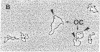Abstract
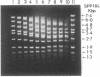
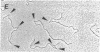
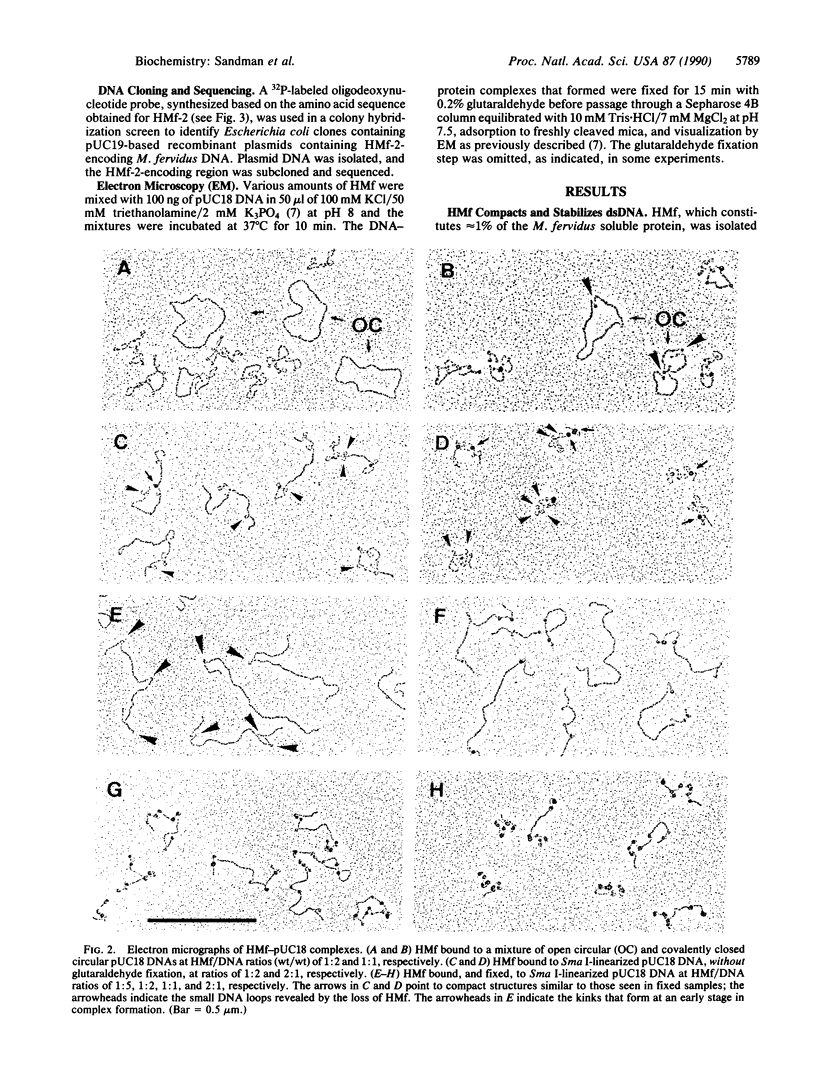
Methanothermus fervidus grows optimally at 83 degrees C. A protein designated HMf (histone M. fervidus) has been isolated from this archaeal hyperthermophile that binds to double-stranded DNA molecules and increases their resistance to thermal denaturation. HMf binding to linear double-stranded DNA molecules of greater than 2 kilobase pairs also increases their electrophoretic mobilities through agarose gels. Visualization of this compaction process by electron microscopy has demonstrated the formation of quasispherical, macromolecular HMf-DNA complexes. HMf is a mixture of approximately equal amounts of two very similar polypeptides designated HMf-1 and HMf-2. Determination of the DNA sequence of the gene encoding HMf-2 (hmfB) has revealed that over 30% of the amino acid residues in HMf-2 are conserved in the consensus sequences derived for eucaryal histones H2A, H2B, H3, and H4. These archaeal polypeptides and eucaryal histones appear therefore to have evolved from a common ancestor and are likely to have related structures and functions.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown J. W., Daniels C. J., Reeve J. N. Gene structure, organization, and expression in archaebacteria. Crit Rev Microbiol. 1989;16(4):287–338. doi: 10.3109/10408418909105479. [DOI] [PubMed] [Google Scholar]
- Chartier F., Crevel G., Laine B., Sautiere P. High-performance liquid chromatographic separation of variants of chromosomal proteins from prokaryotes. J Chromatogr. 1989 Apr 19;466:331–337. doi: 10.1016/s0021-9673(01)84628-8. [DOI] [PubMed] [Google Scholar]
- Chartier F., Laine B., Belaïche D., Touzel J. P., Sautière P. Primary structure of the chromosomal protein MC1 from the archaebacterium Methanosarcina sp. CHTI 55. Biochim Biophys Acta. 1989 Aug 14;1008(3):309–314. doi: 10.1016/0167-4781(89)90021-3. [DOI] [PubMed] [Google Scholar]
- Chartier F., Laine B., Bélaïche D., Sautière P. Primary structure of the chromosomal proteins MC1a, MC1b, and MC1c from the archaebacterium Methanothrix soehngenii. J Biol Chem. 1989 Oct 15;264(29):17006–17015. [PubMed] [Google Scholar]
- Choli T., Henning P., Wittmann-Liebold B., Reinhardt R. Isolation, characterization and microsequence analysis of a small basic methylated DNA-binding protein from the Archaebacterium, Sulfolobus solfataricus. Biochim Biophys Acta. 1988 Jul 13;950(2):193–203. doi: 10.1016/0167-4781(88)90011-5. [DOI] [PubMed] [Google Scholar]
- Choli T., Wittmann-Liebold B., Reinhardt R. Microsequence analysis of DNA-binding proteins 7a, 7b, and 7e from the archaebacterium Sulfolobus acidocaldarius. J Biol Chem. 1988 May 25;263(15):7087–7093. [PubMed] [Google Scholar]
- DeLange R. J., Williams L. C., Searcy D. G. A histone-like protein (HTa) from Thermoplasma acidophilum. II. Complete amino acid sequence. J Biol Chem. 1981 Jan 25;256(2):905–911. [PubMed] [Google Scholar]
- Drlica K., Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987 Sep;51(3):301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabry S., Hensel R. Primary structure of glyceraldehyde-3-phosphate dehydrogenase deduced from the nucleotide sequence of the thermophilic archaebacterium Methanothermus fervidus. Gene. 1988 Apr 29;64(2):189–197. doi: 10.1016/0378-1119(88)90334-4. [DOI] [PubMed] [Google Scholar]
- Fabry S., Lang J., Niermann T., Vingron M., Hensel R. Nucleotide sequence of the glyceraldehyde-3-phosphate dehydrogenase gene from the mesophilic methanogenic archaebacteria Methanobacterium bryantii and Methanobacterium formicicum. Comparison with the respective gene structure of the closely related extreme thermophile Methanothermus fervidus. Eur J Biochem. 1989 Feb 1;179(2):405–413. doi: 10.1111/j.1432-1033.1989.tb14568.x. [DOI] [PubMed] [Google Scholar]
- Haas E. S., Daniels C. J., Reeve J. N. Genes encoding 5S rRNA and tRNAs in the extremely thermophilic archaebacterium Methanothermus fervidus. Gene. 1989 Apr 30;77(2):253–263. doi: 10.1016/0378-1119(89)90073-5. [DOI] [PubMed] [Google Scholar]
- Heilmann H., Reeve J. N. Construction and use of SPP1v, a viral cloning vector for Bacillus subtilis. Gene. 1982 Jan;17(1):91–100. doi: 10.1016/0378-1119(82)90104-4. [DOI] [PubMed] [Google Scholar]
- Kayne P. S., Kim U. J., Han M., Mullen J. R., Yoshizaki F., Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988 Oct 7;55(1):27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Laine B., Chartier F., Imbert M., Lewis R., Sautiere P. Primary structure of the chromosomal protein HMb from the archaebacteria Methanosarcina barkeri. Eur J Biochem. 1986 Dec 15;161(3):681–687. doi: 10.1111/j.1432-1033.1986.tb10493.x. [DOI] [PubMed] [Google Scholar]
- Pettijohn D. E. Histone-like proteins and bacterial chromosome structure. J Biol Chem. 1988 Sep 15;263(26):12793–12796. [PubMed] [Google Scholar]
- Reddy T. R., Suryanarayana T. Archaebacterial histone-like proteins. Purification and characterization of helix stabilizing DNA binding proteins from the acidothermophile Sulfolobus acidocaldarius. J Biol Chem. 1989 Oct 15;264(29):17298–17308. [PubMed] [Google Scholar]
- Searcy D. G., Delange R. J. Thermoplasma acidophilum histone-like protein. Partial amino acid sequence suggestive of homology to eukaryotic histones. Biochim Biophys Acta. 1980 Aug 26;609(1):197–200. doi: 10.1016/0005-2787(80)90212-9. [DOI] [PubMed] [Google Scholar]
- Tanaka I., Appelt K., Dijk J., White S. W., Wilson K. S. 3-A resolution structure of a protein with histone-like properties in prokaryotes. Nature. 1984 Aug 2;310(5976):376–381. doi: 10.1038/310376a0. [DOI] [PubMed] [Google Scholar]
- Weil C. F., Cram D. S., Sherf B. A., Reeve J. N. Structure and comparative analysis of the genes encoding component C of methyl coenzyme M reductase in the extremely thermophilic archaebacterium Methanothermus fervidus. J Bacteriol. 1988 Oct;170(10):4718–4726. doi: 10.1128/jb.170.10.4718-4726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D., McBride C. A comprehensive compilation and alignment of histones and histone genes. Nucleic Acids Res. 1989;17 (Suppl):r311–r346. doi: 10.1093/nar/17.suppl.r311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillig W., Palm P., Reiter W. D., Gropp F., Pühler G., Klenk H. P. Comparative evaluation of gene expression in archaebacteria. Eur J Biochem. 1988 May 2;173(3):473–482. doi: 10.1111/j.1432-1033.1988.tb14023.x. [DOI] [PubMed] [Google Scholar]