Abstract
Posttranscriptional status of messenger RNAs (mRNA) can be affected by many factors, most of which are RNA-binding proteins (RBP) that either bind mRNA in a nonspecific manner or through specific motifs, usually located in the 39 untranslated regions. RBPs can also be recruited by small noncoding RNAs (sncRNA), which have been shown to be involved in posttranscriptional regulations and transposon repression (eg, microRNAs or P-element–induced wimpy testis–interacting RNA) as components of the sncRNA effector complex. Non–sncRNA-binding RBPs have much more diverse effects on their target mRNAs. Some can cause degradation of their target transcripts and/or repression of translation, whereas others can stabilize and/or activate translation. The splicing and exportation of transcripts from the nucleus to the cytoplasm are often mediated by sequence-specific RBPs. The mechanisms by which RBPs regulate mRNA transcripts involve manipulating the 39 poly(A) tail, targeting the transcript to polysomes or to other ribonuclear protein particles, recruiting regulatory proteins, or competing with other RBPs. Here, we briefly review the known mechanisms of posttranscriptional regulation mediated by RBPs, with an emphasis on how these mechanisms might control spermatogenesis in general.
Keywords: Posttranscriptional regulation, RNA stability, ribonuclear particles, translation, splicing, small noncoding RNAs, germ cell, sperm, fertility
Gene expression determines cell function, and it can be regulated in a variety of ways. The most well known regulatory mechanism is through transcriptional regulation by recruiting or blocking transcriptional machinery. Some examples include transcriptional activation or suppression, DNA methylation of the promoter region, or chromatin remodeling. However, a growing field of study focuses on the regulation of gene expression after messenger RNAs (mRNA) are transcribed, and these posttranscriptional mechanisms include regulated splicing, degradation, transportation, and suppression of translation. The different fates of mRNAs are controlled largely by RNA-binding proteins (RBP). RBPs have many roles throughout the body, ranging from embryogenesis to the formation of dendrites to regulating cytokines of the immune system (Crawford et al, 1997; Colegrove-Otero et al, 2005). These functions are achieved by cooperation or competition with other proteins, through mRNA poly(A) tail modification, and/or via targeting transcripts to ribo-nuclear protein particles (RNP), which represent diverse nuclear and cytoplasmic compartments.
Typically, an RBP is expressed in the nucleus and binds to precursor mRNA (pre-mRNA). Following maturation of the transcript, the RBP may remain there for some time before being modified to cause it to move to the cytoplasm, carrying the transcript with it. There, it will be localized to particular cytoplasmic compartments: RNPs. When needed by the cell, the RBP will associate with polysomes and the associated transcript will be translated. There are many variations in this pathway that will be discussed in detail.
Degradation of transcripts is highly variable and thought to be controlled mainly by RBPs. One of the main functions of RNPs is to compartmentalize proteins, effectively localizing transcripts with a subset of cytoplasmic proteins while separating them from others. For example, exosomes serve as sites of general degradation through 3′→5′ exoribonucleases, whereas stress granules have 5′→3′ exoribonucleases, which only degrade decapped transcripts (Mukherjee et al, 2002; Kedersha et al, 2005; Anderson and Kedersha, 2008). In mammals, once transcripts are exported from the nucleus, the half-life generally exceeds 10 hours, but for rapidly degraded transcripts, half-lives can be as short as 15 minutes (Newbury et al, 2006). An important step commonly observed in the regulation of stability is altering the length of the poly(A) tail, which often precedes decapping and 5′→3′ degradation (discussed in the upcoming text; Couttet et al, 1997).
Regulation of translation, likewise, is highly variable and controlled by three main mechanisms: poly(A) tail modification, association with RNPs, and competition with translation machinery. Lengthening the poly(A) tail of a transcript can increase rates of translation up to 100-fold. This process is thought to be mediated by poly(A)-binding protein (PABP), which recruits translation initiation factors (Sheets et al, 1994). However, the most significant mechanism involves directing the transcripts to either RNPs, which usually correlate with translational inhibition, or polyribosomes, which are the sites of translation (Iguchi et al, 2006).
Spermatogenesis provides us with an excellent model to study posttranscriptional fate control of mRNAs because transcriptional machinery is completely shut down when spermatids start the process of nuclear condensation and elongation (Figure 1). De novo transcription is halted permanently during mid spermiogenesis, long before differentiation is complete (Monesi, 1964; Sassone-Corsi, 2002). To compensate for the lack of new mRNAs, transcripts must be synthesized and stored so that their translation can coincide with demand. RBPs have been shown to have a role in this temporal disconnect between transcription and translation (Monesi, 1964; Sassone-Corsi, 2002). In mice, inactivation of many of these RBPs causes infertility and spermatogenic arrest at various steps of germ cell differentiation (Table 1). This highlights the importance of these proteins in regulating the expression of necessary genes, and the necessity for translation to be delayed and regulated. An example of this is observed during chromatin condensation, where histones are replaced by transition proteins, which are subsequently replaced by protamines, resulting in highly compacted chromosomes (Zhao et al, 2004). The mRNAs for protamines can be held inactive for more than a week prior to translation in mammalian spermatogenesis (Kleene et al, 1984). If the protamine mRNA is translated too early, premature nuclear condensation occurs, leading to abnormal spermatogenesis and infertility (Lee et al, 1995).
Figure 1.
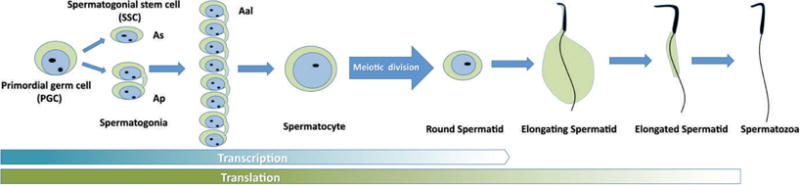
Delayed translation in late spermiogenesis. Spermatogenesis is a process through which spermatogonial stem cells differentiate into spermatocytes, which then undergo meiotic cell divisions and become round spermatids. Round spermatids undergo morphogenesis, a process termed “spermiogenesis,” and differentiate into spermatozoa, which are the male gamete. During this complex differentiation process, transcription completely ceases upon nuclear condensation and elongation in mid spermiogenesis. Therefore, proteins required for the rest of the differentiation program must be produced through translation using the messenger RNAs synthesized and stored prior to the cease of transcription. As indicates single type A spermatogonia; Ap, paired type A spermatogonia; Aal, aligned type A spermatogonia.
Table 1.
Known RNA-binding proteins (RBP) involved in spermatogenesis of the mouse
| RBP | Spermatogenic Arrest | Binding Motif/Transcript Association | Fertility Status | Associated RNP/Organelle | References |
|---|---|---|---|---|---|
| TIAR (TIAL-1) | Defective development of PGCs | A G-U–rich sequence and a C-rich sequence | No sperm. Infertile. | Stress granules | Beck et al, 1998; Kim et al, 2007 |
| NANOS 3 | PGC apoptosis and loss of SSCs | Associates with Pum2 for binding | Infertile, both males and females | … | Tsuda, 2003; Gupta et al, 2009 |
| NANOS 2 | Depleted SSCs | Associates with Pum2 for binding | Infertile, males only | … | Tsuda, 2003; Gupta et al, 2009; Sada et al, 2009 |
| DAZL | Arrest in early meiotic prophase | Minimal binding site: U3G/CU3. More completely: (G/CUn)n. | Infertile, in both male and female | … | Kee et al, 2009; VanGompel and Xu, 2010 |
| PIWIL4 (MIWI 2) | Arrest in early meiotic prophase | piRNA-associated | Infertile, males only | Chromatoid body. Polysomes. | Parvinen, 2005; Grivna et al, 2006; Carmell et al, 2007 |
| PIWIL2 (MILI) | Arrest in early meiotic prophase | piRNA-associated | Infertility, males only | Chromatoid body | Kuramochi-Miyagawa et al, 2004 |
| CPEB | Arrest at pachytene stage | U4–6A1–3U, U12–27 | Infertile, both males and females | … | Tay and Richter, 2001; Charlesworth et al, 2004 |
| DAZAP1 | Arrest around late pachytene | AAAUAG or GU(1–3)AG | Infertile, both male and female | Hsu et al, 2009 | |
| PIWIL1 (MIWI) | Arrest in early round spermatid (steps 1 to 4) | piRNA associated | Infertile, in males only | Chromatoid body. Polysomes. | Deng and Lin, 2002; Parvinen, 2005; Grivna et al, 2006 |
| BOULE | Round spermatid arrest at step 6 | Minimal binding site: U3G/CU3. More completely: (G/CUn)n. | Azoospermia. Infertile, male only. | … | VanGompel and Xu, 2010 |
| CELF1 (CUG-BP1) | Defective elongation. Arrest at step 7 of spermiogenesis. | EDEN motifs. Class III AREs. | Impaired fertility. Males and females. | … | Barreau et al, 2005; Kress et al, 2007 |
| GRTH (Ddx25) | Arrest at step 8 round spermatids and failure to elongate | … | Azoospermia. Infertile, male only. | Chromatoid body. Polysomes. | Tsai-Morris et al, 2004 |
| MSY2 (YBX2) | Misshapen spermatids | Transcripts with Y-box in DNA promoter. AACAUC? | Infertile, both male and female | RNPs. Polysomes. | Yang et al, 2007; Xu et al, 2009 |
| SAM 68 (KHDRBS1) | Aberrant elongated spermatids | AUUAAAA and poly(A) | Immotile sperm. Infertile. | Polysomes | Lin et al, 1997; Paronetto et al, 2009 |
| MSY4 (CADA) | Fewer, but morphologically normal sperm | Transcripts with Y-box in DNA promoter. MSY-RNA. UCCAUCA? | Decreased fertility, male only | … | Bouvet etal,1994; Lu et al, 2006 |
| TIA-1 | Unknown | AREs. 30–37 U-rich element. | Reduced fertility | Stress granules | Kedersha et al, 2000; Piecyk et al, 2000; Lopez de Silanes et al, 2005 |
| PUMILIO | Normal spermatogenesis | 8 copies of UGUANAUA. NREs. UGUANAUARNNNNBBBBSCCS. | Fertile | … | Xu et al, 2007; Gupta et al, 2009 |
| HuR (ELAVL1) | Unknown | 17- to 20-nt–long U/G-rich sequence | Unknown | Chromatoid body. Polysomes. Stress granules. P-bodies. Exosomes. | Nguyen Chi et al, 2009 |
| AUF1 (hnRNP D) | Unknown | Class I and II AREs and a 29- to 39-nt AU-rich sequence containing UUAG | Unknown | Exosomes | Mazan-Mamczarz et al, 2009; Ishimaru et al, 2010 |
| TTP (ZFP36) | Unknown | Class II AREs | Unknown | Stress granules. P-bodies. | Barreau et al, 2005; Kedersha et al, 2005; Newbury et al, 2006 |
| CPSF | Unknown | AAUAAA, 12–30 nt upstream of cleavage site | Unknown | Spliceosomes | Charlesworth et al, 2004 |
Abbreviations: ARE, AU-rich element; AUF1, AU-rich binding factor 1; CELF, CUGBP, ELAV-like family member 1; CPEB, cytoplasmic polyadenylation element–binding protein; CPSF, cleavage and polyadenylation–specific factor; CUG-BP1, CUG repeat binding protein; DAZAP, DAZ-associated protein; DAZL, DAZ-like protein; Ddx, DEAD (Asp-Glu-Ala-Asp) box polypeptide; EDEN, embryo deadenylation element; ELAVL1, embryonic lethal and abnormal vision–like 1; GRTH, gonadotropin-regulated testicular RNA helicase; hnRNP, heterogeneous nuclear ribonucleoprotein; HuR, Hu antigen R; KHDRBS1, KH domain–containing, RNA-binding, signal transduction–associated 1; MILI, MIWI-like; MIWI, murine PIWI homolog; MSY, mouse Y-box protein; NRE, NANOS response element; nt, nucleotide; P-bodies, processing bodies; PGC, primordial germ cell; piRNA, PIWI-interacting RNA; PIWIL, PIWI-like homolog; RNP, ribonucleoprotein particle; SAM68, SRC-associated in mitosis, 68 kd; SSC, spermatogonial stem cell; TIA-1, T-cell–restricted intracellular antigen 1; TIAL-1, TIA-like protein 1; TIAR, TIA-related protein; TTP, tristetraprolin; YBX, Y-box protein; ZFP, zinc finger protein.
Despite the high degree of selective pressure for making functional sperm, male infertility is relatively common. This is likely because of the fact that sperm development is complex, with many opportunities for errors. Primordial germ cells (PGC) migrate to the testis, where they become gonocytes (also called prospermatogonia). These cells then migrate to the basal lamina of the seminiferous epithelium, where they receive signals from somatic Sertoli cells. The gonocytes then divide and differentiate into either spermatogonial stem cells or proliferating spermatogonia (Figure 1). The diploid spermatogonia undergo several rounds of mitosis before entering into the meiotic phase of their development. The resulting spermatocytes then divide twice to become haploid round spermatids, which undergo morphologic changes, including polarization, elongation, chromatin condensation, and development of the acrosome and flagellum to become spermatozoa. Each cell division from spermatogonia to spermatids is incomplete, leaving intercellular bridges that connect the cells. This allows mRNA and proteins from one cell to diffuse or be transported into an adjacent cell, including the movement of chromatoid bodies (CB; discussed in the upcoming text).
It has long been known that motifs in the untranslated regions (UTR) of a transcript are important for regulating nucleus-to-cytoplasm transport, mRNA stability, and translational efficiency, but only in the last 2 decades have the mechanisms that mediate this begun to be uncovered (Barreau et al, 2005). To stabilize/destabilize these transcripts, RBPs must recognize and bind these motifs. There are three basic types of RBPs: those that are non–sequence specific, those that are associated with small noncoding RNAs (sncRNA) in the effector complexes, and those that bind to specific elements in mRNAs. The RBPs that bind mRNAs in a nonspecific manner are unlikely to be responsible for the variable rates of translation of certain subgroups because the regulation of translation is not universal for all transcripts, and it has been experimentally determined that alterations in the 3′ UTR of transcripts can dramatically alter their regulation. Among sncRNA-associated RBPs, microRNA (miRNA)–dependent RBPs bind miRNAs as components of the RNA-induced silencing complex (RISC); miRNAs bind to their specific target sequence, usually in the 3′ UTR, conferring motif-specific activity onto their associated RISC (Agami, 2010). Until recently, this was thought to result only in mRNA degradation or inhibition of translation by Argonaute (AGO)–induced disruption of eukaryotic translation initiation factor 4 (EIF4) and EIF6 (Chendrimada et al, 2007; Kiriakidou et al, 2007). However, new lines of evidence have shown that under certain conditions, mammalian miRNAs can encourage translation by recruiting an Argonaute–fragile X mental retardation-related protein complex, AGO2-FXR1 (Vasudevan and Steitz, 2007).
The much more diverse group of sncRNA-independent RBPs can have a variety of functions. They can have a role in splicing, nuclear export, transportation, targeting, translational regulation, and regulation of stability. Importantly, the same RBP can have different roles depending on the modifications of the RBP, conditions of the cell, or the presence of other proteins (Colegrove-Otero et al, 2005).
The motifs that miRNAs and RBPs bind on a transcript are of vital importance. These are diverse elements that determine which protein complexes will bind to the transcript. The number, location relative to other elements, and nuances in sequence fine-tune the binding of various RBPs and miRNAs, thus determining the fate of the transcript.
General Mechanisms Underlying the Fate Control of mRNA Transcripts
The main mechanisms for the fate control of a transcript involve splicing, cytoplasmic compartmentalization, translational initiation, and stability. Not all compart-mentalized transcripts are translated or stabilized; for example, most transcripts that are localized to the exosome are generally degraded, and some commonly stored transcripts are not translated under normal conditions, such as tumor necrosis factor alpha (TNF-α; Sun et al, 2007). Some RBPs can recognize and bind specific elements in the 3′ UTR and recruit various splicing factors inside the nucleus (Figure 2A). This determines which exons are expressed and which are spliced out. Alternative splicing thus creates variations in proteins that are translated from the same pre-mRNA (Black, 2003). After splicing, pre-mRNAs are modified with a 5′ 7-methylguanosine cap and a poly(A) sequence on their 3′ tails (Figure 2B), which are important modifications for nuclear export (Lei and Silver, 2002). At this point, mRNA is considered a mature transcript and can exit the nucleus through nuclear pore complexes, a process aided by nonspecific and specific binding RBPs (Figure 2B; Lei and Silver, 2002). As a transcript leaves the nucleus, the bound RBPs either remain in the nucleus or accompany the transcript into the cytoplasm. In the cytoplasm, the transcript either remains bound to the same RBPs or recruits others. These RBPs will determine the cytoplasmic compartment to which the transcript will be localized. In Drosophila, an RBP may attach itself to a STAUFEN complex, which then binds to either kinesin or dynein, motor proteins that move unidirectionally along microtubules (Yoon and Mowry, 2004; Weil et al, 2010). In mammals, similar interactions exist, in which STAU1, a homolog of STAUFEN, is necessary for transport to polysomes or exosomes (Tang et al, 2001; Kim et al, 2005). A mammalian testis-specific kinesin family protein, KIF17b, has been identified in nuclear export, but there has been no indication that STAUFEN is necessary for this activity (Kotaja et al, 2006). Using these motor proteins, transcripts can be carried to their destinations within the cytoplasm (Figure 2C).
Figure 2.
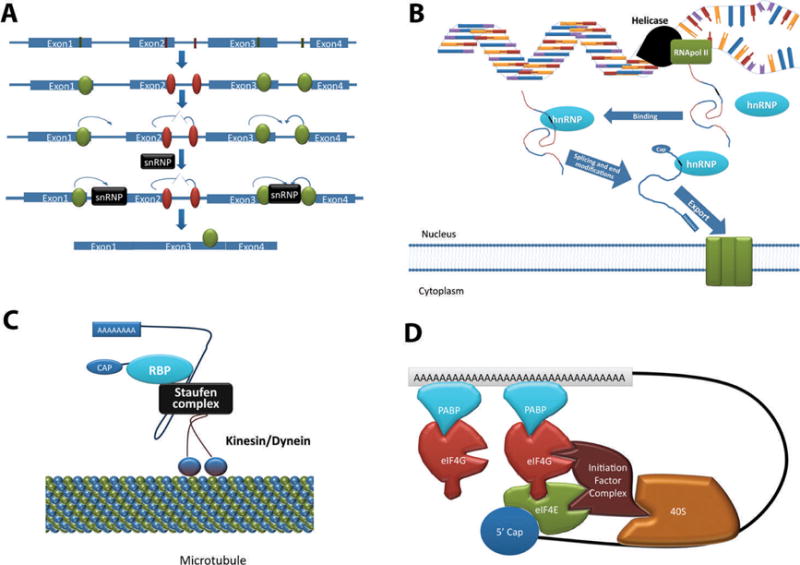
Known mechanisms of posttranscriptional regulation of messenger RNAs (mRNA). (A) Splicing. The top RNA is heterogeneous nuclear RNA (hnRNA) that includes recognition motifs in its exons and introns. Shown in red are enhancers, whereas those shown in green are silencers. Proteins that bind to these sequences are heterogeneous nuclear ribonucleoproteins (hnRNP). Those that encourage exon inclusion are activators, whereas those that discourage exon inclusion are repressors. The mechanism by which they perform this function is by interactions with small nuclear ribonucleoprotein particles (snRNP), particularly U1 and U2. Some hnRNPs remain bound to the transcript after splicing, and will often mediate export from the nucleus. (B) Formation of mature mRNA. Precursor mRNA transcribed from DNA contains motifs that hnRNPs recognize and with which they associate. Splicing occurs simultaneously with cap and tail modifications. This process yields mature mRNA that is exported to the cytoplasm. (C) Transport complex. RNA-binding proteins (RBP) can associate with a STAUFEN complex that associates with kinesin or dynein. This will allow transcripts to be transported along the cytoskeleton to where they are needed. Usually RBPs block the association of the initiation complex, keeping the transcript translationally inactive during transportation. (D) “Closed loop” model of translation initiation. A long poly(A) tail attracts poly(A)-binding protein (PABP), which associates with eukaryotic translation initiation factor 4G (EIF4G). EIF4E has a high affinity for the 59 cap and for EIF4G. An initiation factor complex associates with EIF4G and EIF4E. This complex binds the 40S subunit. The 40S subunit along with several initiation factors (together termed “43S”) will scan the transcript for the start codon, at which point the initiation translational RNA will bind and recruit the large subunit (60S subunit), starting translation.
Interactions between RBPs and other cytoplasmic elements determine whether a transcript will become localized to either polyribosomes or various RNPs (detailed discussion provided shortly). Translation can only occur at ribosomes, so colocalization with polyri-bosomes generally corresponds to active translation, whereas association with RNPs generally is indicative of translational inactivation (Iguchi et al, 2006).
The main regulator of translation is at initiation, although regulation at the elongating and terminating steps can occur as well (DeGracia et al, 2008); however, because regulation at elongation and termination is rare, for the sake of brevity, only initiation will be discussed. The most commonly accepted model of translation initiation is the “closed loop” model (Figure 2D). In this model, the key to initiation of translation is the recruitment of the 43S subunit of ribosomes. The rate-limiting step in translation initiation is the binding of EIF4E to EIF4G. The basic steps are as follows: 1) attachment of EIF4E to the 5′ cap; 2) joining of PABP to the poly(A) tail; 3) binding of EIF4G to PABP and to EIF4E; 4) binding of an initiation factor complex to EIF4G; 5) recruitment of the 40S subunit to the initiation factor complex and to the transcript; and 6) binding of several other proteins, including EIF2 and the initiation methionine translational RNA (tRNAiMet), to the 40S subunit, to create the 43S subunit. This complex searches down the transcript for the initiation codon. At this point, EIF2 recognizes the start codon and stalls so that the tRNAiMet can bind. This allows the large ribosomal subunit (60S) to bind, which causes translation to begin. Many other proteins are involved, and variations exist by which cells initiate translation. For example, histone transcripts use stem-loop–binding proteins that do not require a poly(A) tail or PABP to recruit EIF4G (Sanchez and Marzluff, 2004; Colegrove-Otero et al, 2005).
The association of EIF4E and EIF4G can be inhibited by evolutionarily conserved EIF4E-binding proteins (4E-BPs) or functionally related proteins, such as Maskin or its mammalian homolog TACC3, a cytoplasmic polyadenylation element–binding protein (CPEB) and 4E-BP, conserved in a wide variety of animals (discussed soon; Stebbins-Boaz et al, 1999; Hodgman et al, 2001; Hao et al, 2002; Colegrove-Otero et al, 2005). 4E-BPs are not bound to the transcript, but many functionally similar proteins, such as Bicoid in Drosophila, can bind directly to elements in the 3′ UTR (Niessing et al, 2002; Ottone et al, 2010). Others bind to RBPs, which recognize elements in the 3′ UTR, as is the case with Maskin, which associates with CPEB (Figure 3; Cao and Richter, 2002).
Figure 3.
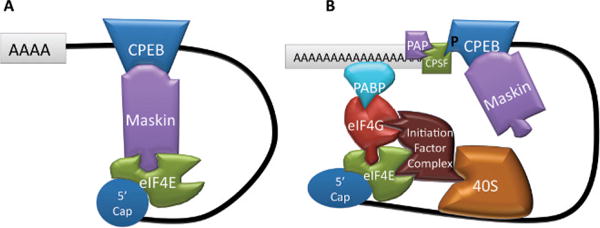
Regulation of cytoplasmic polyadenylation element– binding protein (CPEB) function by phosphorylation. (A) When not phosphorylated, CPEB will hold the transcript inactive by association with Maskin. Maskin binds EIF4E competitively inhibiting binding by eukaryotic translation initiation factor 4G (EIF4G). This activity is encouraged by the maintenance of a short poly(A) tail. (B) Once phosphorylated, CPEB binds to cleavage and polyadenylation–specific factor (CPSF) that associates with polyadenylate polymerase (PAP). This lengthens the poly(A) tail, causing poly(A)-binding protein (PABP) binding. This encourages association with EIF4G. This occurs in conjunction with a loss of affinity of Maskin for EIF4E, although Maskin remains bound to CPEB. Together, this causes the initiation complex to form and encourages the small ribosomal subunit to bind, which is a limiting step for translation initiation.
Control of the length of the poly(A) tails is important for both translation and stability. The poly(A) tails are usually lengthened by recruiting polyadenylate polymerase (PAP) or shortened by recruiting poly(A)-specific ribonuclease (PARN; Copeland and Wormington, 2001; Kashiwabara et al, 2002). The length of the poly(A) tail determines the number of PABPs that can bind. Because PABPs recruit EIF4G, PABP facilitates EIF4G binding to EIF4E and the downstream steps leading to translational initiation. PABP does not act directly with EIF4G; instead, PABP-interacting proteins, namely PAIP1, mediate this interaction (Craig et al, 1998). Transcripts that do not use PABP to bind to EIF4G are not subject to translational up-regulation by polyadenylation. For example, histone transcripts are actually translated at a lower rate if the poly(A) tail is longer (Sanchez and Marzluff, 2004).
During elongation in spermatids, the opposite effect occurs. Translation is accompanied by a rapid shortening of the poly(A) tail (Yanagiya et al, 2010). In late spermiogenesis, there is an increase in levels of PAIP2a, which has low affinity for EIF4G and associates with PABP, competing with PAIP1 for binding sites (Khaleghpour et al, 2001; Yanagiya et al, 2010). PAIP2a knockouts have significantly higher levels of PABP (Yanagiya et al, 2010), leading to the hypothesis that excessive PABP concentration may be responsible for translational inhibition of transcripts with long poly(A) tails. Two possible mechanisms are high PABP levels causing nonspecific binding by PABP or free PABP competing with bound PABP for EIF4G, thus reducing EIF4G binding with EIF4E (Kini et al, 2010). Because there is evidence that high levels of PABP inhibit deadenylation in late spermiogenesis, accompanying translational quiescence, it becomes unclear whether deadenylation is an effect of decreasing levels of PABP or whether deadenylation is necessary for translation at this stage.
Regulation of transcript degradation is vital for the control of translation rates and is of particular importance in spermatogenesis, where transcripts must be stabilized for the temporal disconnect between transcription and translation. The main underlying mechanism is targeting transcripts to RNPs. Different RNPs contain different enzymes; for example, exosomes and processing bodies (P-bodies) have exoribonucleases that degrade mRNAs, whereas most of the others do not (see upcoming text; van Dijk et al, 2007; Anderson and Kedersha, 2008).
A noteworthy point is that deadenylation is usually followed by decapping and rapid degradation. Thus, most RBPs that provide stability for a transcript do so by promoting adenylation or preventing deadenylation. However, this is not always the case. Some RBPs, such as embryonic lethal and abnormal vision–like 1 (ELAVL1), more commonly known as Hu antigen R (HUR), protect a transcript from degradation but have no effect on the length of the poly(A) tail (Espel, 2005).
Ribonuclear Particles: Cytoplasmic Compartments for mRNA Fate Control
RNPs are foci where specific proteins, mRNAs, and sncRNAs are targeted for processing, sorting, storage, or degradation. Aside from polysomes, RNPs are generally associated with translational arrest (Iguchi et al, 2006). Although some are associated with exonucleases and are thus sites for mRNA degradation, others are locations where transcripts can be protected and remain translationally silent until they are needed. A great many contain processing machinery, such as PAPs, PARNs, helicases, splicing factors, RBPs, or RBP-modifying enzymes (Barreau et al, 2005). Although different RNPs contain many common components, they still can be differentiated by unique proteins and by the cell types in which they exist (Table 2). A subtype of RNPs contains groups of functionally related mRNAs and their regulatory proteins, known as RNA operons, which serve to coordinate regulation of associated genes (Keene, 2007). For example, there exist several distinct Hu/ELAV granules (discussed soon) in a cell, each associated with functionally related transcripts (Keene and Lager, 2005; DeGracia et al, 2008).
Table 2.
Ribonuclear protein particles (RNP), their associated proteins, and potential functions
| RNP | Associated Factors | Function |
|---|---|---|
| Stress granules | PABP1, EIF4E (during certain stress), EIF3b, EIF2α, ribosome subunit 48S, TIA-1, TIAR, 5′-3′ exonuclease XRN1, TTP. | General translational arrest. Sorting centers. Aggregate due to EIF2α phosphorylation. |
| Processing-bodies, P-bodies, GW bodies | 5′-3′ exonuclease XRN1, decapping enzymes (DCP1, DCP2, DHH1, DDx6), GW182, RNA-silencing machinery (AGO, RNA helicases), TTP, miRNA machinery. | Decay/storage. Degrades deadenylated mRNAs. miRNA functions. |
| Male germinal granules-intermitochondrial cement, nuage, chromatoid body | Helicases, decapping enzymes, AGO proteins, PABPs, miRNAs, XRN1, GW182, polyadenylated RNAs, MVH (DDX4), HUR. | Male germ cell specific. Storage of long-lived mRNAs, mRNA processing, miRNA activity. Transport through cytoplasmic bridges from one cell to another. |
| Exosome | Initiation factors, KHSRP, 3′–5′ exonucleases, TTP. | Degrades deadenylated and uncapped mRNAs. 3′ →5′ degradation. |
| Spliceosome Heterogeneous nuclear ribonucleoprotein complexes | snRNPs U1, U2, U4, U5, and U6. Nuclear PAP, phosphatase, guanosyl transferase, and (guanine-N7-)-methyltransferase. | Removes introns, ligates exons. Polyadenylation, capping, and general nuclear to cytoplasmic transport. Specific transport complexes also exist. |
| Polyribosomes | Ribosomes. Translation initiation factors. | Site of translation. Form and unform based on cell needs. |
| RNA operon-ELAV/Hu granules | eI4G, EIF4E, PABP, Hu proteins, mRNA. | Coordinate activity of mRNA. Relay center for translation, silencing, or degradation. |
| Transport RNPs | Translationally inactive mRNAs and ribosomes. STAUFEN (mRNA transportation protein). Fragile X mental retardation protein. Do not associate with TIA-1 or TTP. | Transport of mRNAs and ribosomes to distal processes. Aiding in activity- and location-dependent translation. Sequester mRNA from 40S during stress. |
Abbreviations: AGO, Argonaute; DCP, decapping enzyme; DHH1, DExD/H-box helicase 1; EIF, eukaryotic translation initiation factor; GW, glycein tryptophan; KHSRP, KH-type splicing regulatory protein; mRNA, messenger RNA; miRNA, microRNA; MVH, mouse VASA homolog; PABP1, poly(A)-binding protein 1; PAP, poly(A) polymerase; XRN1, exoribonuclease 1. For all other abbreviations, see Table 1 abbreviations footnote.
Polysomes
Polysomes, also known as polyribosomes or ergosomes, are the main sites of translation. Although polysomes are technically RNPs, they are often discussed in the literature as being separate from RNPs (Iguchi et al, 2006). Polysome aggregation is controlled to correspond with cell needs. Polysomes are groups of ribosomes clustered around a single mRNA transcript, allowing for simultaneous translation of the transcript, resulting in rapid protein production. Polysomes have been found ubiquitously in wide evolutionary lineages in all protein-producing cell types.
Stress Granules
Stress granules were originally believed to be created as part of a stress response (Ivanov and Nadezhdina, 2006). Under certain stresses, such as hyperthermia or ischemia, they grow rapidly to a size discernable under a light microscope. Recent revelations have indicated that stress granules exist in nonstressed cells as well but are significantly smaller in size (Kedersha et al, 2005; Anderson and Kedersha, 2008). These smaller stress granules are thought to be sorting centers, constitutively involved in processing and relaying transcripts to other foci. During stressful conditions, transcripts are sequestered in stress granules, resulting in global translational arrest, causing a rapid increase in stress granule size (Kayali et al, 2005; DeGracia et al, 2008). Phosphorylation of EIF2α and association of T-cell–restricted intracellular antigen 1 (TIA-1)–like protein-1 (TIAL-1), an RBP more commonly referred to as TIA-related protein (TIAR), has been shown to be the cause of the rapid increases in the size of stress granules under stressful conditions (Kedersha et al, 1999). Although stress granules have only been observed in somatic cells, they are speculated to exist in germ cells as well (Paronetto and Sette, 2010).
Proteins that have been known to commonly associate with stress granules under certain conditions are TIA-1, TIAR, PABP1, EIF4E (during stress), EIF3b, EIF2α, miRNA machinery, and the small ribosomal subunit (Leung et al, 2006; Anderson and Kedersha, 2008). Even though mammalian stress granules contain 5′→3′ exoribonuclease 1 (XRN1), they do not contain decapping enzymes, so it is unlikely that mRNA degradation occurs in these RNPs (Anderson and Kedersha, 2008). Zinc finger protein 36 (ZFP36), more commonly known as tristetraprolin (TTP), can also be recruited to stress granules when modified (Newbury et al, 2006).
Processing Bodies
Processing bodies, also known as P-bodies, glycein tryptophan (GW) bodies (these may be separate RNPs), or decapping bodies, exist in all somatic cells but have not yet been observed in germ cells. P-bodies are membraneless foci involved in decay or storage of mRNA transcripts (Bettegowda and Wilkinson, 2010). If a transcript has been deadenylated, it will be decapped and degraded (Couttet et al, 1997); thus, both the deadenylase complex, CCR4/NOT, and decapping enzymes DCP1 and DCP2 are required for mRNA degradation in murine P-bodies (Behm-Ansmant et al, 2006). Although technically some components of the CCR4/NOT complex, such as CAF1 and CCR4, are 3′→5′ exoribonucleases, they have a strong preference for poly(A) tails, effectively making them deadenylases (Viswanathan et al, 2004). As their name suggests, P-bodies may also have a role in the processing of transcripts. Although P-bodies have some overlapping functions with stress granules, they are not the same, because they are formed under different conditions and contain some different proteins (Anderson and Kedersha, 2008). An interesting distinction is that P-bodies actively move throughout the cytoplasm, whereas stress granules are relatively static in their location (Kedersha et al, 2005; Newbury et al, 2006). The movement of P-bodies can lead them to physically interact with other foci and organelles.
Associated with this RNP, in mice, are the 5′ to 3′ exoribonuclease XRN1, TTP, and the decapping enzymes, DCP1 and DCP2 (Barreau et al, 2005; Anderson and Kedersha, 2008). In species ranging from yeast to human, P-bodies have DExD/H-box helicase 1 (DHH1) and DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 (DDX6), also known as RC-K8–encoded protein, which are enzymes involved in decapping and 5′–3′ degradation (Colegrove-Otero et al, 2005). MicroRNA and miRNA-associated RBPs, such as GW182 (which is necessary for P-body formation), AGO, and RNA helicases, colocalize with P-bodies, indicating a role in RNA interference (Newbury et al, 2006). MicroRNA-mediated degradation depends on CCR4/NOT and decapping enzymes, but miRNA-mediated repression of transcripts does not (Behm-Ansmant et al, 2006). Recent evidence has suggested that P-bodies are scaffolding centers for miRNA function (Sen and Blau, 2005). Although P-bodies may contain initiation factors, they contain silencing machinery and lack ribosomal subunits, implying that mRNAs associated with them are translationally inactive (DeGracia et al, 2008). Together, this indicates that the dynamic flow of mRNAs between polysomes and P-bodies is one of the main ways by which posttranscriptional fate is regulated (Parker and Sheth, 2007).
Exosomes
Exosomes, which are sites for transcript degradation, are called PM/Scl complexes in humans and exosome complexes in other mammals so as not to confuse them with secreted vesicles in mammals that are also named exosomes (van Dijk et al, 2007). Exosomes contain 3′→5′ exoribonucleases—namely, RRP family proteins, also known as EXOSC family proteins, for degrading deadenylated or uncapped transcripts—and they may contain initiation factors but do not contain ribosomal subunits, implying that they are sites of mRNA decay and translational silencing (Anderson and Kedersha, 2006; Lin-Chao et al, 2007). There is evidence that the CCR4/NOT complex functions within exosomes as well as other exonucleases (Azzouz et al, 2009). RBPs associated with these foci include ZFP36/TTP and KH-type splicing regulatory protein (KHSRP; Chen et al, 2001). Exosomes have been found ubiquitously in eukaryotic and prokaryotic cells (Lin-Chao et al, 2007). A major difference between exosomes and other RNPs is that there is no indication that transcripts are stored in exosomes. Until recently it was unclear whether RNA degradation in the nucleus and the cytoplasm was performed by similar proteins, but recent studies have connected some of the machinery that functions in the cytoplasmic exosomes with nuclear exosomes (Azzouz et al, 2009).
Male Germinal Granules: Intermitochondrial Cement and CBs
Two major types of RNPs, termed “inter-mitochondrial cement” (IMC) and “chromatoid bodies,” respectively, can be readily detected in developing male germ cells in the testis (Figure 4). IMC, also called nuage, exists in the cytoplasm of spermatogonia and spermatocytes, whereas CBs, electron-dense, perinuclear structures, start to appear in late pachytene spermatocytes and become prominent in round spermatids during spermatogenesis (Chuma et al, 2009). Both structures are analogous to processing bodies but only exist in male germ cells.
Figure 4.
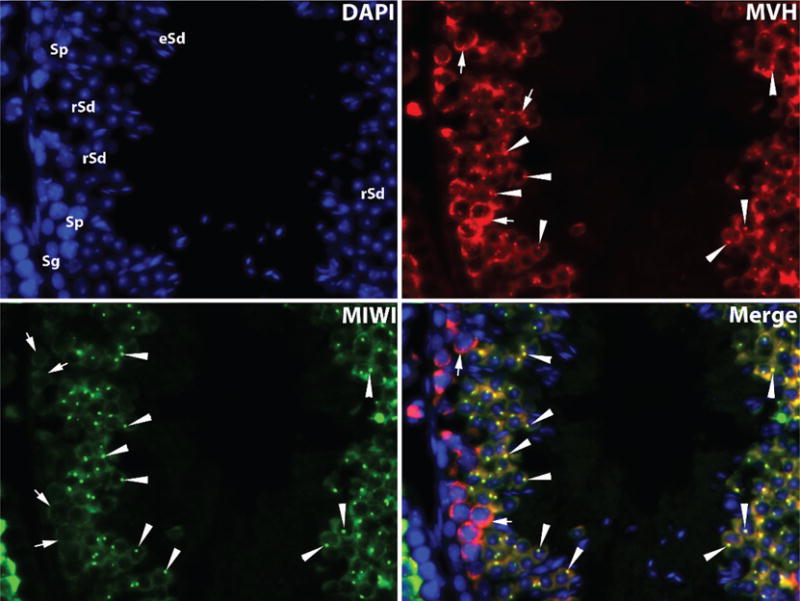
Male germinal granules (intermitochondrial cement/nuage and chromatoid bodies) highlighted by immunofluorescent staining of mouse VASA homolog (MVH [DDX4]) and murine PIWI homolog 1 (MIWI [PIWIL1]) in adult mouse testes. Intermitochondrial cement (arrow) is widely distributed in the cytoplasm of all spermatogenic cells, including spermatogonia (Sg), spermatocytes (Sp), and spermatids (Sd), whereas chromatoid bodies (arrowheads) resemble perinuclear bodies mainly found in round spermatids (rSd). MVH (red fluorescence) is associated with male germinal granules in all types of developing germ cells, including intermitochondrial cement in all spermatogenic cells and chromatoid bodies in round spermatids. MIWI (green fluorescence) is predominantly expressed in intermitochondrial cement in pachytene spermatocytes and chromatoid bodies in round spermatids. eSd, elongated spermatids. Purified human anti-MVH/DDX4 antibody (R11-A3 Fab) was purchased from BD Pharmingen Inc (San Diego, California), and affinity-purified rabbit anti-PIWIL1 polyclonal antibodies were prepared by GenScript Corporation (Piscataway, New Jersey). Both antibodies were used at a dilution of 1:100.
Several reports suggest that the formation of CBs is essential for spermatogenesis (Kotaja and Sassone-Corsi, 2007; Chuma et al, 2009; Nguyen Chi et al, 2009). These membraneless foci have been proposed to function as sites for storing long-lived mRNA transcripts, degrading deadenylated transcripts, regulating miRNA activity, and processing mRNAs (Nguyen Chi et al, 2009; Bettegowda and Wilkinson, 2010). These hypothetical roles were proposed based on the contents of CBs identified thus far, such as DDX4 (more commonly known as mouse VASA homolog [MVH]), miRNAs, AGO proteins, RNA helicases, decapping enzymes, GW182, PABPs, polyadenylated mRNAs, XRN1, and HUR (Kotaja and Sassone-Corsi, 2007; Nguyen Chi et al, 2009). One of the main ways in which transcripts are regulated during spermiogenesis is correlated with movement of HUR and MVH. During steps 1 to 5, HUR, MVH, and associated transcripts accumulate in the CB, and they move to the polysomes after step 6 (Kotaja and Sassone-Corsi, 2007; Nguyen Chi et al, 2009). However, not all long-lived transcripts must be associated with CBs, because a greater proportion of haploid transcripts can be found widely located to other compartments of the cytoplasm, not confined to the CBs. CBs move in a microtubule-dependent fashion and associate with nuclear pores, organelles, and RNPs, suggesting that CBs also have a role in transport (Parvinen and Jokelainen, 1974; Parvinen and Parvinen, 1979; Chuma et al, 2009). KIF17b, the testis-specific kinesin family motor protein, accumulates in CBs, indicating that it may have a role in the nonrandom movement of CBs in the cytoplasm. Because KIF17b is responsible for the transport of RNA, it is also possible that the role of KIF17b is instead to localize other elements to the CB (Kotaja et al, 2006). Interestingly, CBs can move through intercellular bridges from one spermatid to another, although the precise effect of this has yet to be determined (Ventela et al, 2003).
CBs are well conserved over a wide evolutionary lineage, retaining structural and functional similarity, and interacting with homologous proteins. For example, germinal granules in both male and female Drosophila associate with VASA and dead end homolog 1 (DND1), both of which are necessary for germ cell function (Tanaka et al, 2000; Kedde and Agami, 2008).
IMC is a closely related but distinct type of germinal granules that are associated with many of the same proteins as CBs (Chuma et al, 2009). IMC associates with mitochondria and can be seen in small granules in both mitotic (spermatogonia) and meiotic (spermatocytes) male germ cells, whereas CBs are large granules mainly found in the free cytoplasm of round spermatids (Chuma et al, 2009). IMC plays an important role in suppression of retrotransposons through activity of GASZ and P-element–induced wimpy testis (PIWI)-like homolog 2/MIWI-like (PIWIL2/MILI; Ma et al, 2009). PIWIL2 is essential for normal meiosis, possibly through its activity in DNA methylation and transposon suppression, whereas the role that GASZ seems to play is in stabilizing PIWIL2 (Ma et al, 2009). Tudor domain containing 1 (TDRD-1), also known as mouse tudor repeat 1, is an evolutionarily conserved Drosophila TUDOR homolog whose expression is controlled by MVH (Chuma et al, 2006). Tdrd1-deficient mice do not form IMC but still form CBs, indicating that CBs do not arise directly from IMC, and thus CBs and IMC should be considered as separate RNPs (Chuma et al, 2006).
Spliceosomes
Spliceosomes are probably the best-studied RNPs. They are specialized RNPs, generally acting in the nucleus of a cell, that remove introns from a pre-mRNA transcript and ligate the exons, forming a mature mRNA transcript. Key components of the spliceosome contain several small nuclear RNAs (snRNA) and associated proteins. These proteins together with their snRNAs are known as small nuclear ribonucleoprotein particles (snRNP), and are essential for intron removal. Spliceosomes are composed of the snRNPs U1, U2, U4, U5, and U6, along with several other proteins (Kaida et al, 2010).
Commonly, alternative splicing takes place, in which different exons are removed from a transcript; this is responsible for a great deal of the diversity in the eukaryotic genome. Pre-mRNA, often called heterogeneous nuclear RNA (hnRNA), binds to the snRNAs in the spliceosome, forming loops that are excised (Figure 2A). Proteins that bind hnRNA form heterogeneous nuclear ribonucleoproteins (hnRNP) that can encourage or block specific splicing events, resulting in directed alternative splicing. Those that encourage splicing are known as activators and usually have serine/arginine-rich domains (SR splicing factors), and their binding sites on the RNA are known as enhancers, which are required on both the 3′ and 5′ ends of an exon for inclusion (Figure 2A; Wang and Burge, 2008). Those hnRNPs that discourage splicing are known as repressors, and their binding sites are known as silencers. The RNA elements that control splicing events can be located in either the exon or the intron (Wang and Burge, 2008). Cleavage and polyadenylation–specific factor (CPSF) is necessary for processing on the 3′ side through interactions with the snRNP U2 (Li et al, 2001; Kyburz et al, 2006). It is important to note that depending on which proteins are expressed in the cell, the same element can be a silencer or enhancer. Following splicing, hnRNPs may remain bound to the transcript and accompany it into the cytoplasm; thus, many hnRNPs also have cytoplasmic functions (Dreyfuss et al, 2002).
Although hnRNPs usually bind to elements in the coding region of the hnRNA, they can also bind elements in the 3′ UTR, just as cytoplasmic RBPs do. In this way, motifs in the 3′ UTR can also direct splicing events. This modification of 3′ UTR is one way in which the fate of transcripts can be regulated (Thiele et al, 2006). Indeed, alternative splicing events can alter the expression of a protein 3- to 4-fold (Nott et al, 2004; Sanford et al, 2004). There is some debate in the literature on the validity 3′ UTR splicing, because the proposed “nonsense mediated decay” mechanism suggests that attaching splicing factor RBPs to the 3′ UTR of transcripts initiates decay of the transcript (Lykke-Andersen et al, 2001; Dreyfuss et al, 2002).
hnRNP Complexes
The hnRNP binds to hnRNA and forms functional units known as hnRNPs. By recruiting a nuclear PAP and various enzymes such as phosphotase, guanosyl transferase, and (guanine-N7-)-methyltransferase, hnRNPs are responsible for adding the poly(A) tail and the 5′ 7-methylguanosine cap to the pre-mRNA transcripts (Yamada-Okabe et al, 1999; DeGracia et al, 2008). Often hnRNPs are also responsible for nuclear export (Xie et al, 2003). Because most hnRNPs are nonspecific in their binding, their regulation is performed for transcripts in general. Other hnRNPs recognize specific motifs and act only on a small subset of transcripts by modifying and exporting them.
Elav/Hu Granules
Elav/Hu granules appear to be relay centers facilitating the transfer of mRNA to other RNPs. These function as a way to group together and regulate related mRNA transcripts, often referred to as RNA operons (Keene, 2007). Proteins associated with these granules are eI4G, EIF4E, PABP, and Hu proteins (Tenenbaum et al, 2002). Functionally related transcripts simultaneously undergo modifications in these RNPs and then are targeted to other RNPs to be translated, stored, or degraded (Keene and Lager, 2005; DeGracia et al, 2008).
Transport RNPs
Transport RNPs, also referred to as neuronal granules or RNA granules, have been found in oocytes and neurons where localization of translation is of particular importance (Anderson and Kedersha, 2006; Kiebler and Bassell, 2006). They are responsible for sequestering ribosomes, proteins, and mRNA, keeping them inactive until translation is needed (DeGracia et al, 2008). It has been suggested that RNA granules are actually a category of RNP, being split into stress granules, processing bodies, and transport RNPs (Kiebler and Bassell, 2006). Transport RNPs are also responsible for transporting mRNAs and translational machinery to distal processes where they are needed. Common proteins associated with these RNPs are STAUFEN (Villace et al, 2004; Kiebler and Bassell, 2006), a protein involved in the transportation and silencing of transcripts, and FXR (Ferrari et al, 2007), which is a highly regulated protein involved in regulating the posttranscriptional fate of mRNAs. Transport granules actively move throughout the cell, transporting machinery and translationally silenced mRNAs (Kiebler and Bassell, 2006).
Elements: “Secret Codes” in the Untranslated Regions of mRNAs
Elements, also called motifs, are cis-regulatory elements that are involved in the posttranscriptional fate control of mRNAs (Table 3). They exist in a wide variety of transcripts in many different cell types, and are often relatively conserved across diverse evolutionary branches. RBPs and miRNAs recognize specific sequences in mRNAs, and it is thought that this is how transcripts are regulated (Table 3). Generally, these elements are in the 3′ UTR of the transcript. Although these elements generally have such a wide variability that it is nearly impossible to describe all of the variations that can possibly be bound by a particular RBP, altering the code will often disrupt normal regulation. Likewise, the addition of an RNA element into a reporter gene will alter its expression. Because of the extreme difficulty of identifying consistent binding motifs for some proteins, there has been speculation that some RBPs, such as the Y-box family (discussed in the upcoming text), recognize motifs in DNA code and transfer onto RNA as it is transcribed. Interactions between RNA elements have been shown to be vital for proper transcript regulation because RBPs can act cooperatively or competitively. This will be discussed in the RBP section.
Table 3.
Motifs and RNA-binding protein binding sites in order of appearance in the text
| Element and Motif | Sequence |
|---|---|
| CPE | UUUUWWAU {N} AAUAAA, or UUUU{U}{U}AU{U}{U} |
| PRE | VBTYHWWHYNWDHWHRTHHDKBTW |
| EDEN | AUUAUUAUU, or (URUR)n |
| ARE class I | AUUUA |
| ARE class II | A(UUUA)n, or UUAUUUAWW |
| ARE class III | AU-rich sequence without AUUUA |
| SBE | UACUMA |
| TGE | CUCA {N} MMUUUCYU {N} UUUCU, or AAUUUAUU |
| NRE | WCAUWWWWUYGUUGUCSARAAUUGUAYAUAWKCS or GUUGU {N} AUUGUA |
| PBE | UGUANAUARNNNNBBBBSCCS |
| AUF1-binding motif | DKWYDBW{}NBDN{}NHNNRA{}A{N}R{}DM{}DKDVW{}{}WD or UUAG |
| HUR-binding motif | KKNWWD{N}UKNNUUKUWW |
| TIA-1–binding motif | UUNNUHUDYN{N}KWNDRNMVNNBDDKNWRNDRR |
| TIAR-binding motif | UURMMNCCUCCUGYUNS{}{}WRMCC{}{}NNNYNG |
| CPSF-binding motif | AAUAAA |
| DAZL-binding motif | WRUUYAGUAWAANAACUUUKGAAWUR, or UUUSUUU |
| DAZAP1-binding motif | AAAUAG, or GU{U}{U}AG |
| SAM68-binding motif | UCCAUCA |
| MSY2-binding motif | AACAUC |
| MSY4-binding motif | UCCAUCA |
| PUMILIO-binding motif | UGUANAUA |
| hnRNP A1–binding motif | UAGGGW |
Abbreviations: PBE, PUM2 binding element; PRE, polyadenylation response element; SBE, STAR-binding element; TGE, Tra-2 and glioma-associated oncogene homolog (GLI) element. R 5 G A (purine); Y 5 T C (pyrimidine); K 5 G T (keto); M 5 A C (amino); S 5 G C (strong bonds);W5 A T (weak bonds); B 5 G T C (all but A); D 5 G A T (all but C); H 5 A C T (all but G); V 5 G C A (all but T); N 5 A G C T (any); {} 5 possible insert (ie, A{}G 5 AG or ANG); {symbol} 5 possible insert of specified nucleotide (ie, A{U}{U}G 5 AG, AUG, or AUUG); {N} 5 unspecified number of any nucleotide sequence insert (ie, A{N}GT 5 AGT, ANGT, ANNGT, ANNNGT, etc). For all other abbreviations, see Table 1 abbreviations footnote.
Cytoplasmic Polyadenylation Element and Cleavage and Polyadenylation Adenylation Signal
Cytoplasmic poly-adenylation elements (CPE) are conserved in many animal species and are present in 7% to 11% of the 3′ UTRs of transcripts from Caenorhabditis elegans to humans (Charlesworth et al, 2004). There are multiple motifs that represent CPEs, depending on the cell type and developmental stages (eg, embryogenic CPEs have the motif U12–27). CPEs are generally associated with polyadenylation of the poly(A) tail and translation activation. For embryogenic CPEs to function, a CPSF-binding site (AAUAAA), also known as the polyade-nylation signal (PAS), must also exist on the same transcript, where the proximity of the two elements determines activation levels (Simon et al, 1992). Interestingly, in embryos, the rate of polyadenylation of transcripts, rather than the static length of the poly(A) tail, determines translation, although the mechanism for this is unknown (Simon et al, 1992).
An oogenic CPE also exists with the motif of U4–6A1–3U, which is commonly considered the consensus CPE sequence. This sequence is not fixed, because the sequence UUUUAACA has been implicated as a CPE, along with other sequences that lack a terminal U (Charlesworth et al, 2004). Oogenic CPEs are responsible for late-acting maternal mRNA activation in embryos. Oogenic CPEs have to overlap with a CPSF-binding site to function properly. It is hypothesized that CPEB inhibits CPSF from interacting with the transcript, and when CPEB is degraded, CPSF can access the transcript, encouraging adenylation and translation (Colegrove-Otero et al, 2005).
Importantly, several types of CPEBs and CPSFs have been identified in the male germline. The consensus CPE sequence has yet to be identified in spermatogenic cells, but it is likely that the CPE in male germ cells is similar to that of other tissues. However, many essential transcripts have been found to be posttranscriptionally regulated by CPEB in mammalian spermatogenesis, for example, synaptonemal complex proteins Scp1 and Scp2 mRNAs (Tay and Richter, 2001).
Polyadenylation Response Element
Polyadenylation response elements (PRE) are elements that direct polyadenylation in a wide variety of species but have been studied most thoroughly in Xenopus, where they have been shown to direct early polyadenylation and translation in oocytes. This activity is stimulated by progesterone. PREs have been shown to have a conserved sequence in a wide variety of species, including humans, and it has been estimated that 2% to 3% of 3′ UTRs of transcripts from nematodes to mammals have PREs, although transcripts that are regulated by PREs in mammals have not been well elucidated (Pesole et al, 2002; Charlesworth et al, 2004). The consensus sequence for PREs has wide variability (Charlesworth et al, 2004), but despite the seemingly random consensus sequence for PREs, the changes in polyadenylation associated with them can be transferred to other transcripts by adding PREs to their 3′ UTR (Charlesworth et al, 2004). Like CPEs, PREs must also have the PAS nearby to function correctly.
Embryo Deadenylation Elements
Embryo deadenylation elements (EDEN) promote deadenylation and are elements that are formed of U/purine dinucleotide repeats, that is, (UGUA)n. The variable number of repeats, as well as the existence of (AUU)3 just 5′ of the tail and AUUUA elements, coordinate the level of deadenylation (Audic et al, 1998). The end result of deadenylation is destabilization and translational repression of the mRNA transcript. Interestingly, although these elements are conserved in many species, their function is not always the same. For example, in Drosophila, EDEN does not affect the length of the poly(A) tail but does repress translation of the transcript (Castagnetti and Ephrussi, 2003). An EDEN-binding protein (EDEN-BP) is needed for deadenylation by EDEN. In Drosophila, the homolog of EDEN-BP is BRUNO, whereas in mice and humans it is CUG repeat binding protein 1 (CUG-BP1), also known as CUGBP, ELAV-like family member 1(CELF1; Colegrove-Otero et al, 2005). Some examples of transcripts bound by CELF family proteins are Aurora A/(Eg2), Cdk2/(Eg1), c-mos, and c-Jun mRNAs (Paillard et al, 2002, 2003; Graindorge et al, 2008).
AU-Rich Elements
AU-rich elements (ARE) are among the most important and most studied elements involved in posttranscriptional fate control. These elements were initially discovered to be involved in ARE-mediated decay. More recent advances have shown their roles to be more nuanced. Depending on the RBPs that bind to them and the cellular environment, AREs can increase transcript stability, direct splicing events, promote nuclear export, increase or decrease the length of the poly(A) tail, target the transcript to various RNPs, and up- or down-regulate translation (Barreau et al, 2005).
AREs are divided into three classes. Class I has one to three AUUUA motifs dispersed through the 3′ UTR near U-rich regions. Class II has 2 or more overlapping UUAUUUA(U/A)(U/A) motifs inside a U-rich region. Class III is less well characterized, but is composed of AREs that lack an AUUUA motif and contain a U-rich region (Espel, 2005). Examples of ARE-containing transcripts that are active in the mammalian testes are Brd2, Gcnf, and Spam1 (Zhang et al, 2006; Nguyen Chi et al, 2009). AREs often interact with other elements to function correctly. For example, c-Jun contains a class III ARE that encourages EDEN-BP–dependent dead-enylation (Paillard et al, 2002).
STAR-Binding Elements
Signal transduction and activation of RNA (STAR)–binding elements (SBE) are elements that interact with signal transduction and activation of RNA/GRP33, SRC-associated in mitosis, 68 kd (SAM68), GLD-1 (STAR/GSG) family proteins, which themselves are a subset of the KH-type RNA-binding proteins (Colegrove-Otero et al, 2005; Bettegowda and Wilkinson, 2010). STAR/GSG proteins are commonly involved in alternative splicing and nuclear export (Stoss et al, 2001; Ryder and Williamson, 2004). The motif of SBEs, UACU(C/A)A, was first established by binding of protein called defective in germline development of C elegans 1 (GLD-1; Jan et al, 1999; Ryder et al, 2004). However, not all STAR/GSG family proteins bind the UACU(C/A)A motif, because the most well known protein of this family, SAM68, binds to an AUUAAAA motif (Lin et al, 1997). SAM68 has been shown to have a role in alternative splicing of several pre-mRNAs, such as Cd44 and Bcl-X (Matter et al, 2002; Paronetto et al, 2007). It has been shown to regulate the nuclear export of several transcripts, such as Tenr and Pp1γ (Paronetto et al, 2006).
Tra-2 and Glioma-Associated Oncogene Homolog Elements
Tra-2 and glioma-associated oncogene homolog (GLI) elements (TGE), previously known as direct repeat elements (DRE), have 28-nucleotide (nt) sequences separated by 4 nt, given as CUCA{N} (C/A)(C/A)UUUC(C/U)U{N}UUUCU, where {N} represents a string of variable length and composition (Jan et al, 1997). These elements are associated with shorter poly(A) tails, implying a role in deadenylation (Jan et al, 1997). There is evidence that transcripts that have TGEs also have 3 SBEs in their 3′ UTRs (Jan et al, 1997). Although these elements were first identified in C elegans, the evidence suggests that these elements are conserved in mammals (Jan et al, 1999).
NANOS Response Elements and PUMILIO Response Elements
NANOS response elements (NRE) are 32-nt–long sequences that have 2 completely constant regions labeled box-A (GUUGU) and box-B (AUU-GUA; Gupta et al, 2009). NANOS itself does not bind NREs; instead, boxes A and B refer to the actual binding sites for PUMILIO and fem-3–binding factor protein family (Puf) domain proteins—PUMILIO in Drosophila and fem-3–binding factor (FBF) in C elegans (Colegrove-Otero et al, 2005; Gupta et al, 2009). The entire 32-nt sequence, not simply the PUF domain– binding sites, is necessary for NRE regulation, implying other factors may be active on the same transcript (Gupta et al, 2009). On most transcripts that have NREs, there are 2 separate NREs, indicating that PUF domain proteins may function as dimers (Gupta et al, 2009). The NRE sequence is distinct from the PUMILIO response element sequence given as UGUA-NAUA(G/A)NNNN(G/A/T)(G/A/T)(G/A/T) (G/A/T)(G/C)CC(G/C), which is essential for PUM2 regulation (a murine PUMILIO homolog; White et al, 2001).
RBPs: The Ultimate Players in the Control of mRNA Fate
RBPs can either exist in the nucleus or the cytoplasm, but most often they are able to shuttle in between. Because they often exist in the nucleus as hnRNPs, when they relocate to the cytoplasm they generally facilitate nuclear export of their associated transcripts. Once in the cytoplasm they can regulate transcript fate by associating with other proteins, complexes, or RNPs. The control of transcript fate is generally achieved by sequestering the transcript from other machinery or altering the length of the poly(A) tail. Many RBPs can be modified (phosphorylation, methylation, acetylation, etc) to alter their activity and their location in the cell; thus, their effects on posttranscriptional fate can be controlled.
RBPs can either bind RNA in a nonspecific manner or recognize specific motifs. There is also a subset of RBPs that recognize stem-loop structures. These are semispecific because although they do not bind to specific motifs, the tertiary structures that they recognize are dependent on the sequences of the transcript. Here, RBPs will be divided into two groups: small noncoding RNA (sncRNA)–associated and sncRNA-independent RBPs. The sncRNA-independent RBPs will be subdivided into AU-rich element–binding proteins (ARE-BP) and non–ARE-binding proteins. This is not because AREs are particularly unique elements, but instead because of the fact that the group is prolific and diverse to the extent that it warrants its own separate discussion. Indeed, it is estimated that 5% to 8% of mRNAs contain AREs in their 3′ UTRs, and 7% testicular transcripts possess AREs (Barreau et al, 2005; Newbury et al, 2006; Nguyen Chi et al, 2009). Furthermore, AREs have been shown to play a vital role in spermatogenesis.
sncRNA-Associated RBPs
More and more distinct sncRNA species are being discovered. Given limited knowledge of these sncRNAs, here we only discuss 2 well-characterized sncRNAs: miRNAs and PIWI-interacting RNAs (piRNAs). Mi-croRNAs are generally ~22 nt long and are derived from precursor sequences that form stem-loop structures. MicroRNAs are associated with RISCs. The piRNAs are longer RNAs (24–31 nt) that associate with PIWILs 1, 2, and 4. MicroRNAs are created in a DICER-dependent pathway, whereas piRNA production is independent of DICER (Figure 5).
Figure 5.
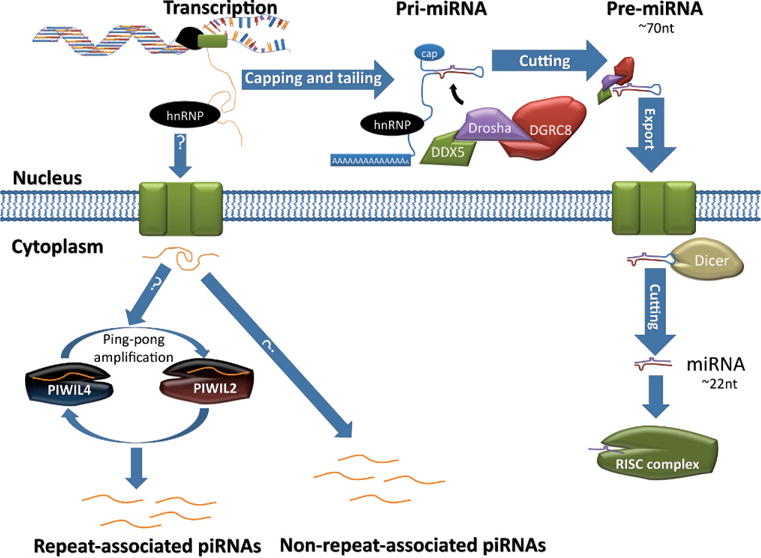
Biogenesis of microRNAs (miRNA) and PIWI-interacting RNAs (piRNA). DNA is transcribed into RNA by RNA polymerase II/III, and the resulting primary transcript (pri-miRNA) contains a hairpin structure that is recognized and cleaved by a microprocessor complex consisting of DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 (DDX5), DGRC8, and DROSHA. This results in a ~70-nucleotide (nt) strand, called precursor miRNA (pre-miRNA), which retains its hairpin structure. Pre-miRNA is exported, and in the cytoplasm it is recognized and processed by an RNase III enzyme, DICER. This results in two ~22-nt mature miRNAs, which can then be incorporated into the RNA-induced silencing complex (RISC). The pathways forming piRNAs are not yet elucidated. But piRNAs are likely derived from long single-stranded RNAs transcribed in the nucleus. Once in the cytoplasm the precursor piRNAs are then processed into ~30-nt piRNAs. It is known that repeat-associated piRNAs are derived from transcripts of repetitive elements (eg, transposons), and PIWIL2 and PIWIL4 mediate the so-called “ping-pong” amplification loop to generate a large quantity of repeat-associated piRNAs, which can then suppress the expression of those “mother” repetitive elements through methylation of these loci. How non–repeat-associated piRNAs are produced remains unknown.
RBPs that are associated with miRNAs include those that synthesize miRNAs and those that are recruited by miRNAs as components of miRNA effector complexes. It has been estimated that ~1000 miRNAs exist in the human genome, regulating approximately one-third of protein coding genes (Lewis et al, 2005). MicroRNAs recognize motifs that are generally located in the 3′ UTRs of their target transcripts. Although miRNAs and their associated proteins were originally thought to be responsible only for mRNA degradation and repression, it has been recently discovered that miRNA causes translational activation under certain conditions, and there is some evidence to indicate that miRNAs are also involved in transport of some mRNA transcripts (Vasudevan et al, 2007).
Most miRNAs begin by being transcribed by RNA polymerase II into long single-stranded RNA strands, which are then capped, polyadenylated, and spliced by spliceosomes and hnRNPs, in the same way that mRNAs are produced (Figure 5). The resulting RNAs are known as primary miRNAs, in which the miRNA-containing region forms a hairpin/stem-loop structure that can be recognized by RBPs, particularly by DiGeorge syndrome critical region gene 8 (DGCR8; Wang et al, 2007). In mammals, DROSHA, DGCR8, DDX5, and DDX17 form the microprocessor complex, which cleaves out the ~70-nt precursor miRNAs (pre-miRNAs) that retain their hairpin structure (Davis et al, 2008; Agami, 2010). Nuclear export of pre-miRNAs is accomplished by EXPORTIN 5. Once in the cytoplasm, DICER binds to pre-miRNAs and cleaves the hairpin structure, resulting in 2 mature miRNAs (~22 nt; Hammond, 2005). Mature miRNAs are then incorporated into RISCs. MicroRNAs have partially complementary sequences for their target elements located in the 3′ UTR of transcripts. This allows the RISC to bind to specific subsets of transcripts that contain the correct motif, causing transcript degradation or translational inhibition. Important components of RISCs are the Argonaute family proteins (AGOs 1–3). AGO proteins assist in degradation or compete with initiation factors, EIF4 and EIF6, and thus repress the initiation of translation (Chendrimada et al, 2007; Kiriakidou et al, 2007). Interestingly, miRNAs themselves may regulated by upstream elements, because a conserved miRNA-specific motif has been observed in many human miRNAs (Inouchi et al, 2007). A mostly unknown aspect of miRNAs is that under certain conditions they can actually encourage translation of associated transcripts. For example, AGO2 forms a complex with FXR1 in human cells under stressful conditions. This complex appears to encourage associated transcripts to be translated, particularly with respect to a subset of ARE-mRNAs to which miRNA binds (Vasudevan et al, 2007).
An important subset of AGOs has a domain known as the PIWI domain, which is vital in spermatogenesis (Tushir et al, 2009). PIWIL1, PIWIL4, and PIWIL2, also known as murine PIWI homologs 1 and 2 (MIWI and MIWI2) and MILI, respectively, are the murine orthologs of Drosophila PIWI. PIWIL4 and PIWIL2 are known for their roles in the suppression of transposable elements (Tushir et al, 2009). Rather than associating with RISCs and miRNAs, PIWI family proteins associate with piRNAs, ~24- to 3-nt–long small RNAs involved in transposable element repression through methylation and posttranscriptional regulation, probably through RNA interference. PIWI-interfering RNAs often come from cleaved transposon transcripts. Sense piRNAs bind to PIWIL2, which causes cleavage of transcripts to make antisense piRNAs, which then bind to PIWIL4, causing the formation of sense piRNA (Figure 5); this amplification cycle is known as “ping-pong” amplification (Tushir et al, 2009). PIWIL proteins have been shown to associate with CBs and with polysomes, indicating a role in temporal regulation of translation (Parvinen, 2005; Grivna et al, 2006). PIWIL2, PIWIL1, and PIWIL4 are necessary for fertility in male mice. PIWIL1 knockout mice have spermiogenic arrest at steps 1 to 4 of round spermatids (Deng and Lin, 2002). Both PIWIL2 and PIWIL4 knockouts have spermatocyte arrest in early meiotic prophase (Kuramochi-Miyagawa et al, 2004; Carmell et al, 2007). The phenotype of PIWIL2 or PIWIL4 knockout mice reflects their role in silencing transposable elements during embryonic male germ cell development, because DNMT3L has a similar role and Dnmt3l knockout mice display a similar phenotype (Webster et al, 2005; Tushir et al, 2009).
ARE-BPs
ARE-BPs, also abbreviated AU-BPs, were initially thought only to induce deadenylation, degradation, and transcriptional repression. They are now known to be involved in a wide variety of activities, such as splicing, export, and targeting (Barreau et al, 2005; Izquierdo et al, 2005; Newbury et al, 2006). These proteins are particularly important in spermatogenesis, because 78% of ARE-containing mRNAs (ARE-mRNAs) are expressed in the testis; 6% are exclusively expressed in the testis and another 13% are preferentially expressed in the testis (Nguyen Chi et al, 2009). One-third of the exclusively and preferentially expressed groups is expressed in spermatocytes (meiotic germ cells) and spermatids (postmeiotic germ cells; Nguyen Chi et al, 2009).
AU-Rich Binding Factor 1/hnRNP D
The first discovered ARE-BP was hnRNP D, more commonly called AU-rich binding factor 1 (AUF1). It has been shown to bind and regulate both class I and class II ARE-mRNAs in mammalian cells (Barreau et al, 2005; Mazan-Mamczarz et al, 2009), but its signature motif has recently been determined to be a 29- to 39-nt AU-rich sequence. AUF1 has 4 isoforms caused by alternative splicing, and each retains 2 RNA-recognition motifs (RRM; Lu and Schneider, 2004). AUF1 can bind both DNA and RNA, and although its expression is primarily nuclear, under certain conditions it can be exported into the cytoplasm (Brennan and Steitz, 2001). AUF1 has been shown to have a role in nuclear export (Sarkar et al, 2003; Barreau et al, 2005). It has been shown to compete with HUR and TIAR for binding sites (Kharraz et al, 2010). Its competition with HUR is highlighted by their opposing actions on mRNA stability and their colocalization to the same tissues. HUR is shown to increase stability, whereas AUF1 generally decreases stability, which is likely related to the association of AUF1 with exosomes (Ishimaru et al, 2010); however, in some instances AUF1 has been shown to stabilize mRNAs (Barreau et al, 2005). This may be due to phosphorylation of AUF1, which alters the transcript from a closed conformation to an open conformation (Wilson et al, 2003). This is not cell type specific, but rather transcript specific, because overexpression and immune-suppression studies show that AUF1 is responsible for stabilizing one subset of transcripts and destabilizing another subset within the same tissue (Barreau et al, 2005). AUF1’s competition with TIAR likely contributes to translation of a transcript, because TIAR is involved in translational suppression. It should also be noted that AUF1 associates with both PABP and EIF4G, both of which encourage translation.
AUF1 is expressed in a wide range of tissues, including the thymus, spleen, lungs, brain, and germ cells. In the testis, AUF1 is expressed in spermatocytes and round spermatids, but not in elongated spermatids (Lu and Schneider, 2004). The literature is lacking in loss-of-function studies of AUF1 on spermatogenesis.
Hu Antigen/ELAV Family
ELAVL1, more commonly referred to as HUR, also known as HuA, is a murine homolog of the Drosophila embryonic lethal, abnormal vision 1 (ELAV1) protein. HUR has been shown to bind and regulate all three classes of ARE-mRNAs (Barreau et al, 2005). HUR has 3 RRMs, 2 of which recognize AREs in the 3′ UTR, and the third binds to the poly(A) tail of transcripts (Brennan and Steitz, 2001). This third RRM is responsible for the increased stabilization of ARE-mRNAs (Fan and Steitz, 1998). Although HUR is an ARE-BP, it can associate with many transcripts that do not have the AUUUA region generally associated with AREs in their 3′ UTRs (Barreau et al, 2005; Espel, 2005). Hu proteins are generally associated with increasing the stability of associated transcripts and regulating their translation. It has been suggested that Hu antigen (HU) family proteins are the only RBPs associated with increases in mRNA stability, but this does not seem to be correct, because several others can be responsible for protecting mRNAs from degradation, such as the αCP protein in mammals (Wang and Kiledjian, 2000). Interestingly, unlike other RBPs that have similar functions, HU family proteins are not associated with alterations in the length of the poly(A) tail (Peng et al, 1998; Brennan and Steitz, 2001). During stressful conditions, such as heat shock or ischemia, HUR has a role in stimulating global arrest of translation and in the up-regulation of recovery proteins (DeGracia et al, 2008). Another function of HUR, which is not well known, is that it can relieve miRNA repression under stressful conditions (Bhattacharyya et al, 2006; Agami, 2010). HUR is also involved in nuclear export and may have other roles as an hnRNP. It is noteworthy that nuclear export is dependent on modifications of HUR, such as methylation or phosphorylation (Kim et al, 2008), and this exists as a possible mechanism for regulating cytoplasmic associations as well (Nguyen Chi et al, 2009).
HUR is expressed in many tissues, and its expression patterns seem to match AUF1 expression patterns (Lu and Schneider, 2004). It has been suggested that the balance between AUF1 and HUR levels has an important role in the regulation of transcripts. HUR has been found to colocalize with many RNPs under certain conditions, including polysomes, stress granules, and P-bodies (Barreau et al, 2005). In developing spermatids, HUR has also been shown to target transcripts to the CB (Nguyen Chi et al, 2009).
Although many HU family proteins are necessary for development and have pivotal roles in neurons and lymphocytes, only HUR is expressed in the testis, where it is expressed in meiotic and postmeiotic germ cells and in Sertoli cells (Lu and Schneider, 2004). Its overexpression leads to impaired transcript turnover, and in the testis overexpression of HUR causes impaired spermiogenesis (Nguyen Chi et al, 2009). HUR is primarily expressed in the nucleus of spermatocytes and round spermatids (Nguyen Chi et al, 2009). It is exported along with its associated transcripts to CBs from step 1 to 5 spermatids (Nguyen Chi et al, 2009). It is thought to protect transcripts from degradations and keep them translationally inactive in CBs until step 6. At this point, it moves from the CBs along with MVH and the associated transcripts to the polysomes. Following the rapid translation of HUR-associated transcripts, HUR is degraded and does not appear in detectable levels in elongated spermatids (Nguyen Chi et al, 2009).
Tristetrapolin
Tristetraprolin (TTP) binds to class II AREs (Barreau et al, 2005). The binding of 2 TTP molecules to a transcript is necessary for maximum deadenylation (Lai et al, 2005). TTP is expressed both in the nucleus and the cytoplasm; however, its role in the nucleus has not been explored. In the cytoplasm it is associated with PARN-dependent deadenylation and degradation. Mitogen-activated protein kinase–activated protein kinase-2 is responsible for TTP phosphorylation, which leads to inhibition of the degradation activity of TTP and export of TTP from the nucleus (Newbury et al, 2006). Presumably, this accumulation of TTP in the nucleus during ARE-mRNA expression is to facilitate the degradation and rapid arrest of translation following the translational period of ARE-mRNAs (Newbury et al, 2006).
TTP has been known to associate with stress granules and P-bodies. Its association with stress granules is modification specific. It will only associate with stress granules in its active form (nonphosphorylated; Newbury et al, 2006). Interestingly, overexpression of TTP induces the P-bodies to physically associate with stress granules (Kedersha et al, 2005). The most well known role of TTP is the silencing and degradation of the TNF-α transcript and other cytokine RNAs (Lu and Schneider, 2004). In mice, knockouts of TTP have been shown to have systemic TNF-α overexpression, even though transcription of the gene remains constant (Lai et al, 1999). TTP has been shown to be expressed in many tissues, including the testis, although its role in the testis has not yet been discovered (Lu and Schneider, 2004).
TIA-1 and TIAR
TIA-1 and TIAR are both AREBPs that have been shown to bind and regulate class II ARE-containing transcripts (Barreau et al, 2005); however, not all TIA-1–regulated or TIAR-regulated mRNAs have an AUUUA sequence in their 3′ UTRs (Lopez de Silanes et al, 2005; Kim et al, 2007). They exist in the nucleus as hnRNPs and in the cytoplasm as RBPs. In the nucleus these proteins have a role in alternative splicing and nuclear export (Forch et al, 2000; Zhang et al, 2005). In the cytoplasm they are associated with translational repression (Lopez de Silanes et al, 2005). They promote the association of the preinitiation complex but repress the association of tRNAiMet (Anderson and Kedersha, 2002; Paronetto and Sette, 2010). TIA-1 and TIAR also have a role in global translational repression under stressful conditions, which is likely due to association with stress granules (Kedersha et al, 2000; Kharraz et al, 2010).
Both proteins are expressed in the testis, but only TIAR is necessary for spermatogenesis (Beck et al, 1998). TIA-1 knockout mice are less viable, but survivors are still fertile (Piecyk et al, 2000). Inactivation of TIAR caused embryonic lethality, with only 14% of embryos surviving to adulthood, but all male mice were sterile (Beck et al, 1998). These mice do not make any sperm and do not have any germ cells in the gonads. TIAR appears to be important for the survival of migrating PGCs to the testis, because the number of PGCs reaching the gonadal ridges was significantly reduced, and the surviving PGCs in the testis quickly disappeared (Beck et al, 1998).
CUG-Binding Protein 1/CUGBP, ELAV-like Family Member 1
CUGBP, ELAV-like family member 1 (CELF1), also referred to as CUG-binding protein 1 (CUG-BP1), the mammalian version of EDEN-BP, is an ELAV homolog generally associated with rapid dead-enylation (Paillard et al, 2003; Newbury et al, 2006). There are several different CELF proteins, which have diverse binding motifs, including class III AREs (Barreau et al, 2005). CELF proteins have a role in the nucleus regulating alternative splicing, although their expression is primarily cytoplasmic, where they repress translation (Kress et al, 2007). Translational repression is due to their role in recruiting PARN, which causes deadenylation (Moraes et al, 2006). The activity of CELF1 is thought to be regulated by the CELF phosphorylating protein, dystrophia myotonica-protein kinase, which itself is alternatively spliced by CELF1 (Llamusi and Artero, 2008).
CELF1 is expressed in many tissues, including the testis (Kress et al, 2007). CELF1 knockout mice are generally not viable, but some that do survive show variability in phenotype in spermatogenesis (Kress et al, 2007). A mild phenotype is associated with defective elongation of spermatids and greatly reduced fertility, whereas a strong phenotype displays complete spermio-genic arrest at step 7 and infertility (Kress et al, 2007). Defects may begin earlier, because increased apoptosis is detected in late spermatocytes and early spermatids (Kress et al, 2007).
KHSRP/Vg mRNA-Binding Protein 71 kd (KHSRP/KSRP/VGRBP71)
KHSRP has 4 RNA-binding domains and has been shown to bind to vegetal pole localization elements, VLEs (a type of vegetative pole protein [Vg1] element), and all 3 classes of AREs (Barreau et al, 2005; Colegrove-Otero et al, 2005). There are 2 common splice variants for KHSRP, ~75 kd and ~52 kd; the ~75-kd KHSRP is primarily nuclear, whereas the ~52-kd splice variant is primarily cytoplasmic (Xu et al, 2008). KHSRP is expressed in many different tissues, including the testis, where both splice variants are detected (Lu and Schneider, 2004; Xu et al, 2008). KHSRP is an hnRNP involved in regulating alternative splicing, localization, and degradation of associated transcripts, and, interestingly, also has a role in the formation of miRNA (Min et al, 1997; Gherzi et al, 2004; Trabucchi et al, 2009). In mammals, the activity in alternative splicing occurs in the nucleus, where KHSRPs recruit hnRNP F (Min et al, 1997). KHSRP activity in degradation is due to its association with exosomes and with PARN, the canonical dead-enylase (Gherzi et al, 2004). KHSRP homologs affect localization by association with STAUFEN; thus, it is likely that KHSRP functions in a similar way, but this has not yet been explored (Allison et al, 2004; Colegrove-Otero et al, 2005). KHSRP activity in the creation of miRNAs is due to its activity in DICER and DROSHA complexes (Trabucchi et al, 2009).
CPEB
CPEB, an evolutionarily conserved protein in many lineages from yeast to humans, has several variations in its recognition motifs. This protein is a class III ARE-BP because the motif is described as a highly U-rich sequence inside an AU-rich region (Charlesworth et al, 2004). Although many variants in the CPE sequence exist, there is no known functional correlation between CPE variants and temporal or spatial regulation of transcripts (Charlesworth et al, 2004).
CPEB controls posttranscriptional fate through transcriptional repression by competition for EIF-binding sites, changes in regulation controlled by modification and competition with other RBPs, and localization by association with STAUFEN and motor proteins (Bestman and Cline, 2008). In vertebrates, the activity of CPEB is largely controlled by its phosphorylation by AURORA and dephosphorylation by the phosphatase PP1 (Tay et al, 2003). It has been shown in Xenopus that CPEB, when unphosphorylated, binds the transcript and Maskin, which competes with EIF4G for binding to EIF4E, thus holding the transcript translationally inactive (Figure 3). When phosphorylated, CPEB remains bound to Maskin but decreases its affinity for EIF4E (Stebbins-Boaz et al, 1999). CPEB also recruits CPSF and PAP, lengthening the poly(A) tail, allowing PABP to recruit EIF4G, stimulating translation (Cao and Richter, 2002). Thus, although CPEB is generally involved in translational repression, phosphorylation of this protein causes it to encourage translation (Mendez et al, 2000a; Richter, 2007). Because of the conserved nature of CPEB activity, a similar mechanism probably exists in mammals, with the Maskin homolog, TACC3.
In Xenopus oocytes, it has been shown that the overlapping of CPE and a CPSF-binding site can determine temporal regulation (Charlesworth et al, 2004). Levels of CPEB fluctuate temporally, which allows CPSF to compete with CPEB for the binding site (Mendez et al, 2002). There are several different CPEBs, but only CPEBs 1 and 2 are expressed in the testis (Colegrove-Otero et al, 2005). Knocking out CPEB1 results in infertility in both sexes, with males having spermatogenic arrest at pachytene spermatocytes (Tay and Richter, 2001).
CPSF
CPSF is an evolutionarily conserved protein found in nearly all eukaryotic cell types. The number and proximity of CPE sites around the CPSF-binding site (AAUAAA) alter its regulation of associated transcripts (Colegrove-Otero et al, 2005). A CPE motif is generally found to be within 25 nt of the CPSF-binding site (Charlesworth et al, 2004), and a CPE element is required somewhere on the transcript for CPSF to cause cytoplasmic polyadenylation (Colegrove-Otero et al, 2005). By its binding to CPEB and PAP, CPSF encourages the growth of the poly(A) tail (Mendez et al, 2000b). CPSF also has a role in nuclear cleavage (Li et al, 2001; Kyburz et al, 2006). Because both global knockout and overexpression studies result in embryonic lethality, it is unclear what the effect on spermatogenesis would be (Xu et al, 2006).
Non–ARE-BPs
Deleted in Azoospermia Family Proteins and DAZ-Associated Protein-1
Deleted in azoospermia (Daz) refers to genes in the azoospermic factor region on the Y chromosome that is deleted in 10% to 15% of azoospermic men (VanGompel and Xu, 2010). In the Catarrhine lineage (Old World monkeys and the great apes), encoded in this region are 4 DAZ proteins; however, the more ancestral DAZ family proteins— DAZ-like protein (DAZL) and BOULE—are encoded on autosomal chromosomes (Bielawski and Yang, 2001). The DAZ family proteins—DAZ, DAZL, and BOULE—are all expressed solely in germ cells (Yen, 2004). BOULE is the ancestor of the DAZ family proteins, with conserved homologs existing from C elegans to humans, although the C elegans homolog is involved in oogenesis rather than spermatogenesis (Karashima et al, 2000). DAZL homologs exist in vertebrates only and are thought to have given rise to the DAZ protein, which exists only in Catarrhine primates (Bielawski and Yang, 2001). Although some studies on the evolution of the Daz gene have suggested that it is not acted on by selection, indicating that it is not necessary for spermatogenesis (Agulnik et al, 1998), more recent studies have since shown that it is acted on by positive and negative selection (Bielawski and Yang, 2001).
Although they are essential for normal spermatogenesis, the exact function of DAZ family proteins has yet to be elucidated. DAZ and BOULE are necessary for late-stage meiosis and development of haploid spermatids, whereas DAZL functions primarily in PGC formation (Kee et al, 2009). Other studies have differed, indicating that BOULE does not play a role in meiosis, as problems are first noticeable at steps 2 to 4 of round spermatids, and development continues to step 6 (VanGompel and Xu, 2010). It has been speculated that DAZL is necessary for PGCs to respond to meiosis-inducing factors because in males, development is arrested at prophase of meiosis I, indicating a possible role in stimulating metaphase by DAZL (Paronetto and Sette, 2010; VanGompel and Xu, 2010).
In PGCs and spermatogonia, both DAZ and DAZL are expressed in the nucleus but are primarily cytoplasmic in spermatocytes (Xu et al, 2001). Because of their expression, it is suggested that DAZ and DAZL also have a role in the nucleus (Paronetto and Sette, 2010). Interestingly, DAZL can alleviate repression by disrupting miRNA-mediated repression of transcripts in both the nucleus and the cytoplasm (Takeda et al, 2009).
DAZ-associated protein-1 (DAZAP1), also known as mouse proline rich RNA-binding protein, is a nuclear hnRNP and cytoplasmic RBP with 2 RRMs that commonly associates with DAZL (Kurihara et al, 2004). The consensus sequences for DAZAP1 binding are AAAUAG and GU(1–3)AG (Yang et al, 2009). It is involved in alternative splicing in the nucleus, and in the cytoplasm it has a role in transcript localization and translational regulation (Martinez-Contreras et al, 2007). Despite its association with DAZ family proteins, the expression of DAZAP1 is different from that of DAZ family proteins, indicating it has a role aside from its association with this family of proteins (Kurihara et al, 2004). It further has been shown that the binding targets of DAZAP1 and DAZL differ, indicating that they regulate a different subset of transcripts (Kurihara et al, 2004). Regulation of DAZAP1 requires cooperation with several other RBPs, such as KHSRP, hnRNP A1, AUF1, and DDX20/GEMIN3—a member of miRNP (an RNP involved in miRNA-targeted suppression of translation) and the let-7 programmed RISC (Hutvagner and Zamore, 2002; Mourelatos et al, 2002; Meister and Tuschl, 2004; Yang et al, 2009). The expression of DAZAP1 is primarily nuclear in spermatocytes and round spermatids, but shifts to cytoplasmic just before elongation (Lin and Yen, 2006).
DAZAP1 hypomorphic mice are infertile in both males and females. Male DAZAP1-suppressed mice have higher rates of germ cell apoptosis, and development does not proceed beyond postpachytene spermatocytes (Hsu et al, 2008).
Gonadotropin-Regulated Testicular RNA Helicase/Ddx25
DDX25, more commonly referred to as gonadotropin-regulated testicular RNA helicase (GRTH), is a helicase that up-regulates translation of its associated transcripts, which are often associated with cell survival (Bettegowda and Wilkinson, 2010). GRTH is expressed in both the nucleus and the cytoplasm, and is thought to have a role in selective nuclear export and shuttling between RNPs, controlled by phosphorylation (Sheng et al, 2006). The expression of GRTH is limited to the testes in germ cells and Leydig cells (Tsai-Morris et al, 2004). GRTH associates with both CBs and polyribosomes, and, because of its helicase activity, may encourage translation by removing inhibitory secondary structures of transcripts (Tang et al, 1999). Because the role of CBs is often transcript storage, this implies GRTH may have a role in temporal regulation of translation (Tang et al, 1999). This is further evidenced by the fact that GRTH is expressed in pachytene spermatocytes to round spermatids, and it is responsible for translation of many transcripts, including protamine transcripts, which are transcribed long before they are translated (Tsai-Morris et al, 2004). Among other transcripts regulated by GRTH are transition proteins and antiapoptotic mRNAs, such as Bcl-2, Bcl-xL, Bid, Bak, and Bad (Tsai-Morris et al, 2004; Gutti et al, 2008). An interesting aspect of GRTH regulation is that the transcript of GRTH has 2 possible AUG start codons, the selection of which is androgen dependent (Tsai-Morris et al, 2010).
Knockout of GRTH causes infertility due to azoospermia, with increased apoptosis occurring at stage XII of spermatocytes, or metaphase of meiosis, implying that errors begin prior to spermatid formation (Tsai-Morris et al, 2004). The final stage of development seen in knockouts of GRTH is step 8 spermatids, indicating that GRTH is necessary for elongation of spermatids (Tsai-Morris et al, 2004). GRTH knockout mice also have smaller CBs and significantly smaller testis (Tsai-Morris et al, 2004).
SRC-Associated in Mitosis, 68 kd/KH Domain–Containing, RNA-Binding, Signal Transduction–Associated 1
KH domain–containing, RNA-binding, signal transduction–associated 1 (KHDRBS1), more commonly referred to as SRC-associated in mitosis, 68 kd (SAM68), a protein conserved from insects to mammals, is a member of the Star/GSG family of proteins, a subfamily of the KH domain proteins. SAM68 has a unique binding motif of AUUAAA and associates with transcripts that have this motif and long poly(A) tails (Lin et al, 1997). It is primarily a nuclear protein that moves into the cytoplasm after phosphorylation (Haegebarth et al, 2004), which is one of the functions it can have in signal transduction (Najib et al, 2005). It is involved in alternative splicing, nuclear export, and translational up-regulation (Paronetto et al, 2007; Bettegowda and Wilkinson, 2010). SAM68 up-regulates transcripts both by targeting them to polysomes and by increasing transcript stability (Paronetto et al, 2009). It is expressed ubiquitously in multiple organs (Haegebarth et al, 2004). In spermatogenesis, SAM68 is expressed during two periods—from PGCs to spermatogonia, and from pachytene spermatocytes to round spermatids (Paronetto and Sette, 2010). Knockouts of SAM68 are infertile because of increased apoptosis of germ cells and aberrant elongated sperm (Richard et al, 2005; Bettegowda and Wilkinson, 2010; Paronetto and Sette, 2010).
Mouse Y-Box Protein 2/Y-Box Protein 2 and Mouse Y-Box Protein 4/Cold Shock Domain Protein A
Mouse Y-box protein 2 (MSY2) and MSY4 are murine members of the cold shock domain (CSD) protein family, which can bind to both RNA and DNA (Matsumoto and Wolffe, 1998). Y-box proteins (YBX) bind to a Y-box motif in the promoter region of a gene, the consensus sequence of which is given as CTGATTGG(C/T)(C/T)AA (Matsumoto and Wolffe, 1998). Homologs of MSY2, namely, FRGY2, the Xenopus homolog, must bind to a separate AACAUC motif in two separate locations in the 3′ UTR of mRNA (Bouvet et al, 1994), and MSY4 has been shown to bind to a similar sequence (UCCAUCA; Giorgini et al, 2001). However, a more recent study of the transcripts bound by YBXs showed no consensus sequence in the 3′ UTR; instead, transcripts had a Y-box region in the promoter of the DNA, indicating that transcripts are marked during transcription rather than by elements in their 3′ UTRs (Yang et al, 2005a). While in the nucleus, these proteins are known to stimulate transcription, but once in the cytoplasm they generally repress translation and increase transcript stability (Giorgini et al, 2002; Lu et al, 2006; Xu and Hecht, 2008). A novel aspect of MSY2 is its association with sncRNAs that are ~25 to 31 nt in length, referred to as MSY-RNAs. The exact function of this association has not been elucidated (Xu et al, 2009).
MSY4 is expressed throughout embryogenesis; however, there is no noticeable effect in embryogenesis of knockouts. Some evidence suggests the lack of phenotypic difference is due to redundancy of function with other CSD family proteins (Lu et al, 2006). Both MSY4 and MSY2 are expressed in both meiotic and early postmeiotic spermatocytes (Lu et al, 2006). In MSY4 knockouts there is an increase in apoptosis of meiotic and postmeiotic germ cells, leading to infertility in some mice and reduced fertility in others, where surviving cells completely differentiate without noticeable problems, resulting in fewer but functionally normal sperm (Lu et al, 2006). Accompanying this is a reduction in testis size, degradation of seminiferous tubules, and appearance of vacuoles in spermatocytes (Lu et al, 2006).
MSY2 knockout mice experience infertility in both males and females (Yang et al, 2005b). The first noticeable defect in male MSY2 knockouts is an increase in apoptosis of spermatocytes, and in the surviving cells, spermiogenesis proceeds to step 8/9, but many defective spermatids are detected, such as misshapen, multinucleated, or vacuolated spermatids with reduced chromatin condensation (Yang et al, 2007). All developing cells undergo apoptosis before complete differentiation, resulting in a complete absence of spermatozoa in the epididymis and a reduction in the size of seminiferous tubules (Yang et al, 2007). This phenotype has been shown to be due to MSY2’s role in the cytoplasm because transcription and nuclear export of associated transcripts are unchanged, whereas association with polysomes was reduced (Yang et al, 2007).
NANOS2 and NANOS3
The NANOS family consists of RBPs that repress translation. Although they are most well known for their role in early embryo development of Drosophila, NANOS2 and NANOS3 are required for spermatogenesis in mice (Tsuda et al, 2003). For NANOS to function properly, association with PUMILIO family proteins is required (Gupta et al, 2009). NANOS2 knockouts are infertile in males only, because knockouts show depletion of spermatogonial stem cells (Sada et al, 2009). NANOS3 is expressed in undifferentiated spermatogonia and is essential for maintaining an undifferentiated state (Lolicato et al, 2008).
RNA-Binding Motif Protein, Y Chromosome, and hnRNP G/T
Mammalian RNA-binding motif protein, Y chromosome (RBMY) and hnRNP G/T are members of the RNA-binding motif (RBM) family of proteins that recognize certain stem-loop structures of transcripts that form as part of the secondary structure of mRNA rather than specific motifs (Skrisovska et al, 2007). RBMY is encoded by 6 genes on the Y chromosome that are frequently deleted in infertile men (Skaletsky et al, 2003; Elliott, 2004). In humans, RBMY is expressed only in the nucleus from spermatogonia to round spermatids (Elliott et al, 1998), whereas in mice it is only expressed up to the early spermatocyte stage (Saunders et al, 2003).
The hnRNP G/T is an RBM protein, X chromosome (RBMX) homolog that is closely related to RBMY. It is expressed in the nucleus of meiotic spermatocytes and early spermatids, where it has a role in splicing (Ehrmann et al, 2008). Male knockouts of hnRNP G/T are generally infertile because of low sperm count (Ehrmann et al, 2008).
Miscellaneous
Adenosine deaminase domain–containing 1 (ADAD1), also known as testis nuclear RNA-binding protein (TENR), is expressed in the nucleus of mammals from mid pachytene spermatocytes to elongating spermatids (Connolly et al, 2005). It is thought to be involved in RNA editing (Connolly et al, 2005). Knockouts of ADAD1 are infertile, with low numbers of often defective spermatozoa (Connolly et al, 2005).
TAR (HIV) RNA-binding protein 2 (TARBP2) in mammals, also known as the protamine RNA-binding protein (PRBP), is a double-stranded RNA-binding protein that is associated with translational activation (Zhong et al, 1999). TARBP2 encourages translation of transition proteins and protamines in developing germ cells (Zhong et al, 1999). Male mouse Tarbp2 knockouts have defects in spermiogenesis, generally in packaging of DNA; thus, although mature spermatozoa are formed, mice without PBRP are infertile because of oligospermia (Zhong et al, 1999). This may be due to the role of TARBP2 in the DICER complex (Melo et al, 2009).
DND1, a protein conserved from zebrafish to humans, interferes with miRNA-mediated repression of specific miRNAs, and thus encourages stability of their associated transcripts. Inactivation of this gene results in infertility due to a loss of germ cells (Kedde et al, 2007).
Summary and Concluding Remarks
A typical pathway for transcript-RBP interactions involves RBPs binding to the transcript in the nucleus, and thus directing splicing events; in this case, the RBP would also be an hnRNP. The RBP and its associated transcript may remain in the nucleus, separating the transcript from cytoplasmic particles until the transcript is needed (Figure 6). The transcript then moves from the nucleus along with its associated RBP. The RBP directs movement to stress granules that modify the associated RBPs, directing it to one of many other RNPs. If the transcript is fated for immediate translation, it will be directed to polysomes. If it is fated for delayed translation, it will be directed to processing bodies (or CBs in male gametes), where either it is held in translational arrest or marked by attaching translation-suppressive RBPs (eg, HUR) within the processing bodies followed by exit to cytoplasmic RNP compartments (eg, HUR granules) for long-term storage. If the transcript is fated for degradation, it is either directed toward exosomes or degraded in processing bodies (Figure 6).
Figure 6.
An overview of posttranscriptional fate control of messenger RNAs (mRNA). Splicing and the addition of the 59 cap and 39 poly(A) tail occur simultaneously and are executed by spliceosomes and heterogeneous nuclear ribonucleoprotein (hnRNP) complexes in the nucleus. Transcripts exported from the nucleus are often associated with hnRNPs. Once in the cytoplasm, the transcripts and their recruited RNA-binding proteins (RBP) are usually first directed to processing or sorting ribonuclear protein particles (RNP; eg, stress granules, p-granules, chromatoid bodies, etc). From there, the transcripts can be either transported and loaded onto polyribosomes for translation, or directed to storage RNPs (RNA operons like Hu antigen R [HUR]/Elav1 granules) for future translation. Transcripts that are no longer needed are directed to the exosomes for degradation.
There are many variations in this pathway. RBPs may begin or end associations with their RNAs at any point during their life cycle. Some are only nuclear or only cytoplasmic; some are only associated with particular RNPs or with specific functions. RBPs can interact with each other through cooperation or competition. RBPs can be modified, typically by phosphorylation, inducing altered behavior. Other modifications occur but are less common.
The mechanisms by which RBPs function are variable, but there are some commonalities. The two main ways in which RBPs hold the transcript translationally inactive are by sequestering the transcript away from translational machinery and by competition with factors needed for translation initiation. Likewise, the two main means by which transcripts become translationally active are by association with polyribosomes and by encouraging binding of translation initiation factors. A very common way to encourage or discourage binding of initiation factors is by altering the length of the poly(A) tail. A long poly(A) tail binds many PABPs, which have a high affinity for EIF4G. This encourages EIF4G to bind to EIF4E, forming a closed loop, and attracts the preinitiation complex to bind. This complex recruits the small subunit to bind to the transcript—the main limiting factor in translation initiation.
In the future, the motifs to which RBPs bind should be explored. It seems strange that there should be so much variability in their sequences. Do proteins recognize sequences, or is it secondary structures that are bound? Even the most widely comprehensive sequence variations rarely describe all possible sequences to which a particular RBP can bind (Mazan-Mamczarz et al, 2009). Perhaps low-affinity binding is advantageous, yielding wide variability in sequences. It should also be discovered what ways RBPs regulate the activity of each other and miRNA complexes. Finding other RBPs, the effects of various modifications, and the methods by which they interact with each other will further our understanding.
RNPs have very much in common regarding their activity and contents. It should also be determined whether several RNPs (eg, IMC and CBs) are unique structures or merely the temporarily modified versions of the same structure. It may be that the differences among RNPs are mostly superficial, where the addition of a selective few proteins can completely alter their behavior. A vital advancement to our understanding is to actually catalog the various RNPs and their functions. This paper was a very rough attempt at that, but to have a discussion of RNPs, there must first be a consensus on what they are. An important question that must be addressed is whether these motifs determine the fate of the transcript, whether the presence of particular binding proteins or modifying proteins determines fate, or whether some other undefined mechanism is responsible.
It can be speculated that translational fate is likely controlled by a combination of mechanisms, with general trends being determined by protein levels and more nuanced control being determined by motifs. For spermatogenesis, it must also be determined whether or not a hierarchy of control exists, such as exists in embryo development. If it were found that master control switches exist for translational regulation it would be a breakthrough in this area.
Our understanding of posttranscriptional control is in its infancy. Because regulation of transcripts has been shown to have a pivotal role in differentiation and in rapid signal response, advancements in this area should greatly increase our knowledge of cell physiology and development.
Clarification on Abbreviations and Usage
Most of the proteins discussed exist in Mus musculus. Where the protein is studied (or does not exist in mice), the model organism was mentioned. If no organism is mentioned, it may be inferred that the relevant model organism is M musculus.
In the Appendix, the first name given is the proper name, whereas the bolded name is the name used in this paper, which usually is the name used more commonly in the literature.
RNP is sometimes used in the literature to mean ribonuclear protein (ie, a protein/RNA complex), and in other cases to mean ribonuclear protein particle (ie, distinct foci within the cell). In this paper, RNP is used only in the second way. However, when using hnRNP, this refers to the protein/pre-mRNA complex within the nucleus (similar to the first usage of RNP). After splicing and tail-end modification, this same protein complex is technically referred to as “ribonuclear protein,” but for clarity’s sake, sometimes the term “hnRNP” was still used. When discussing the ribonuclear protein particle formed by hnRNP and other proteins, the term “hnRNP complex” was used. When using snRNP, this refers to the protein/snRNA complex that generally occurs within the nucleus.
For many proteins, the more proper name given in the Mouse Genome Informatics website is rarely used. In these cases, the proper name was mentioned when introducing the protein; however, throughout the paper, the more commonly used name rather than the proper name was used. This is true for AGO/F-box and WD-40 domain protein 7 (AGO/FBXW7), HUR/ELAVL1, TIAR/TIAL-1, MVH/DDX4, TTP/TFP36, SAM68/KHDRBS1, GRTH/DDX25, AUF1/hnRNPD, and MSY/YBX.
Acknowledgments
We thank Amy Mauseth and Randy R. Idler for critical reading of the manuscript. We apologize to colleagues whose work is relevant to this topic but not mentioned in the review due to page limits for the Journal.
Supported by National Institutes of Health grants HD048855 and HD050281 (W.Y.).
Appendix
4E-BP: EIF4E-binding protein
ADAD1/TENR: adenosine deaminase domain containing 1 (testis specific)/testis nuclear RNA-binding protein
ARE: AU-rich element
ARE-BP/AU-BP: ARE-binding protein
CB: chromatoid body
CELF1/CUG-BP1/EDEN-BP: CUGBP, ELAV-like family member 1/EDEN-binding protein
CPE: cytoplasmic polyadenylase element
CPEB: CPE-binding protein
CPSF: cleavage and polyadenylation–specific factor
CSDA/MSY4: cold shock domain protein A/mouse Y-box protein 4
DAZ: deleted in azoospermia
DAZAP1/mPrrP: DAZ-associated protein 1/mouse pro-line rich RNA-binding protein
DAZL: DAZ-like protein
DCP: decapping enzyme
DDX25/GRTH: DEAD (Asp-Glu-Ala-Asp) box polypeptide 25/gonadotropin-regulated testicular RNA helicase
DDX4/MVH: DEAD (Asp-Glu-Ala-Asp) box polypeptide 4/mouse VASA homolog
DDX5: DEAD (Asp-Glu-Ala-Asp) box polypeptide 5
DDX6/RCK: DEAD (Asp-Glu-Ala-Asp) box polypeptide 6/RC-K8 encoded protein
DGCR8: DiGeorge syndrome critical region gene 8
DMPK: dystrophia myotonica-protein kinase
DND1: dead end homolog 1
dsRNA: double-stranded RNA
EDEN: embryo deadenylation element
EIF: eukaryotic translation initiation factor
ELAVL1/HUR/HUA: embryonic lethal and abnormal vision–like 1/Hu antigen R
FBF: fem-3–binding factor
FBXW7/AGO: F-box and WD-40 domain protein 7/Argonaute
GDF1/VG1: growth and differentiation factor/vegetative pole protein
GLI: glioma-associated oncogene homolog
hnRNA/pre-mRNA: heteronuclear RNA/precursor messenger RNA
hnRNP: heterogeneous nuclear ribonucleoprotein
hnRNP D/AUF1: heterogeneous nuclear ribonucleopro-tein D/AU-rich binding factor
KHDRBS1/SAM68: KH domain–containing, RNA-binding, signal transduction–associated 1/SRC-associated in mitosis, 68 kd
KHRSP/KSRP/VGRBP71: KH-type splicing regulatory protein
MAPK: mitogen-activated protein kinase
MAPKAPK2/MK2: MAPK-activated protein kinase-2
ME31B/DHH1: maternal expression at 31B/DExD/H-box helicase
miRNA: microRNA
MSY-RNAs: MIWI-independent small RNAs
NR1H4/FXR: nuclear receptor subfamily 1, group H, member 4/fragile X retardation protein
NRE: NANOS response element
PABP: poly(A)-binding protein
PAP: polyadenylate polymerase
PARN: poly(A)-specific ribonuclease
PGC: primordial germ cell
piRNAs: PIWI-interacting RNAs
PIWI: P-element–induced wimpy testis
PIWIL1/MIWI: PIWI-like homolog 1/murine PIWI gene 1
PIWIL2/MILI: PIWI-like homolog 2/MIWI-like
PIWIL4/MIWI2: PIWI-like homolog 4/murine PIWI gene 2
pre-miRNA: precursor miRNA
PRE: polyadenylation response element
pri-miRNA: primary miRNA
Puf: PUMILIO and fem-3–binding factor protein family
RBMX: RNA-binding motif protein, X chromosome
RBMY: RNA-binding motif protein, Y chromosome
RBP: RNA-binding proteins
RISC: RNA-induced silencing complex
RNP: ribonuclear particle
RRM: RNA-recognition motif
SBE: STAR-binding element
SLBP: stem-loop–binding protein sncRNA small noncoding RNA
snRNA: small nuclear RNA
snRNP: small nuclear ribonucleoprotein
SR: serine/arginine-rich protein
SSC: spermatogonial stem cell
STAR-GSG: signal transduction and activation of RNA/GRP33, SAM68, GLD-1
TARBP2/PBRP: TAR (HIV) RNA-binding protein 2
TGE/DRE: tra-2 and GLI elements/direct repeat element
TIA-1: T-cell–restricted intracellular antigen 1/cytotoxic granule–associated RNA-binding protein 1
TIAL-1/TIAR: TIA1-like 1/TIA-related protein
TNF-α: tumor necrosis factor α
TNRC6A/GW182: trinucleotide repeat containing 6a/glycerin (G) tryptophan (W) repeat protein
tRNAiMet: initiator methionyl tRNA
UTR: untranslated region
XRN1: 5′-3′ exoribonuclease 1
YBX2/MSY2: Y-box protein 2/mouse Y-box protein 2
ZFP36/TTP: zinc finger protein 36/tristetraprolin
References
- Agami R. microRNAs, RNA binding proteins and cancer. Eur J Clin Invest. 2010;40:370–374. doi: 10.1111/j.1365-2362.2010.02279.x. [DOI] [PubMed] [Google Scholar]
- Agulnik AI, Zharkikh A, Boettger-Tong H, Bourgeron T, McElreavey K, Bishop CE. Evolution of the DAZ gene family suggests that Y-linked DAZ plays little, or a limited, role in spermatogenesis but underlines a recent African origin for human populations. Hum Mol Genet. 1998;7:1371–1377. doi: 10.1093/hmg/7.9.1371. [DOI] [PubMed] [Google Scholar]
- Allison R, Czaplinski K, Git A, Adegbenro E, Stennard F, Houliston E, Standart N. Two distinct Staufen isoforms in Xenopus are vegetally localized during oogenesis. RNA. 2004;10:1751–1763. doi: 10.1261/rna.7450204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones. 2002;7:213–221. doi: 10.1379/1466-1268(2002)007<0213:vstroe>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci. 2008;33:141–150. doi: 10.1016/j.tibs.2007.12.003. [DOI] [PubMed] [Google Scholar]
- Audic Y, Omilli F, Osborne HB. Embryo deadenylation element-dependent deadenylation is enhanced by a cis element containing AUU repeats. Mol Cell Biol. 1998;18:6879–6884. doi: 10.1128/mcb.18.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouz N, Panasenko OO, Colau G, Collart MA. The CCR4-NOT complex physically and functionally interacts with TRAMP and the nuclear exosome. PLoS One. 2009;4:e6760. doi: 10.1371/journal.pone.0006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AR, Miller IJ, Anderson P, Streuli M. RNA-binding protein TIAR is essential for primordial germ cell development. Proc Natl Acad Sci U S A. 1998;95:2331–2336. doi: 10.1073/pnas.95.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestman JE, Cline HT. The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc Natl Acad Sci U S A. 2008;105:20494–20499. doi: 10.1073/pnas.0806296105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettegowda A, Wilkinson MF. Transcription and posttranscriptional regulation of spermatogenesis. Philos Trans R Soc Lond B Biol Sci. 2010;365:1637–1651. doi: 10.1098/rstb.2009.0196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z. Positive and negative selection in the DAZ gene family. Mol Biol Evol. 2001;18:523–529. doi: 10.1093/oxfordjournals.molbev.a003831. [DOI] [PubMed] [Google Scholar]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- Bouvet P, Omilli F, Arlot-Bonnemains Y, Legagneux V, Roghi C, Bassez T, Osborne HB. The deadenylation conferred by the 39 untranslated region of a developmentally controlled mRNA in Xenopus embryos is switched to polyadenylation by deletion of a short sequence element. Mol Cell Biol. 1994;14:1893–1900. doi: 10.1128/mcb.14.3.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Richter JD. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 2002;21:3852–3862. doi: 10.1093/emboj/cdf353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Castagnetti S, Ephrussi A. Orb and a long poly(A) tail are required for efficient oskar translation at the posterior pole of the Drosophila oocyte. Development. 2003;130:835–843. doi: 10.1242/dev.00309. [DOI] [PubMed] [Google Scholar]
- Charlesworth A, Cox LL, MacNicol AM. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J Biol Chem. 2004;279:17650–17659. doi: 10.1074/jbc.M313837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Ong SE, Chan EL, Raijmakers R, Pruijn GJ, Stoecklin G, Moroni C, Mann M, Karin M. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci U S A. 2006;103:15894–15899. doi: 10.1073/pnas.0601878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Tanaka T, Nakatsuji N. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: germinal granules in mammals. Mol Cell Endocrinol. 2009;306:17–23. doi: 10.1016/j.mce.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Colegrove-Otero LJ, Minshall N, Standart N. RNA-binding proteins in early development. Crit Rev Biochem Mol Biol. 2005;40:21–73. doi: 10.1080/10409230590918612. [DOI] [PubMed] [Google Scholar]
- Connolly CM, Dearth AT, Braun RE. Disruption of murine Tenr results in teratospermia and male infertility. Dev Biol. 2005;278:13–21. doi: 10.1016/j.ydbio.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Copeland PR, Wormington M. The mechanism and regulation of deadenylation: identification and characterization of Xenopus PARN. RNA. 2001;7:875–886. doi: 10.1017/s1355838201010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc Natl Acad Sci U S A. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig AW, Haghighat A, Yu AT, Sonenberg N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature. 1998;392:520–523. doi: 10.1038/33198. [DOI] [PubMed] [Google Scholar]
- Crawford EK, Ensor JE, Kalvakolanu I, Hasday JD. The role of 39 poly(A) tail metabolism in tumor necrosis factor-alpha regulation. J Biol Chem. 1997;272:21120–21127. doi: 10.1074/jbc.272.34.21120. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGracia DJ, Jamison JT, Szymanski JJ, Lewis MK. Translation arrest and ribonomics in post-ischemic brain: layers and layers of players. J Neurochem. 2008;106:2288–2301. doi: 10.1111/j.1471-4159.2008.05561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Ehrmann I, Dalgliesh C, Tsaousi A, Paronetto MP, Heinrich B, Kist R, Cairns P, Li W, Mueller C, Jackson M, Peters H, Nayernia K, Saunders P, Mitchell M, Stamm S, Sette C, Elliott DJ. Haploinsufficiency of the germ cell-specific nuclear RNA binding protein hnRNP G-T prevents functional spermatogenesis in the mouse. Hum Mol Genet. 2008;17:2803–2818. doi: 10.1093/hmg/ddn179. [DOI] [PubMed] [Google Scholar]
- Elliott DJ. The role of potential splicing factors including RBMY, RBMX, hnRNPG-T and STAR proteins in spermatogenesis. Int J Androl. 2004;27:328–334. doi: 10.1111/j.1365-2605.2004.00496.x. [DOI] [PubMed] [Google Scholar]
- Elliott DJ, Oghene K, Makarov G, Makarova O, Hargreave TB, Chandley AC, Eperon IC, Cooke HJ. Dynamic changes in the subnuclear organisation of pre-mRNA splicing proteins and RBM during human germ cell development. J Cell Sci. 1998;111:1255–1265. doi: 10.1242/jcs.111.9.1255. [DOI] [PubMed] [Google Scholar]
- Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Semin Cell Dev Biol. 2005;16:59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F, Mercaldo V, Piccoli G, Sala C, Cannata S, Achsel T, Bagni C. The fragile X mental retardation protein-RNP granules show an mGluR-dependent localization in the post-synaptic spines. Mol Cell Neurosci. 2007;34:343–354. doi: 10.1016/j.mcn.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Forch P, Puig O, Kedersha N, Martinez C, Granneman S, Seraphin B, Anderson P, Valcarcel J. The apoptosis-promoting factor TIA-1 is a regulator of alternative pre-mRNA splicing. Mol Cell. 2000;6:1089–1098. doi: 10.1016/s1097-2765(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Gherzi R, Lee KY, Briata P, Wegmuller D, Moroni C, Karin M, Chen CY. A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol Cell. 2004;14:571–583. doi: 10.1016/j.molcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Giorgini F, Davies HG, Braun RE. MSY2 and MSY4 bind a conserved sequence in the 39 untranslated region of protamine 1 mRNA in vitro and in vivo. Mol Cell Biol. 2001;21:7010–7019. doi: 10.1128/MCB.21.20.7010-7019.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgini F, Davies HG, Braun RE. Translational repression by MSY4 inhibits spermatid differentiation in mice. Development. 2002;129:3669–3679. doi: 10.1242/dev.129.15.3669. [DOI] [PubMed] [Google Scholar]
- Graindorge A, Le Tonqueze O, Thuret R, Pollet N, Osborne HB, Audic Y. Identification of CUG-BP1/EDEN-BP target mRNAs in Xenopus tropicalis. Nucleic Acids Res. 2008;36:1861–1870. doi: 10.1093/nar/gkn031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A. 2006;103:13415–13420. doi: 10.1073/pnas.0605506103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta YK, Lee TH, Edwards TA, Escalante CR, Kadyrova LY, Wharton RP, Aggarwal AK. Co-occupancy of two Pumilio molecules on a single hunchback NRE. RNA. 2009;15:1029–1035. doi: 10.1261/rna.1327609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutti RK, Tsai-Morris CH, Dufau ML. Gonadotropin-regulated testicular helicase (DDX25), an essential regulator of spermatogenesis, prevents testicular germ cell apoptosis. J Biol Chem. 2008;283:17055–17064. doi: 10.1074/jbc.M708449200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegebarth A, Heap D, Bie W, Derry JJ, Richard S, Tyner AL. The nuclear tyrosine kinase BRK/Sik phosphorylates and inhibits the RNA-binding activities of the Sam68-like mammalian proteins SLM-1 and SLM-2. J Biol Chem. 2004;279:54398–54404. doi: 10.1074/jbc.M409579200. [DOI] [PubMed] [Google Scholar]
- Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- Hao Z, Stoler MH, Sen B, Shore A, Westbrook A, Flickinger CJ, Herr JC, Coonrod SA. TACC3 expression and localization in the murine egg and ovary. Mol Reprod Dev. 2002;63:291–299. doi: 10.1002/mrd.90012. [DOI] [PubMed] [Google Scholar]
- Hodgman R, Tay J, Mendez R, Richter JD. CPEB phosphorylation and cytoplasmic polyadenylation are catalyzed by the kinase IAK1/Eg2 in maturing mouse oocytes. Development. 2001;128:2815–2822. doi: 10.1242/dev.128.14.2815. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Chen HY, Lin YW, Chu WC, Lin MJ, Yan YT, Yen PH. DAZAP1, an hnRNP protein, is required for normal growth and spermatogenesis in mice. RNA. 2008;14:1814–1822. doi: 10.1261/rna.1152808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Iguchi N, Tobias JW, Hecht NB. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc Natl Acad Sci U S A. 2006;103:7712–7717. doi: 10.1073/pnas.0510999103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouchi A, Shinohara S, Inoue H, Kita K, Itakura M. Identification of specific sequence motifs in the upstream region of 242 human miRNA genes. Comput Biol Chem. 2007;31:207–214. doi: 10.1016/j.compbiolchem.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Ishimaru D, Zuraw L, Ramalingam S, Sengupta TK, Bandyopadhyay S, Reuben A, Fernandes DJ, Spicer EK. Mechanism of regulation of bcl-2 mRNA by nucleolin and A+U-rich element-binding factor 1 (AUF1) J Biol Chem. 2010;285:27182–27191. doi: 10.1074/jbc.M109.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov PA, Nadezhdina ES. Stress granules: RNP-containing cytoplasmic bodies springing up under stress: the structure and mechanism of organization [in Russian] Mol Biol (Mosk) 2006;40:937–944. [PubMed] [Google Scholar]
- Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R, Bilbao D, Valcarcel J. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Jan E, Motzny CK, Graves LE, Goodwin EB. The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenowd-habditis elegans. EMBO J. 1999;18:258–269. doi: 10.1093/emboj/18.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Yoon JW, Walterhouse D, Iannaccone P, Goodwin EB. Conservation of the C. elegans tra-2 39UTR translational control. EMBO J. 1997;16:6301–6313. doi: 10.1093/emboj/16.20.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karashima T, Sugimoto A, Yamamoto M. Caenorhabditis elegans homologue of the human azoospermia factor DAZ is required for oogenesis but not for spermatogenesis. Development. 2000;127:1069–1079. doi: 10.1242/dev.127.5.1069. [DOI] [PubMed] [Google Scholar]
- Kashiwabara S, Noguchi J, Zhuang T, Ohmura K, Honda A, Sugiura S, Miyamoto K, Takahashi S, Inoue K, Ogura A, Baba T. Regulation of spermatogenesis by testis-specific, cytoplasmic poly(A) polymerase TPAP. Science. 2002;298:1999–2002. doi: 10.1126/science.1074632. [DOI] [PubMed] [Google Scholar]
- Kayali F, Montie HL, Rafols JA, DeGracia DJ. Prolonged translation arrest in reperfused hippocampal cornu Ammonis 1 is mediated by stress granules. Neuroscience. 2005;134:1223–1245. doi: 10.1016/j.neuroscience.2005.05.047. [DOI] [PubMed] [Google Scholar]
- Kedde M, Agami R. Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle. 2008;7:899–903. doi: 10.4161/cc.7.7.5644. [DOI] [PubMed] [Google Scholar]
- Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, Lund AH, Perrakis A, Raz E, Agami R. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol. 2000;151:1257–1268. doi: 10.1083/jcb.151.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee K, Angeles VT, Flores M, Nguyen HN, Reijo Pera RA. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Keene JD, Lager PJ. Post-transcriptional operons and regulons coordinating gene expression. Chromosome Res. 2005;13:327–337. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- Khaleghpour K, Svitkin YV, Craig AW, DeMaria CT, Deo RC, Burley SK, Sonenberg N. Translational repression by a novel partner of human poly(A) binding protein, Paip2. Mol Cell. 2001;7:205–216. doi: 10.1016/s1097-2765(01)00168-x. [DOI] [PubMed] [Google Scholar]
- Kharraz Y, Salmand PA, Camus A, Auriol J, Gueydan C, Kruys V, Morello D. Impaired embryonic development in mice overexpressing the RNA-binding protein TIAR. PLoS One. 2010;5:e11352. doi: 10.1371/journal.pone.0011352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Kim HH, Abdelmohsen K, Lal A, Pullmann R, Jr, Yang X, Galban S, Srikantan S, Martindale JL, Blethrow J, Shokat KM, Gorospe M. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008;22:1804–1815. doi: 10.1101/gad.1645808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kuwano Y, Zhan M, Pullmann R, Jr, Mazan-Mamczarz K, Li H, Kedersha N, Anderson P, Wilce MC, Gorospe M, Wilce JA. Elucidation of a C-rich signature motif in target mRNAs of RNA-binding protein TIAR. Mol Cell Biol. 2007;27:6806–6817. doi: 10.1128/MCB.01036-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 39UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- Kini HK, Vishnu MR, Liebhaber SA. Too much PABP, too little translation. J Clin Invest. 2010;120:3090–3093. doi: 10.1172/JCI44091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Kleene KC, Distel RJ, Hecht NB. Translational regulation and deadenylation of a protamine mRNA during spermiogenesis in the mouse. Dev Biol. 1984;105:71–79. doi: 10.1016/0012-1606(84)90262-8. [DOI] [PubMed] [Google Scholar]
- Kotaja N, Lin H, Parvinen M, Sassone-Corsi P. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J Cell Sci. 2006;119:2819–2825. doi: 10.1242/jcs.03022. [DOI] [PubMed] [Google Scholar]
- Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- Kress C, Gautier-Courteille C, Osborne HB, Babinet C, Paillard L. Inactivation of CUG-BP1/CELF1 causes growth, viability, and spermatogenesis defects in mice. Mol Cell Biol. 2007;27:1146–1157. doi: 10.1128/MCB.01009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, Nakano T. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe H, Kawaguchi A, Hori T, Mishiro K, Ono M, Sawada H, Uesugi S. Dynamic changes in intranuclear and subcellular localizations of mouse Prrp/DAZAP1 during spermatogenesis: the necessity of the C-terminal proline-rich region for nuclear import and localization. Arch Histol Cytol. 2004;67:325–333. doi: 10.1679/aohc.67.325. [DOI] [PubMed] [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W. Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 39 end processing and splicing. Mol Cell. 2006;23:195–205. doi: 10.1016/j.molcel.2006.05.037. [DOI] [PubMed] [Google Scholar]
- Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Carrick DM, Blackshear PJ. Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. J Biol Chem. 2005;280:34365–34377. doi: 10.1074/jbc.M506757200. [DOI] [PubMed] [Google Scholar]
- Lee K, Haugen HS, Clegg CH, Braun RE. Premature translation of protamine 1 mRNA causes precocious nuclear condensation and arrests spermatid differentiation in mice. Proc Natl Acad Sci U S A. 1995;92:12451–12455. doi: 10.1073/pnas.92.26.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Silver PA. Protein and RNA export from the nucleus. Dev Cell. 2002;2:261–272. doi: 10.1016/s1534-5807(02)00134-x. [DOI] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Li Y, Chen ZY, Wang W, Baker CC, Krug RM. The 39-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA. 2001;7:920–931. doi: 10.1017/s1355838201010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Taylor SJ, Shalloway D. Specificity and determinants of Sam68 RNA binding: implications for the biological function of K homology domains. J Biol Chem. 1997;272:27274–27280. doi: 10.1074/jbc.272.43.27274. [DOI] [PubMed] [Google Scholar]
- Lin YT, Yen PH. A novel nucleocytoplasmic shuttling sequence of DAZAP1, a testis-abundant RNA-binding protein. RNA. 2006;12:1486–1493. doi: 10.1261/rna.42206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Chao S, Chiou NT, Schuster G. The PNPase, exosome and RNA helicases as the building components of evolutionarily-conserved RNA degradation machines. J Biomed Sci. 2007;14:523–532. doi: 10.1007/s11373-007-9178-y. [DOI] [PubMed] [Google Scholar]
- Llamusi B, Artero R. Molecular effects of the CTG repeats in mutant dystrophia myotonica protein kinase gene. Curr Genomics. 2008;9:509–516. doi: 10.2174/138920208786847944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lolicato F, Marino R, Paronetto MP, Pellegrini M, Dolci S, Geremia R, Grimaldi P. Potential role of Nanos3 in maintaining the undifferentiated spermatogonia population. Dev Biol. 2008;313:725–738. doi: 10.1016/j.ydbio.2007.11.011. [DOI] [PubMed] [Google Scholar]
- Lopez de Silanes I, Galban S, Martindale JL, Yang X, Mazan-Mamczarz K, Indig FE, Falco G, Zhan M, Gorospe M. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol Cell Biol. 2005;25:9520–9531. doi: 10.1128/MCB.25.21.9520-9531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Schneider RJ. Tissue distribution of AU-rich mRNA-binding proteins involved in regulation of mRNA decay. J Biol Chem. 2004;279:12974–12979. doi: 10.1074/jbc.M310433200. [DOI] [PubMed] [Google Scholar]
- Lu ZH, Books JT, Ley TJ. Cold shock domain family members YB-1 and MSY4 share essential functions during murine embryogenesis. Mol Cell Biol. 2006;26:8410–8417. doi: 10.1128/MCB.01196-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA. Communication of the position of exon-exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science. 2001;293:1836–1839. doi: 10.1126/science.1062786. [DOI] [PubMed] [Google Scholar]
- Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, Yan W, Matzuk MM. GASZ is essential for male meiosis and suppression of retro-transposon expression in the male germline. PLoS Genet. 2009;5:e1000635. doi: 10.1371/journal.pgen.1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R, Cloutier P, Shkreta L, Fisette JF, Revil T, Chabot B. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–147. doi: 10.1007/978-0-387-77374-2_8. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Wolffe AP. Gene regulation by Y-box proteins: coupling control of transcription and translation. Trends Cell Biol. 1998;8:318–323. doi: 10.1016/s0962-8924(98)01300-2. [DOI] [PubMed] [Google Scholar]
- Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Kuwano Y, Zhan M, White EJ, Martindale JL, Lal A, Gorospe M. Identification of a signature motif in target mRNAs of RNA-binding protein AUF1. Nucleic Acids Res. 2009;37:204–214. doi: 10.1093/nar/gkn929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, Ferreira B, Liu CG, Villanueva A, Capella G, Schwartz S, Jr, Shiekhattar R, Esteller M. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mendez R, Barnard D, Richter JD. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 2002;21:1833–1844. doi: 10.1093/emboj/21.7.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature. 2000a;404:302–307. doi: 10.1038/35005126. [DOI] [PubMed] [Google Scholar]
- Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol Cell. 2000b;6:1253–1259. doi: 10.1016/s1097-2765(00)00121-0. [DOI] [PubMed] [Google Scholar]
- Min H, Turck CW, Nikolic JM, Black DL. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- Monesi V. Ribonucleic acid synthesis during mitosis and meiosis in the mouse testis. J Cell Biol. 1964;22:521–532. doi: 10.1083/jcb.22.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. RNA. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourelatos Z, Dostie J, Paushkin S, Sharma A, Charroux B, Abel L, Rappsilber J, Mann M, Dreyfuss G. miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs. Genes Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D, Gao M, O’Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najib S, Martin-Romero C, Gonzalez-Yanes C, Sanchez-Margalet V. Role of Sam68 as an adaptor protein in signal transduction. Cell Mol Life Sci. 2005;62:36–43. doi: 10.1007/s00018-004-4309-3. [DOI] [PubMed] [Google Scholar]
- Newbury SF, Muhlemann O, Stoecklin G. Turnover in the Alps: an mRNA perspective: workshops on mechanisms and regulation of mRNA turnover. EMBO Rep. 2006;7:143–148. doi: 10.1038/sj.embor.7400628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Chi M, Chalmel F, Agius E, Vanzo N, Khabar KS, Jegou B, Morello D. Temporally regulated traffic of HuR and its associated ARE-containing mRNAs from the chromatoid body to polysomes during mouse spermatogenesis. PLoS One. 2009;4:e4900. doi: 10.1371/journal.pone.0004900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niessing D, Blanke S, Jackle H. Bicoid associates with the 59-cap-bound complex of caudal mRNA and represses translation. Genes Dev. 2002;16:2576–2582. doi: 10.1101/gad.240002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott A, Le Hir H, Moore MJ. Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev. 2004;18:210–222. doi: 10.1101/gad.1163204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottone C, Galasso A, Gemei M, Pisa V, Gigliotti S, Piccioni F, Graziani F, Verrotti di Pianella A. Diminution of eIF4E activity suppresses parkin mutant phenotypes. Gene. 2010;470:12–19. doi: 10.1016/j.gene.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Paillard L, Legagneux V, Beverley Osborne H. A functional dead-enylation assay identifies human CUG-BP as a deadenylation factor. Biol Cell. 2003;95:107–113. doi: 10.1016/s0248-4900(03)00010-8. [DOI] [PubMed] [Google Scholar]
- Paillard L, Legagneux V, Maniey D, Osborne HB. c-Jun ARE targets mRNA deadenylation by an EDEN-BP (embryo deadenylation element-binding protein)-dependent pathway. J Biol Chem. 2002;277:3232–3235. doi: 10.1074/jbc.M109362200. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Messina V, Bianchi E, Barchi M, Vogel G, Moretti C, Palombi F, Stefanini M, Geremia R, Richard S, Sette C. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J Cell Biol. 2009;185:235–249. doi: 10.1083/jcb.200811138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Sette C. Role of RNA-binding proteins in mammalian spermatogenesis. Int J Androl. 2010;33:2–12. doi: 10.1111/j.1365-2605.2009.00959.x. [DOI] [PubMed] [Google Scholar]
- Paronetto MP, Zalfa F, Botti F, Geremia R, Bagni C, Sette C. The nuclear RNA-binding protein Sam68 translocates to the cytoplasm and associates with the polysomes in mouse spermatocytes. Mol Biol Cell. 2006;17:14–24. doi: 10.1091/mbc.E05-06-0548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvinen M. The chromatoid body in spermatogenesis. Int J Androl. 2005;28:189–201. doi: 10.1111/j.1365-2605.2005.00542.x. [DOI] [PubMed] [Google Scholar]
- Parvinen M, Jokelainen PT. Rapid movements of the chromatoid body in living early spermatids of the rat. Biol Reprod. 1974;11:85–92. doi: 10.1095/biolreprod11.1.85. [DOI] [PubMed] [Google Scholar]
- Parvinen M, Parvinen LM. Active movements of the chromatoid body: a possible transport mechanism for haploid gene products. J Cell Biol. 1979;80:621–628. doi: 10.1083/jcb.80.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G, Liuni S, Grillo G, Licciulli F, Mignone F, Gissi C, Saccone C. UTRdb and UTRsite: specialized databases of sequences and functional elements of 59 and 39 untranslated regions of eukaryotic mRNAs: update 2002. Nucleic Acids Res. 2002;30:335–340. doi: 10.1093/nar/30.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M, Wax S, Beck AR, Kedersha N, Gupta M, Maritim B, Chen S, Gueydan C, Kruys V, Streuli M, Anderson P. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S, Torabi N, Franco GV, Tremblay GA, Chen T, Vogel G, Morel M, Cleroux P, Forget-Richard A, Komarova S, Tremblay ML, Li W, Li A, Gao YJ, Henderson JE. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet. 2005;1:e74. doi: 10.1371/journal.pgen.0010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Ryder SP, Frater LA, Abramovitz DL, Goodwin EB, Williamson JR. RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat Struct Mol Biol. 2004;11:20–28. doi: 10.1038/nsmb706. [DOI] [PubMed] [Google Scholar]
- Ryder SP, Williamson JR. Specificity of the STAR/GSG domain protein Qk1: implications for the regulation of myelination. RNA. 2004;10:1449–1458. doi: 10.1261/rna.7780504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada A, Suzuki A, Suzuki H, Saga Y. The RNA-binding protein NANOS2 is required to maintain murine spermatogonial stem cells. Science. 2009;325:1394–1398. doi: 10.1126/science.1172645. [DOI] [PubMed] [Google Scholar]
- Sanchez R, Marzluff WF. The oligo(A) tail on histone mRNA plays an active role in translational silencing of histone mRNA during Xenopus oogenesis. Mol Cell Biol. 2004;24:2513–2525. doi: 10.1128/MCB.24.6.2513-2525.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JR, Gray NK, Beckmann K, Caceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–768. doi: 10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar B, Lu JY, Schneider RJ. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J Biol Chem. 2003;278:20700–20707. doi: 10.1074/jbc.M301176200. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P. Unique chromatin remodeling and transcriptional regulation in spermatogenesis. Science. 2002;296:2176–2178. doi: 10.1126/science.1070963. [DOI] [PubMed] [Google Scholar]
- Saunders PT, Turner JM, Ruggiu M, Taggart M, Burgoyne PS, Elliott D, Cooke HJ. Absence of mDazl produces a final block on germ cell development at meiosis. Reproduction. 2003;126:589–597. doi: 10.1530/rep.0.1260589. [DOI] [PubMed] [Google Scholar]
- Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. The 39-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- Sheng Y, Tsai-Morris CH, Gutti R, Maeda Y, Dufau ML. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is a transport protein involved in gene-specific mRNA export and protein translation during spermatogenesis. J Biol Chem. 2006;281:35048–35056. doi: 10.1074/jbc.M605086200. [DOI] [PubMed] [Google Scholar]
- Simon R, Tassan JP, Richter JD. Translational control by poly(A) elongation during Xenopus development: differential repression and enhancement by a novel cytoplasmic polyadenylation element. Genes Dev. 1992;6:2580–25891. doi: 10.1101/gad.6.12b.2580. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Skrisovska L, Bourgeois CF, Stefl R, Grellscheid SN, Kister L, Wenter P, Elliott DJ, Stevenin J, Allain FH. The testis-specific human protein RBMY recognizes RNA through a novel mode of interaction. EMBO Rep. 2007;8:372–379. doi: 10.1038/sj.embor.7400910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol Cell. 1999;4:1017–1027. doi: 10.1016/s1097-2765(00)80230-0. [DOI] [PubMed] [Google Scholar]
- Stoss O, Olbrich M, Hartmann AM, Konig H, Memmott J, Andreadis A, Stamm S. The STAR/GSG family protein rSLM-2 regulates the selection of alternative splice sites. J Biol Chem. 2001;276:8665–8673. doi: 10.1074/jbc.M006851200. [DOI] [PubMed] [Google Scholar]
- Sun L, Stoecklin G, Van Way S, Hinkovska-Galcheva V, Guo RF, Anderson P, Shanley TP. Tristetraprolin (TTP)-14-3-3 complex formation protects TTP from dephosphorylation by protein phosphatase 2a and stabilizes tumor necrosis factor-alpha mRNA. J Biol Chem. 2007;282:3766–3777. doi: 10.1074/jbc.M607347200. [DOI] [PubMed] [Google Scholar]
- Takeda Y, Mishima Y, Fujiwara T, Sakamoto H, Inoue K. DAZL relieves miRNA-mediated repression of germline mRNAs by controlling poly(A) tail length in zebrafish. PLoS One. 2009;4:e7513. doi: 10.1371/journal.pone.0007513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- Tang PZ, Tsai-Morris CH, Dufau ML. A novel gonadotropin-regulated testicular RNA helicase: a new member of the dead-box family. J Biol Chem. 1999;274:37932–37940. doi: 10.1074/jbc.274.53.37932. [DOI] [PubMed] [Google Scholar]
- Tang SJ, Meulemans D, Vazquez L, Colaco N, Schuman E. A role for a rat homolog of staufen in the transport of RNA to neuronal dendrites. Neuron. 2001;32:463–475. doi: 10.1016/s0896-6273(01)00493-7. [DOI] [PubMed] [Google Scholar]
- Tay J, Hodgman R, Sarkissian M, Richter JD. Regulated CPEB phosphorylation during meiotic progression suggests a mechanism for temporal control of maternal mRNA translation. Genes Dev. 2003;17:1457–1462. doi: 10.1101/gad.1071403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay J, Richter JD. Germ cell differentiation and synaptonemal complex formation are disrupted in CPEB knockout mice. Dev Cell. 2001;1:201–213. doi: 10.1016/s1534-5807(01)00025-9. [DOI] [PubMed] [Google Scholar]
- Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–198. doi: 10.1016/S1046-2023(02)00022-1. [DOI] [PubMed] [Google Scholar]
- Thiele A, Nagamine Y, Hauschildt S, Clevers H. AU-rich elements and alternative splicing in the beta-catenin 39UTR can influence the human beta-catenin mRNA stability. Exp Cell Res. 2006;312:2367–2378. doi: 10.1016/j.yexcr.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai-Morris CH, Sheng Y, Gutti R, Li J, Pickel J, Dufau ML. Gonadotropin-regulated testicular RNA helicase (GRTH/DDX25) gene: cell-specific expression and transcriptional regulation by androgen in transgenic mouse testis. J Cell Biochem. 2010;109:1142–1147. doi: 10.1002/jcb.22493. [DOI] [PubMed] [Google Scholar]
- Tsai-Morris CH, Sheng Y, Lee E, Lei KJ, Dufau ML. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is essential for spermatid development and completion of spermatogenesis. Proc Natl Acad Sci U S A. 2004;101:6373–6378. doi: 10.1073/pnas.0401855101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Tushir JS, Zamore PD, Zhang Z. SnapShot: mouse piRNAs, PIWI proteins, and the ping-pong cycle. Cell. 2009;139:830–830.e1. doi: 10.1016/j.cell.2009.10.042. [DOI] [PubMed] [Google Scholar]
- van Dijk EL, Schilders G, Pruijn GJ. Human cell growth requires a functional cytoplasmic exosome, which is involved in various mRNA decay pathways. RNA. 2007;13:1027–1035. doi: 10.1261/rna.575107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanGompel MJ, Xu EY. A novel requirement in mammalian spermatid differentiation for the DAZ-family protein Boule. Hum Mol Genet. 2010;19:2360–2369. doi: 10.1093/hmg/ddq109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Ventela S, Toppari J, Parvinen M. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: mechanisms of haploid gene product sharing. Mol Biol Cell. 2003;14:2768–2780. doi: 10.1091/mbc.E02-10-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villace P, Marion RM, Ortin J. The composition of Staufen-containing RNA granules from human cells indicates their role in the regulated transport and translation of messenger RNAs. Nucleic Acids Res. 2004;32:2411–2420. doi: 10.1093/nar/gkh552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan P, Ohn T, Chiang YC, Chen J, Denis CL. Mouse CAF1 can function as a processive deadenylase/39–59-exonuclease in vitro but in yeast the deadenylase function of CAF1 is not required for mRNA poly(A) removal. J Biol Chem. 2004;279:23988–23995. doi: 10.1074/jbc.M402803200. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Burge CB. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kiledjian M. The poly(A)-binding protein and an mRNA stability protein jointly regulate an endoribonuclease activity. Mol Cell Biol. 2000;20:6334–6341. doi: 10.1128/mcb.20.17.6334-6341.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster KE, O’Bryan MK, Fletcher S, Crewther PE, Aapola U, Craig J, Harrison DK, Aung H, Phutikanit N, Lyle R, Meachem SJ, Antonarakis SE, de Kretser DM, Hedger MP, Peterson P, Carroll BJ, Scott HS. Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis. Proc Natl Acad Sci U S A. 2005;102:4068–4073. doi: 10.1073/pnas.0500702102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil TT, Xanthakis D, Parton R, Dobbie I, Rabouille C, Gavis ER, Davis I. Distinguishing direct from indirect roles for bicoid mRNA localization factors. Development. 2010;137:169–176. doi: 10.1242/dev.044867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EK, Moore-Jarrett T, Ruley HE. PUM2, a novel murine puf protein, and its consensus RNA-binding site. RNA. 2001;7:1855–1866. [PMC free article] [PubMed] [Google Scholar]
- Wilson GM, Lu J, Sutphen K, Suarez Y, Sinha S, Brewer B, Villanueva-Feliciano EC, Ysla RM, Charles S, Brewer G. Phosphorylation of p40AUF1 regulates binding to A + U-rich mRNA-destabilizing elements and protein-induced changes in ribonucleoprotein structure. J Biol Chem. 2003;278:33039–33048. doi: 10.1074/jbc.M305775200. [DOI] [PubMed] [Google Scholar]
- Xie J, Lee JA, Kress TL, Mowry KL, Black DL. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc Natl Acad Sci U S A. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu EY, Chang R, Salmon NA, Reijo Pera RA. A gene trap mutation of a murine homolog of the Drosophila stem cell factor Pumilio results in smaller testes but does not affect litter size or fertility. Mol Reprod Dev. 2007;74:912–921. doi: 10.1002/mrd.20687. [DOI] [PubMed] [Google Scholar]
- Xu EY, Moore FL, Pera RA. A gene family required for human germ cell development evolved from an ancient meiotic gene conserved in metazoans. Proc Natl Acad Sci U S A. 2001;98:7414–7419. doi: 10.1073/pnas.131090498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Hecht NB. MSY2 and polypyrimidine tract binding protein 2 stabilize mRNAs in the mammalian testis. Int J Androl. 2008;31:457–461. doi: 10.1111/j.1365-2605.2008.00885.x. [DOI] [PubMed] [Google Scholar]
- Xu M, McCarrey JR, Hecht NB. A cytoplasmic variant of the KH-type splicing regulatory protein serves as a decay-promoting factor for phosphoglycerate kinase 2 mRNA in murine male germ cells. Nucleic Acids Res. 2008;36:7157–7167. doi: 10.1093/nar/gkn800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Medvedev S, Yang J, Hecht NB. MIWI-independent small RNAs (MSY-RNAs) bind to the RNA-binding protein, MSY2, in male germ cells. Proc Natl Acad Sci U S A. 2009;106:12371–12376. doi: 10.1073/pnas.0903944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Zhao H, Dinkins RD, Cheng X, Carberry G, Li QQ. The 73 kD subunit of the cleavage and polyadenylation specificity factor (CPSF) complex affects reproductive development in Arabidopsis. Plant Mol Biol. 2006;61:799–815. doi: 10.1007/s11103-006-0051-6. [DOI] [PubMed] [Google Scholar]
- Yamada-Okabe T, Mio T, Kashima Y, Matsui M, Arisawa M, Yamada-Okabe H. The Candida albicans gene for mRNA 5-cap methyltransferase: identification of additional residues essential for catalysis. Microbiology. 1999;145:3023–3033. doi: 10.1099/00221287-145-11-3023. [DOI] [PubMed] [Google Scholar]
- Yanagiya A, Delbes G, Svitkin YV, Robaire B, Sonenberg N. The poly(A)-binding protein partner Paip2a controls translation during late spermiogenesis in mice. J Clin Invest. 2010;120:3389–3400. doi: 10.1172/JCI43350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HT, Peggie M, Cohen P, Rousseau S. DAZAP1 interacts via its RNA-recognition motifs with the C-termini of other RNA-binding proteins. Biochem Biophys Res Commun. 2009;380:705–709. doi: 10.1016/j.bbrc.2009.01.166. [DOI] [PubMed] [Google Scholar]
- Yang J, Medvedev S, Reddi PP, Schultz RM, Hecht NB. The DNA/RNA-binding protein MSY2 marks specific transcripts for cytoplasmic storage in mouse male germ cells. Proc Natl Acad Sci U S A. 2005a;102:1513–1518. doi: 10.1073/pnas.0404685102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Medvedev S, Yu J, Tang LC, Agno JE, Matzuk MM, Schultz RM, Hecht NB. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc Natl Acad Sci U S A. 2005b;102:5755–5760. doi: 10.1073/pnas.0408718102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Morales CR, Medvedev S, Schultz RM, Hecht NB. In the absence of the mouse DNA/RNA-binding protein MSY2, messenger RNA instability leads to spermatogenic arrest. Biol Reprod. 2007;76:48–54. doi: 10.1095/biolreprod.106.055095. [DOI] [PubMed] [Google Scholar]
- Yen PH. Putative biological functions of the DAZ family. Int J Androl. 2004;27:125–129. doi: 10.1111/j.1365-2605.2004.00469.x. [DOI] [PubMed] [Google Scholar]
- Yoon YJ, Mowry KL. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development. 2004;131:3035–3045. doi: 10.1242/dev.01170. [DOI] [PubMed] [Google Scholar]
- Zhang H, Barnoski BL, Sol-Church K, Stabley DL, Martin-Deleon PA. Murine Spam1 mRNA: involvement of AU-rich elements in the 39UTR and antisense RNA in its tight post-transcriptional regulation in spermatids. Mol Reprod Dev. 2006;73:247–255. doi: 10.1002/mrd.20400. [DOI] [PubMed] [Google Scholar]
- Zhang T, Delestienne N, Huez G, Kruys V, Gueydan C. Identification of the sequence determinants mediating the nucleo-cytoplasmic shuttling of TIAR and TIA-1 RNA-binding proteins. J Cell Sci. 2005;118:5453–5463. doi: 10.1242/jcs.02669. [DOI] [PubMed] [Google Scholar]
- Zhao M, Shirley CR, Mounsey S, Meistrich ML. Nucleoprotein transitions during spermiogenesis in mice with transition nuclear protein Tnp1 and Tnp2 mutations. Biol Reprod. 2004;71:1016–1025. doi: 10.1095/biolreprod.104.028191. [DOI] [PubMed] [Google Scholar]
- Zhong J, Peters AH, Lee K, Braun RE. A double-stranded RNA binding protein required for activation of repressed messages in mammalian germ cells. Nat Genet. 1999;22:171–174. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]


