Significance
Chronic pancreatitis is a progressive inflammatory disease that inflicts hundreds of thousands of people annually in the United State alone, and it has no effective treatment. In the present paper we develop a mathematical model of the progression of the disease and use the model to evaluate the effect of two potential drugs, anti-TNF- and anti-IL-6. The model is represented by a system of partial differential equations with carefully estimated parameters. The simulation of the model shows that both drugs can reduce significantly the progressive fibrosis of the pancreas. The model has the potential to generate an interesting clinically testable hypothesis.
Keywords: mathematical model, chronic pancreatitis, drug studies
Abstract
Chronic pancreatitis (CP) is a progressive inflammatory disease of the pancreas, leading to its fibrotic destruction. There are currently no drugs that can stop or slow the progression of the disease. The etiology of the disease is multifactorial, whereas recurrent attacks of acute pancreatitis are thought to precede the development of CP. A better understanding of the pathology of CP is needed to facilitate improved diagnosis and treatment strategies for this disease. The present paper develops a mathematical model of CP based on a dynamic network that includes macrophages, pancreatic stellate cells, and prominent cytokines that are present at high levels in the CP microenvironment. The model is represented by a system of partial differential equations. The model is used to explore in silico potential drugs that could slow the progression of the disease, for example infliximab (anti-TNF-) and tocilizumab or siltuximab (anti-IL-6/IL-6R).
Chronic pancreatitis (CP) is a continuous or recurrent inflammatory disease of the pancreas characterized by progressive and irreversible morphological changes (scarring). Extensive fibrosis and inflammation often lead to exocrine and endocrine insufficiency (1). Symptoms include persistent abdominal pain, steatorrhea, nausea, and vomiting (2, 3). In the United States, CP results in 56,000 hospitalizations each year and 122,000 outpatient visits. The disease usually begins in adulthood and is more common among men. No curative treatment for CP exists. Medical treatment of this condition includes pain control with analgesic agents and replacement of pancreatic enzymes (1–5).
The etiology of CP is complex and multifactorial (2, 5). Recurrent attacks of acute pancreatitis (AP) are thought to precede the development of CP. Factors that may increase risk of recurrent AP or CP include alcohol abuse, gallstones, genetic mutations, autoimmunity, hypercalcemia, the use of certain prescription medications, steroids, trauma, and others. Still, a large number of cases of CP are considered idiopathic. A better understanding of the pathology of CP is needed to facilitate improved diagnosis and treatment strategies for this disease (6–8).
One cell type known to contribute to CP pathology is the pancreatic stellate cell (PSC). PSCs are myofibroblast-like cells that reside in the periacinar regions of the pancreas in a quiescent state (4, 9). Quiescent PSCs contribute to maintenance of proper tissue architecture by regulating the synthesis of extracellular matrix (ECM) (10). Upon stimulation, these PSCs become activated, migrate to injured locations, promote inflammation, and secrete ECM proteins. During CP, PSCs remain persistently activated and are hypothesized to play a pivotal role in the development of CP by facilitating sustained inflammation and fibrosis in the organ (11–13).
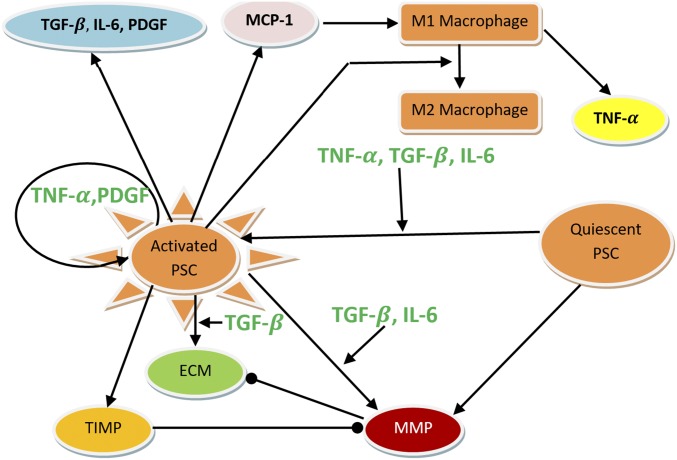
These pathological processes are orchestrated by PSCs, in conjunction with parenchymal and immune cells, via the secretion of soluble signaling factors, including cytokines and chemokines. Interactions between these mediators and the cells of the pancreatic microenvironment are complex and dynamic. During pancreatic inflammation, endothelial cells secrete monocyte chemoattractant protein (MCP-1) (14–19), triggering recruitment of monocytes from the blood into the inflamed area (20, 21). Upon entering the pancreas, monocytes differentiate into macrophages, which secrete tumor necrosis factor alpha (TNF-) (22, 23) and interleukin 6 (IL-6) (24, 25). TNF- and IL-6 from macrophages, along with other soluble factors, are then capable of causing PSC activation. Once active, PSCs and other inflammatory subsets of fibroblasts secrete proinflammatory cytokines, including platelet-derived growth factor (PDGF), transforming growth factor beta (TGF-) and IL-6 (9, 10, 24–27) as well as MCP-1 (14–19). Furthermore, a recent study (28) shows that IL-4 and IL-13 secreted by PSCs preferentially change classically activated macrophages M1 to alternatively activated macrophages M2, which secrete IL-10, TGF-, and PDGF. The resultant mixture of cytokines and chemokines in the inflamed pancreas is quite dynamic and characterized by other interactions between these factors. For example, PSC activation may be enhanced by autocrine and paracrine of TGF- (in addition to IL-6 and TNF-) (9, 10, 27). TNF- and PDGF increase proliferation of active PSCs (APSCs) (9, 10, 12, 29–33), whereas TGF- and IL-6 decrease the proliferation of APSCs (9, 10). Conversely, the production of matrix metalloproteinases (MMPs) (34) and tissue inhibitors of metalloproteinase (TIMP) was enhanced by these same cytokines (34). Taken together, the activation of PSCs is enhanced by TGF-, PDGF, and TNF- (10).
The above processes are summarized in Fig. 1. We did not include in Fig. 1 IL-1 and IL-10, both of which are produced by macrophages. IL-1 and IL-10 are known to affect both the activation and proliferation of PSCs (10), similar to IL-6. For simplicity, however, we do not account for them explicitly and use IL-6 to implicitly represent their activity by adjusting parameter values associated with IL-6.
Fig. 1.
Interaction of cells and cytokines in CP. Arrows represent activation or secretion, and circular heads represent inhibition. Circular arrow (about the activated PSC) means increased proliferation, and circular head means decreased proliferation.
Due to the complexity of these interactions, as well as the synergistic and pleotropic effects of such signaling molecules, it is necessary to better understand—in a quantitative manner—the way each signaling node may affect PSC activation, inflammation, and fibrosis to contribute to CP pathology. Mathematical modeling of these interactions would identify the most relevant targets for future therapies, including neutralizing antibodies against specific cytokines. Such therapies have not been tested in the context of CP.
In the present report, we have developed a mathematical model of cytokine interactions that influence PSCs in the setting of CP. Simulations of the model show significant increases in TNF-, TGF-, IL-6, and PDGF in the pancreatic tissue of patients. These predictions were in agreement with clinical data. The model was then used to explore the impact of treatment with anti-IL-6 and anti- TNF- drugs on this physiological system.
Mathematical Model
The mathematical model for CP includes quiescent PSCs and APSCs. Based on the interactive network in Fig. 1, we include in the model the following densities of cells and concentrations of cytokines:
 |
All terms are in units of grams, centimeters, and days.
Equations for Quiescent PSC () and Activated PSC (P).
The evolution of the densities of quiescent and activated PSCs is modeled by the equations
| [1] |
| [2] |
where , , , and are the half-saturation values of TNF-, PDGF, TGF-, and IL-6, respectively. The first term on the right-hand side of Eq. 1 is the migration of PSCs by chemotaxis induced by PDGF (31). The proliferation of APSCs is enhanced by TNF- (9, 10, 35) and PDGF (12, 29–33). As reported in ref. 10, IL-6, at the very large concentration of g/mL, inhibited the proliferation of PSCs, but because the concentration of IL-6 in tissue is of the much smaller order of g/ml, we do not include such inhibition. The PSCs are activated by TNF-, TGF-, and IL-6 (9, 10, 27).
Equations for MCP-1 (C).
The MCP-1 equation is given by
| [3] |
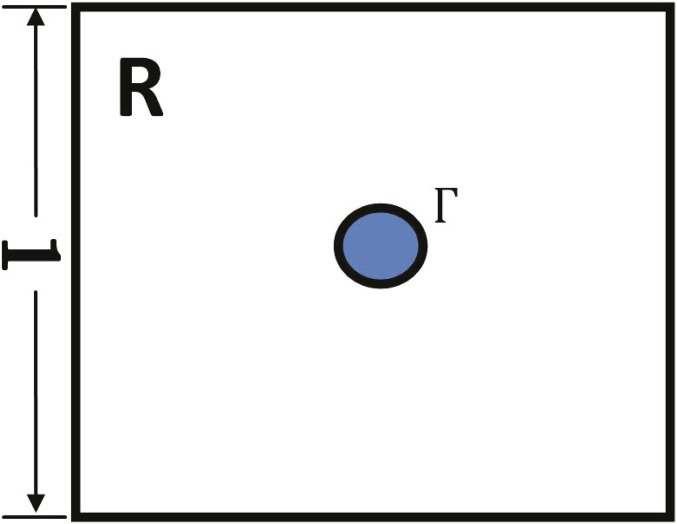
where is the characteristic function of the injured area in the pancreas, which gives rise to the initial inflammation (Fig. 2). The second term on the right-hand side accounts for the production of MCP-1 by activated PSCs (14–19). MCP-1 is a chemoattractant to M1 macrophages and is therefore internalized by macrophages (the third term) (36, 37).
Fig. 2.
Domain with an injured (initially inflamed) area .
Equations for Macrophages ( and ).
As reported in ref. 28, in chronic pancreatitis IL-4 () and IL-13 () produced by active PSCs preferentially change M1 to M2 macrophages. The velocity of change can be modeled by , where
Because the cytokine dynamic is much faster than cell dynamics, we use the steady-state approximation to conclude that IL-4 and IL-13 concentrations are proportional to . Hence the velocity of change from M1 to M2 is modeled by , where .
The evolution of macrophages outside blood vessels is modeled by
Here the first term on the right-hand side accounts for recruitment of M1 macrophages by MCP-1 (20), and is the chemotactic parameter, and the second term is the transition from M1 to M2 macrophages.
Because macrophages are the monocytes that migrated from the blood vessels into the tissue, we assume that satisfies the following boundary condition at the blood vessel’s membrane,
where is monocyte density in the blood, and depends on the MCP-1 concentration, . We assume that the blood vessels (in a cross-section) are distributed according to Fig. 3, and they occupy 5% of the total area.
Fig. 3.
Cross-section of parallel blood vessels. (A) From above. (B) From the side.
Assuming that all of the small circles in Fig. 3 are arranged approximately in a periodic manner and that their diameter is small, we can use the homogenization theory (38) to write down the “effective” equation for over the entire tissue,
| [4] |
where ( is the saturation level of MCP-1).
Indeed the coefficient arises from the 5% assumption above, and the coefficient arises from the effective diffusion coefficient where is replaced by . The coefficient is computed by
where satisfies the equation
and is periodic, with period 1, in the horizontal and vertical directions; here is a unit square from which a circle located at the center is removed; the area of the circle is 0.5 and its boundary is denoted by . (Fig. 4) Computing by finite-element discretization, we find that and .
Fig. 4.
Homogenization domain.
M2 macrophages satisfy the equation
| [5] |
Equation for ECM ().
ECM is produced by both quiescent and activated PSCs (10). The production by is enhanced by TGF- (12); it is also enhanced by TNF- and PDGF and is inhibited by IL-6 (10, 12). However, TNF-, PDGF, and IL-6 affect very little the production of ECM in comparison with TGF-, and we therefore neglect them in modeling the ECM equation. Finally the ECM is degraded by MMP and remodeled at rate , which, for simplicity, is assumed to be a constant. Hence the equation of the density of ECM is given by
| [6] |
Alterations in ECM structure during CP result in enhanced fibrosis and glandular stiffness that may affect PSC activation and cytokine activity within the pancreatic milieu. In one study, Asaumi et al. (39) demonstrate that activation of PSCs is enhanced by externally applied pressure. Several papers have demonstrated the ability of ECM stiffness to promote the activation of hepatic stellate cells, liver fibroblasts that closely resemble PSCs (40, 41). ECM-associated forces have also been shown to alter the activation of latent TGF-, which is secreted from active PSCs and promotes the differentiation of myofibroblasts (42, 43). Thus, it is important to note that the effects of ECM structure and pressure are not represented in these models.
Equations for TGF- (), TNF- (), IL-6 (), and PDGF (G).
The equations for TGF-, TNF-, IL-6, and PDGF are
| [7] |
| [8] |
| [9] |
| [10] |
For simplicity, we assumed that the cytokines are produced (by PSCs or macrophages) at a constant rate, and they undergo a natural decay with constant rates (9, 10, 24, 25, 27). However, in the production of IL-6, we also included enhancement of IL-6 production by TGF- (24).
Equations for MMP (Q) and TIMP ().
We have the following sets of reaction diffusion equations for MMP and TIMP ( and ):
| [11] |
| [12] |
and are produced by PSCs. The production of MMP by APSCs is enhanced by TGF- and IL-6 (34). In Eq. 11, MMP is depleted by binding with TIMP (second to last term); this entails also depletion of TIMP by MMP in Eq. 12.
Equation for Scar (S).
Fibrotic diseases are characterized by excessive scarring due to excessive production and deposition of ECM and disruption of normal healthy protein cross-linking. MMP disrupts collagen cross-linking and increases scarring in cases of excessive collagen concentrations (44, 45). We accordingly represent the state of the scar by the equation
| [13] |
Boundary Conditions.
We simulate the model in a square domain with an injured area (Fig. 2). We take and . We assume periodic boundary conditions for all variables.
Initial Conditions.
We assume that initially , , , and , where , , and are determined by steady states of Eqs. 6, 11, and 12. All other variables are initially equal to zero.
Results
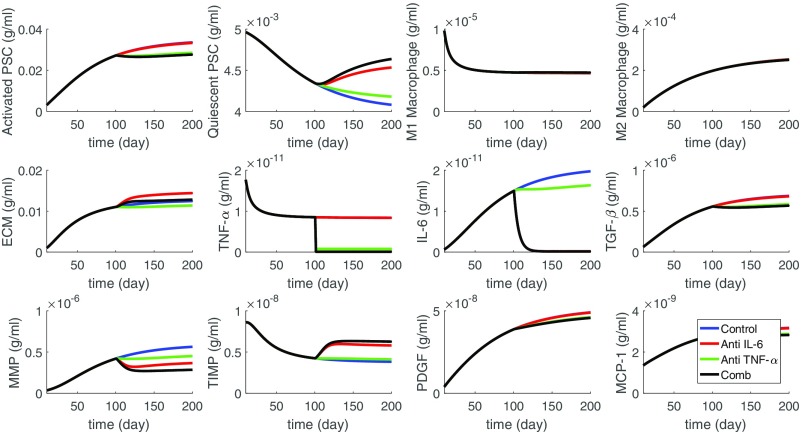
From Fig. 5 (the blue profiles) we see that almost all of the PSCs became activated. As a result, the concentrations of cytokines IL-6, TGF-, PDGF, and MCP-1 have significantly increased, by a factor of 5–15 compared with their concentration in healthy normal pancreatic tissue given by the values of , , , and in SI Appendix, Table S2. Also, with the increase in migration of macrophages, the concentration of TNF- in tissue increased by a factor of 5 compared with . The increase in IL-6, TGF-, TNF-, and PDGF in tissue of CP patients is documented in refs. 46–49 whereas the increase of MCP-1 in serum of CP patients is reported in ref. 47.
Fig. 5.
Simulation of the average densities of cells and concentrations of cytokines for the first 200 d since the start of the diseases (blue). Anti-IL-6 reduces the production rate of IL-6 by 90% (red) and anti-TNF- reduces the production rate of TNF- by 90% (green). All of the parameters are from SI Appendix, Tables S1 and S2.
We also see that more than 90% of the PSCs became activated, and the average ECM concentration increased 70-fold, from g/mL to g/mL. The disruption of proteins cross-linking in the ECM may be indirectly inferred from the significant increase in MMP and decrease in TIMP.
CP is a chronic inflammatory disease characterized by progressive fibrotic destruction of the pancreas. The current treatment options of CP are limited to supportive and palliative care, and there is an urgent demand to develop drugs that can stop or, at least, slow the progress of the disease (50). As mentioned in ref. 50, several animal studies have been conducted, but they have not been translated into the clinical setting.
We use our model to explore, in silico, the efficacy of two potential drugs proposed in ref. 50: anti-TNF- (infliximab, golimruab) and anti-IL-6 (tocilizumab).
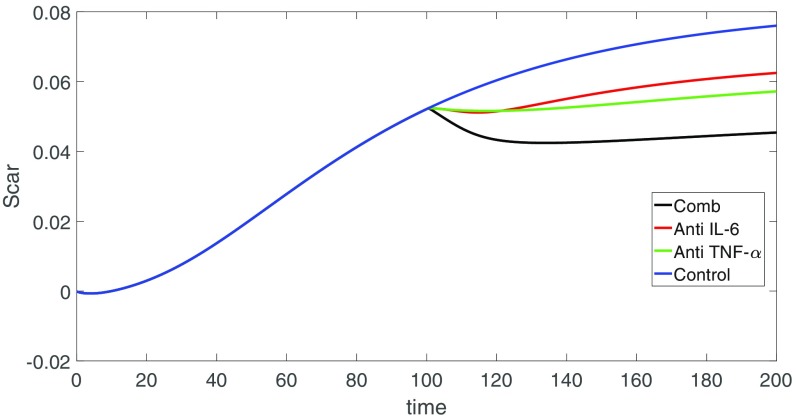
We implemented the treatment by anti-IL-6 and by anti-TNF- by reducing the production coefficients of IL-6 and TNF- by a factor of 10. Figs. 5 and 6 show the results of the drugs. The control curves are in the color blue. Some of these profiles are changed only very little by the drugs. When the changes are significant, the profiles corresponding to anti-IL-6 appear in the color red, and the profiles corresponding to anti-TNF- appear in the color green. We see that anti-TNF- was more effective than anti-IL-6 in reducing TGF- and PDGF and this results in decreases in ECM. Although anti-IL-6 increases the ECM, it also decreases MMP and thus perhaps does better than anti-TNF- in preserving the architecture of the tissue. When we used the combination of anti-TNF- and anti-IL-6, both at the same level as in Fig. 5, we found (not shown here) that there was only slight improvement in the reduction of MMP.
Fig. 6.
Scar concentration for different drugs vs. control. All of the parameters are from SI Appendix, Tables S1 and S2.
Discussion
CP is a progressive inflammatory disease of the pancreas, leading to its fibrotic destruction. There is currently no drug that can stop or slow the progression of the disease. Therefore, animal models are needed for advancing our understanding of the pathophysiological processes associated with CP (51). Animal models with pigs, dogs, opossums, rats, and other animals are reported in refs. 4 and 51–53. But the models, so far, have their limitations and have not reproduced the aspects of histopathology, endocrine/exocrine insufficiency, and pain that characterize human CP (51). In the present paper we developed a mathematical model of the disease. The model is based on a dynamical network of immune cells and cytokines that affect the pancreatic tissue. Simulations of the model indicated that TNF-, TGF-, IL-6, and PDGF are likely to increase in the pancreatic microenvironment during chronic pancreatitis. These results are consistent with those observed from prior clinical data (46–49). The model can be used to explore, in silico, the efficacy of treatments that target some of the overexpressed cytokines. We conducted treatments by anti-IL-6 and anti-TNF- drugs. Assuming that each of these drugs reduces the production of the cytokines by the same percentage, it appears that anti-TNF- is more effective.
For simplicity we did not include in our model T cells and associated cytokines such as IL-2, IL-10, and IL-12 and IFN-. When clinical data become available, the model could then be further extended, and some of the parameters better adjusted to fit the data, to make the model predictions more reliable. In addition, we have not included in the analysis soluble factors from parenchymal cells or other infiltrating immune cells, such as neutrophils. Furthermore, we have not included in the analysis subclassification of individual fibroblast populations (IL-6 high vs. IL-6 low) that have been identified in the setting of pancreatic cancer (26). Certainly the biologic complexity within the pancreatic microenvironment is appreciated and could be modeled more rigorously as information becomes available on the abundance of other relevant cell populations and soluble factors. Due to the complexity of CP and the lack of standardized data available in serum factor analysis, several shortcomings exist in the data on which we based our analyses. First, data gathered from the literature to use in developing this model may have inherent inconsistencies in the experimental methods and patient inclusion criteria used, and some data defining cytokine circuits may have been derived in part from early studies of stellate cell biology conducted from rat sources. In addition, serum analysis we performed has inherently high levels of variability within disease groups for any given analyte. This may be due to natural human variability or differences in disease severity or staging or may be based on etiology. The spectrum of disease from AP to recurrent AP to CP and the various associated etiologies may significantly impact the cytokine profile of each individual patient. Thus, each stage or etiology could have one or several specific cytokine signaling circuits that predominate. In the future, it may be necessary to analyze these differences in circulating levels of soluble factors for the various subtypes of CP to further refine this model.
Supplementary Material
Acknowledgments
W.H. and A.F. have been supported by the Mathematical Biosciences Institute and the National Science Foundation under Grant DMS 0931642. This work was also supported in part through funding from the National Institutes of Health, including Grants 1 R01 CA208253-01 (to G.B.L.), 1 R21 AI124687-01 (to G.B.L.), and 1 U01 DK108327-01 (to D.L.C. and P.A.H.) and a grant from Chi Rho Clin, Inc. (to G.B.L.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620264114/-/DCSupplemental.
References
- 1.Lee P, Stevens T. 2014 Chronic pancreatitis. Cleveland Clin. http://www.clevelandclinicmeded.com/medicalpubs/diseasemanagement/gastroenterology/chronic-pancreatitis/. Accessed April 17, 2017.
- 2.Huffman LC, Reed MF, Howington JA. Video-assisted thoracoscopic splanchnicectomy for pain control in chronic pancreatitis. Innovations. 2006;1:171–174. doi: 10.1097/01.IMI.0000220917.68537.2f. [DOI] [PubMed] [Google Scholar]
- 3.Zaheer A, Singh VK, Qureshi RO, Fishman EK. The revised Atlanta classification for acute pancreatitis: Updates in imaging terminology and guidelines. Abdom Imaging. 2013;38:125–136. doi: 10.1007/s00261-012-9908-0. [DOI] [PubMed] [Google Scholar]
- 4.Aghdassi AA, et al. Animal models for investigating chronic pancreatitis. Fibrogenesis Tissue Repair. 2011;4:26. doi: 10.1186/1755-1536-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman S, Whitcomb D, Grover S. 2016 Treatment of chronic pancreatitis. UpToDate. https://www.uptodate.com/contents/treatment-of-chronic-pancreatitis. Accessed April 17, 2017.
- 6.Ammann RW, Muellhaupt B. Progression of alcoholic acute to chronic pancreatitis. Gut. 1994;35:552–556. doi: 10.1136/gut.35.4.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brock C, Nielsen LM, Lelic D, Drewes AM. Pathophysiology of chronic pancreatitis. World J Gastroenterol. 2013;19:7231–7240. doi: 10.3748/wjg.v19.i42.7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens T, Conwell DL, Zuccaro G. Pathogenesis of chronic pancreatitis: An evidence-based review of past theories and recent developments. Am J Gastroenterol. 2004;99:2256–2270. doi: 10.1111/j.1572-0241.2004.40694.x. [DOI] [PubMed] [Google Scholar]
- 9.Marzoq AJ, Giese N, Hoheisel JD, Alhamdani MS. Proteome variations in pancreatic stellate cells upon stimulation with proinflammatory factors. J Biol Chem. 2013;288:32517–32527. doi: 10.1074/jbc.M113.488387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mews P, et al. Pancreatic stellate cells respond to inflammatory cytokines: Potential role in chronic pancreatitis. Gut. 2002;50:535–541. doi: 10.1136/gut.50.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Apte MV, Pirola RC, Wilson JS. Pancreatic stellate cells: A starring role in normal and diseased pancreas. Front Physiol. 2012;3:344. doi: 10.3389/fphys.2012.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Apte MV, et al. Pancreatic stellate cells are activated by proinflammatory cytokines: Implications for pancreatic fibrogenesis. Gut. 1999;44:534–541. doi: 10.1136/gut.44.4.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haber PS, et al. Activation of pancreatic stellate cells in human and experimental pancreatic fibrosis. Am J Pathol. 1999;155:1087–1095. doi: 10.1016/S0002-9440(10)65211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masamune A, et al. Ligands of peroxisome proliferator-activated receptor-gamma block activation of pancreatic stellate cells. J Biol Chem. 2002;277:141–147. doi: 10.1074/jbc.M107582200. [DOI] [PubMed] [Google Scholar]
- 15.Masamune A, Shimosegawa T. Signal transduction in pancreatic stellate cells. J Gastroenterol. 2009;44:249–260. doi: 10.1007/s00535-009-0013-2. [DOI] [PubMed] [Google Scholar]
- 16.Andoh A, et al. Cytokine regulation of chemokine (IL-8, MCP-1, and RANTES) gene expression in human pancreatic periacinar myofibroblasts. Gastroenterology. 2000;119:211–219. doi: 10.1053/gast.2000.8538. [DOI] [PubMed] [Google Scholar]
- 17.Vonlaufen A, et al. Bacterial endotoxin: A trigger factor for alcoholic pancreatitis? Evidence from a novel, physiologically relevant animal model. Gastroenterology. 2007;133:1293–1303. doi: 10.1053/j.gastro.2007.06.062. [DOI] [PubMed] [Google Scholar]
- 18.Gibo J, et al. Camostat mesilate attenuates pancreatic fibrosis via inhibition of monocytes and pancreatic stellate cells activity. Lab Invest. 2005;85:75–89. doi: 10.1038/labinvest.3700203. [DOI] [PubMed] [Google Scholar]
- 19.Marra F. Renaming cytokines: MCP-1, major chemokine in pancreatitis. Gut. 2005;54:1679–1681. doi: 10.1136/gut.2005.068593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Detlefsen S, Sipos B, Feyerabend B, Kloppel G. Fibrogenesis in alcoholic chronic pancreatitis: The role of tissue necrosis, macrophages, myofibroblasts and cytokines. Mod Pathol. 2006;19:1019–1026. doi: 10.1038/modpathol.3800613. [DOI] [PubMed] [Google Scholar]
- 21.Hao W, Friedman A. The LDL-HDL profile determines the risk of atherosclerosis: A mathematical model. PLoS One. 2014;9:e90497. doi: 10.1371/journal.pone.0090497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beidelschies MA, et al. Stimulation of macrophage TNFalpha production by orthopaedic wear particles requires activation of the ERK1/2/Egr-1 and NF-kappaB pathways but is independent of p38 and JNK. J Cell Physiol. 2008;217:652–666. doi: 10.1002/jcp.21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pereda J, et al. Effect of simultaneous inhibition of TNF-alpha production and xanthine oxidase in experimental acute pancreatitis: The role of mitogen activated protein kinases. Ann Surg. 2004;240:108–116. doi: 10.1097/01.sla.0000129343.47774.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoki H, et al. Existence of autocrine loop between interleukin-6 and transforming growth factor-beta1 in activated rat pancreatic stellate cells. J Cell Biochem. 2006;99:221–228. doi: 10.1002/jcb.20906. [DOI] [PubMed] [Google Scholar]
- 25.Omary MB, Lugea A, Lowe AW, Pandol SJ. The pancreatic stellate cell: A star on the rise in pancreatic diseases. J Clin Invest. 2007;117:50–59. doi: 10.1172/JCI30082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohlund D, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214:579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wakefield LM, et al. Recombinant latent transforming growth factor beta 1 has a longer plasma half-life in rats than active transforming growth factor beta 1, and a different tissue distribution. J Clin Invest. 1990;86:1976–1984. doi: 10.1172/JCI114932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue J, et al. Alternatively activated macrophages promote pancreatic fibrosis in chronic pancreatitis. Nat Commun. 2015;6:7158. doi: 10.1038/ncomms8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebert M, et al. Overexpression of platelet-derived growth factor (PDGF) B chain and type beta PDGF receptor in human chronic pancreatitis. Dig Dis Sci. 1998;43:567–574. doi: 10.1023/a:1018867209170. [DOI] [PubMed] [Google Scholar]
- 30.Mahadevan D, Von Hoff DD. Tumor-stroma interactions in pancreatic ductal adenocarcinoma. Mol Cancer Ther. 2007;6:1186–1197. doi: 10.1158/1535-7163.MCT-06-0686. [DOI] [PubMed] [Google Scholar]
- 31.Masamune A, Satoh M, Kikuta K, Suzuki N, Shimosegawa T. Activation of JAK-STAT pathway is required for platelet-derived growth factor-induced proliferation of pancreatic stellate cells. World J Gastroenterol. 2005;11:3385–3391. doi: 10.3748/wjg.v11.i22.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Apte MV, Pirola RC, Wilson JS. Battle-scarred pancreas: Role of alcohol and pancreatic stellate cells in pancreatic fibrosis. J Gastroenterol Hepatol. 2006;21(Suppl 3):S97–S101. doi: 10.1111/j.1440-1746.2006.04587.x. [DOI] [PubMed] [Google Scholar]
- 33.Schneider E, et al. Identification of mediators stimulating proliferation and matrix synthesis of rat pancreatic stellate cells. Am J Physiol Cell Physiol. 2001;281:C532–C543. doi: 10.1152/ajpcell.2001.281.2.C532. [DOI] [PubMed] [Google Scholar]
- 34.Phillips PA, et al. Rat pancreatic stellate cells secrete matrix metalloproteinases: Implications for extracellular matrix turnover. Gut. 2003;52:275–282. doi: 10.1136/gut.52.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erkan M, et al. StellaTUM: Current consensus and discussion on pancreatic stellate cell research. Gut. 2012;61:172–178. doi: 10.1136/gutjnl-2011-301220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mantovani A, Locati M. Housekeeping by chemokine scavenging. Blood. 2008;112:215–216. doi: 10.1182/blood-2008-04-149237. [DOI] [PubMed] [Google Scholar]
- 37.Schiavo R, et al. Chemokine receptor targeting efficiently directs antigens to MHC class I pathways and elicits antigen-specific CD8+ T-cell responses. Blood. 2006;107:4597–4605. doi: 10.1182/blood-2005-08-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goel P, Sneyd J, Friedman A. Homogenization of the cell cytoplasm: The calcium bidomain equations. Multiscale Model Simul. 2006;5:1045–1062. [Google Scholar]
- 39.Asaumi H, Watanabe S, Taguchi M, Tashiro M, Otsuki M. Externally applied pressure activates pancreatic stellate cells through the generation of intracellular reactive oxygen species. Am J Physiol Gastrointest Liver Physiol. 2007;293:G972–G978. doi: 10.1152/ajpgi.00018.2007. [DOI] [PubMed] [Google Scholar]
- 40.Olsen AL, et al. Hepatic stellate cells require a stiff environment for myofibroblastic differentiation. Am J Physiol Gastrointest Liver Physiol. 2011;301:G110–G118. doi: 10.1152/ajpgi.00412.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells RG. The role of matrix stiffness in hepatic stellate cell activation and liver fibrosis. J Clin Gastroenterol. 2005;39(Suppl 2):S158–S161. doi: 10.1097/01.mcg.0000155516.02468.0f. [DOI] [PubMed] [Google Scholar]
- 42.Klingberg F, et al. Prestress in the extracellular matrix sensitizes latent TGF-Î21 for activation. J Cell Biol. 2014;207:283–297. doi: 10.1083/jcb.201402006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wipff PJ, Rifkin DB, Meister JJ, Hinz B. Myofibroblast contraction activates latent TGF-beta1 from the extracellular matrix. J Cell Biol. 2007;179:1311–1323. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hadi MF, Sander EA, Ruberti JW, Barocas VH. Simulated remodeling of loaded collagen networks via strain-dependent enzymatic degradation and constant-rate fiber growth. Mech Mater. 2012;44:72–82. doi: 10.1016/j.mechmat.2011.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee B, et al. A three-dimensional computational model of collagen network mechanics. PLoS One. 2014;9:e111896. doi: 10.1371/journal.pone.0111896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fukumura Y, Suda K, Mitani K, Takase M, Kumasaka T. Expression of transforming growth factor beta by small duct epithelium in chronic, cancer-associated, obstructive pancreatitis: An in situ hybridization study and review of the literature. Pancreas. 2007;35:353–357. doi: 10.1097/mpa.0b013e318073cf98. [DOI] [PubMed] [Google Scholar]
- 47.Jablonowska M, et al. Immunohistochemical localization of interleukin-6 in human pancreatitis. Appl Immunohistochem Mol Morphol. 2008;16:40–43. doi: 10.1097/PAI.0b013e318031ffe8. [DOI] [PubMed] [Google Scholar]
- 48.Koninger J, et al. The ECM proteoglycan decorin links desmoplasia and inflammation in chronic pancreatitis. J Clin Pathol. 2006;59:21–27. doi: 10.1136/jcp.2004.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pavan Kumar P, et al. Interferon and glycemic status in diabetes associated with chronic pancreatitis. Pancreatology. 2012;12:65–70. doi: 10.1016/j.pan.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 50.Gasiorowska A, et al. Subclinical inflammation and endothelial dysfunction in patients with chronic pancreatitis and newly diagnosed pancreatic cancer. Dig Dis Sci. 2016;61:1121–1129. doi: 10.1007/s10620-015-3972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reed AM, Gorelick FS. Animal models of chronic pancreatitis. Pancreapedia: Exocrine Pancreas Knowledge Base. 2014 doi: 10.3998/panc.2014.1. [DOI] [Google Scholar]
- 52.Otsuki M, Yamamoto M, Yamaguchi T. Animal models of chronic pancreatitis. Gastroenterol Res Pract. 2010;2010:403295. doi: 10.1155/2010/403295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhan X, Wang F, Bi Y, Ji B. Animal models of gastrointestinal and liver diseases. Animal models of acute and chronic pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2016;311:G343–G355. doi: 10.1152/ajpgi.00372.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.