Abstract
Mitochondria are the main consumers of molecular O2 in a cell as well as an abundant source of reactive oxygen species (ROS). Both, molecular oxygen and ROS are powerful regulators of the hypoxia-inducible factor-1α-subunit (HIF-α). While a number of mechanisms in the oxygen-dependent HIF-α regulation are quite well known, the view with respect to mitochondria is less clear. Several approaches using pharmacological or genetic tools targeting the mitochondrial electron transport chain (ETC) indicated that ROS, mainly formed at the Rieske cluster of complex III of the ETC, are drivers of HIF-1α activation. However, studies investigating non-ETC located mitochondrial defects and their effects on HIF-1α regulation are scarce, if at all existing. Thus, in the present study we examined three cell lines with non-ETC mitochondrial defects and focused on HIF-1α degradation and transcription, target gene expression, as well as ROS levels. We found that cells lacking the key enzyme 2-enoyl thioester reductase/mitochondrial enoyl-CoA reductase (MECR), and cells lacking manganese superoxide dismutase (MnSOD) showed a reduced induction of HIF-1α under long-term (20 h) hypoxia. By contrast, cells lacking the mitochondrial DNA depletion syndrome channel protein Mpv17 displayed enhanced levels of HIF-1α already under normoxic conditions. Further, we show that ROS do not exert a uniform pattern when mediating their effects on HIF-1α, although all mitochondrial defects in the used cell types increased ROS formation. Moreover, all defects caused a different HIF-1α regulation via promoting HIF-1α degradation as well as via changes in HIF-1α transcription. Thereby, MECR- and MnSOD-deficient cells showed a reduction in HIF-1α mRNA levels whereas the Mpv17 lacking cells displayed enhanced HIF-1α mRNA levels under normoxia and hypoxia. Altogether, our study shows for the first time that mitochondrial defects which are not related to the ETC and Krebs cycle contribute differently to HIF-1α regulation by affecting HIF-1α degradation and HIF-1α transcription where ROS play not a major role.
Keywords: Hypoxia, HIF-1α, Mitochondrial defects, MFAS II, Mpv17, MnSOD
Graphical abstract
Highlights
-
•
non-ETC located mitochondrial defects promote HIF-1α degradation.
-
•
differently affected HIF-1α transcription accounts for the effects of non-ETC defects.
-
•
enhanced ROS are not primarily involved in the regulation of HIF-1α by non-ETC mitochondrial defects.
1. Introduction
Oxygen homeostasis is essential for survival and normal function of all aerobic living cells and organisms. Adaptation to low oxygen levels involves a wide range of responses at different cellular levels. Among the first line responses to hypoxia is the reprogramming of gene expression by activation of hypoxia-inducible factors (HIFs). HIFs are heterodimers, containing an oxygen-dependent α-subunit (HIF-1α, HIF-2α and HIF-3α) and a constitutively expressed β-subunit (HIF-1β or aryl hydrocarbon receptor nuclear translocator (ARNT) [1]. From the three α-subunits known to date, HIF-1α is the best characterized. Its regulation occurs primarily at the level of protein stability; under normal oxygen tension HIF-1α is hydroxylated at two specific residues (P402 and P564) which allows binding of a VHL-containing ubiquitin ligase complex that mediates degradation via the ubiquitin/ proteasome pathway [2], [3]. The hydroxylation is carried out by a family of proline hydroxylases which are dependent on the availability of molecular oxygen, Fe2+ and α-ketoglutarate [4]. Under low oxygen levels the HIF-1α-subunit is stabilized and translocates to the nucleus where it regulates more than 800 target genes [5], [6]. Interestingly, the levels of HIF-α subunits are not only affected by oxygen, but also by reactive oxygen species (ROS). One major source of ROS are mitochondria which are important organelles involved in a variety of cellular processes, including ATP production, calcium homeostasis, as well as cell survival and cell death [7], [8].
Mitochondria consume oxygen in order to synthesize ATP via oxidative phosphorylation (OXPHOS). The OXPHOS machinery consists of five multi-subunit enzyme complexes (complex I, II, III, IV and V), which are located in the inner mitochondrial membrane [9]. The electrons generated in the TCA cycle and which are donated by the coenzymes, NADH and FADH2 are passing through the electron transport chain (ETC) where oxygen functions as a final acceptor. However, if oxygen does not become fully reduced to water, ROS are generated as a byproduct of this electron transport process [10]. Several reports using pharmacological or genetic approaches targeting the ETC indicated that ROS, mainly formed at the Rieske cluster of complex III of the ETC, are drivers of HIF-1α activation [11], [12], [13], [14], [15], [16]. However, the approaches targeting the ETC are expected not only to interfere with ROS production, but also to interfere with mitochondrial oxygen consumption and thus, make it difficult to estimate to which extent ROS production or as proposed a shift in oxygen distribution [17] accounts for the effects on HIF-1α.
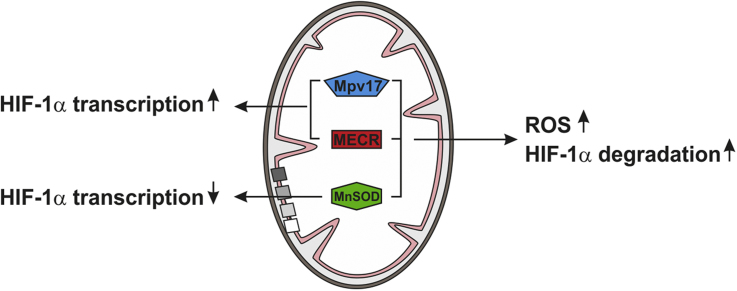
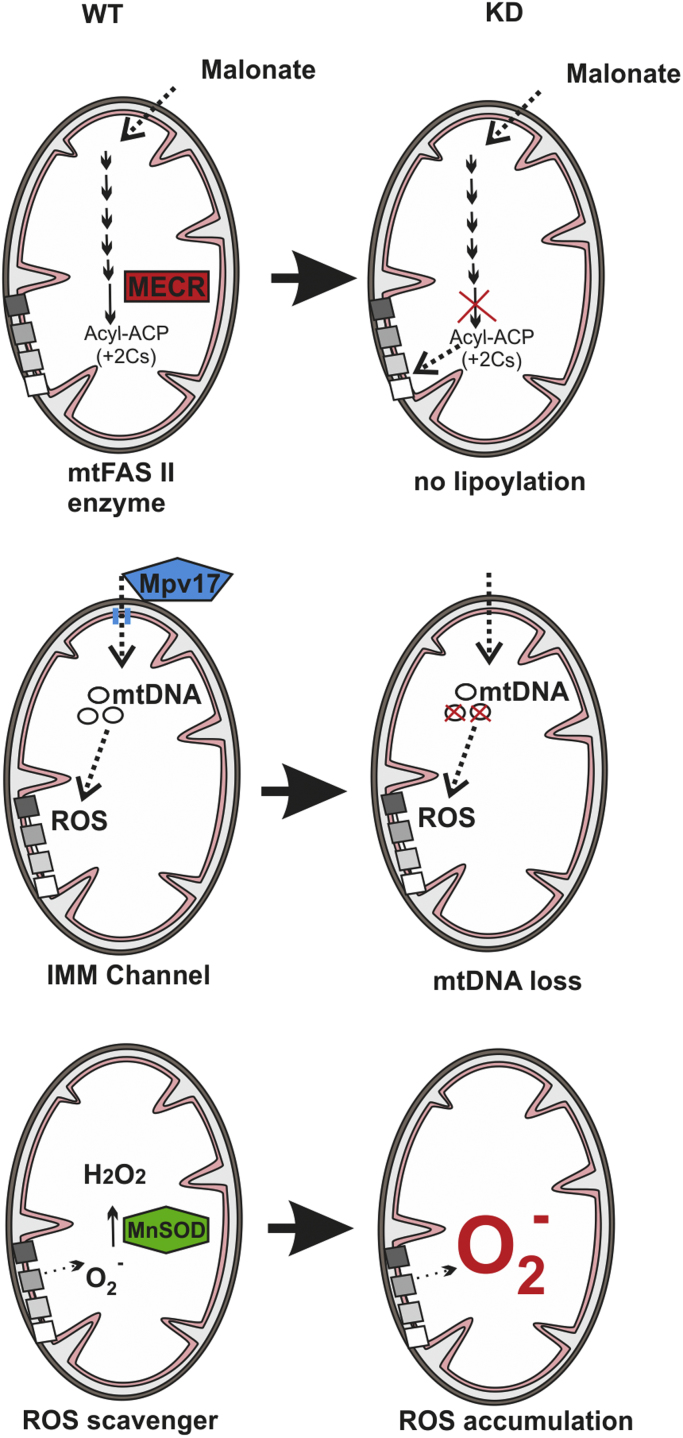
To better understand the mitochondria´s contribution to HIF-1α regulation we hypothesized that non-ETC and non-Krebs cycle mitochondrial defects may have different or even no effect on HIF-1α levels. Therefore, we investigated in the present work three cell lines with such mitochondrial defects with respect to HIF-1α regulation, mitochondrial membrane potential, ROS production, and HIF target gene expression (Fig. 1). The first cell line under study was devoid of 2-enoyl thioester reductase/mitochondrial enoyl-CoA reductase (MECR). MECR is a component of the fatty acid synthesis type II (FAS II) complex and catalyzes the NADPH-dependent reduction of trans-2-enoyl thioesters. Loss-of-function mutations in the MECR gene cause childhood-onset dystonia and optic atrophy [18]. The second cell line used was lacking Mpv17, a mitochondrial inner membrane channel forming protein whose dysfunction causes hepatocerebral mitochondrial DNA depletion syndrome including Navajo neurohepatopathy [19], [20]. The third cell line under study was depleted of manganese superoxide dismutase (MnSOD). MnSOD is the second (SOD2) of three superoxide dismutases; in contrast to the cytosolic (SOD1), and the extracellular (SOD3), it is found in the mitochondrial matrix. All SODs are responsible for the dismutation of superoxide anion radicals to hydrogen peroxide and oxygen. Importantly, deficiency of MnSOD in several cell types and in organs of mice was shown to contribute to carcinogenesis [21], [22].
Fig. 1.
Scheme of investigated mitochondrial defects and functional consequences. Acyl-ACP, acetyl-(acyl-carrier-protein); MECR, mitochondrial trans-E-enoyl-CoA reductase; mtFASII, mitochondrial fatty acid synthase system; MnSOD, manganese superoxide dismutase; ROS, reactive oxygen species.
Our study shows for the first time that mitochondrial defects which are not related to the ETC and Krebs cycle enzymes contribute differently to HIF-1α regulation by affecting HIF-1α degradation and HIF-1α transcription with ROS being rather indirectly involved.
2. Material and methods
All biochemicals and enzymes were of analytical grade and were obtained from commercial suppliers.
2.1. Cell culture
NIH 3T3 were stably transfected with a MECR-shRNA expressing vector and a respective scrambled control vector and cultured under normoxia (16% O2, 79% N2 and 5% CO2 [by volume]) in Dulbecco´s modified Eagle´s medium (DMEM) supplemented with 10% fetal calf serum (FCS) in an Invivo2 400 hypoxia workstation (Ruskin Technologies). HepG2-scr and HepG2-MnSOD-KD cells were previously described and cultured in minimal essential medium (MEM) supplemented with 10% FCS [21]. Mouse embryonic fibroblasts (MEFs) Mpv17+/+ and Mpv17-/- cells were cultured in DMEM supplemented with 10% FCS as described previously [20]. For protein extraction, cells were seeded onto 10 cm dishes and cultured under either normoxia or hypoxia (1% O2, 94% N2 and 5% CO2 [by volume]) in an Invivo2 400 hypoxia workstation (Ruskin Technologies) for 20 h and then harvested.
2.2. Western blot analyses and HIF-1α protein half-life studies
Western blot analyses were carried out as described previously [23]. In brief, lysates from NIH 3T3-scr, NIH 3T3-MECR-KD, HepG2-scr, HepG2-MnSOD-KD, Mpv17+/+ and Mpv17-/- cells were collected and 100 µg of total protein was loaded onto a 7.5% or 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel. After electrophoresis and electroblotting onto a nitrocellulose membrane, proteins were detected with primary antibodies against human HIF-1α (1:2000; BD Bioscience), mouse HIF-1α (1:1000; Novus Biologicals), MnSOD (SOD2) (1:1000; Calbiochem), MECR (1:1000; Proteintech), and against α-tubulin (1:10.000; Sigma). The secondary antibody was either an anti-mouse, an anti-rabbit or an anti-sheep immunoglobulin G conjugated to horseradish peroxidase (1:5000; Bio-Rad Laboratories). The ECL system (Amersham) was used for detection.
For half-life studies, NIH 3T3-scr, NIH 3T3-MECR-KD, HepG2-scr, HepG2-MnSOD-KD, Mpv17+/+ and Mpv17-/- cells were cultured under normoxic or hypoxic conditions. After 20 h, cycloheximide (10 µg/ml; Sigma) was added to the cell culture medium, cells were scraped in lysis buffer (50 mM Tris/HCL, pH 7.5, 150 mM NaCl, 1% Triton X-100, 2 mM EDTA, 2 mM EGTA, 1 mM PMSF and complete protease inhibitor cocktail tablet (Roche)) at indicated time points and protein levels were measured by immunoblot analysis.
2.3. TMRE mitochondrial staining
NIH 3T3-scr, NIH 3T3-MECR-KD, HepG2-scr, HepG2-MnSOD-KD, Mpv17+/+ and Mpv17-/- cells were seeded on a 96-well plate at a density of 15,000 cells/well and cultured under normoxic or hypoxic conditions. After 20 h, the cells were washed with 1×PBS and stained with 1 µM of tetramethyl rhodamine ester (TMRE, Abcam) for 30 min. Although these conditions of TMRE usage are considered as being the quenching mode, dinitrophenol (DNP) reduced the signal indicating validity of the assay. After incubation, cells were washed twice with 0.2% BSA in 1 x PBS in the hypoxia workstation where the 96-well plates were sealed with parafilm and the fluorescence was immediately measured using a Tecan infinite M1000Pro microplate reader at an excitation wavelength of 549 nm and an emission wavelength of 575 nm.
2.4. ROS detection
The total ROS content of cells was assessed using 2′,7′-dichlorodihydrofluorescein diacetate (H2DCF-DA), whereas DHE was used to measure superoxide levels. Cells were seeded onto a 24-well plate at a density of 10,000 cells per well and cultured either under normoxia or hypoxia. After 20 h, cells were washed with 1 x PBS and incubated with 10 μM H2DCF-DA (Molecular Probes) or 10 μM DHE (Sigma) in 1 x PBS for 15 min at 37 °C. After replacement of the reactive agents with 1 x PBS in the hypoxia workstation and sealing of the plate with parafilm, DCF formation was recorded at λex/λem: 495/529 nm and DHE fluorescence at λex/λem: 530/610 nm using a Tecan infinite M1000Pro microplate reader. Values were corrected for H2DCF-DA and DHE auto-oxidation in 1 x PBS.
2.5. RNA preparation and reverse transcription
Isolation of total RNA was performed by using the RNeasy Mini Kit (Qiagen) following the manufacturer's instructions. Total RNA concentration and purity were measured on a NanoDrop spectrophotometer. One μg of total RNA was used for cDNA synthesis using SuperScript II Reverse Transcriptase (Invitrogen).
2.6. Quantitative real-time PCR
qRT-PCR was performed with the iTaq SYBR Green Supermix in an Applied Biosystems 7500 Real-Time PCR System (Life Technologies, Finland). The following primer sets were used: hLDHA-F (5´- TTGACCTACGTGGCTTGGAAG-3´), hLDHA-R (5´-GGTAACGGAATCGGGCTGAAT-3´), mLDHA-F (5´-ACATTGTCAAGTACAGTCCACAC-3´), mLDHA-R (5´-TTCCAATTACTCGGTTTTTGGGA-3´), hGLUT1-F (5´-TCTGGCATCAACGCTGTCTTC-3´), hGLUT1-R (5´-CGATACCGGAGCCAATGGT-3´), mGLUT1-F (5´-CTCTGTCGGCCTCTTTGTTAAT-3´), mGLUT1-R (5´- CCAGTTTGGAGAAGCCCATAAG-3´), hHIF-1α-F (5´-ATCCATGTGACCATGAGGAAATG-3´), hHIF-1α-R (5´-TCGGCTAGTTAGGGTACACTTC-3´), mHIF-1α-F (5´-GGGGAGGACGATGAACATCAA-3´), mHIF-1α-R (5´-GGGTGGTTTCTTGTACCCACA-3´), hPGC1-F (5´-TGACTGGCGTCATTCAGGAG-3´), hPGC1-R (5´-CCAGAGCAGCACACTCGAT-3), mPGC1-F (5´-TGACTGGCGTCATTCGGGAG-3´), mPGC1-R (5´-CCAGAGCAGCACACTCTAT-3´) hHPRT-F (5´-CCTGGCGTCGTGATTAGTGAT-3´), hHPRT-R (5´-AGACGTTCAGTCCTGTCCATAA -3´), mHPRT-F (5´-CGAAGTGTTGGATACAGGCC-3´), mHPRT-R (5´-GGCAACATCAACAGGACTCC-3´). The experiments for each data point were carried out in triplicate. The relative quantification of gene expression was determined using the ΔΔCt method [24].
2.7. Statistical analysis
Densitometry data were plotted as fold induction of relative density units, with the zero value absorbance in each figure set arbitrarily to 1 or 100%. Statistical comparisons of absorbance differences were performed by the Mann-Whitney test (Statview 4.5, Abacus Concepts, Berkeley, CA), and p values p<0.05 were considered significant. All data are shown as means ±SD of at least three independent experiments.
3. Results
3.1. MECR, Mpv17, and MnSOD deficiency influence mitochondrial membrane potential, and ROS formation
In order to investigate whether the three different non-ETC mitochondrial defects such as loss of MECR, Mpv17, and MnSOD (Fig. 1) affect mitochondrial function and ROS levels we cultured the respective deficient cells along with their normal counterparts under normoxia and hypoxia and measured the mitochondrial membrane potential as a parameter of mitochondrial function, and ROS levels.
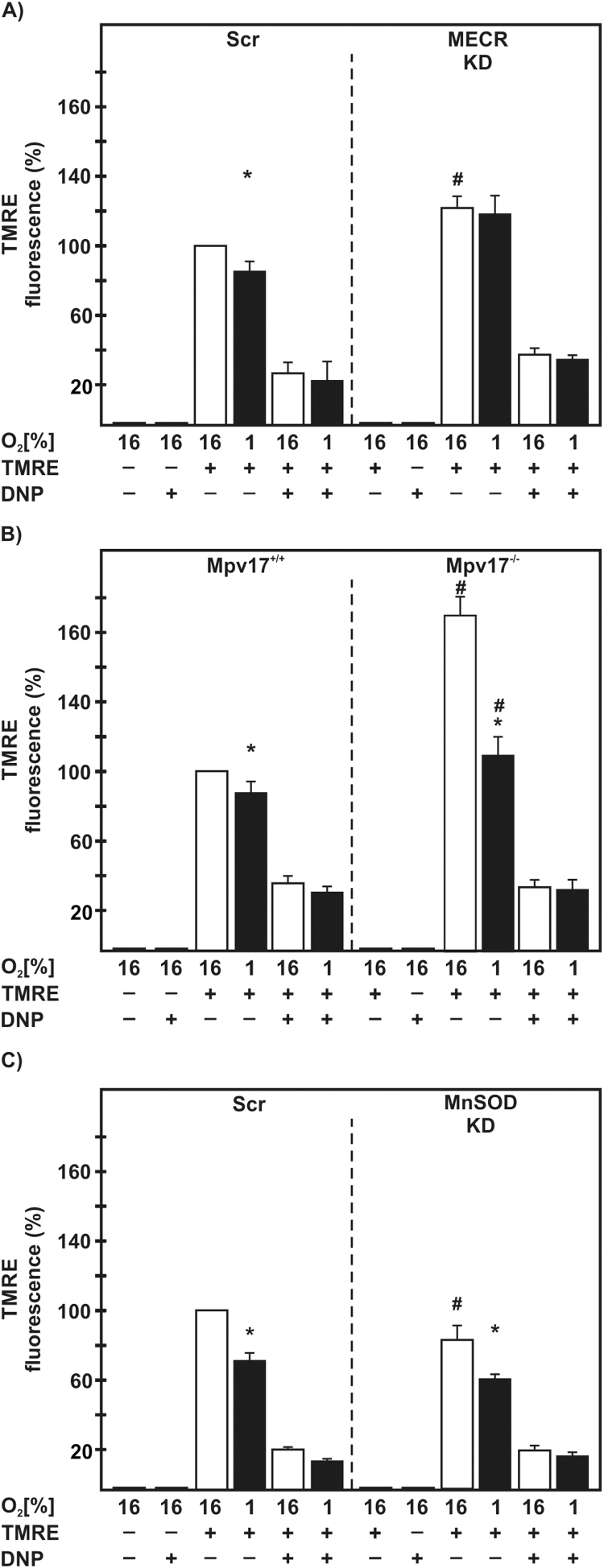
To assess the mitochondrial membrane potential, we first labeled the intact and active mitochondria with the fluorescent dye tetramethylrhodamine ethyl ester (TMRE) and found that hypoxia reduced the TMRE signal in all control cells (Fig. 2). Interestingly, loss of MECR induced the TMRE signal and abolished the difference caused by hypoxia. Loss of Mpv17 also increased the TMRE signal; by about 60% under normoxia in line with previous reports [20] and by about 20% under hypoxia. In contrast, cells with a loss of MnSOD showed a similar reduction of the TMRE signal under both normoxia and hypoxia (Fig. 2).
Fig. 2.
MECR-, Mpv17-, and MnSOD-deficiency influences the mitochondrial membrane potential. (A) NIH 3T3-scr, MECR-KD, (B) Mpv17+/+, Mpv17-/-, (C) HepG2-scr and MnSOD-KD cells were cultured under normoxia (16% O2) and hypoxia (1% O2) for 20 h. Mitochondrial membrane potential (ΔΨm) was assessed by TMRE fluorescence; dinitrophenol (DNP 50 µM) was added when indicated to uncouple the respiratory chain. Results are shown as percentage normalized to the control (16% O2). * significant difference 16% O2 versus 1% O2, # significant difference NIH 3T3-scr versus MECR-KD, Mpv17+/+ versus Mpv17-/-, and HepG2-scr versus MnSOD-KD at 16% O2; p <0.05.
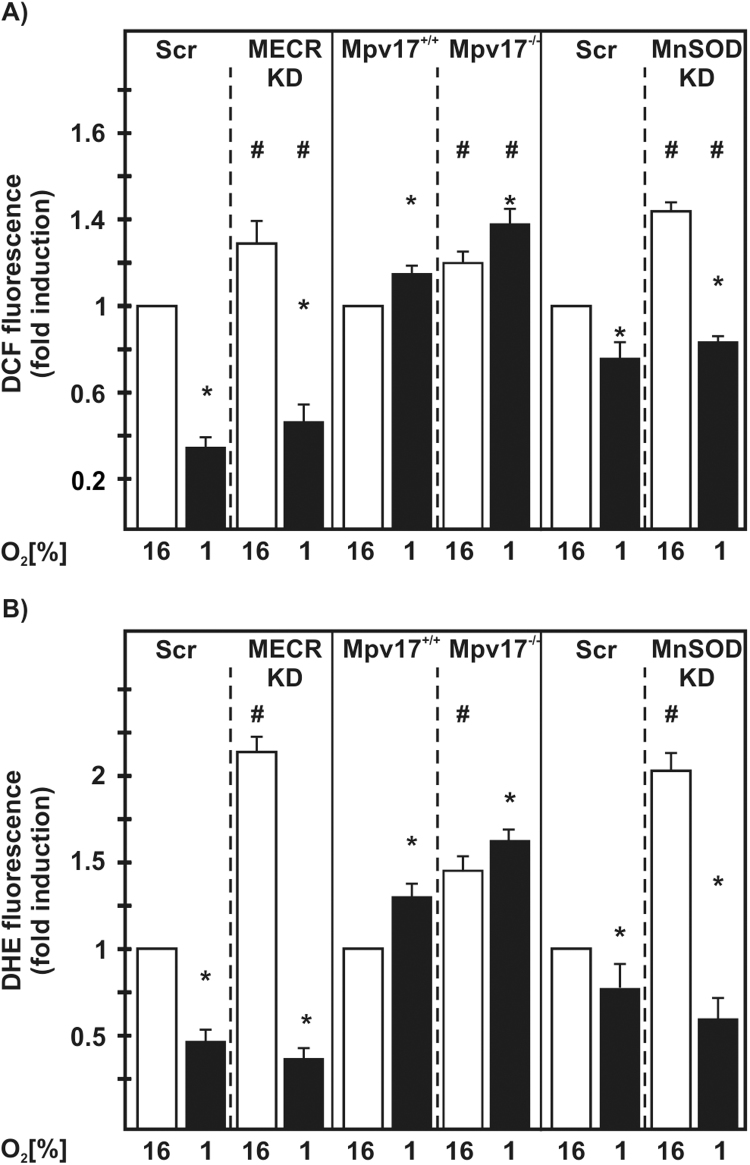
To assess whether the loss of MECR, Mpv17 or MnSOD affect the mitochondrial ROS levels, we labeled the cells with either dichlorodihydrofluorescein diacetate (DCF), to measure overall ROS levels, or dihydroethidium (DHE) to measure superoxide. When we compared ROS levels under normoxia with ROS levels under hypoxia, we found that hypoxia reduced ROS levels in MECR-Scr and MECR-KD cells as well as in MnSOD-Scr and MnSOD-KD cells. By contrast, hypoxia increased ROS in Mpv17+/+ and Mpv17-/- cells. However, all cells with mitochondrial defects displayed higher ROS levels than the respective normal control cells either under normoxia or hypoxia (Fig. 3A).
Fig. 3.
Mitochondrial ROS production is affected by hypoxia and mitochondrial defects. NIH 3T3-scr, MECR-KD, HepG2-scr, MnSOD-KD, Mpv17+/+, and Mpv17-/- cells were cultured under normoxia (16% O2) and hypoxia (1% O2) for 20 h. The redox state was determined by measuring (A) DCF fluorescence, and (B) DHE fluorescence. * significant difference 16% O2 versus 1% O2, # significant difference NIH 3T3-scr versus MECR-KD, Mpv17+/+ versus Mpv17-/-, and HepG2-scr versus MnSOD-KD at 16% O2; p<0.05.
Similar to the ROS levels, we found that hypoxia reduced DHE fluorescence indicating superoxide in MECR-Scr and MECR-KD cells as well as in MnSOD-Scr and MnSOD-KD cells. Again, hypoxia increased DHE fluorescence in Mpv17+/+ and Mpv17-/- cells under normoxia and hypoxia. Further, the cells harboring the mitochondrial defects showed enhanced DHE fluorescence under normoxia but not under hypoxia when compared to the respective control cells. (Fig. 3B). Together, these results suggest that all the mitochondrial defects affect ROS formation under normoxia and hypoxia.
3.2. Loss of MECR, Mpv17 and MnSOD affect HIF-1α protein levels with ROS playing different roles
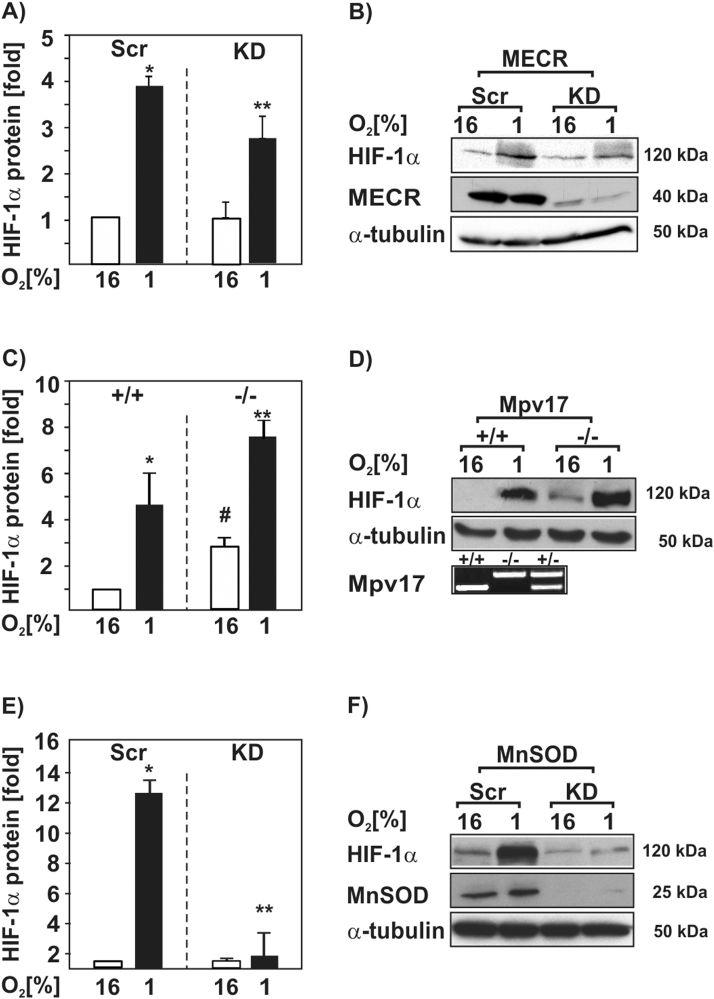
We next wanted to know whether loss of MECR, Mpv17, and MnSOD have an influence on HIF-1α protein levels and whether ROS may be involved in this regulation. To do this, we cultured the respective deficient cells along with their normal counterparts under normoxia and hypoxia for 20 h and measured HIF-1α protein levels by Western blot analysis. We found that hypoxia induced the HIF-1α protein levels in all three control cell lines (Fig. 4). Cells with a knockdown of MECR as well as cells with a knockdown of MnSOD showed reduced HIF-1α protein levels under hypoxia (Fig. 4A,B,E,F). By contrast, knockout of Mpv17 increased HIF-1α protein levels by about 2.5-fold under normoxia and by about 7.5-fold under hypoxia (Fig. 4C,D). Together, these results indicate that lack of MECR, Mpv17, and MnSOD can modulate HIF-1α protein levels.
Fig. 4.
Loss of MECR, Mpv17, and MnSOD affects HIF-1α protein levels. (A,C,E) The shRNA-depleted NIH 3T3 MECR (MECR KD) or HepG2 MnSOD (MnSOD KD) cells and their respective scrambled controls (Scr) as well as Mpv17+/+ and Mpv17-/- mouse embryonic fibroblasts were cultured under normoxia (16% O2) and hypoxia (1% O2) for 20 h. The HIF-1α protein levels measured by Western blot under normoxia (16% O2) were set to 1. *, significant difference between control 16% O2 versus control 1% O2; #, significant difference between control 16% O2 versus KD 16% O2; **, significant difference between control 1% O2 versus KD 1% O2; p <0.05 (B,D,F) Representative Western blot analysis. 100 µg of total protein lysates were analyzed with antibodies against HIF-1α, MECR, MnSOD and α-tubulin. Genotyping of isolated genomic DNA from mouse embryonic fibroblasts indicate lack of Mpv17.
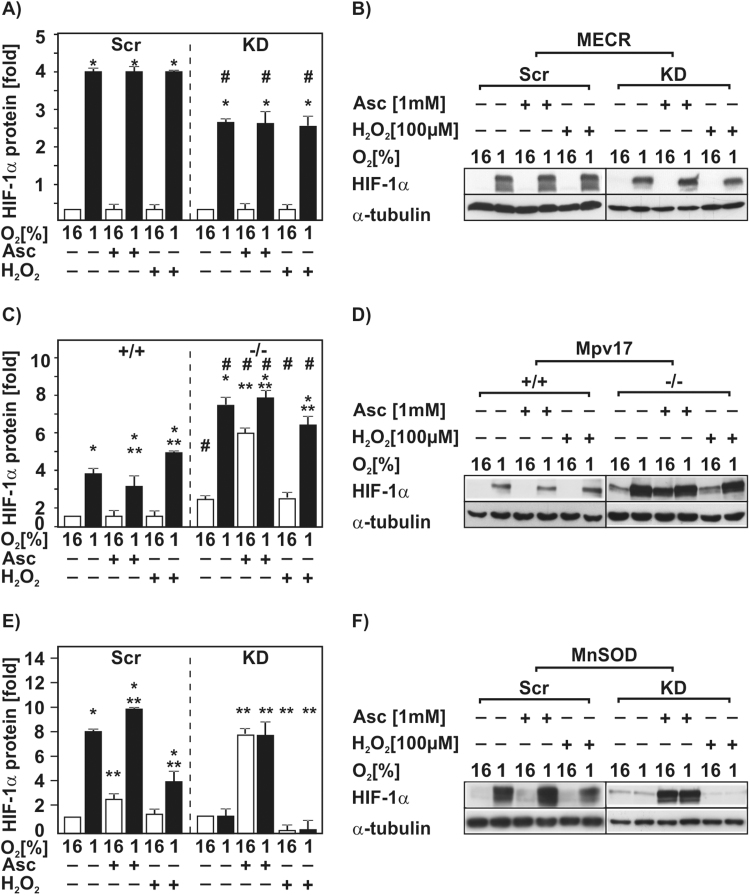
To further investigate whether the changed ROS levels in these cells affect HIF-1α protein levels, we treated the cells with the antioxidant ascorbate, and the oxidant hydrogen peroxide (H2O2). In the MECR-Scr and MECR-KD cells neither treatment with ascorbate nor with H2O2 had an effect on HIF-1α protein levels under both normoxia and hypoxia (Fig. 5A). In Mpv17+/+ cells treatment with ascorbate let to a slight reduction in the HIF-1α protein levels by about 30% under hypoxia compared to the untreated cells under hypoxia, whereas treatment with H2O2 induced the HIF-1α protein levels by about 1.3-fold, but only under hypoxia (Fig. 5C). In contrast, ascorbate induced the HIF-1α protein levels in Mpv17-/- cells only under normoxia. Treatment of the Mpv17-/- cells with H2O2 had no effect on HIF-1α protein levels under normoxic and hypoxic conditions compared to the untreated control (Fig. 5C). In MnSOD-Scr cells ascorbate treatment induced HIF-1α under normoxia by about 2-fold and under hypoxia by about 10-fold. Treatment with H2O2 let to a reduction by about 50% under hypoxia (Fig. 5E). Importantly, knockdown of MnSOD abolished the hypoxia-dependent HIF-1α induction. Ascorbate treatment of the MnSOD-KD cells strongly increased the HIF-1α protein levels by about 7-fold under both normoxia and hypoxia. Upon exposure of these cells to H2O2 almost no HIF-1α protein could be detected (Fig. 5E). Thus, these findings indicate that H2O2 and ascorbate can influence the HIF-1α protein levels differently in the used cells with non-ETC defects.
Fig. 5.
H2O2 and ascorbate influence HIF-1α protein levels. NIH 3T3-scr, MECR-KD, Mpv17+/+, Mpv17-/-, HepG2-scr and MnSOD-KD cells were cultured under normoxic (16% O2) or hypoxic (1% O2) conditions for 20 h and then treated with ascorbate (Asc) or H2O2 for 4 h. (A,C,E) In each experiment the untreated HIF-1α protein levels were set to 1. * significant difference 16% O2 versus 1% O2, ** significant difference respective 16% O2 or 1%O2 control versus respective treatment with H2O2 or ascorbate, # significant difference NIH 3T3-scr versus MECR-KD, Mpv17+/+ versus Mpv17-/-, and HepG2-scr versus MnSOD-KD; p<0.05. (B,D,F) Representative Western blot analysis. One hundred micrograms of total protein were analyzed with antibodies against HIF-1α and α-tubulin.
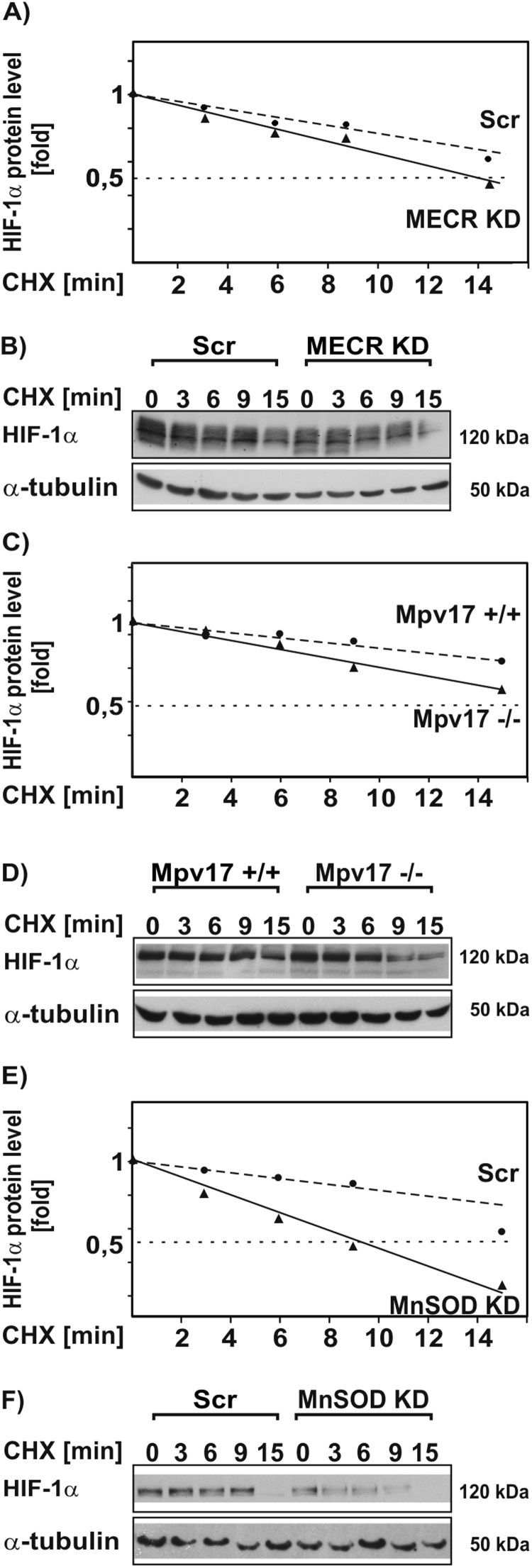
3.3. Loss of MECR, Mpv17, and MnSOD reduces HIF-1α half-life
Since HIF-1α is primarily regulated at the level of protein stability, we were interested to see whether MECR, Mpv17, and MnSOD deficiency affects the half-life of HIF-1α. Therefore, we cultured cells for 20 h under hypoxic conditions (1% O2) to stabilize HIF-1α. After treatment with cycloheximide (CHX) for several indicated time points, we collectively found that deficiency of MECR, Mpv17, and MnSOD reduced the HIF-1α protein levels and shortened the half-life with the strongest effect in the MnSOD-KD cells (Fig. 6A-F). Together, these data show that all non-ETC mitochondrial defects reduce the half-life of HIF-1α.
Fig. 6.
Loss of MECR, Mpv17, and MnSOD affect HIF-1α protein stability. (A,C,E) NIH 3T3-scr, MECR-KD, HepG2-scr, MnSOD-KD, Mpv17+/+, and Mpv17-/- cells were cultured for 20 h under hypoxia (1% O2). After inhibition of protein synthesis with cycloheximide (CHX; 10 µg/ml) HIF-1α protein levels were measured by Western blot analysis at the indicated time points. The HIF-1α protein levels without CHX were set to 1. *, significant difference, p≤0.05. (B,D,F) Representative immunoblots. One hundred micrograms of total protein were analyzed with antibodies against HIF-1α and α-tubulin.
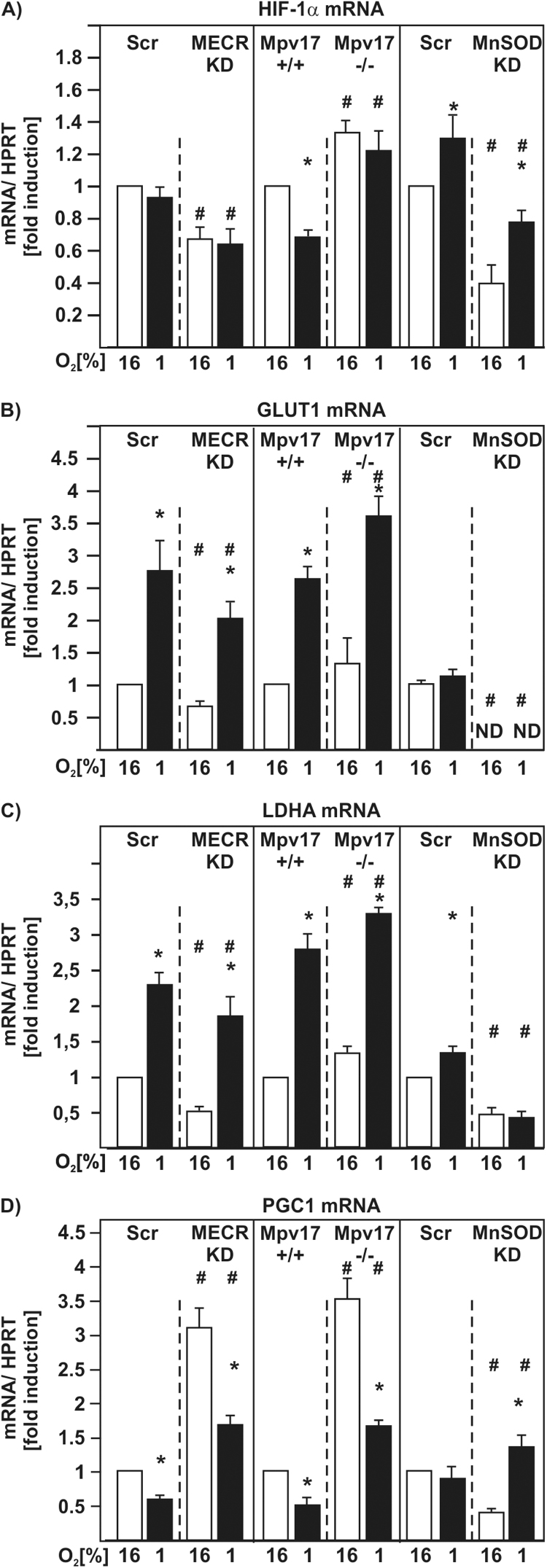
3.4. Loss of MECR, Mpv17, and MnSOD affect HIF-1α mRNA levels and HIF-1α target gene expression
The above made observations and the unexpected regulatory pattern achieved with respect to ROS-dependent HIF-1α regulation made us asking to what extent HIF-1α mRNA levels would be affected by the loss of MECR, Mpv17, and MnSOD. Accordingly, we analyzed HIF-1α mRNA levels by quantitative real-time PCR (qRT-PCR). The obtained data indicated that there were no significant changes in the HIF-1α mRNA levels in MECR-Scr cells under hypoxia. However, HIF-1α mRNA levels were reduced in MECR-KD cells by about 40% under both normoxia and hypoxia (Fig. 7A). In Mpv17+/+ cells hypoxia let to a reduction in the HIF-1α mRNA levels by about 40%. In contrast, loss of Mpv17 let to an increase of HIF-1α mRNA levels by about 30% under normoxia and by about 25% under hypoxia (Fig. 7A). The MnSOD-Scr cells had about 30% induced HIF-1α mRNA levels in response to hypoxia. In contrast, MnSOD-KD cells showed a significant reduction in HIF-1α mRNA levels by about 60% under normoxia and by 20% under hypoxia (Fig. 7A). Next we investigated the mRNA levels of two specific HIF-1α target genes, glucose transporter 1 (GLUT1) and lactate dehydrogenase A (LDHA), as well as those of master regulator for mitochondrial biogenesis peroxisome proliferator-activated receptor-gamma coactivator (PGC)−1alpha (PGC-1α). As expected hypoxia induced the mRNA levels of GLUT1 and LDHA in MECR-Scr, Mpv17 wild-type- and MnSOD-Scr cells. Surprisingly, there were no significant changes detectable in the mRNA levels of GLUT1 under hypoxia in the MnSOD-Scr cells (Fig. 7B,C). In line with the reduced HIF-1α levels, knockdown of MECR significantly reduced GLUT1 as well as LDHA mRNA levels under both normoxia and hypoxia (Fig. 7B,C). The Mpv17-/- cells showed an induction of GLUT1 mRNA by about 25% under normoxia and by about 40% under hypoxia; LDHA mRNA levels showed a similar pattern (Fig. 7B,C). In the MnSOD-KD cells no GLUT1 mRNA was detectable under normoxia and hypoxia. The LDHA mRNA was reduced in the MnSOD-KD cells by about 50% under both normoxia and hypoxia (Fig. 7B,C).
Fig. 7.
MECR, Mpv17, and MnSOD deficiency affect HIF-1α and HIF-1α target gene expression as well as expression of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1). NIH 3T3-scr, MECR-KD, HepG2-scr, MnSOD-KD, Mpv17+/+, and Mpv17-/- cells were cultured under normoxic (16% O2) or hypoxic (1% O2) conditions for 24 h. Total RNA was extracted, and relative HIF-1α, GLUT1, LDHA and PGC1 mRNA levels were evaluated by quantitative real-time PCR. *, significant difference 16% O2 versus 1% O2, #, significant difference NIH 3T3-scr versus MECR-KD, Mpv17+/+ versus Mpv17-/-, and HepG2-scr versus MnSOD-KD at the respective pO2; p≤0.05.
In all cells hypoxia reduced PGC1 mRNA levels. Interestingly loss of MECR and Mpv17 increased the PGC1 mRNA levels under normoxic conditions by about 3-fold and 3.5-fold, respectively. Again, after exposure of these cells to hypoxia the PGC1 mRNA levels decreased (Fig. 7D). Further, a reduction of PGC1 mRNA by about 50% could be detected in the MnSOD-KD cells when compared to the MnSOD-Scr cells. Surprisingly, hypoxia induced the PGC1 mRNA levels in the MnSOD-KD cells (Fig. 7D).
Together, these data show that non-ETC mitochondrial defects regulate HIF-1α at the degradation and transcriptional level in processes where ROS may play a role.
4. Discussion
Over the years, a number of studies have addressed the involvement of mitochondria, as well as ROS, in HIF-1α regulation. Although all previous reports, even if being conflicting, underlined that mitochondria contribute to HIF-1α regulation and oxygen sensing a complete picture about the mechanisms involved has not been reached. This is in part likely due to the fact that the majority of studies focused either on the depletion of mitochondrial DNA (rho(0)-cells), or on the inhibition of the ETC via pharmacological approaches as well as by using cells with major ETC defects (for review see [25], [26], [27], [28]). To the best of our knowledge, no studies considering other mitochondrial defects outside the ETC were carried out yet. Although descriptive, the current study is the first to show that non-ETC and non-Krebs cycle mitochondrial defects contribute to HIF-1α regulation in a different manner.
Thereby, we describe three entirely new aspects with respect to the involvement of mitochondria in hypoxia-dependent HIF-1α regulation. First, the data show that three different non-ETC and non-Krebs cycle mitochondrial defects contribute to HIF-1α regulation in a different manner. Thereby, cells lacking the key enzyme of mitochondrial fatty acid synthesis MECR, and cells lacking MnSOD showed a reduced induction of HIF-1α under hypoxia. By contrast, cells lacking the mitochondrial DNA depletion syndrome channel protein Mpv17 displayed enhanced levels of HIF-1α already under normoxic conditions. Second, our study shows that ROS do not exert a uniform pattern when mediating their effects on HIF-1α, although all mitochondrial defects in the used cell types increased ROS formation under normoxia. Third, the current data indicate that the mitochondrial defects cause the different HIF-1α regulation via promoting HIF-1α degradation as well as via changes in HIF-1α transcription. Thereby, MECR- and MnSOD-deficient cells showed a reduction in HIF-1α mRNA levels whereas the Mpv17 lacking cells displayed enhanced HIF-1α mRNA levels under normoxia and hypoxia.
The MECR deficiency leading to a reduced mitochondrial fatty acid synthesis (mtFAS) affects the function of other important metabolic enzymes such as α-ketoglutarate dehydrogenase (KGD), branched-chain keto-acid dehydrogenase, 2-oxoadipate dehydrogenase, pyruvate dehydrogenase (PDH), and enzymes of the glycine cleavage system. This is due to the fact that mtFAS is involved in the synthesis of lipoic acid (LA) which is as a covalently bound cofactor essential for the activity of the above mentioned enzymes. Hence, a loss of MECR leads to a loss of lipoylated proteins [29], [30]. With respect to HIFα regulation this may be especially relevant if considering KGD activity and the accumulation of α-ketoglutarate in MECR-deficient cells which could feed the HIF hydroxylation reaction and consequently lead to an enhanced degradation of HIF-1α. This is indeed reflected in our study where MECR deficiency reduces the half-life of HIF-1α. Moreover, the reduced HIF-1α mRNA levels add to this effect and may be seen in conjunction with the role of MECR in the regulation of pentose metabolism. Together, the findings with respect to MECR and its effect on HIF-1α regulation are in line with its role in causing an inborn error of metabolism which resembles typical mitochondrial disorders with the involvement of organs being susceptible to oxygen deficiency such as basal ganglia and the optic nerve [18].
Our finding that HIF-1α is upregulated in cells lacking Mpv17 was at the first glance surprising. This was based on the fact that lack of Mpv17 causes mitochondrial DNA depletion, a feature also found in cells chemically modified to lack mitochondrial DNA (rho(0)-cells). These rho(0)- Hep3B and HEK293 cells were earlier shown to fail to induce HIF-1α and to generate ROS in response to hypoxia [12], [15], [31], hence, we expected that Mpv17 deficiency would abolish HIF-1α induction. Although we observed the opposite on the overall level of HIF-1α, we do not believe that cell-type specific effects as previously discussed in a report investigating the rho(0) effect in a human rho(0) osteosarcoma cell line with normal induction of HIF-1α [32] account for this regulation. We think that, in this particular case, the Mpv17 Δψm- and ROS-modulating channel activity [20] and its subsequent, so far unknown, signaling account for these effects.
In line, we showed in our recent report that the gating properties of the Mpv17 channel can be affected by the redox state and that it participates in the modulation of the membrane potential (Δψm) to preserve mitochondrial homeostasis [20]. The data of the current and other reports [33] support this view. In particular, we found that hypoxia decreases Δψm in all cell types investigated. Uncoupling of the ETC by dinitrophenol (DNP) elicited the expected dye responses indicating that the observed changes mirror the activity of the respiratory chain, although TMRE was used in a quenching mode. Interestingly, lack of MECR and Mpv17 increased Δψm likely due to enhanced ROS formation especially under normoxia and although to a lesser extent, under hypoxia; by contrast, MnSOD-deficient cells displayed reduced Δψm but also enhanced ROS formation. Overall, these data suggest a mutual relation between Δψm and ROS where ROS levels can change Δψm and vice versa changes in Δψm may affect ROS production. This may, to a certain extent reflect also the severity of the mitochondrial defect and in this regard, the total lack of channel-forming activity in Mpv17 deficient cells causes mitophagy, a process ultimately linked to hypoxic cell death and autophagy [20]. Thus, the increase in HIF-1α in Mpv17 lacking cells is in line with the findings showing that HIF-1α acts as an inducer of the Bcl2 family members, BCL2/adenovirus E1B 19 kDa interacting protein 3 (BNIP3) or BINP3L which both promote mitochondrial elimination via autophagosome formation [34].
Under these conditions, one can expect some quite similar aberrations in ROS levels and by avoiding a reoxygenation phase due to the use of an Invivo2 400 hypoxia workstation before ROS measurements we found, in line with previous investigations, that hypoxia affects ROS levels [35] and thus, the oxidative status of the cell. Interestingly, none of the mutations affected the difference in ROS levels obtained upon transition of cells from normoxia to hypoxia, and although not entirely clear, these differences could simply reflect the different metabolic conditions obtained during the 20 h exposure of the cells to hypoxia. However and as discussed earlier [26], [27], [28], ROS production under hypoxia appeared to be cell-type specific and appeared to decrease in the respective MECR NIH3T3 fibroblasts and MnSOD HepG2 control cells, whereas ROS levels increased in hypoxic Mpv17+/+ mouse embryonic fibroblasts. Interestingly, the loss of function of all the genes led to an additional increase in ROS levels under normoxia. By contrast, an increase in ROS was only observed with Mpv17-/- cells under hypoxia. Thus, the pattern of ROS, neither H2O2 nor. O2- as indicated by DCF or DHE fluorescence [21], respectively, did not entirely reflect the HIF-1α levels. However, our data also showed that H2O2 or the ROS scavenger ascorbate contributed to HIF regulation, although with different effects with respect to cell type and mitochondrial defect. This is in particular evident in the MnSOD deficient cells which do not show an upregulation of HIF-1α under hypoxia but where the ascorbate exposure strongly induces HIF-1α under both normoxia and hypoxia.
These findings fit with previous reports from MnSOD overexpressing human breast carcinoma MCF-7 cells, where a highly increased MnSOD activity promoted HIF-1α appearance and overexpression of H2O2 metabolizing enzymes prevented HIF-1α induction under hypoxia [22]. Since the efficacies of superoxide dismutase and ascorbic acid for catalyzing the decay of superoxide radicals in animal tissues are similar [36] our data fit with the proposal that an increase in HIF-1α could be mediated by mitochondrial hydrogen peroxide. Our earlier findings [37], [38] and reports from other groups [39], [40], [41] support that this may be partially the case and, as discussed earlier [26], depend on the cell type. Our data also support this, since addition of H2O2 to Mpv17+/+ cells increased HIF-1α levels. However, when control and deficient MECR NIH 3T3 fibroblasts as well as MnSOD HepG2 cells were exposed to H2O2, HIF-1α was not found to be induced, neither under normoxia nor under hypoxia. Although these data fit with the idea that ROS can regulate HIF-1α, they appear not to follow the same regulatory mechanism influencing PHD-mediated HIF-1α degradation. This is based on the observation that the HIF-1α half-life is decreased in all cells with mitochondrial defects investigated here, although they all displayed enhanced ROS levels. Thus, these findings prompted us to look into the HIF-1α mRNA expression pattern. Importantly, we found that the HIF-1α mRNA levels correlated with the protein levels; decreased HIF-1α mRNA and protein levels in MECR- and MnSOD-deficient cells and the opposite in Mpv17-lacking cells. Thus, these data suggest that, redox sensitive transcription factors could contribute to the changes in HIF-1α mRNA. In particular, NFκB [38], [42] and Nrf2 [35] may account for an induction of HIF-1α expression. However, it is unlikely that these factors also directly mediate the negative effects on HIF-1α mRNA abundance. Thus, it is tempting to speculate that either other transcription factors or indirect regulators of NFκB and Nrf2 may be involved.
In addition to other interacting transcription factors, indirect regulators may also be miRNAs. Indeed, previous reports suggested that several miRNAs, being regulated by hypoxia, ROS, or regulating antioxidant responses can contribute to the regulation of oxidative phosphorylation, TCA cycle activity as well as transcription factors such as HIFs, NFκB, and Nrf2 [43], [44], [45]. Thereby, miRNAs such as miR210, which during hypoxia represses the iron-sulfur cluster assembly proteins ISCU1/2, affect TCA cycle activity and oxidative phosphorylation [46]. Further, miR9 that controls for example the NFκB1/p50 subunit, or miR200a that affects the Nrf2 regulator Keap1, or miR126 [47] may affect HIF-1α expression and stabilization. Again, cell-type specific effects may account for differences. For example, miR126 was found to be up-regulated by hypoxia and oxidative stress in nonmalignant mesothelial cell lines. By contrast, in malignant H28 mesothelioma cells the induction of miR126 by hypoxia and oxidative stress was lost [47]. Thus, certain miRNAs, identification of which was not the scope of this study, may serve in important feedback loops and future work will provide us with the details about those.
The survival of cells under hypoxia is also accompanied by active changes in mitochondrial function. For example, HIF-1α directs processes leading to downregulation of complex I activity and complex IV activity [48], [49], [50]. In addition to limiting the entry of metabolites into the TCA cycle, the HIF system was also shown to shut down O2-demanding processes such as fatty acid β-oxidation. Indeed, hypoxia reduced the expression of carnitine palmitoyltransferase 1 (CPT-1) [51], which catalyzes the rate-limiting step in the mitochondrial import of fatty acids for β-oxidation. Moreover, hypoxia exposure reduced expression of the medium- and long-chain acyl-CoA dehydrogenases (MCAD and LCAD) [52] and decreased ROS as well as an important regulator of β-oxidation proliferator-activated receptor-γ coactivator-1α (PGC-1) protein levels [51], [52]. These findings are concurrent with the present study. However, when PGC-1 expression was analyzed under normoxia we found that its expression was increased in all cells harboring a mitochondrial defect. Since PGC-1 is considered to be one of the most, if not the most, important driver of mitochondrial biosynthesis these effects constitute likely a feedback mechanism by which the cells aim to compensate for their respective deficit in mitochondrial function. Mitochondrial number and mtDNA, as well as mitochondrial protein composition can vary with the mutations and interventions, and PGC-1 variations support this. Further, this also correlates well with the enhanced ROS levels under normoxia which are known to be activators of PGC-1 [53].
Altogether, the current study substantiates that mitochondrial integrity is an important modulatory factor of HIF-1α regulation. In line with the present findings and the knowledge that HIF-1α can be regulated by ROS in different ways (reviewed in [28], [54], [55]), we show that, in addition to the complex network controlling HIF-1α at the level of protein stability, mitochondrial defects regulate HIF-1α mRNA expression. Thus, the identification of transcriptional and post-transcriptional HIF regulatory networks may provide an additional way of controlling HIF-1α levels and may open new therapeutic options to modulate the HIF pathway.
References
- 1.Wang G.L., Jiang B.H., Rue E.A., Semenza G.L. Proc. Natl. Acad. Sci. USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 3.Jaakkola P., Mole D.R., Tian Y.-, Wilson M.I., Gielbert J., Gaskell S.J. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 4.Kaelin W.G., Jr., Ratcliffe P.J. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Manalo D.J., Rowan A., Lavoie T., Natarajan L., Kelly B.D., Ye S.Q. Blood. 2005;105:659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 6.Semenza G.L. Trends Mol. Med. 2012;18:534–543. doi: 10.1016/j.molmed.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J.A., Wei Y., Sowers J.R. Circ. Res. 2008;102:401–414. doi: 10.1161/CIRCRESAHA.107.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhatti J.S., Bhatti G.K., Reddy P.H. Biochim. Biophys. Acta. 2016 [Google Scholar]
- 9.Dallner G., Sindelar P.J. Free Radic. Biol. Med. 2000;29:285–294. doi: 10.1016/s0891-5849(00)00307-5. [DOI] [PubMed] [Google Scholar]
- 10.Turrens J.F. J. Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon M.C. Adv. Exp. Med. Biol. 2006;588:165–170. doi: 10.1007/978-0-387-34817-9_15. [DOI] [PubMed] [Google Scholar]
- 12.Chandel N.S., Maltepe E., Goldwasser E., Mathieu C.E., Simon M.C., Schumacker P.T. Proc. Natl. Acad. Sci. Usa. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guzy R.D., Schumacker P.T. Exp. Physiol. 2006;91:807–819. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- 14.Bell E.L., Klimova T.A., Eisenbart J., Moraes C.T., Murphy M.P., Budinger G.R. J. Cell Biol. 2007;177:1029–1036. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mansfield K.D., Guzy R.D., Pan Y., Young R.M., Cash T.P., Schumacker P.T. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunelle J.K., Bell E.L., Quesada N.M., Vercauteren K., Tiranti V., Zeviani M. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Hagen T. Biochem. Res. Int. 2012;2012:436981. doi: 10.1155/2012/436981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heimer G., Kerätär J.M., Riley L.G., Balasubramaniam S., Eyal E., Pietikäinen L.P. Am. J. Hum. Genet. 2016;99:1229–1244. doi: 10.1016/j.ajhg.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dalla Rosa I., Camara Y., Durigon R., Moss C.F., Vidoni S., Akman G. PLoS Genet. 2016;12:e1005779. doi: 10.1371/journal.pgen.1005779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antonenkov V.D., Isomursu A., Mennerich D., Vapola M.H., Weiher H., Kietzmann T. J. Biol. Chem. 2015;290:13840–13861. doi: 10.1074/jbc.M114.608083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konzack A., Jakupovic M., Kubaichuk K., Gorlach A., Dombrowski F., Miinalainen I. Antioxid. Redox Signal. 2015;23:1059–1075. doi: 10.1089/ars.2015.6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang M., Kirk J.S., Venkataraman S., Domann F.E., Zhang H.J., Schafer F.Q. Oncogene. 2005;24:8154–8166. doi: 10.1038/sj.onc.1208986. [DOI] [PubMed] [Google Scholar]
- 23.Immenschuh S., Hinke V., Ohlmann A., Gifhorn-Katz S., Katz N., Jungermann K. Biochem. J. 1998;334(Pt 1):141–146. doi: 10.1042/bj3340141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak K.J., Schmittgen T.D. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Taylor C.T. Biochem. J. 2008;409:19–26. doi: 10.1042/BJ20071249. [DOI] [PubMed] [Google Scholar]
- 26.Kietzmann T., Gorlach A. Semin. Cell Dev. Biol. 2005;16:474–486. doi: 10.1016/j.semcdb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 27.Dehne N., Brüne B. Antioxid. Redox Signal. 2014;20:339–352. doi: 10.1089/ars.2012.4776. [DOI] [PubMed] [Google Scholar]
- 28.Gorlach A., Dimova E.Y., Petry A., Martinez-Ruiz A., Hernansanz-Agustin P., Rolo A.P. Redox Biol. 2015;6:372–385. doi: 10.1016/j.redox.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wada H., Shintani D., Ohlrogge J. Proc. Natl. Acad. Sci. USA. 1997;94:1591–1596. doi: 10.1073/pnas.94.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng D., Witkowski A., Smith S. J. Biol. Chem. 2009;284:11436–11445. doi: 10.1074/jbc.M806991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandel N.S., McClintock D.S., Feliciano C.E., Wood T.M., Melendez J.A., Rodriguez A.M. J. Biol. Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 32.Vaux E.C., Metzen E., Yeates K.M., Ratcliffe P.J. Blood. 2001;98:296–302. doi: 10.1182/blood.v98.2.296. [DOI] [PubMed] [Google Scholar]
- 33.Frezza C., Zheng L., Tennant D.A., Papkovsky D.B., Hedley B.A., Kalna G. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang H., Bosch-Marce M., Shimoda L.A., Tan Y.S., Baek J.H., Wesley J.B. J. Biol. Chem. 2008;283:10892–10903. doi: 10.1074/jbc.M800102200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Taleb Z. al, Petry A., Chi T.F., Mennerich D., Görlach A., Dimova E.Y. J. Mol. Med. 2016;94:1153–1166. doi: 10.1007/s00109-016-1439-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Som S., Raha C., Chatterjee I.B. Acta Vitaminol. Enzymol. 1983;5:243–250. [PubMed] [Google Scholar]
- 37.BelAiba R.S., Bonello S., Zahringer C., Schmidt S., Hess J., Kietzmann T. Mol. Biol. Cell. 2007;18:4691–4697. doi: 10.1091/mbc.E07-04-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonello S., Zahringer C., BelAiba R.S., Djordjevic T., Hess J., Michiels C. Arterioscler. Thromb. Vasc. Biol. 2007;27:755–761. doi: 10.1161/01.ATV.0000258979.92828.bc. [DOI] [PubMed] [Google Scholar]
- 39.Fang J., Ding M., Yang L., Liu L.Z., Jiang B.H. Cell. Signal. 2007;19:2487–2497. doi: 10.1016/j.cellsig.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhong H., Chiles K., Feldser D., Laughner E., Hanrahan C., Georgescu M.M. Cancer Res. 2000;60:1541–1545. [PubMed] [Google Scholar]
- 41.Richard D.E., Berra E., Pouyssegur J. J. Biol. Chem. 2000;275:26765–26771. doi: 10.1074/jbc.M003325200. [DOI] [PubMed] [Google Scholar]
- 42.Frede S., Stockmann C., Freitag P., Fandrey J. Biochem. J. 2006;396:517–527. doi: 10.1042/BJ20051839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crosby M.E., Devlin C.M., Glazer P.M., Calin G.A., Ivan M. Curr. Pharm. Des. 2009;15:3861–3866. doi: 10.2174/138161209789649367. [DOI] [PubMed] [Google Scholar]
- 44.Contreras J., Rao D.S. Leukemia. 2012;26:404–413. doi: 10.1038/leu.2011.356. [DOI] [PubMed] [Google Scholar]
- 45.Espinosa-Diez C., Miguel V., Mennerich D., Kietzmann T., Sanchez-Perez P., Cadenas S. Redox Biol. 2015;6:183–197. doi: 10.1016/j.redox.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chan S.Y., Zhang Y., Hemann C., Mahoney C.E., Zweier J.L., Loscalzo J. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomasetti M., Nocchi L., Staffolani S., Manzella N., Amati M., Goodwin J. Antioxid. Redox Signal. 2014;21:2109–2125. doi: 10.1089/ars.2013.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tello D., Balsa E., Acosta-Iborra B., Fuertes-Yebra E., Elorza A., Ordóñez Á. Cell Metab. 2011;14:768–779. doi: 10.1016/j.cmet.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 49.Soro-Arnaiz I., Li Q., Torres-Capelli M., Meléndez-Rodríguez F., Veiga S., Veys K. Cell Rep. 2016;16:2991–3002. doi: 10.1016/j.celrep.2016.08.041. [DOI] [PubMed] [Google Scholar]
- 50.Fuhrmann D.C., Brüne B. Redox Biol. 2017;12:208–215. doi: 10.1016/j.redox.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu Y., Ma Z., Zhao C., Wang Y., Wu G., Xiao J. Toxicol. Lett. 2014;226:117–123. doi: 10.1016/j.toxlet.2014.01.033. [DOI] [PubMed] [Google Scholar]
- 52.Huang D., Li T., Li X., Zhang L., Sun L., He X. Cell. Rep. 2014;8:1930–1942. doi: 10.1016/j.celrep.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 53.LaGory E.L., Wu C., Taniguchi C.M., Ding C.-, Chi J.-, von Eyben R. Cell Rep. 2015;12:116–127. doi: 10.1016/j.celrep.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Samoylenko A., Hossain J.A., Mennerich D., Kellokumpu S., Hiltunen J.K., Kietzmann T. Antioxid. Redox Signal. 2013;19:2157–2196. doi: 10.1089/ars.2012.4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gorlach A., Kietzmann T. Methods Enzymol. 2007;435:421–446. doi: 10.1016/S0076-6879(07)35022-2. [DOI] [PubMed] [Google Scholar]