Abstract
Single-chain derivatives of JRFL gp120 linked to the first two domains of human CD4 (gp120-CD4D12) or to the CD4 miniprotein analog CD4M9 (gp120-M9), have been constructed. Biacore studies revealed that gp120-CD4D12 and gp120-M9 bound to antibody 17b with dissociation constants of 0.8 and 25 nM, respectively, at pH 7.0, while gp120 alone did not bind. The binding of gp120-CD4D12 to 17b is not affected by the addition of excess soluble CD4D12, while the binding of gp120-M9 is enhanced. This finding indicates that the M9 component of the single chain interacts relatively weakly with gp120 and can be displaced by soluble CD4D12. Immunogenicity studies of gp120, gp120-CD4D12, and gp120-M9 were carried out with guinea pigs. All three molecules were highly immunogenic. The resulting antisera were examined for neutralizing activities against various human immunodeficiency virus type 1 isolates. Broadly neutralizing activity was observed only with sera generated against gp120-CD4D12. These antisera were depleted of anti-CD4D12 antibodies by being passed over a column containing immobilized CD4D12. The depleted sera showed a loss of broadly neutralizing activity. Sera that were affinity purified over a column containing immobilized gp120-M9 also lacked such neutralizing activity. This finding suggests that the broadly neutralizing response observed is exclusively due to anti-CD4 antibodies. Competition experiments showed that only antisera generated against gp120-CD4D12 competed with the CD4i antibody 17b and that this activity was not affected by depletion of anti-CD4 antibodies. The data indicate that although antibodies targeting the CD4i epitope were generated by the gp120-CD4D12 immunogen, these antibodies were nonneutralizing.
One of the major goals of human immunodeficiency virus (HIV) vaccine research is to find an immunogen that will elicit broadly cross-reactive neutralizing antibodies against HIV. Most antibodies in HIV type 1 (HIV-1)-infected individuals are dircted against the Env surface glycoprotein of the virus. The gp120 subunit of Env binds to the cellular receptor CD4 (10). CD4 binding results in a conformational change which enables subsequent binding of gp120 to the coreceptor CCR5 and/or CXCR4. The conformational change results in the exposure of previously buried (cryptic) epitopes known as CD4-induced (CD4i) epitopes (1, 2, 7, 11, 16, 31, 32, 36-39, 42). Previous attempts to use gp120 as a vaccine failed to elicit antibodies capable of neutralizing primary isolates of the virus (5, 9, 14, 23, 24, 40). Antibody responses in vaccinated individuals were often found to be directed against linear epitopes accessible in denatured gp120 that are not exposed in correctly folded gp120 (40). A recent phase III vaccine trial that used monomeric gp120 as an immunogen also failed to demonstrate any efficacy for this molecule (VaxGen press release, 12 November 2003 [http://www.vaxgen.com]).
Several different innovative strategies have been employed to obtain Env-based immunogens capable of generating a broadly neutralizing response. Immunogens can be subdivided into protein- and peptide-based immunogens. In the former category, strategies include (i) attempts to stabilize gp120 by filling in a part of the CD4 binding site (44); (ii) attempts to construct immunogens that display cryptic epitopes that are normally not exposed, such as the coreceptor binding site (examples are the use of cross-linked gp120:CD4 complexes as immunogens [12], the use of cross-linked complexes of gp120 with antibody A32, which induces exposure of CD4i epitopes on gp120 [22], and the use of gp120 from CD4-independent viruses that have enhanced exposure of the coreceptor binding site [17]); (iii) use of hyperglycosylated derivatives of gp120 that attempt to focus the immune response to conserved epitopes that form part of the CD4 binding site (29); and (iv) design of Env derivatives that mimic the gp120:gp41 native trimer on the virus (including gp140 derivatives with cleavage site mutations and with [4, 46] or without [34, 35] artificial C-terminal trimerization sequences, as well as gp140 derivatives with engineered disulfides between the gp120 and gp41 components [3, 33]).
Alternative approaches have attempted to generate peptides which bind known broadly neutralizing antibodies such as immunoglobulin Gb12 (IgGb12) (6) or 2F5 (49). In one such study, a peptide that bound the broadly neutralizing antibody IgGb12 (48) was isolated by phage display, though there have been no subsequent reports of the ability of the peptide to yield b12-like antibodies when used as an immunogen. Similar difficulties were encountered in attempts to generate 2F5-like antibodies by using constrained peptide epitope mimics (25).
Of the immunogens described above, some of the gp140-based trimeric immunogens have yielded neutralizing responses of greater breadth than monomeric gp120 (4). However, the best neutralizing responses observed to date were obtained in a recent study that employed cross-linked complexes of gp120 with the four extracellular domains of human CD4 as an immunogen in rhesus macaques (12). The study suggested that antibodies against CD4i epitopes had broadly neutralizing activity and hence that antigens that expose such epitopes are potentially important immunogens.
In the present work we report on the biophysical and immunological characterization of JRFL gp120 and two of its single-chain derivatives, one linked to CD4D12 (gp120-CD4D12) and one linked to M9 (gp120-M9). These constructs are described in greater detail in Materials and Methods. Similar single chains have been constructed previously by using gp120 from the Bal isolate (13). In the present work, gp120, gp120-CD4D12, and gp120-M9 were injected into guinea pigs, and the resulting antisera were characterized. As in an earlier macaque study that used cross-linked complexes of gp120 with the four extracellular domains of human CD4 (12), broadly cross-reactive neutralizing responses were observed only in guinea pigs injected with the gp120-CD4D12 single chain. However, in these animals there was a very high proportion of anti-CD4 antibodies. Sera depleted of these antibodies did not show a broadly neutralizing response. Furthermore, sera that were affinity purified by binding to immobilized gp120-M9 also showed no neutralization. The data suggest that the broadly neutralizing response observed was due to anti-CD4 antibodies. Although antibodies against CD4i epitopes in gp120 were also found in sera from animals immunized with gp120-CD4D12, there was no evidence to suggest that these antibodies had significant neutralizing activity. In contrast to the results of earlier work (12), the present results suggest that gp120 antigens displaying the CD4i epitope are unlikely to be able to generate a gp120-directed broad neutralizing response.
MATERIALS AND METHODS
Construct descriptions.
The gp120 (amino acids 1 to 505) used in this work is from the JRFL isolate. The gene sequence was codon optimized for expression in human cells. CD4D12 refers to the first two domains (amino acids 1 to 183) of human CD4. All amino acid residues that contact gp120 are located in domain D1 of CD4 (20). M9 is a 27-residue CD4 analog based on a scyllatoxin scaffold. It is a structural mimic of beta-strands C′ and C" of domain D1 of human CD4. M9 binds gp120 about 100-fold less strongly than CD4 (41). Single chains similar to gp120-M9 and gp120-CD4D12 have been constructed previously by using gp120 from the Bal isolate (13).
Cell lines and antibodies.
The 293 cell line was obtained from the National Centre for Cell Sciences, Pune, India. Monoclonal antibody (MAb) 17b was obtained from the National Institutes of Health AIDS Reagent Program. The anti-gp120 MAb NEA-9301 was purchased from NEN. Polyclonal antisera against CD4M9 and CD4D12 were generated in rabbits. Lyophilized protein was reconstituted in water and mixed with Freund's complete adjuvant at a 1:1 (vol/vol) ratio of protein to emulsion. Rabbits were immunized subcutaneously with 100 μg of protein. For booster injections, protein was mixed with Freund's incomplete adjuvant at a 1:1 (vol/vol) ratio of protein to emulsion. Boosters with 50 μg of the immunogen were given after every 15 days. Bleeds were collected on the second day after each injection following the second immunization.
CD4M9 gene synthesis.
The CD4M9 gene was synthesized from a series of six overlapping oligonucleotides. These six oligonucleotides collectively encode the synthetic version of 81-bp CD4M9. Complementary oligonucleotides overlap by 20 nucleotides. All the oligonucleotides were combined in a 50-μl PCR mixture to a final concentration of 1 μM and subjected to PCR amplification. The gene assembly reaction mixture was then further diluted 20-fold in 50 μl of a PCR mixture containing 3 U of Taq DNA polymerase and outside primers at 1 μM. This mixture was subjected to 25 cycles of PCR. The amplified product was gel purified and cloned into the pTZ57R/T vector, provided with the InsT/A clone PCR product cloning kit (MBI), according to the manufacturer's protocol.
Construction of gp120-CD4D12 and gp120-M9.
Plasmids carrying gp120-CD4D12 and gp120-M9 were constructed as follows. The gene encoding gp120 was PCR amplified from the V1jnstpaoptgp140 vector. This vector contains the codon-optimized sequence for gp140 from the JRFL isolate with a tissue plasminogen activator leader under the control of a cytomegalovirus promoter. A 20-amino-acid linker sequence similar to that described previously (13) was generated by overlap PCR using four oligonucleotides in a manner similar to that described above for CD4M9 gene synthesis. The CD4D12 or CD4M9 gene was amplified with a 5′ overhang of the linker sequence. The three PCR products (gp120, linker, and CD4 or CD4M9) were mixed together and subjected to overlap PCR. The resultant product was digested with the ApaI and KpnI enzymes and cloned into V1jnstpaoptgp140 digested with the same enzymes to produce the final constructs. These are designated V1Jnstpagp120-CD4D12 and V1Jnstpagp120-M9. The nucleotide sequence of the linker used was GGAAGCGCTGGATCTGCTGGTTCCGCCGGCAGCGCAGGCAGTGCGGGCAGCGCAGGTTCT.
The corresponding peptide sequence is GSAGSAGSAGSAGSAGSAGS.
Expression of gp120, gp120-CD4D12, and gp120-M9 proteins.
Plasmids V1Jnstpagp120-CD4D12, V1Jnstpagp120-M9, and V1Jnstpagp120 were transiently transfected into 293 cells by using Ca3(PO4)2 (Amersham Pharmacia Biotech) according to the manufacturer's protocol. Eight hours after transfection, the medium was replaced with Optimem reduced serum medium (Gibco). After an additional 48 to 72 h, supernatants were collected and analyzed for protein expression by anti-gp120 and anti-CD4D12 immunoblot assays.
Immunoblot analysis.
Samples were treated by boiling for 5 min in 2% sodium dodecyl sulfate (SDS) and 1% β-mercaptoethanol and were then electrophoresed over a 10% polyacrylamide gel. Protein was then electrophoretically transferred to polyvinylidene difluoride membranes. The membrane was then treated with 5% nonfat milk (Carnation) in phosphate-buffered saline (PBS) for 2 h at room temperature. The membrane was washed three times with PBS containing 3% milk-0.05% Tween and then incubated with either MAb NEA-9301 (anti-gp120) or rabbit polyclonal anti-CD4. The membrane was washed three times with PBS containing 3% milk-0.05% Tween and then incubated with an appropriate secondary antibody conjugated to horseradish peroxidase in 3% milk-PBS at a dilution of 1:1,000. Bound antibody was detected with an ECL-Plus kit (Amersham Pharmacia Biotech).
Purification of gp120, gp120-CD4D12, and gp120-M9 proteins.
Protein was purified by affinity chromatography using Lentil-Lectin Sepharose 4B (Amersham-Pharmacia Biotech). Bound protein was eluted with 500 mM α-D-methyl mannopyranoside in PBS. Protein concentration was determined by a BCA assay (SIGMA) according to the manufacturer's protocol using BSA as a standard.
Expression and purification of CD4D12.
Plasmid pET-9a-pELB-rsCD4183, encoding human CD4D12 (two domains), was a gift from Marcia Osborne (28). In this construct, the first two domains of human CD4 (amino acids 1 to 183) are expressed C-terminal to a pelB signal sequence under the transcriptional control of the T7 promoter in plasmid pET9a. The fragment encoding the PelB signal peptide and the CD4D12 region was isolated by digestion of plasmid pET-9a-pELB-rsCD4183 with XbaI and XhoI and then cloned into the corresponding sites of the pET-20b vector to generate pET20b-CD4D12. In this construct a C-terminal His tag has been added to CD4D12 to facilitate protein purification. Subsequently, the CD4D12 gene, along with the pelB signal sequence and His tag, was cloned into the arabinose-inducible vector pBAD24 (13a) to generate pBAD24CD4D12. The recombinant plasmid was transformed into Escherichia coli strain DH5α. The culture was induced with 1% arabinose during mid-logarithmic growth. Cells were lysed by sonication for 10 min. Protein was purified from the soluble fraction by affinity purification over a nickel-nitrilotriacetic acid (Ni-NTA) column (QIAGEN). The yield of purified protein was 0.7 mg/liter.
The CD4D12 gene without the pelB signal sequence but with the C-terminal His tag was subcloned from pBAD24CD4D12 into the pET9a vector to generate pET9a-CD4D12. pET9a CD4D12 was expressed in E. coli strain BL21(DE3). After cell growth and lysis, inclusion bodies were resuspended in 6 M guanidine hydrochloride (GdnCl) in PBS for 4 h at room temperature. The solution was centrifuged at 9,000 × g for 30 min. The supernatant was bound on Ni-NTA beads and eluted with 6 M GdnCl in PBS containing 300 mM imidazole at room temperature. The eluted sample was extensively dialyzed against PBS at 4°C. The dialyzed sample was spun at 9,000 × g for 30 min. at 4°C. The supernatant was collected and subjected to SDS-polyacrylamide gel electrophoresis (PAGE). Protein was found to be >95% pure.
Gel filtration analysis of CD4D12, gp120, gp120-CD4D12, and gp120-CD4M9.
Approximately 50 μg of each protein was analyzed under nondenaturing conditions by gel filtration chromatography in PBS buffer at room temperature by using either a Superdex-75 (for CD4D12) or a Superdex-200 (for gp120, gp120-CD4D12, and gp120-M9) analytical gel filtration column. A standard curve of elution volume versus molecular weight was generated by using a set of protein standards. This curve was then used to calculate the apparent molecular weights of the proteins.
Far-UV CD spectroscopy.
Circular dichroism (CD) spectra were recorded on a Jasco J-715 C spectropolarimeter flushed with nitrogen gas. The spectra were recorded with a 0.1-cm-path length cuvette using a scan rate of 10 nm/min and a time constant of 8 s. All data were averaged over a minimum of six scans. Ellipticities were calculated as follows:
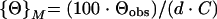 |
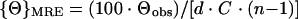 |
where {Θ}M is the calculated molar ellipticity (expressed as degrees per square centimeter per decimole), {Θ}MRE is the calculated mean residue ellipticity (expressed as degrees per square centimeter per decimole), {Θ}obs is the observed ellipticity (expressed in degrees), d is the path length (in centimeters), C is the molar protein concentration, and n is the total number of amino acids in the protein or protein-protein complex. Far-UV CD spectra were taken for gp120, gp120-CD4D12, gp120-M9, CD4D12, and gp120 preincubated with a threefold molar excess of CD4D12. Molar ellipticities were calculated for all the proteins. To calculate the molar ellipticity of the gp120:CD4D12 complex, the following formula was used:
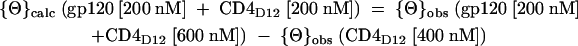 |
This {Θ}calc was used to calculate the {Θ}MRE of the gp120:CD4D12 complex. To compare the secondary structure of gp120 in free gp120, gp120-CD4D12, and gp120-M9, the {Θ}M of CD4D12 was subtracted from the {Θ}M of gp120-CD4D12, the {Θ}M of M9 was subtracted from the {Θ}M of gp120-M9, and {Θ}MRE was calculated from {Θ}M as described above.
Biacore experiments.
All Biacore experiments were performed with a Biacore 2000 (Biacore, Uppsala, Sweden) optical biosensor at 25°C. In the first assay, 500 resonance units (RU) of the full-length human MAb 17b was attached by amine coupling to the surface of a research-grade CM5 chip. The binding of gp120 and its derivatives to this surface was examined in the presence and absence of CD4D12. A naked sensor surface without antibody served as a negative control for each binding interaction. gp120 and its derivatives, which had been serially diluted, were run across each sensor surface at four different concentrations in a running buffer of PBS plus 0.05% Tween 20. Protein concentrations ranged from 50 to 500 nM for gp120 and from 12.5 to 100 nM for gp120-CD4D12. In addition, the binding of the noncovalent gp120:CD4D12 complex to 17b was also examined. CD4D12 was added at a fivefold molar excess to gp120 at least 15 min before binding was measured. The binding of gp120-CD4D12 and gp120-M9 to 17b in the presence of additional CD4D12 was also examined. Binding and dissociation were measured for 75 and 300 s, respectively, at a flow rate of 30 μl/min. In all cases, the sensor surface was regenerated between binding reactions by one to two washes with 10 mM NaOH for 60 s at 10 μl/min. Each binding curve was corrected for nonspecific binding by subtraction of the signal obtained from the negative-control flow cell. Kinetic constants for association and dissociation were derived from linear transformations of the exported binding data using at least four concentrations of the analyte. The kinetic parameters obtained were compared with those estimated by fitting the data to the simple 1:1 Langmuir interaction model by using BIA EVALUATION 3.1 software. The pH dependence of the binding of gp120 and its derivatives to 17b was determined by using CGH25 buffer (25 mM citrate, 25 mM glycine, 25 mM HEPES) adjusted to the desired pH.
Denaturation of gp120, gp120-CD4 D12, and CD4D12.
A 500-μl volume of a 10 μM solution of protein was unfolded in 6 M GdnCl-PBS containing 400 μM of dithiothreitol (DTT) for 1 h. To this solution, iodoacetamide was added to a final concentration of 2 mM and incubated in the dark for 30 min. The reaction mixture was rapidly desalted on a PD10 column equilibrated in PBS.
Immunization protocol.
All animal experiments were approved by Merck Research Laboratories Institutional Animal Care and Use Committee. Three different protocols, with DNA alone, DNA primed with a protein boost, and protein alone, respectively, were used. For DNA alone, guinea pigs were injected intramuscularly with 2 mg of plasmid DNA expressing the desired antigen in phosphate buffer and were boosted with 2 mg of plasmid DNA 4, 8, and 24 weeks after the initial immunization. Sera were collected 3 weeks after each injection. Terminal bleeds were collected 44 weeks after the first injection. For DNA primed with a protein boost, two DNA injections at weeks 0 and 4 were followed by injection of a purified protein antigen (40 μg of protein with 40 μg of QS21 adjuvant). For protein alone, each of the four injections contained 40 μg of protein along with 40 μg of QS21, and the schedule described above was followed.
Neutralization assays.
Two different assay formats were employed. The first was a peripheral blood mononuclear cell (PBMC) assay. PBMCs at 106/ml were infected at a multiplicity of infection of ∼0.01, incubated overnight, then washed extensively, and plated into 96-well plates. Test sera were diluted by twofold serial dilutions and mixed with the cells. Cultures were incubated for an additional 72 h and then assayed for viral production by a commercial simian immunodeficiency virus viral core p27 assay kit (Coulter Immunology). End point titers were recorded as the reciprocal of the serum dilution in which 90% or more of the viral antigen production was inhibited relative to production in untreated viral growth control wells.
The second format was a single-cycle infectivity assay. P4/R5 cells containing a stably integrated long terminal repeat-lacZ reporter were seeded in 96-well plates at 2.5 × 103/well, incubated overnight, and infected with different strains of HIV-1 at a multiplicity of infection of ∼0.01in the presence of varying dilutions of antisera. After incubation for an additional 48 h, cells were lysed, and β-galactosidase was detected by using GalScreen chemilumniscent substrate (Tropix).
Depletion of anti-CD4 antibodies from sera collected from vaccinated animals.
Collected sera were depleted of anti-CD4 antibodies by using CD4D12 immobilized on Ni-NTA agarose as follows. One milligram of CD4D12 was incubated with 200 μl of Ni-NTA slurry overnight at 4°C in PBS. The mixture was centrifuged at 1,000 × g. Beads were washed three times with PBS. For quantitation of bound CD4D12, CD4-Ni-NTA beads were treated by boiling for 15 min in 2% SDS and 1% β-mercaptoethanol and were then electrophoresed over a 12% polyacrylamide gel.
Affinity purification of antisera by using immobilized gp120-M9.
One hundred micrograms of biotinylated gp120-M9 (1 μg/ml) was incubated with 40 μl of streptavidin-agarose beads (Sigma) for 3 h at room temperature. The suspension was spun at 2,000 × g for 2 min, and unbound gp120-M9 was removed. The beads were washed three times with 300 μl of PBS. To quantitate the amount of bound gp120-M9, beads were boiled for 15 min in 2% SDS-1% β-mercaptoethanol and then electrophoresed over a 10% polyacrylamide gel. Approximately 80% of the protein was found to be bound. Sera from terminal bleeds were incubated with gp120-M9-streptavidin beads for 2 h at room temperature. Beads were spun at 2,000 × g for 2 min, and the unbound fraction was collected. Beads were washed three times with PBS. Bound antibodies were eluted at pH 3 in 10 mM glycine-HCl, and the pH was immediately adjusted to 7 with 1 M phosphate buffer. Increasing either the amount of immobilized gp120-M9 or the total volume of beads had no effect on the titer of bound antibodies.
Determination of titers of antibody against native and rcm gp120, gp120-CD4D12, and CD4D12.
Enzyme-linked immunosorbent assays (ELISAs) against native gp120 or native gp120-CD4D12 were performed in 96-well plates to which D7324 (1 μg/ml) (Cliniqa Corporation, Fallbrook, Calif.) had been adsorbed after overnight incubation at 4°C in capture buffer (100 mM NaHCO3 [pH 9.5]). D7324 is a sheep antibody against the carboxy-terminal 15 amino acids of gp120. Plates were washed three times with PBS-0.5% Tween20 (PBST) and blocked with 200 μl of 5% nonfat milk in PBST. After a wash, 70 μl of native gp120 (100 ng/ml) was captured over the plate by incubating for 2 h. Serial dilutions of serum in a total volume of 70 μl were added in separate wells and incubated for 2 h at room temperature. For the negative control, PBST was added without any serum. Bound sera were detected by using a peroxidase-conjugated goat anti-guinea pig antibody (Jackson ImmunoResearch) at a dilution of 1:5,000 and the chromogenic substrate o-phenylenediamine dihydrochloride. The reciprocal of the serum dilution showing an optical density reading more than twice that of the negative control and greater than 0.1 was taken as the antibody titer. For corresponding ELISAs with reduced, carboxymethylated (rcm) gp120, rcm gp120 was desalted on a PD10 column to remove DTT and GdnCl before incubation with D7324. To determine the titer of serum against CD4D12, microtiter wells were coated with either native or rcm CD4D12 (2 μg/ml) overnight at 4°C. After three washes, the wells were blocked with 5% nonfat milk in PBST. Wells were washed three times with PBST, and 70 μl of serum was added in different dilutions. Incubation was carried out for 2 h at room temperature. Bound antibodies were detected with a peroxidase-conjugated goat anti-guinea pig antibody as described above.
Competition ELISAs of sera with 17b and 447-52d.
ELISA plates were coated with D7324 as described above. A 60-μl volume of gp120-CD4D12 (5 μg/ml) was added per well and incubated for 2 h. After three washes with PBST, serial dilutions of sera in 3% milk were added. Wells were washed three times, and 70 μl of 17b or 447-52d (5 μg/ml)was added and incubated for 2 h. Wells were washed three times, and 70 μl of a peroxidase-conjugated anti-human antibody was added per well and detected by using o-phenylenediamine dihydrochloride as described above.
RESULTS
Construction, expression, and purification of gp120 and its single-chain derivatives
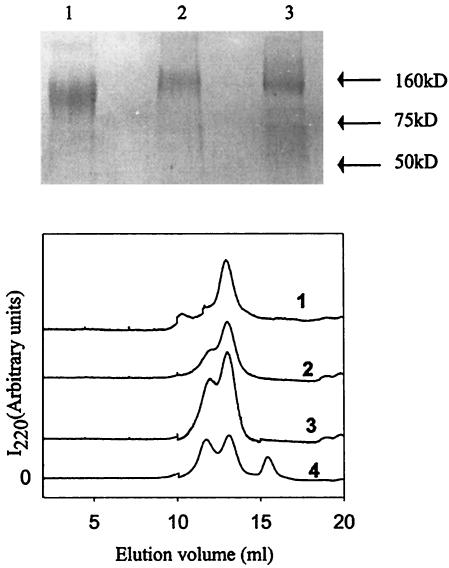
The gp120 used in the present work comprises codons 1 to 505 from the JRFL isolate. The gene sequence was codon optimized for expression in human cell lines. Plasmid DNA was transiently transfected into 293 cells, and protein was purified from tissue culture supernatants by affinity chromatography as described in Materials and Methods. Yields of purified proteins were approximately 1 to 2 mg/liter of cell culture supernatants. No cleavage in the linker region was observable in either of the single-chain constructs (Fig. 1, top). Gel filtration studies (Fig. 1, bottom) showed that all three proteins are monomeric, though in the case of gp120-M9 an appreciable fraction of a higher-molecular-weight aggregate was also present. The observed elution volume of gp120 (Fig. 1, bottom) was 13.05 ml, while the calculated elution volume based on the column calibration was 13.2 ml.
FIG. 1.
Characterization of gp120, gp120-CD4D12, and gp120-M9. (Top) SDS-PAGE analysis. Three micrograms of each purified protein was loaded onto a 10% gel. Samples were boiled with 2% SDS and 100 mM DTT prior to loading. The gel was stained with Coomassie blue. Lane 1, gp120; lane 2, gp120-CD4D12; lane 3, gp120-M9. (Bottom) Gel filtration analysis of native gp120, gp120-CD4D12, and gp120-M9 polypeptides on a Superdex 200 analytical gel filtration column in PBS buffer at room temperature. For comparison, gp120 was also preincubated with a molar excess of CD4D12 for 30 min. at room temperature and then loaded onto the gel filtration column. The absorbance at 220 nm is shown as a function of the elution volume for gp120-CD4D12 (trace 1), gp120-M9 (trace 2), gp120 (trace 3), and gp120 preincubated with CD4D12 (trace 4).
Expression and purification of CD4D12 and its derivatives.
CD4D12 was cloned into the E. coli expression vector pET9a to generate the pET9aCD4D12 construct with a C-terminal His tag. BL21(DE3) cells transformed with this plasmid were used to express CD4D12. The protein was purified from inclusion bodies under denaturing conditions using Ni-NTA agarose and was refolded as described above. The refolded protein was found to be monomeric by gel filtration chromatography on Superdex-75 columns and to have gp120 binding activity comparable to that of either commercially available four-domain CD4 expressed in mammalian cells or two-domain CD4 expressed in soluble form by using a construct with a signal peptide (data not shown). The yields of purified protein were approximately 5 to 10 mg/liter of E. coli culture.
Characterization of gp120 constructs by SPR.
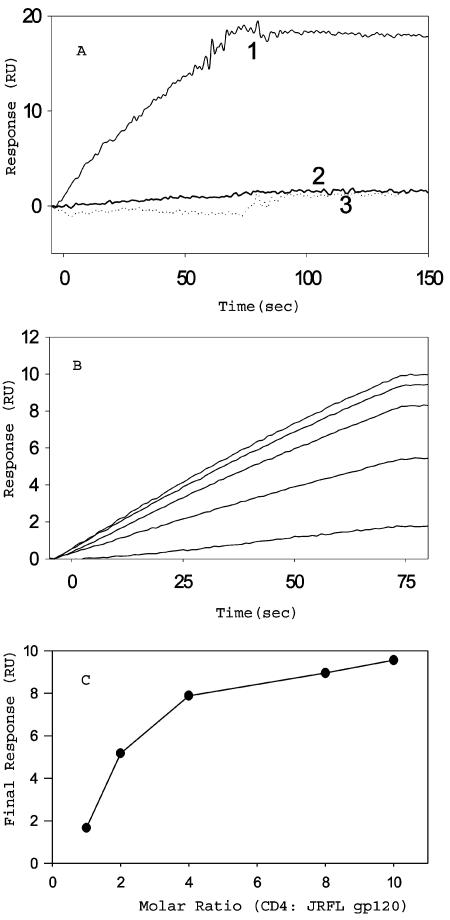
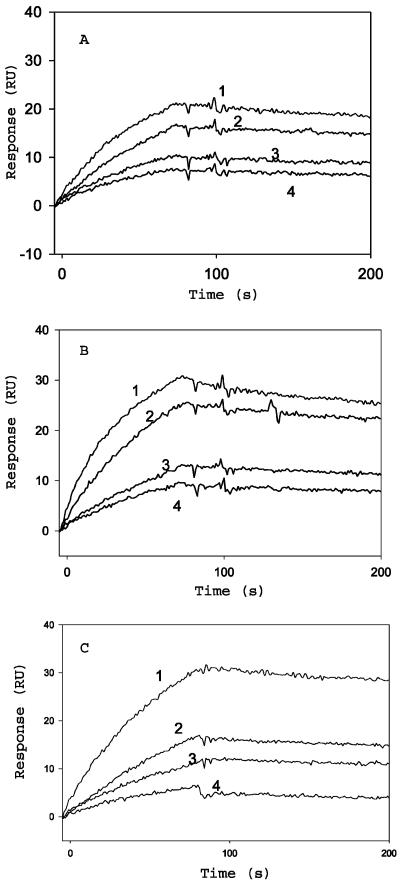
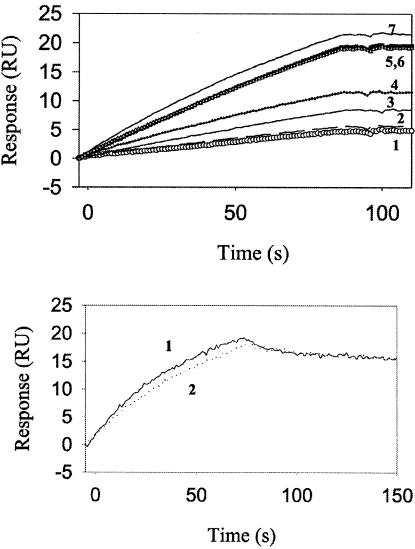
CD4 binding to gp120 results in a conformational change in gp120 that exposes cryptic epitopes known as CD4i epitopes. One MAb specific for the CD4i conformation of gp120 is 17b (38). The binding of all three gp120 constructs to 17b, in the presence or absence of soluble CD4D12, was characterized by surface plasmon resonance (SPR). 17b was immobilized on the surface of a CM5 chip, and a solution of the gp120 derivative was passed over the chip surface. Real-time binding to surface-immobilized 17b was assayed by using the BIACORE 2000 SPR instrument. Binding was assayed both in the presence and in the absence of a fivefold excess of soluble CD4D12. JRFL gp120 in the absence of CD4D12 showed no detectable binding to 17b. Binding increased in the presence of added CD4D12 and became saturated in the presence of a fivefold molar excess of CD4D12 (Fig. 2). In contrast, the gp120-CD4D12 single chain bound to 17b in the absence of soluble CD4D12, and no enhancement of binding was observed after addition of a fivefold molar excess of CD4D12. The extent of 17b binding for gp120-CD4D12 was similar to that of an identical concentration of the noncovalent gp120:CD4D12 complex, suggesting that all of the single chain is properly folded (Fig. 3). The gp120-M9 complex bound to 17b, but with much lower affinity than gp120-CD4D12. Furthermore, 17b binding to gp120-M9 was appreciably enhanced in the presence of excess CD4D12, in contrast to 17b binding to gp120-CD4D12 (Fig. 4). The data suggest that the gp120-M9 single chain may be relatively flexible and may contain both molecules in which the M9 component is complexed to gp120 (CD4i conformation) and a large fraction of molecules which more closely resemble free gp120. CD4D12 would be able to bind to the latter molecules and convert them to a CD4i conformation.
FIG. 2.
Sensorgram overlays to show the binding of JRFL gp120 to the immobilized 17b surface. Surface density, 500 RU; buffer, 10 mM phosphate (pH 7.4)-150 mM NaCl-0.0005% Tween 20; flow rate, 30 μl/min. (A) JRFL gp120 at 100 nM does not bind to 17b (curve 2), but when it is preincubated with 500 nM CD4D12, there is a marked increase in binding (curve 1). CD4D12 at 500 nM does not show any detectable binding to 17b (curve 3). (B) Binding of gp120 to 17b as a function of CD4D12 concentration. A 40 nM concentration of gp120 was incubated with 400, 320, 160, 80, or 40 nM CD4D12 for 30 min, and binding to 17b was monitored (curves shown from top to bottom of the graph in order of decreasing CD4D12 concentration). (C) The final response at the end of the association phase was plotted against the molar ratio of CD4D12:gp120.
FIG. 3.
Sensorgram overlays for the binding of gp120 (preincubated with CD4D12), gp120-CD4D12, and gp120-M9 to surface-immobilized 17b. Surface density, 500 RU; buffer, 10 mM phosphate (pH 7.4)-150 mM NaCl-0.0005% Tween 20; flow rate, 30 μl/min. (A) Binding of 100, 50, 25, or 12.5 nM gp120, preincubated with a fivefold molar excess of CD4D12 for 30 min, to surface-immobilized 17b (curves 1, 2, 3, and 4, respectively). (B) Binding of 100, 50, 25, or 12.5 nM gp120-CD4D12 to surface-immobilized 17b (curves 1, 2, 3, and 4, respectively). (C) Binding of 5,500, 250, 125, or 62.5 nM gp120-M9 to surface immobilized 17b (curves 1, 2, 3, and 4, respectively).
FIG. 4.
Effect of added CD4D12 on binding of gp120-M9 and gp120-CD4D12 to 17b. (Top) Binding of gp120-M9 to surface-immobilized 17b increases upon preincubation with increasing concentrations of CD4D12 (curves 1 to 6). Binding saturates at a fivefold molar excess of CD4D12. This binding is comparable to that of the same concentration of gp120 preincubated with a fivefold molar excess of CD4D12 (curve 7). Shown is the binding of 100 nM gp120-M9 to surface immobilized 17b (curve 1), with 50 nM (curve 2), 100 nM (curve 3), 250 nM (curve 4), 500 nM (curve 5), and 750 nM (curve 6) CD4D12. (Bottom) Binding of gp120-CD4D12 to surface-immobilized 17b does not increase upon preincubation with a fivefold molar excess of CD4D12. Shown is the binding of 100 nM gp120-CD4D12 to surface-immobilized 17b with (curve 1) and without (curve 2) CD4D12. Surface density, 500 RU; buffer, 10 mM phosphate (pH 7.4)-150 mM NaCl-0.0005% Tween 20; flow rate, 30 μl/min.
SPR studies were carried out as a function of protein concentration and pH to obtain values of the rate constants for association and dissociation (ka and kd, respectively) and the equilibrium dissociation constant (KD) value (kd/ka) for binding of the various constructs to 17b (Table 1). Binding is largely independent of pH in the pH range 5 to 9. The gp120-CD4D12 single chain and the noncovalent gp120:CD4D12 complex have similar KDs for 17b, while the corresponding KD of the gp120-M9 single chain is 10-fold higher. This difference results primarily from a lower ka for this protein. The binding constant of the noncovalent gp120:CD4D12 complex to 17b is similar to that reported previously (17). In addition, the binding of CD4D12 to gp120 was also measured by us, and the binding parameters (9.5 × 104 M−1 s−1 for ka and 1.7 × 10−3 s−1 for kd) were very similar to published values (6.7 × 104 M−1 s−1 for ka and 1.5 × 10−3 s−1 for kd) (27). All the above studies suggest that the gp120 and gp120 derivatives used in the present work are properly folded. In contrast to some previous studies (19, 47), we did not observe binding of gp120 to 17b in the absence of CD4D12 for the full-length gp120 constructs used in the present work, though such binding was observed in gp120 constructs in which the loop was deleted (K. Chakraborty and R. Varadarajan, unpublished data). The reasons for this difference are not clear, but we speculate that it may be due to differences in the glycosylation pattern of JRFL gp120, used in these studies (expressed by transient transfection in 293 cells), and the gp120 molecules used in the earlier studies, which were expressed in Drosophila S2 cells.
TABLE 1.
Rate constants and equilibrium constants for the binding of the different constructs to 17b as a function of pH as measured by BIAcore SPR
| Protein | Valuea at:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| pH 9
|
pH 7
|
pH 5
|
|||||||
| ka (M−1 s−1) (10−5) | kd (s−1) (104) | KD (M) (109) | ka (M−1 s−1) (10−5) | kd (s−1) (104) | KD (M) (109) | ka (M−1 s−1) (10−5) | kd (s−1) (104) | KD (M) (109) | |
| gp120:CD4D12 (1:5) | 1.7 | 5.6 | 3.4 | 1.8 | 3.0 | 1.6 | 3.1 | 4.6 | 1.5 |
| gp120-CD4D12 | 2.2 | 2.2 | 1.0 | 3.0 | 2.3 | 0.8 | 2.2 | 4.9 | 2.2 |
| gp120-M9 | 0.2 | 6.0 | 32.0 | 0.3 | 7.9 | 25.0 | 0.2 | 4.5 | 20 |
ka and kd, rate constants for association and dissociation, respectively. KD, equilibrium dissociation constant.
Spectroscopic characterization of single-chain complexes.
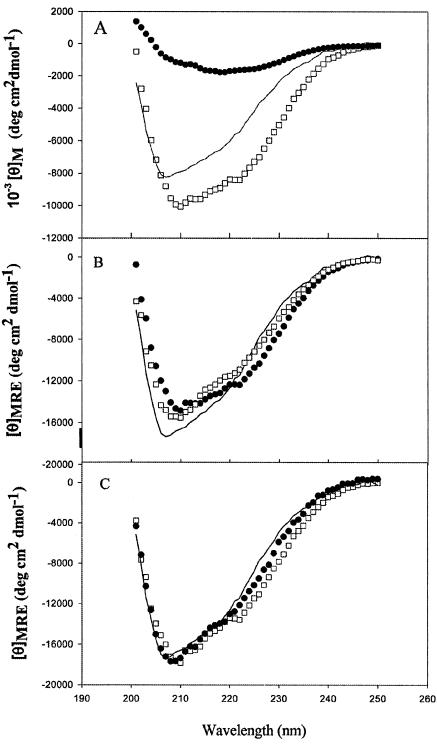
Far-UV CD spectra of gp120, gp120-CD4D12, and CD4D12 are shown in Fig. 5A. The spectrum for gp120-M9 (data not shown) has molar ellipticity values intermediate between those for gp120 and gp120-CD4D12. Figure 5B shows mean residue ellipticities (MRE) for gp120, gp120-CD4D12, and the noncovalent gp120:CD4D12 complex. For the complex, the contribution of excess unbound CD4D12 has been subtracted. Figure 5C shows MRE for gp120, gp120-M9, and gp120-CD4D12. For the latter two molecules, the contributions of M9 and CD4D12 have been subtracted. In all cases the subtractions were carried out before the raw data were converted to MRE (see Materials and Methods). The subtractions make the reasonable assumption that the secondary structures of these two ligands (CD4D12 and M9) are the same in the free form and the single chain. The figure shows that the secondary structure of gp120 is identical in all three constructs. This implies that the conformational changes induced by CD4 or M9 in gp120 primarily involve rigid body motions of domains, without any overall change in secondary structure.
FIG. 5.
Far-UV CD spectra of gp120 and its derivatives. Spectra were obtained with 0.61 μM gp120, 0.76 μM gp120-CD4D12, 0.80 μM gp120-M9, and 2 μM CD4D12 in CGH5 buffer (pH 7.4) with a 0.1-cm-path length cuvette, a scan rate of 10 nm/min, and a time constant of 8 s. Data reported are averaged over a minimum of six scans. (A) Molar ellipticities for gp120 (solid line), gp120-CD4D12 (open squares), and CD4D12 (solid circles). (B) MRE for gp120 (solid line), gp120-CD4D12 (solid circles), and gp120 preincubated with a threefold molar excess of CD4D12 after subtraction of the contribution from excess CD4D12 (open squares). (C) MRE for gp120 (solid line), gp120-M9 after subtraction of the M9 contribution (solid circles), and gp120-CD4D12 after subtraction of the CD4D12 contribution (open squares).
Immunogenicity and neutralization studies.
Immunization of guinea pigs with the three antigens (gp120, gp120-CD4D12, and gp120-M9) was carried out using the three different immunization protocols described in Materials and Methods. Table 2 shows ELISA and neutralization titers obtained after the fourth immunization. ELISA titers to all antigens in all groups increased progressively for injections 1 to 3, reached a plateau for injections 3 and 4, and decreased somewhat for the terminal bleeds (data not shown). Values reported are means for each group. The DNA-alone and DNA-protein injection protocols yielded similar titers, while the animals receiving protein alone had markedly lower titers (data not shown). All animals receiving the gp120-CD412 antigen had very high titers against CD412. These were 5- to 10-fold higher than the corresponding gp120 titers. Surprisingly, low but consistent and reproducible titers against CD4 were also seen in all animals receiving gp120 alone, suggesting that some antibodies against gp120 cross-react with CD4D12. Neutralization studies were carried out for all animals receiving DNA alone or DNA-protein by using a standard PBMC assay as described in Materials and Methods. The data show that broadly neutralizing activity is present only in sera derived from animals receiving the gp120-CD4D12 immunogen. Neutralization was also examined by using a more-sensitive single-cycle infectivity assay. Here, too, neutralization was observed only with sera from animals receiving the gp120-CD4D12 immunogen (Table 2, footnote c). No differences in neutralization profiles were observed between animals receiving the gp120 and gp120-M9 antigens. Hence the gp120-M9 antisera were not subjected to further detailed characterization.
TABLE 2.
ELISA and neutralization titersa for sera obtained after the fourth immunization
| Group | Vaccine antigen | ELISA titer
|
Neutralization titer (IC90)b
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| gp120 | M9 | CD4D12 | MN | JRFL | Bal | IIIB | 1357-0 | ||
| 1 | gp120 (DNA) | 237,500 | 100 | 3,200 | 72 | <10 | <10c | <10c | <10 |
| 2 | gp120-CD4D12 (DNA) | 175,000 | 100 | 2,532,600 | 600 | 70 | 50c | 40c | 120 |
| 3 | gp120-M9 (DNA) | 64,000 | 256,000 | 100 | 120 | <10 | <10c | <10c | <10 |
| 4 | gp120 (DNA-protein) | 1,024,000 | 100 | 1,000 | 240 | <10 | <10 | <10 | <10 |
| 5 | gp120-CD4D12 (DNA-protein) | 2,867,200 | 4,800 | 4,915,200 | 1,000 | 120 | 45 | <10 | 10 |
| 6 | gp120-M9 (DNA-protein) | 409,600 | 307,200 | 100 | 45 | <10 | <10 | <10 | <10 |
Values are means for each group.
IC90, 90% inhibitory concentration, expressed as the reciprocal of the serum dilution in which 90% or more of the viral antigen production was inhibited relative to production in untreated viral growth control wells.
Neutralization titers were also measured by using a more-sensitive single-cycle infectivity assay. Mean titers (IC50) for animals vaccinated with gp120-CD4D12 were 1,765 and 4,982 for HIV strains IIIB and Bal, respectively. No neutralization was observed in sera from animals receiving gp120 or gp120-M9 as the immunogen.
CD4D12 depletion and affinity purification studies.
Since the gp120-CD4D12 antisera contained very high titers of anti-CD4 antibodies, it is likely that these are primarily responsible for the broad neutralizing response. In order to test this possibility, antisera from gp120-CD4D12 animals were passed over a column containing immobilized CD4D12. In a control experiment, corresponding gp120 antisera were also subjected to the same procedure. Table 3 shows ELISA and neutralization titers for these sera before and after depletion. The data clearly show that the depletion procedure completely depletes sera of anti-CD4D12 antibodies without significantly affecting the titer against gp120. There is also a corresponding loss of neutralization activity upon depletion.
TABLE 3.
Comparison of ELISA and neutralization titers before and after depletion of anti-CD4 antibodies from sera collected after the fourth immunization
| Group | Vaccine antigen | ELISA titer
|
Neutralization titer (IC90)a
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| CD4
|
gp120
|
MN
|
1357-0b
|
||||||
| Undepleted | Depleted | Undepleted | Depleted | Undepleted | Depleted | Undepleted | Depleted | ||
| 1 | gp120 (DNA) | 3,200 | 3,200 | 100,000 | 100,000 | 80 | 46 | 10 | <46 |
| 1,600 | 1,600 | 400,000 | 400,000 | 160 | 46 | <10 | <46 | ||
| 1,600 | 1,600 | 50,000 | 50,000 | <10 | <40 | 20 | <40 | ||
| 6,400 | 6,400 | 400,000 | 400,000 | 40 | 44 | <10 | <44 | ||
| 2 | gp120-CD4D12 (DNA) | 300,000 | 1,000 | 400,000 | 400,000 | >1,280 | 51 | 160 | <51 |
| 1,638,400 | 1,000 | 100,000 | 100,000 | 320 | <57 | 160 | <57 | ||
| 6,553,600 | 1,000 | 100,000 | 100,000 | 640 | <67 | 320 | <67 | ||
| 1,638,400 | 1,000 | 100,000 | 100,000 | 160 | <48 | 20 | <48 | ||
IC90, 90% inhibitory concentration, expressed as the reciprocal of the serum dilution in which 90% or more of the viral antigen production was inhibited relative to production in untreated viral growth control wells.
CCR5-utilizing primary isolate.
Antisera were affinity purified by being passed over gp120-M9 immobilized on agarose beads. Table 4 shows that both the CD4 ELISA titers and the neutralization titers were present exclusively in the unbound fraction. A substantial fraction of anti-gp120 antibodies bound to the column and were eluted. However, these antibodies did not show any neutralization. A control experiment showed that the CD4i antibody 17b could be efficiently captured and eluted from the beads and that the elution procedure did not result in any loss of bound gp120-M9 from the beads. Both the depletion and the affinity purification experiments clearly indicate that the broad neutralizing response observed in animals receiving the gp120-CD4D12 immunogen is wholly due to anti-CD4D12 antibodies.
TABLE 4.
Characterization of neutralization properties of guinea pig sera before and after affinity purification on immobilized gp120-M9 beads
| Serial no. | Material | ELISA titer
|
Neutralization titer
|
||
|---|---|---|---|---|---|
| CD4a | gp120a | MNb | 1357-0b | ||
| 1 | Untreated sera | 1,000,000 | 150,000 | 640 | 80 |
| 2 | Column flowthrough | 1,000,000 | 150,000 | 704 | 88 |
| 3 | Eluted fraction | 3,600 | 150,000 | <44 | <44 |
ELISA antigen.
Viral strain used for neutralization assay.
Immunological characterization of antisera.
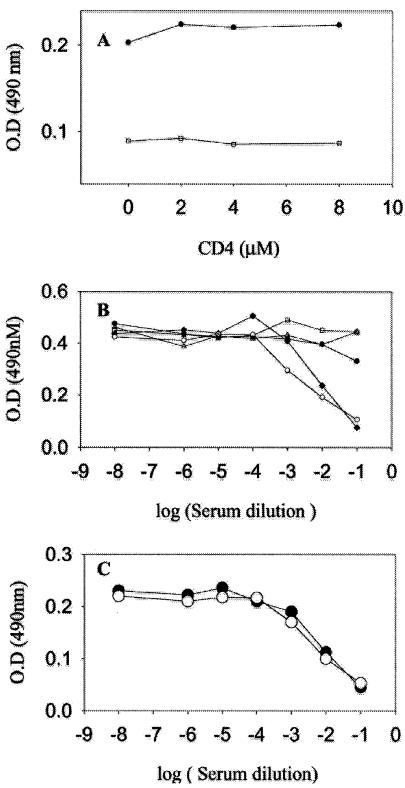
Antisera were further characterized for their relative titers against native and denatured gp120. It has been reported previously (40) that individuals vaccinated with gp120 have a high proportion of antibodies directed aginst linear epitopes that are present only in the denatured protein. Table 5 demonstrates that animals inoculated with gp120 or gp120-CD4D12 had equivalent titers to native and rcm gp120. There was little or no change in these titers after depletion of anti-CD4 antibodies. Thus, the presence of the stabilizing CD4D12 ligand in the single chain does not appear to have resulted in a very large increase in the proportion of antibodies that recognize conformational epitopes present only in intact gp120. In contrast, the anti-CD4 antibodies were directed primarily against conformational epitopes present only in intact CD4. Antisera were also characterized for their abilities to compete with the CD4i antibody 17b and the V3 loop-directed antibody 447-52d. gp120-CD4D12 and gp120 antisera competed equally well with the anti-V3 loop antibody 447-52d. This was expected, because this loop should be accessible in both immunogens. Figure 6 shows that only antisera to gp120-CD4D12 were able to compete with 17b. Interestingly, this activity was not affected by depletion of anti-CD4D12 antibodies. Preincubation of the gp120-CD4D12 sera with CD4D12 also had no effect on the ability of the sera to compete with 17b. Both these experiments suggest that the desired CD4i antibodies have been generated in response to the gp120-CD4D12 immunogen. However, these antibodies either have a low titer relative to other anti-gp120 antibodies or are weakly neutralizing. A recent study suggests that CD4i antibodies are likely to be poorly neutralizing because they may be sterically excluded after attachment of the virus to the CD4 receptor (21).
TABLE 5.
ELISA titers against native and rcm protein for terminal bleed sera
| Serial no. | Vaccine antigen (DNA) | ELISA antigen | ELISA titer |
|---|---|---|---|
| 1 | gp120 | Native gp120 | 17,000 |
| 2 | gp120 | rcm gp120 | 20,000 |
| 3 | gp120-CD4D12 | Native gp120 | 25,000 |
| 4 | gp120-CD4D12 | rcm gp120 | 25,000 |
| 5 | gp120 | Native CD4D12 | 600 |
| 6 | gp120 | rcm CD4D12 | 600 |
| 7 | gp120-CD4D12 | Native CD4D12 | 1,00,000 |
| 8 | gp120-CD4D12 | rcm CD4D12 | 500 |
FIG. 6.
Characterization of immune sera by a competition ELISA. (A) A 1:10 dilution of the serum obtained from a gp120-CD4D12-vaccinated animal preincubated with varying amounts of CD4D12 (□) was able to compete out 17b binding to gp120-CD4D12. This competition was not due to anti-CD4 antibodies, because it was independent of the CD4D12 concentration. The preimmune serum control (•) showed no competition. (B) Sera obtained from animals vaccinated with gp120-CD4D12 (○), gp120-M9 (□), gp120 (▵), or the preimmune serum (•) were examined for their abilities to compete out 17b binding to gp120-CD4D12 in a competition ELISA (details in Materials and Methods). Only gp120-CD4D12 antisera competed with 17b. The gp120-CD4D12 antisera depleted of anti-CD4D12 (♦) retained the ability to compete out 17b binding to gp120-CD4D12. (C) Sera obtained from animals vaccinated with gp120-CD4D12 (○) or gp120 (•) competed out 447-52D binding to gp120-CD4D12 to equal extents (for details, see Materials and Methods).
DISCUSSION
Despite intensive efforts over the past decade, there has been little success in obtaining an immunogen that elicits a broadly neutralizing antibody response against HIV-1. The virus has evolved a number of mechanisms to avoid a neutralizing response. While the gp120 and gp140 proteins are obvious targets for the immune system, the protein surface is heavily glycosylated. Recent work (18) has shown that mutations that alter the glycosylation pattern contribute significantly to immune evasion. It is believed that conserved epitopes such as the receptor or coreceptor binding sites are either recessed (CD4 binding site) or sterically inaccessible (coreceptor binding site) in the trimeric form of the Env protein on the virus. Upon receptor binding, gp120 undergoes a conformational change that exposes its coreceptor binding sites. Epitopes exposed at this stage are known as CD4i epitopes. Several MAbs that target CD4i epitopes, such as 17b and 48d, have been isolated (38). However, none of these antibodies has been shown to have broadly neutralizing activity. Recently a Fab fragment named X5, isolated by phage display against a gp120:CD4:CCR5 complex, was reported to have broadly neutralizing activity (26). However, subsequent work showed that when X5 was converted to a full-length antibody, there was a substantial decrease in its neutralizing activity (21). The neutralizing activities of X5, 17b, and 48d were studied for full-length IgG, Fab, and scFv forms. In all three cases the neutralizing activity was found to decrease as molecular size increased, i.e., the activity of scFv was highest, followed by Fab and IgG, in that order.
The data were consistent with a neutralization mechanism in which the CD4i epitope is exposed only after attachment of the virus to CD4 on the host cell. After attachment, it is likely that the larger IgG molecules are sterically excluded from binding to CD4i epitopes, and this is a probable explanation for the inverse correlation of antibody neutralization efficiency with antibody size. However, all these experiments were carried out with a limited set of MAbs. Given the high degree of flexibility and diversity in antibody structure, it is still unclear if antigens that present the CD4i epitope are capable of generating a neutralizing response. A recent study (8) demonstrated that the sulfated antibody 412d, derived from an infected individual, neutralized the primary R5 ADA isolate better than the well-studied unsulfated CD4i antibodies 17b and 48d. Another similarly derived sulfated antibody, E51, was also shown (45) to neutralize primary isolates JRFL and SA32 better than 17b, though the enhancement in neutralization efficacy was relatively modest. Both 412d and E51 are CD4i antibodies. A CD4-independent virus was previously shown to exhibit partially exposed CD4i epitopes (17), but there is so far no evidence demonstrating that the corresponding envelope is a useful immunogen. It was also recently reported that cross-linked complexes of gp120 with four-domain soluble human CD4 generated a potent and broadly neutralizing immune response. Although anti-CD4 antibodies were present in these sera, the sera were reported to show appreciable neutralization activity after being affinity purified on a column containing the immobilized gp120-M9 single chain (12).
This finding suggested that gp120:CD4 complexes or other, related molecules, which exposed the CD4i epitope, might be useful immunogens. Chemically cross-linked protein complexes are relatively inhomogeneous molecular species which are difficult to produce reproducibly on a large scale. An earlier report described the construction of gp20-CD4D12 and gp120-M9 single chains in which the two molecules were linked by a 20-amino-acid linker (13). These constructs used gp120 derived from the Bal isolate. In the present work we have described the biophysical and immunological characterization of gp120-CD4D12 and gp120-M9 single chains derived from the JRFL isolate. In both cases, folded and largely monomeric proteins were obtained. gp120-CD4D12 was found to bind 17b, a prototypical CD4i antibody, with a 10-fold lower KD than gp120-M9. The binding of 17b to gp120-CD4D12 was not enhanced by addition of exogenous CD4D12, demonstrating that all single-chain molecules were in a properly folded conformation that exposed the CD4i epitope. In contrast, exogenous CD4D12 considerably enhanced the ability of gp120-M9 to bind 17b, suggesting that the CD4D12 was able to displace the M9 component of the single chain from its intramolecular binding site in gp120. Free M9 is known to bind approximately 100-fold less strongly than CD4D12 to gp120 (41). Evidently, even linking M9 and gp120 in a single chain to decrease the entropic penalty for binding is not sufficient to prevent CD4D12 from displacing M9. The present work indicates that the broadly neutralizing response observed in guinea pig sera, associated with the gp120-CD4D12 single-chain construct, is exclusively due to anti-CD4D12 antibodies. While CD4i antibodies are clearly induced by this immunogen, they do not appear to be responsible for neutralization. This result is quite different from that observed in the earlier study that utilized cross-linked gp120-CD4D12 complexes in rhesus macaques (12). There are several possible explanations for this discrepancy.
First, cross-linked complexes behave differently from single chains. It is possible that the single-chain molecules may be more susceptible to proteolysis and/or degradation than the cross-linked complexes. However, we found no evidence that the linker region in our constructs was particularly sensitive to proteolysis. Indeed, incubation of our purified single-chain constructs with a serum-containing medium for more than 72 h showed no cleavage of the single chain into its individual components.
Second, the earlier animal studies (12) were carried out with macaques, while the present data used guinea pigs. Rodent immune responses are known to differ in important respects from those of primates. In particular, rodents lack the ability to produce long CDR3 loops (30); these loops are an important feature of the broadly neutralizing antibody b12, which targets the recessed CD4 binding site of gp120 (30). It was recently reported that sulfated CD4i antibodies derived from HIV-1-infected individuals and able to neutralize R5 isolates are also associated with long CDR3 loops.
Third, in the previous study (12), efforts were made to affinity purify the neutralizing activity by passing the material over an affinity matrix composed of gp120-M9 immobilized onto agarose beads. The ratios of antiserum titers to gp120 and CD4, respectively, were approximately 10 and 20 before and after the affinity purification (Fig. 2A and C) (12). It was suggested that the observed CD4 titer after affinity purification was due to promiscuous binding of anti-gp120 antibodies. However, the small change in relative titer after affinity purification also indicates that all CD4 antibodies may not have been removed by this treatment. Hence it is possible that some nontrivial fraction of the neutralization observed in the earlier study may have been due to anti-CD4 antibodies. A similar affinity purification procedure in our case resulted in a complete removal of anti-CD4 antibodies as well as a lack of observable neutralizing activity. In a recent study (15), the Bal gp120-CD4 single chain was injected into mice. While the molecule was highly immunogenic, the corresponding antisera did not show any neutralizing activity. MAbs generated from the immunized mouse bound to the single chain but, surprisingly, did not bind to the noncovalent gp120:CD4 complex or to free gp120. Since the present work clearly demonstrates that the single chain and the noncovalent complex are structurally very similar, this result is quite puzzling. It suggests that the MAbs may be binding to the linker region of the single chain. Another recent study (22) examined the immunogenicity of HIV gp120 cross-linked to MAb A32. A32 is an antibody that induces 17b and 48d epitopes upon binding on gp120, although it binds at a site distinct from CD4 (43). In agreement with the results reported here, no increase in the breadth of neutralization, relative to that observed by using gp120 alone as an immunogen, was seen with the A32-gp120 immunogen.
In summary, the present work shows that the broad neutralization activity observed in guinea pigs receiving the gp120-CD4D12 single-chain immunogen is due exclusively to anti CD4D12 antibodies. Although antibodies against CD4i epitopes were present in these sera, they were not associated with any significant neutralizing activity. These studies illustrate the difficulties involved in targeting CD4i epitopes and also suggest that CD4i epitopes may not be a good target for HIV-1 vaccine design.
Acknowledgments
This work was supported in part by a grant from the NIH AIDS Innovation grant program (NIH-AI-46984) to R.V. R.V. is a recipient of the Swarnajayanthi Fellowship (Government of India) and a Senior Research Fellowship from the Wellcome Trust.
We thank J. Robinson for providing antibody 17b and the reviewers for suggestions.
REFERENCES
- 1.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 2.Bandres, J. C., Q. F. Wang, J. O'Leary, F. Baleaux, A. Amara, J. A. Hoxie, S. Zolla-Pazner, and M. K. Gorny. 1998. Human immunodeficiency virus (HIV) envelope binds to CXCR4 independently of CD4, and binding can be enhanced by interaction with soluble CD4 or by HIV envelope deglycosylation. J. Virol. 72:2500-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binley, J. M., R. W. Sanders, B. Clas, N. Schuelke, A. Master, Y. Guo, F. Kajumo, D. J. Anselma, P. J. Maddon, W. C. Olson, and J. P. Moore. 2000. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J. Virol. 74:627-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bower, J. F., X. Yang, J. Sodroski, and T. M. Ross. 2004. Elicitation of neutralizing antibodies with DNA vaccines expressing soluble stabilized human immunodeficiency virus type 1 envelope glycoprotein trimers conjugated to C3d. J. Virol. 78:4710-4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bures, R., A. Gaitan, T. Zhu, C. Graziosi, K. M. McGrath, J. Tartaglia, P. Caudrelier, R. El Habib, M. Klein, A. Lazzarin, D. M. Stablein, M. Deers, L. Corey, M. L. Greenberg, D. H. Schwartz, and D. C. Montefiori. 2000. Immunization with recombinant canarypox vectors expressing membrane-anchored glycoprotein 120 followed by glycoprotein 160 boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 16:2019-2035. [DOI] [PubMed] [Google Scholar]
- 6.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 7.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 8.Choe, H., W. Li, P. L. Wright, N. Vasilieva, M. Venturi, C. C. Huang, C. Grundner, T. Dorfman, M. B. Zwick, L. Wang, E. S. Rosenberg, P. D. Kwong, D. R. Burton, J. E. Robinson, J. G. Sodroski, and M. Farzan. 2003. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 114:161-170. [DOI] [PubMed] [Google Scholar]
- 9.Connor, R. I., B. T. Korber, B. S. Graham, B. H. Hahn, D. D. Ho, B. D. Walker, A. U. Neumann, S. H. Vermund, J. Mestecky, S. Jackson, E. Fenamore, Y. Cao, F. Gao, S. Kalams, K. J. Kunstman, D. McDonald, N. McWilliams, A. Trkola, J. P. Moore, and S. M. Wolinsky. 1998. Immunological and virological analyses of persons infected by human immunodeficiency virus type 1 while participating in trials of recombinant gp120 subunit vaccines. J. Virol. 72:1552-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 11.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 12.Fouts, T., K. Godfrey, K. Bobb, D. Montefiori, C. V. Hanson, V. S. Kalyanaraman, A. DeVico, and R. Pal. 2002. Crosslinked HIV-1 envelope-CD4 receptor complexes elicit broadly cross-reactive neutralizing antibodies in rhesus macaques. Proc. Natl. Acad. Sci. USA 99:11842-11847. [Online.] doi:10.1073/pnas.182412199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouts, T. R., R. Tuskan, K. Godfrey, M. Reitz, D. Hone, G. K. Lewis, and A. L. DeVico. 2000. Expression and characterization of a single-chain polypeptide analogue of the human immunodeficiency virus type 1 gp120-CD4 receptor complex. J. Virol. 74:11427-11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haynes, B. F. 1996. HIV vaccines: where we are and where we are going. Lancet 348:933-937. [DOI] [PubMed] [Google Scholar]
- 15.He, Y., P. D'Agostino, and A. Pinter. 2003. Analysis of the immunogenic properties of a single-chain polypeptide analogue of the HIV-1 gp120-CD4 complex in transgenic mice that produce human immunoglobulins. Vaccine 21:4421-4429. [DOI] [PubMed] [Google Scholar]
- 16.Hill, C. M., H. Deng, D. Unutmaz, V. N. Kewalramani, L. Bastiani, M. K. Gorny, S. Zolla-Pazner, and D. R. Littman. 1997. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J. Virol. 71:6296-6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman, T. L., C. C. LaBranche, W. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. USA 96:6359-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koch, M., M. Pancera, P. D. Kwong, P. Kolchinsky, C. Grundner, L. Wang, W. A. Hendrickson, J. Sodroski, and R. Wyatt. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313:387-400. [DOI] [PubMed] [Google Scholar]
- 19.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 20.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure Fold Des. 8:1329-1339. (Erratum, 9:I, 2201.) [DOI] [PubMed] [Google Scholar]
- 21.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao, H. X., S. M. Alam, J. R. Mascola, J. Robinson, B. Ma, D. C. Montefiori, M. Rhein, L. L. Sutherland, R. Scearce, and B. F. Haynes. 2004. Immunogenicity of constrained monoclonal antibody A32-human immunodeficiency virus (HIV) Env gp120 complexes compared to that of recombinant HIV type 1 gp120 envelope glycoproteins. J. Virol. 78:5270-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, D. S. Burke, et al. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 24.Matthews, T. J. 1994. Dilemma of neutralization resistance of HIV-1 field isolates and vaccine development. AIDS Res. Hum. Retrovir. 10:631-632. [DOI] [PubMed] [Google Scholar]
- 25.McGaughey, G. B., M. Citron, R. C. Danzeisen, R. M. Freidinger, V. M. Garsky, W. M. Hurni, J. G. Joyce, X. Liang, M. Miller, J. Shiver, and M. J. Bogusky. 2003. HIV-1 vaccine development: constrained peptide immunogens show improved binding to the anti-HIV-1 gp41 MAb. Biochemistry 42:3214-3223. [DOI] [PubMed] [Google Scholar]
- 26.Moulard, M., S. K. Phogat, Y. Shu, A. F. Labrijn, X. Xiao, J. M. Binley, M. Y. Zhang, I. A. Sidorov, C. C. Broder, J. Robinson, P. W. Parren, D. R. Burton, and D. S. Dimitrov. 2002. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proc. Natl. Acad. Sci. USA 99:6913-6918. [Online.] doi:10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osburne, M. S., E. A. Neidhardt, J. E. Godoy, M. R. van Schravendijk, and T. H. Grossman. 1999. Production of secreted, soluble human two-domain CD4 protein in Escherichia coli. J. Immunol. Methods 224:19-24. [DOI] [PubMed] [Google Scholar]
- 29.Pantophlet, R., I. A. Wilson, and D. R. Burton. 2003. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J. Virol. 77:5889-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 31.Sattentau, Q. J., and J. P. Moore. 1991. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J. Exp. Med. 174:407-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sattentau, Q. J., J. P. Moore, F. Vignaux, F. Traincard, and P. Poignard. 1993. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 67:7383-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srivastava, I. K., L. Stamatatos, E. Kan, M. Vajdy, Y. Lian, S. Hilt, L. Martin, C. Vita, P. Zhu, K. H. Roux, L. Vojtech, D. C. Montefiori, J. Donnelly, J. B. Ulmer, and S. W. Barnett. 2003. Purification, characterization, and immunogenicity of a soluble trimeric envelope protein containing a partial deletion of the V2 loop derived from SF162, an R5-tropic human immunodeficiency virus type 1 isolate. J. Virol. 77:11244-11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Srivastava, I. K., K. VanDorsten, L. Vojtech, S. W. Barnett, and L. Stamatatos. 2003. Changes in the immunogenic properties of soluble gp140 human immunodeficiency virus envelope constructs upon partial deletion of the second hypervariable region. J. Virol. 77:2310-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sullivan, N., Y. Sun, J. Binley, J. Lee, C. F. Barbas III, P. W. Parren, D. R. Burton, and J. Sodroski. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J. Virol. 72:6332-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 40.VanCott, T. C., F. R. Bethke, D. S. Burke, R. R. Redfield, and D. L. Birx. 1995. Lack of induction of antibodies specific for conserved, discontinuous epitopes of HIV-1 envelope glycoprotein by candidate AIDS vaccines. J. Immunol. 155:4100-4110. [PubMed] [Google Scholar]
- 41.Vita, C., E. Drakopoulou, J. Vizzavona, S. Rochette, L. Martin, A. Menez, C. Roumestand, Y. S. Yang, L. Ylisastigui, A. Benjouad, and J. C. Gluckman. 1999. Rational engineering of a miniprotein that reproduces the core of the CD4 site interacting with HIV-1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 96:13091-13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 43.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiang, S. H., P. D. Kwong, R. Gupta, C. D. Rizzuto, D. J. Casper, R. Wyatt, L. Wang, W. A. Hendrickson, M. L. Doyle, and J. Sodroski. 2002. Mutagenic stabilization and/or disruption of a CD4-bound state reveals distinct conformations of the human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 76:9888-9899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiang, S. H., L. Wang, M. Abreu, C. C. Huang, P. D. Kwong, E. Rosenberg, J. E. Robinson, and J. Sodroski. 2003. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology 315:124-134. [DOI] [PubMed] [Google Scholar]
- 46.Yang, X., J. Lee, E. M. Mahony, P. D. Kwong, R. Wyatt, and J. Sodroski. 2002. Highly stable trimers formed by human immunodeficiency virus type 1 envelope glycoproteins fused with the trimeric motif of T4 bacteriophage fibritin. J. Virol. 76:4634-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, W., A. P. Godillot, R. Wyatt, J. Sodroski, and I. Chaiken. 2001. Antibody 17b binding at the coreceptor site weakens the kinetics of the interaction of envelope glycoprotein gp120 with CD4. Biochemistry 40:1662-1670. [DOI] [PubMed] [Google Scholar]
- 48.Zwick, M. B., L. L. Bonnycastle, A. Menendez, M. B. Irving, C. F. Barbas III, P. W. Parren, D. R. Burton, and J. K. Scott. 2001. Identification and characterization of a peptide that specifically binds the human, broadly neutralizing anti-human immunodeficiency virus type 1 antibody b12. J. Virol. 75:6692-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zwick, M. B., H. K. Komori, R. L. Stanfield, S. Church, M. Wang, P. W. Parren, R. Kunert, H. Katinger, I. A. Wilson, and D. R. Burton. 2004. The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5. J. Virol. 78:3155-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]