xylose consumption was higher in the DS68616-Cyc8-Y353C mutant strain compared with that of the DS68616-Cyc8-Y353 wild-type strain (Fig. S4B). By contrast, d-glucose consumption remained unaltered (Fig. S4A). This shows that in a glucose-consuming strain, xylose consumption is also stimulated by the CYC8(Y353C) mutation.
Overexpression of the upregulated silent HXT genes.
To examine the role of individual upregulated HXTs in the phenotype of the DS69473Evo strain, the genes encoding several HXTs were amplified from genomic DNA of the DS69473Evo strain and cloned into the yeast expression vector, pRS313-P7T7 (22). It should be noted that we assumed that the DS69473 and DS69473Evo strains display a deletion of the last 18.9 kbp of the telomeric region of the right arm of chromosome XIV, including HXT17 (accession no. YNR072W). This is partially in contrast to the de novo genome sequencing of the progenitor strain CEN.PK113-7D, where only the genes designated YNR070W, YNR071C, YNR074C, YNR075C, and YNR077C, which cover different parts of the telomeric region, were found to be deleted (24). However, the genes reported to be present in this study, designated YNR072W (HXT17), YNR073C (DSF1), and YNR076W (PAU6) all have homologs elsewhere in the genome with 97, 99, and 100% similarity, respectively. Therefore, it appears that these were mapped incorrectly and that instead, the entire 18-kbp DNA fragment was deleted in the CEN.PK113-7D strain. PCR amplification of genomic DNA of the yeast strains S288C, CEN.PK113-7D, and DS69473 showed that 3 of 3 fragments failed to be amplified in the latter two strains, whereas in the S288C strain, this amplification was possible (data not shown). Since HXT13 contains a stop codon at position 842 (W281stop), it was not further examined for overexpression and complementation. Moreover, due to the very high nucleotide sequence homology (99%) between HXT15 and HXT16, the individual genes could not be separated by sequencing. In the DS69473 strain, HXT10 and HXT15-HXT16 are not expressed. However, these genes are upregulated in the DS69473Evo strain and were therefore also overexpressed individually in the hexose transporter deletion strain, in which HXT10 and HXT15-HXT16 are silent (see Table S3). Both HXT10 and HXT15 were able to complement growth on 2% d-glucose (see Fig. S5A). However, only HXT10 showed growth complementation on 2% d-xylose (Fig. S5B), but growth was relatively poor compared with that from strains with other HXT genes (e.g., HXT2 and HXT7) (data not shown). Thus, HXT10 was not further analyzed. These data suggest that HXTs are not individually responsible for the observed phenotype. Rather, the CYC8 mutation causes an increased expression of a set of HXTs, thereby changing the transporter landscape and concomitantly elevating the rates of d-glucose and d-xylose transport.
DISCUSSION
In the yeast Saccharomyces cerevisiae, HXTs (encoded by HXT1 to HXT7) function as facilitators for d-glucose uptake, allowing cells to grow efficiently with medium containing high concentrations of this sugar. In an industrial setting, where a xylose-fermenting S. cerevisiae strain is used to convert d-xylose and d-glucose from lignocellulosic biomass into bioethanol, coconsumption of these sugars is essential to shorten fermentation times and to prevent inhibitory effects of toxic metabolites on d-xylose metabolism that is usually slower in these engineered strains. Therefore, cometabolism is desired for the development of a robust fermentation process. Although the S. cerevisiae strain DS68616 used in this study metabolizes d-xylose efficiently, the uptake, and therefore consumption, of d-xylose is strongly inhibited by d-glucose. This is because d-xylose is transported via HXTs that prefer d-glucose over d-xylose (9, 21, 22). To overcome this glucose transport inhibition and to select for mutants with an increased and more specific d-xylose uptake, a d-xylose-metabolizing DS69473 strain was used which lacks the four hexokinase genes and is thus unable to grow on d-xylose in the presence of high concentrations of d-glucose. This strain does not grow on d-glucose because of the inability to metabolize this sugar. The evolutionary engineering process yielded the DS69473Evo strain, which is able to grow on a high ratio of d-glucose to d-xylose. Results from uptake experiments indeed showed an improved d-xylose uptake in the presence of an increasing concentration of d-glucose, albeit d-xylose uptake remained inhibited. Genome sequencing of the evolved DS69473Evo strain identified the Y353C mutation for the CYC8 gene, and by transcriptomics, the subsequently increased expression of a large number of genes encoding HXTs was demonstrated causing both an increased Vmax of d-xylose and increased d-glucose uptake. Further mutagenesis of the Y353 position showed that other substitutions can result in improved growth, but the original cysteine mutant appears to exhibit the strongest phenotype. A previous study also described a point mutation in CYC8 in an evolutionary engineering experiment which optimized d-xylose consumption in a d-xylose-fermenting S. cerevisiae strain (26). Either because of that mutation or because of two other mutations found in the evolved strain (26), the expression levels of the genes involved in xylose metabolism (e.g., XYL1 and XYL2) were increased, causing improved d-xylose consumption but not increased growth on d-xylose in the presence of d-glucose. The Y353C mutation for the CYC8 gene is located in the next-to-last (number 9 out of 10) tetratricopeptide (TPR), which is a functional domain required for the interaction with Tup1p. A distinct subset of TPR motifs is needed for the repression of different classes of genes affected by the Cyc8p-Tup1p corepressor complex, especially TPRs 8 and 9, and possibly 10. These are shown to be critical for glucose repression (27, 28). During glucose repression, the Cyc8p-Tup1p corepressor complex interacts with Mig1p and inhibits Mig1p activation (29, 30). Transcriptome data of a S. cerevisiae wild-type strain in which the TUP1 gene was deleted showed the 2-fold upregulation of 225 genes, including 15 genes involved in flocculation and encoding serine-rich cell wall mannoproteins, seripauperin, and members of the hexose transporter family (31). The latter group includes all genes encoding HXTs except for HXT5, HXT10, and HXT14. Although there is extensive overlap among upregulated HXT genes in our data set, the CYC8 mutation does not cause the upregulation of GAL2, HXT4, HXT8, HXT9, HXT11, or HXT14, and thus the phenotype differs from that of the tup1 deletion. Of the 93 genes, which are upregulated in the CYC8(Y353C) mutant strain, 34 genes are 2-fold upregulated in the tup1 deletion strain. Our data further demonstrate that a CYC8 deletion strain does not have the same phenotype as the CYC8(Y353C) mutant strain. This CYC8 deletion strain showed a tendency to flocculate, which was not observed with the CYC8(Y353C) mutant strain. Flocculation was also observed previously in a cyc8 (32) and tup1 (33, 34) deletion strain. However, since Hxk2p is a bifunctional enzyme, both a catalyst and an important regulator in glucose repression, the phenotype of a strain with 2 mutations in the glucose catabolite repression mechanism (CYC8[Y353C] and Δhxk2) might differ from that with only the CYC8(Y353C) mutation. Overall, we conclude that the phenotype of the Y353C mutation for the CYC8 gene has substantial overlap with the phenotype of strains carrying a deletion of CYC8 or TUP1. It should also be noted that in addition to the aforementioned functional classes of genes, including the HXTs, a number of individual genes were upregulated. In particular, the almost 6-fold upregulation of TKL2 is of interest as this gene encodes a transketolase that catalyzes the conversion of xylulose-5-phosphate and ribose-5-phosphate to sedoheptulose-7-phosphate and glyceraldehyde-3-phosphate in the pentose phosphate pathway, and thus is involved in the metabolism of d-xylose. The lowly expressed TKL2 is a paralog of TKL1. Although TKL1 is 1.4-fold downregulated in the CYC8 mutant strain, its expression is still 26-fold higher than TKL2. Based on the expression levels, it appears unlikely that an increase in TKL2 expression causes a major change in d-xylose metabolism. The upregulated HXT15 and HXT16 genes have recently been identified as encoding mannitol and sorbitol transporters (35). They cluster with the genes encoding sorbitol dehydrogenases, SOR2 and SOR1. The latter genes were also upregulated in the DS69473-Y353C mutant strain (see Table S2 in the supplemental material). However, the upregulation of SOR1 could also be caused by the increased influx of d-xylose into the cell, since d-xylose induces the expression of SOR1 (36). This may also apply to SOR2. Furthermore, HXT15 is able to transport xylitol, which may have a negative effect on biomass and/or ethanol production, since xylitol inhibits the xylose isomerase (37). It should be noted that in the DS69473-derived strains, the aldose reductase encoded by GRE3 was deleted to prevent the accumulation of xylitol. Although the upregulation of silent hexose transporters, HXT10 and HXT15, occurred in the CYC8(Y353C) mutant, these HXTs do not substantially contribute to the overall phenotype of an increased growth rate on d-xylose in the presence of d-glucose. The genes are lowly expressed and individually could not fully complement a transporter deletion strain for xylose transport and metabolism. We conclude that the phenotype relates to the increased expression of the “main” hexose transporters HXT2, HXT36, and HXT7 and, to a lesser extent, to expression of HXT5 and HXT1, causing elevated rates of uptake of both d-glucose and d-xylose. Under these conditions, d-glucose remains the most favorable transported sugar, but because of the higher transport capacity, an increased rate of d-xylose transport is also observed, driving improved d-xylose metabolism. Thus, mutation of CYC8 might be a general means to increase the expression of HXTs to improve hexose/pentose cometabolism.
Conclusions.
A mutation (Y353C) in the general transcriptional corepressor Cyc8 causes the altered transcription of a large group of genes involved in sugar metabolism and cell wall biogenesis, including the upregulation of almost all genes encoding HXTs. This leads to an increased uptake of d-xylose in the presence of d-glucose, providing a general means to increase the sugar transport flux in strains that cometabolize d-glucose and d-xylose.
MATERIALS AND METHODS
Molecular biology techniques and chemicals.
DNA polymerase, restriction enzymes, and T4 DNA ligase were acquired from Fermentas. Oligonucleotides used for strain constructions were purchased from Sigma-Aldrich (Zwijndrecht, the Netherlands). Yeast genomic DNA for genome sequencing was isolated using the YeaStar genomic DNA kit (ZymoResearch, Irvine, CA, USA) according to the manufacturer's instructions. Total RNA was isolated and cDNA was prepared from S. cerevisiae cells as described before (22). Antibiotics, hygromycin and Geneticin (G418), for the selection of introduced constructs in yeast were acquired from Invitrogen (Toulouse, France); nourseothricin was acquired from Werner Bioagents (Jena, Germany).
Strains and growth conditions.
The construction of DS68625 (38) and DS68616 (23) have been described elsewhere. S. cerevisiae strains used in this study (see Table S3 in the supplemental material) were provided by DSM Bio-based Products & Services. Xylose-fermenting S. cerevisiae strains were provided by DSM and made available for academic research under a material transfer agreement with DSM. Fed-batch cultures were grown in minimal medium supplemented with vitamin solution and trace elements (39) in a laboratory fermentor with a working volume of 500 ml (Applikon, Schiedam, the Netherlands) at a temperature of 30°C and pH 4.5. The dissolved oxygen (DO) was set at 5%, the solution was stirred at 400 rpm, and the starting OD600 was 0.2. Shake flask experiments at 200 rpm were also conducted in minimal medium supplemented with 2% d-maltose, 2% d-xylose-0.05% d-maltose, and 2% d-glucose. The 0.05% d-maltose was added to circumvent an elongated lag phase in minimal medium with only 2% d-xylose. Cell growth was monitored by optical density (OD) at 600 nm using a UV-visible spectrophotometer (Novaspec Plus).
In vivo evolution.
The quadruple hexokinase deletion mutant DS69473 was evolved in batch cultivation to grow with 1% d-xylose in the presence of increasing concentrations of d-glucose (from 3 to 8%). Growth of the DS69473 strain was followed over time by CO2 measurements, whereas the levels of d-xylose and d-glucose were monitored by high-performance liquid chromatography (HPLC) to confirm that the cells were growing solely on d-xylose. The d-glucose-to-d-xylose ratio at the start of the evolutionary engineering was kept low at a 1:3 ratio but was increased during the experiment, eventually reaching 1% d-xylose and 8% d-glucose. In the setup used, the DS69473 strain consumes only the d-xylose, which leads to higher glucose-to-xylose ratios over time and therefore a drop in the growth rate. When the CO2 production was reduced, additional xylose (5 ml of 50% d-xylose added to a 500-ml fermentor volume) was added to maintain growth. On average, after 6 to 7 days, the culture was diluted into fresh medium with a higher d-glucose-to-d-xylose ratio (Fig. S1). After 40 days, the evolved DS69473 strain was plated on 1% d-xylose and 8% d-glucose. A single colony (DS69473Evo) was used for further analysis.
Analytical methods.
HPLC was performed using an Aminex HPX-87H column (Bio-Rad) at 65°C, and a refractive index detector (Shimadzu, Kyoto, Japan) was used to measure the concentrations of d-glucose and d-xylose. The mobile phase was 0.005 N H2SO4 at a flow rate of 0.55 ml/min.
Genome sequencing.
Strains DS69473 and DS69473Evo were sequenced using Illumina HiSeq with mate-pair (50-bp-long reads; insert size, 3.2 to 6.3 kbp) and paired-end libraries (100-bp-long reads; insert size, 200 to 400 bp) at Baseclear B.V. (Leiden). For DS69473, 31 million read pairs were obtained and for DS69473Evo, 37 million read pairs were obtained. Data were of high quality (average Phred score >35). CLC genomics workbench 7.5.1 (Qiagen) was used to analyze the data. Reads were trimmed at a Phred score of 30, allowing only 2 ambiguous nucleotides per read. Both data sets retained more than 95% of the data after trimming. Mapping was done versus the CEN.PK113-7D public genome (24) downloaded as a GenBank file from NCBI (accession no. PRJNA52955; 70 scaffolds, 12 Mbp assembly) using strict alignment settings. This resulted in 78% (DS69473) and 75% (DS69473Evo) of the data mapped amounting to 300× coverage of the CEN.PK113-7D genome. The public CEN.PK113-7D genome assembly does not contain mitochondrial DNA, which explains the relatively low mapping frequency. The low frequency variant detection from the CLC genomics workbench was used to detect variants using a minimum count of 10 (at least 10 reads needed to support the variant). Since these are haploid strains, the variant frequency (number of reads supporting variant/total coverage at that position) was set to 80%. Variants from DS69473 were compared with DS69473Evo, only variants present in the evolved DS69473Evo were kept. This resulted in 75 variants (31 single nucleotide variants [SNV], 12 MNVs, 17 insertion variants, and 15 deletion variants), of which 33 are in assembly gaps (nucleotide unknown in reference) (Table S1). Of the remaining 42 variants, the mapping was inspected visually; only one variant remained that could potentially cause the improved phenotype of DS69473evo, namely, Tyr353Cys in Cyc8p.
Saturation mutagenesis of CYC8(Y353).
The Cas9 expression vector p414-TEF1p-Cas9-CYCt (Addgene) was cut using the restriction enzymes AdeI and MunI to remove the auxotrophic tryptophan marker. The kanMX marker was amplified (primers listed in Table S4) from a genomic DNA (gDNA) template from the DS68625 strain, was cut with the same restriction enzymes, and was subsequently ligated into plasmid p414-TEF1p-Cas9-CYCt, yielding plasmid p414-KanMX-TEF1p-Cas9-CYCt. The p414-KanMX-TEF1p-Cas9-CYCt plasmid was transformed into the DS69473 strain, and the expression of the CAS9 gene was analyzed using qPCR. Herein, we used the CRISPR/Cas9 protocol described by Mans et al. (40) and pMEL16 to express the guide RNA targeting the CYC8 gene at the Y353 position (Fig. S4) and targeting a PAM site 6 bp in front of the Y353 position. Two repair fragments with degenerated codons were used (Table S4) in which the 353 position was replaced by a WNN or SNN, yielding codons starting with an A/T or C/G, respectively. Transformation to the DS69473 strain yielded all amino acids at position 353 except for E, K, M, W, and F that were obtained separately using specific repair fragments. All mutations were verified after sequencing of the CYC8 gene.
Transcriptome analysis.
Total RNA was isolated in triplicates from both strains after 29 h of growth on 2% d-xylose in the presence of 6% d-glucose. RNA-seq was analyzed on an Ion Proton sequencer (PrimBio, USA) and high-quality read data were obtained for both strains in triplicates with an average read length of 114 bp and an average number of reads of 11.2 M per sample. The FastQ files were run through a BowTie2-TopHat-SamTools pipeline, and the resulting BAM files were analyzed in SeqMonk v0.27.0. The CEN.PK113-7D strain was used as a reference genome. All genes were quantified and run in an intensity difference statistical test in which a statistical difference of below 0.05 was used (P < 0.05).
CYC8 replacement.
To obtain the same Y353C mutation in the original DS69473 strain, CYC8 in the DS69473 strain was replaced with the mutant gene with a kanMX resistance marker in front of the CYC8 promoter region. A fragment of 52 bp was used as a 5′ flanking region and a major part of the CYC8(Y353C) gene was used as a 3′ flanking region (Table S4). The kanMX resistance marker was amplified via PCR with the Phusion high-hidelity PCR master mix in HF buffer, using a forward primer (F kanMX 5′tail) with a 52-bp deletion of the 5′ flanking region (667 to 719 upstream of CYC8) and a reverse primer (R KanMX + CYC8) with a small 22-bp amplification at the 3′ flanking region (645 to 667 upstream of CYC8). The 3′ flanking region was amplified using a forward primer (F kanMX + CYC8), which is the reverse complement of the R kanMX + CYC8 primer, and a reverse primer (R CYC8) which anneals 60 bp behind the Y353C mutation. Both fragments were used in an overlap PCR using only the outside primers (F kanMX 5′tail and R CYC8) to fuse the two fragments together. After transformation of the fused fragment into the DS69473 strain, cells were plated on minimal medium containing 2% d-xylose and G418 (200 mg/liter). Colonies were tested via PCR using primers F CYC8 and R CYC8 (Table S4) and were subsequently sequenced. Sequences were verified for the presence or absence of the Y353C mutation in the CYC8 sequence.
CYC8 deletion.
The kanMX resistance marker was amplified from genomic DNA of the DS68625 strain using the F 5′FR CYC8 kanMX and the R 3′FR CYC8 kanMX primers (Table S4) using the Phusion high-fidelity PCR master mix in HF buffer. The F 5′FR CYC8 kanMX primer contains a 60-bp flanking region homologous to the 5′ upstream region of CYC8, whereas in the R 3′FR CYC8 kanMX primer, this region is homologous to 59 bp downstream of CYC8. DS69473 and DS68616 strains were transformed with the PCR amplicons and colonies were selected on plates containing minimal medium supplemented with vitamin solution, trace elements, G418, and 2% d-xylose (DS69473) or 2% d-maltose (DS68616).
Real-time PCR and primers.
Real-time PCR (qPCR) analysis of the expression of HXT1 to HXT17, GAL2, ADH1, FIT1, and CAS9 was performed with the primers indicated in Table S5 in the supplemental material using the SensiMix SYBR and fluorescein kit (Quantace, Ltd.) and the iCYCLER iQ real-time PCR instrument (Bio-Rad). In all experiments, the gene for actin was used as a reference to normalize fold changes. The SYBR green master mix was used as described before (22).
Cloning of HXT10 and HXT15.
HXT10 and HXT15 were amplified from genomic DNA of the DS69473Evo strain using the primers listed in Table S4 in the supplemental material with the Phusion high-fidelity PCR master mix with HF buffer. The full-length open reading frames (ORFs) of HXT10 and HXT15 were amplified using primers F HXT10 XbaI and R HXT10 Cfr9I and F HXT15 XbaI and HXT15 Cfr9I, respectively, and were cloned into pRS313-P7T7. The vector pRS313-P7T7 was described before (22) and used for the expression of HXTs under the control of the HXT7 promoter. The vector was derived from pRS313 (kindly supplied by DSM Biotechnology Center, the Netherlands) as the backbone containing the histidine selection marker and the CEN/ARS low-copy origin for cloning in yeast.
Uptake measurements.
Uptake experiments were performed as follows. Cells were grown for 24 h at 30°C in shake flasks in minimal medium containing 2% d-xylose and were washed (via centrifugation at 3,000 rpm for 3 min at 20°C) and resuspended in minimal medium without a carbon source. d-[14C]Xylose or d-[14C]glucose stocks (ARC, USA) were added to the cell suspension, and the reaction was stopped after various time intervals by the addition of 5 ml of ice-cold 0.1 M lithium chloride. Samples were filtered through 0.45-μm HV membrane filters (Millipore, France), were washed once with 5 ml of an ice-cold lithium chloride solution, and were counted using a liquid scintillation counter (Perkin-Elmer, USA). To determine the uptake kinetics, the d-xylose and d-glucose concentrations were varied from 0.5 to 500 mM and from 0.1 to 500 mM, respectively. For competition experiments, the uptake of 100 mM d-[14C]xylose was analyzed in the presence of 0 to 800 mM unlabeled d-glucose.
Supplementary Material
ACKNOWLEDGMENTS
The research was financially supported by an EOS long-term grant from the Dutch Ministry of Economical Affairs, Agriculture and Innovation, and by the research program of the biobased ecologically balanced sustainable industrial chemistry (BE-BASIC).
L.G.M.B., P.P.D.W., and P.K. work for the company DSM.
J.G.N., H.Y.S., P.P.D.W., and A.J.M.D. conceived and designed the research, J.G.N. performed the experiments, P.P.D.W. constructed the strains, and P.P.D.W., P.K., and A.J.M.D. supervised the project. All authors contributed to the writing of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00095-17.
REFERENCES
- 1.Solomon BD. 2010. Biofuels and sustainability. Ann N Y Acad Sci 1185:119–134. doi: 10.1111/j.1749-6632.2009.05279.x. [DOI] [PubMed] [Google Scholar]
- 2.Zaldivar J, Nielsen J, Olsson L. 2001. Fuel ethanol production from lignocellulose: a challenge for metabolic engineering and process integration. Appl Microbiol Biotechnol 56:17–34. [DOI] [PubMed] [Google Scholar]
- 3.Carroll A, Somerville C. 2009. Cellulosic biofuels. Annu Rev Plant Biol 60:165–182. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- 4.Kotter P, Ciriacy M. 1993. Xylose fermentation by Saccharomyces cerevisiae. Appl Microbiol Biotechnol 38:776–783. doi: 10.1007/BF00167144. [DOI] [Google Scholar]
- 5.Tantirungkij M, Seki T, Yoshida T. 1994. Genetic improvement of Saccharomyces cerevisiae for ethanol production from xylose. Ann N Y Acad Sci 721:138–147. doi: 10.1111/j.1749-6632.1994.tb47386.x. [DOI] [PubMed] [Google Scholar]
- 6.Kuyper M, Harhangi HR, Stave AK, Winkler AA, Jetten MS, de Laat WT, den Ridder JJ, Op den Camp HJ, van Dijken JP, Pronk JT. 2003. High-level functional expression of a fungal xylose isomerase: the key to efficient ethanolic fermentation of xylose by Saccharomyces cerevisiae? FEMS Yeast Res 4:69–78. doi: 10.1016/S1567-1356(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 7.Kuyper M, Toirkens MJ, Diderich JA, Winkler AA, van Dijken JP, Pronk JT. 2005. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res 5:925–934. doi: 10.1016/j.femsyr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Kuyper M, Winkler AA, van Dijken JP, Pronk JT. 2004. Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res 4:655–664. doi: 10.1016/j.femsyr.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Hamacher T, Becker J, Gardonyi M, Hahn-Hagerdal B, Boles E. 2002. Characterization of the xylose-transporting properties of yeast hexose transporters and their influence on xylose utilization. Microbiology 148(Pt 9):2783–2788. doi: 10.1099/00221287-148-9-2783. [DOI] [PubMed] [Google Scholar]
- 10.von Sivers M, Zacchi G, Olsson L, Hahn-Hagerdal B. 1994. Cost analysis of ethanol production from willow using recombinant Escherichia coli. Biotechnol Prog 10:555–560. doi: 10.1021/bp00029a017. [DOI] [PubMed] [Google Scholar]
- 11.Young EM, Comer AD, Huang H, Alper HS. 2012. A molecular transporter engineering approach to improving xylose catabolism in Saccharomyces cerevisiae. Metab Eng 14:401–411. doi: 10.1016/j.ymben.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Ren C, Chen T, Zhang J, Liang L, Lin Z. 2009. An evolved xylose transporter from Zymomonas mobilis enhances sugar transport in Escherichia coli. Microb Cell Fact 8:66. doi: 10.1186/1475-2859-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du J, Li S, Zhao H. 2010. Discovery and characterization of novel d-xylose-specific transporters from Neurospora crassa and Pichia stipitis. Mol Biosyst 6:2150–2156. doi: 10.1039/c0mb00007h. [DOI] [PubMed] [Google Scholar]
- 14.Hector RE, Qureshi N, Hughes SR, Cotta MA. 2008. Expression of a heterologous xylose transporter in a Saccharomyces cerevisiae strain engineered to utilize xylose improves aerobic xylose consumption. Appl Microbiol Biotechnol 80:675–684. doi: 10.1007/s00253-008-1583-2. [DOI] [PubMed] [Google Scholar]
- 15.Leandro MJ, Goncalves P, Spencer-Martins I. 2006. Two glucose/xylose transporter genes from the yeast Candida intermedia: first molecular characterization of a yeast xylose-H+ symporter. Biochem J 395:543–549. doi: 10.1042/BJ20051465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Runquist D, Hahn-Hagerdal B, Radstrom P. 2010. Comparison of heterologous xylose transporters in recombinant Saccharomyces cerevisiae. Biotechnol Biofuels 3:5. doi: 10.1186/1754-6834-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farwick A, Bruder S, Schadeweg V, Oreb M, Boles E. Engineering of yeast hexose transporters to transport d-xylose without inhibition by d-glucose. Proc Natl Acad Sci U S A 111:5159–5164. doi: 10.1073/pnas.1323464111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young EM, Tong A, Bui H, Spofford C, Alper HS. 2014. Rewiring yeast sugar transporter preference through modifying a conserved protein motif. Proc Natl Acad Sci U S A 111:131–136. doi: 10.1073/pnas.1311970111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kruckeberg AL. 1996. The hexose transporter family of Saccharomyces cerevisiae. Arch Microbiol 166:283–292. doi: 10.1007/s002030050385. [DOI] [PubMed] [Google Scholar]
- 20.Boles E, Hollenberg CP. 1997. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev 21:85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 21.Saloheimo A, Rauta J, Stasyk OV, Sibirny AA, Penttila M, Ruohonen L. 2007. Xylose transport studies with xylose-utilizing Saccharomyces cerevisiae strains expressing heterologous and homologous permeases. Appl Microbiol Biotechnol 74:1041–1052. doi: 10.1007/s00253-006-0747-1. [DOI] [PubMed] [Google Scholar]
- 22.Nijland JG, Shin HY, de Jong RM, de Waal PP, Klaassen P, Driessen AJ. 2014. Engineering of an endogenous hexose transporter into a specific d-xylose transporter facilitates glucose-xylose co-consumption in Saccharomyces cerevisiae. Biotechnol Biofuels 7:168. doi: 10.1186/s13068-014-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reznicek O, Facey SJ, de Waal PP, Teunissen AWRH, de Bont JAM, Nijland JG, Driessen AJM, Hauer B. 2015. Improved xylose uptake in Saccharomyces cerevisiae due to directed evolution of galactose permease Gal2 for sugar co-consumption. J Appl Microbiol 119:99–111. doi: 10.1111/jam.12825. [DOI] [PubMed] [Google Scholar]
- 24.Nijkamp JF, van den Broek M, Datema E, de Kok S, Bosman L, Luttik MA, Daran-Lapujade P, Vongsangnak W, Nielsen J, Heijne WHM, Klaassen P, Paddon CJ, Platt D, Kötter P, van Ham RC, Reinders MJT, Pronk JT, de Ridder D, Daran J-M. 2012. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113-7D, a model for modern industrial biotechnology. Microb Cell Fact 11:36. doi: 10.1186/1475-2859-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleming AB, Beggs S, Church M, Tsukihashi Y, Pennings S. 2014. The yeast Cyc8-Tup1 complex cooperates with Hda1p and Rpd3p histone deacetylases to robustly repress transcription of the subtelomeric FLO1 gene. Biochim Biophys Acta 1839:1242–1255. doi: 10.1016/j.bbagrm.2014.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zha J, Shen M, Hu M, Song H, Yuan Y. 2014. Enhanced expression of genes involved in initial xylose metabolism and the oxidative pentose phosphate pathway in the improved xylose-utilizing Saccharomyces cerevisiae through evolutionary engineering. J Ind Microbiol Biotechnol 41:27–39. doi: 10.1007/s10295-013-1350-y. [DOI] [PubMed] [Google Scholar]
- 27.Tzamarias D, Struhl K. 1995. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev 9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 28.Smith RL, Johnson AD. 2000. Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci 25:325–330. doi: 10.1016/S0968-0004(00)01592-9. [DOI] [PubMed] [Google Scholar]
- 29.Nehlin JO, Ronne H. 1990. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J 9:2891–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Treitel MA, Carlson M. 1995. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci U S A 92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeRisi JL, Iyer VR, Brown PO. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 32.Chujo M, Yoshida S, Ota A, Murata K, Kawai S. 2015. Acquisition of the ability to assimilate mannitol by Saccharomyces cerevisiae through dysfunction of the general corepressor Tup1-Cyc8. Appl Environ Microbiol 81:9–16. doi: 10.1128/AEM.02906-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipke PN, Hull-Pillsbury C. 1984. Flocculation of Saccharomyces cerevisiae tup1 mutants. J Bacteriol 159:797–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong KH, Struhl K. 2011. The Cyc8-Tup1 complex inhibits transcription primarily by masking the activation domain of the recruiting protein. Genes Dev 25:2525–2539. doi: 10.1101/gad.179275.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordan P, Choe J-Y, Boles E, Oreb M. 2016. Hxt13, Hxt15, Hxt16 and Hxt17 from Saccharomyces cerevisiae represent a novel type of polyol transporters. Sci Rep 6:23502. doi: 10.1038/srep23502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toivari MH, Salusjärvi L, Ruohonen L, Penttilä M. 2004. Endogenous xylose pathway in Saccharomyces cerevisiae. Appl Environ Microbiol 70:3681–3686. doi: 10.1128/AEM.70.6.3681-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Traff KL, Otero Cordero RR, van Zyl WH, Hahn-Hagerdal B. 2001. Deletion of the GRE3 aldose reductase gene and its influence on xylose metabolism in recombinant strains of Saccharomyces cerevisiae expressing the xylA and XKS1 genes. Appl Environ Microbiol 67:5668–5674. doi: 10.1128/AEM.67.12.5668-5674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin HY, Nijland JG, de Waal PP, de Jong RM, Klaassen P, Driessen AJM. 2015. An engineered cryptic Hxt11 sugar transporter facilitates glucose-xylose co-consumption in Saccharomyces cerevisiae. Biotechnol Biofuels 8:176. doi: 10.1186/s13068-015-0360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luttik MA, Kötter P, Salomons FA, van der Klei IJ, van Dijken JP, Pronk JT. 2000. The Saccharomyces cerevisiae ICL2 gene encodes a mitochondrial 2-methylisocitrate lyase involved in propionyl-coenzyme A metabolism. J Bacteriol 182:7007–7013. doi: 10.1128/JB.182.24.7007-7013.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mans R, van Rossum HM, Wijsman M, Backx A, Kuijpers NGA, van den Broek M, Daran-Lapujade P, Pronk JT, van Maris AJA, Daran J-MG. 2015. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res 15:fov004–fov004. doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


