Significance
The mutualistic association between legumes and rhizobia has ecological and agronomical relevance because of its contribution to the global nitrogen cycle by biological nitrogen fixation. Legumes from the Inverted Repeat Lacking Clade (IRLC) impose irreversible differentiation to their endosymbionts through nodule-specific cysteine-rich (NCR) peptides. This study indicates that NCR gene families evolved via different pathways in IRLC species, in which their size and composition directly impacted the morphotype of their bacterial partners. The positive correlation between the diversity of NCRs with their physiological effects on bacteria provides a better understanding of the multiple roles played by this large family in nodule functioning.
Keywords: nodule-specific cysteine-rich peptides, symbiosis, legume, nodule, nitrogen-fixing bacteroids
Abstract
In legume nodules, rhizobia differentiate into nitrogen-fixing forms called bacteroids, which are enclosed by a plant membrane in an organelle-like structure called the symbiosome. In the Inverted Repeat-Lacking Clade (IRLC) of legumes, this differentiation is terminal due to irreversible loss of cell division ability and is associated with genome amplification and different morphologies of the bacteroids that can be swollen, elongated, spherical, and elongated–branched, depending on the host plant. In Medicago truncatula, this process is orchestrated by nodule-specific cysteine-rich peptides (NCRs) delivered into developing bacteroids. Here, we identified the predicted NCR proteins in 10 legumes representing different subclades of the IRLC with distinct bacteroid morphotypes. Analysis of their expression and predicted sequences establishes correlations between the composition of the NCR family and the morphotypes of bacteroids. Although NCRs have a single origin, their evolution has followed different routes in individual lineages, and enrichment and diversification of cationic peptides has resulted in the ability to impose major morphological changes on the endosymbionts. The wide range of effects provoked by NCRs such as cell enlargement, membrane alterations and permeabilization, and biofilm and vesicle formation is dependent on the amino acid composition and charge of the peptides. These effects are strongly influenced by the rhizobial surface polysaccharides that affect NCR-induced differentiation and survival of rhizobia in nodule cells.
In rhizobial-infected nodule cells, the symbiotic bacteria differentiate to nitrogen-fixing bacteroids. Depending on the legume host, bacteroids can retain their morphology and their capacity to divide and regrow into free-living cells or can become irreversibly differentiated, which is associated with the loss of reproductive capacity, genome amplification, and cell enlargement (1, 2). This differentiation can be manifested with different morphotypes such as swollen (S), elongated (E), spherical (SP), and elongated–branched (EB), all of which are host-dependent (3–6). Swollen (most probably, terminally differentiated) bacteroids were found in five of the six major papilionoid subclades (7), but terminal differentiation of bacteroids has been studied only in certain Inverted Repeat-Lacking Clade (IRLC) legumes and Aeschynomene species (8). The differentiation involves nodule-specific cysteine-rich (NCR) and NCR-like plant peptides (4, 6, 8, 9). In the IRLC legume Medicago truncatula, there are more than 600 NCR genes encoding mature peptides of 30–50 amino acids with four to six cysteine residues at conserved positions. The mature peptides are targeted to the bacteroids, in which they can interact with the membrane or various components of the cytosol (1, 9–13). Species of the IRLC develop indeterminate nodules, which contain a persistent meristem (zone I) and a central region composed of an infection zone (II), interzone (IZ), nitrogen-fixing zone (III), and senescent zone (IV) where NCRs are expressed in waves in differentiating or fully functioning symbiotic cells (9, 14). The large number (>600) of M. truncatula NCR genes suggests redundant functions. However, recent findings have revealed the existence of NCR peptides essential for symbiosis because the deletion of NCR169 or NCR211 impaired differentiation and survival of bacteroids in M. truncatula nodules (12, 13). Cationic NCRs perform in vitro antimicrobial activities (15, 16) partly by disturbing the integrity of the microbial membranes (17) or by interacting with a wide range of proteins involved in transcription, translation, and cell division in Sinorhizobium meliloti, the rhizobial partner of M. truncatula (9, 10, 18). Terminal differentiation of bacteroids is a complex process that occurs in coordination with host-cell development and requires various subsets and balanced production of cationic and noncationic NCRs. NCR genes have also been identified in other IRLC legumes accommodating in their nodules bacteroids that are swollen (S), elongated (E), spherical (SP), or elongated-branched (EB) (4, 6, 19–25). Such variability in the morphotype of bacteroids indicates that the range of NCR peptides produced by legumes is able to shape the microsymbionts with various outcomes. Such differences in the morphotypes may reflect distinct pathways in the evolution of the NCR family. Our previous results, indicating many fewer NCR genes in chickpea than in Medicago (6), and our failure to identify pea orthologs of the symbiotically essential NCR genes (13), prompted us to question how these NCR genes evolved and how large the gene families are in different genera of IRLC legumes. These questions were addressed in this study by analyzing the nodule transcriptome of 10 legumes representing the main IRLC subclades and different bacteroid morphotypes.
Results
The Numbers of NCR Genes Are Highly Variable and Expanded Independently in Different Lineages of IRLC Legumes.
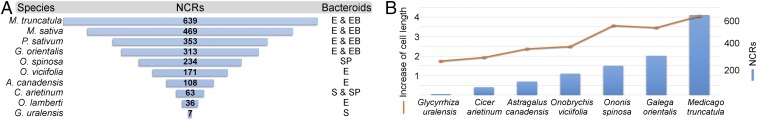
To assess the number of NCR genes in different IRLC legumes, RNA pools of mature nodules from Glycyrrhiza uralensis, Oxytropis lamberti, Astragalus canadensis, Onobrychis viciifolia, Galega orientalis, and Ononis spinosa plants were sequenced; the publicly available genome and/or transcriptome sequencing data from Cicer arietinum, Pisum sativum, Medicago sativa, and Medicago truncatula (26–29) were downloaded; and the predicted mRNA consensus sequences were assembled. These 10 legumes provide an overview of the IRLC, from the most basal species (G. uralensis) to members of the three main subclades: Hedysaroid (O. viciifolia), Astragalean (O. lamberti and A. canadensis) and Vicioid (G. orientalis, C. arietinum, O. spinosa, P. sativum, M. sativa, and M. truncatula) (Fig. S1). These species have different bacteroid morphotypes such as S, E, SP, and EB bacteroids (Fig. S1). Putative NCRs were identified in all of the species using Position-Specific Iterated BLAST searches, which revealed highly variable numbers of NCRs in the species, ranging from 7 in G. uralensis to over 600 in M. truncatula (Fig. 1A and Dataset S1). It is possible that the number of NCRs is underestimated, particularly in those species where genome information is not available. Furthermore, in the M. truncatula A17 genome in which over 700 NCR genes were predicted (Dataset S1), expression of only 639 NCRs was detected (29), and only those NCRs were included in this analysis.
Fig. 1.
Numbers of NCR peptides in different IRLC legumes are correlated with bacteroid morphology. Numbers of NCR peptides predicted from nodule transcriptomes or genome sequences of 10 IRLC legumes are shown in relation to the morphotype of the bacteroids (A). There is a positive correlation between average bacteroid length and the size of the NCR family (B). Pearson correlation coefficient: 0.90 (P value > 0.001).
Although the amino acid composition of the individual NCR peptides is highly variable, the average frequency of a given amino acid per peptide (“amino acid usage”) in the NCR family is very similar in all species investigated (Fig. S2A). The average length of the mature peptides is also very similar (i.e., 36–41 amino acids) in these species (Fig. S2B). Over 30% of the NCR peptides in Vicioid plants (excluding C. arietinum) had two potential disulfide bridges, whereas, in the other IRLC species, the vast majority of NCRs contained three potential disulfide bridges (Fig. S2C).
To investigate how the NCR families evolved, we compared the NCR sequences from the different IRLC lineages. Using protein BLAST (BLASTP) alignments, we found that G. uralensis, O. lamberti, C. arietinum, A. canadensis, and O. viciifolia shared several NCRs with amino acid identities ≥80% (Fig. S3 and Dataset S2). In fact, each of the seven G. uralensis NCRs had at least one highly similar homolog in another legume. Similarly, 20 of the 63 C. arietinum NCRs had at least one putative ortholog in one of these four legumes: G. uralensis, O. lamberti, A. canadensis, and O. viciifolia (Fig. S3), whereas no NCRs with such high similarity were found in the other five Vicioid legume species. Indeed, these Vicioid legumes, containing the most numerous NCRs, show a very different pattern. Except for two G. orientalis NCRs that display high homology to G. uralensis NCRs, all of the peptides shared less than 80% identity with peptides produced in other genera (Dataset S2). These results suggest common origin and conservation of a few NCRs, coupled with the emergence of many species-specific peptides. To explore this possibility, NCRs from different pairs of legumes were compared by phylogenetic analyses using the predicted mature peptide sequences (Fig. S4). Putative orthologs could be recognized based on their distribution in closely related species such as O. lamberti and A. canadensis, and even more so when comparing M. truncatula and M. sativa. The expansion of the NCR families might be driven by local gene duplications followed by diversification as can be observed in the M. truncatula genome where, in agreement with a previous analysis (2), both recent and more ancient duplications could be detected.
Bacteroid Size and Morphotype Correlate with Size and Composition of NCRs and Expression Level of Cationic Peptides.
We showed previously that bacteroids in all tested IRLC legumes were larger and had more DNA than cultured cells, but the degree of cell elongation was rather variable in different species (6). Therefore, we investigated whether the numbers and/or the type of expressed NCRs correlate with the morphotype of bacteroids (Fig. 1A). G. uralensis with 7 NCRs and C. arietinum with 63 NCRs produced swollen and swollen/spherical bacteroids, respectively. O. lamberti with 36 NCRs, A. canadensis with 108 NCRs, and O. viciifolia with at least 171 NCRs formed E bacteroids. The nodules of O. spinosa expressed 234 NCRs and hosted SP bacteroids that were preceded with a brief elongated phase. G. orientalis, P. sativum, M. sativa, and M. truncatula expressed the most NCRs (313, 353, 469, and 639, respectively), and their nodules contain remarkably elongated and EB-type bacteroids. There was a positive correlation between the degree of bacteroid elongation and the number of the expressed NCRs (correlation coefficient 0.90) (Fig. 1B). These results strongly suggest that NCR peptides have a direct impact on bacteroid elongation and that the profile of NCRs produced by the legume affects the morphotype of the bacteroids.
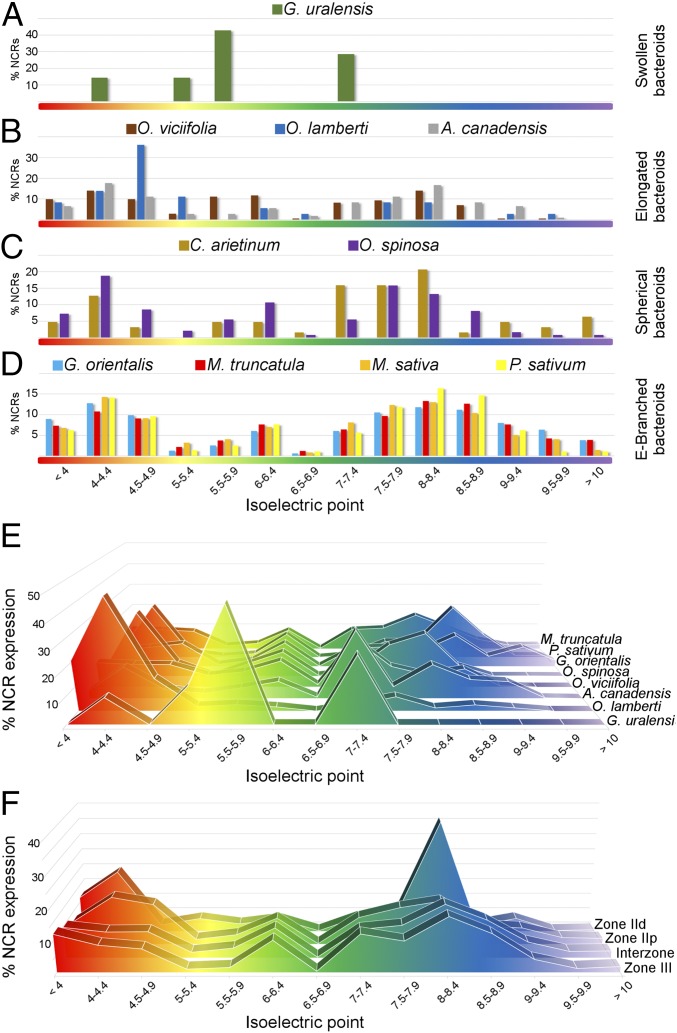
It was reported that the isoelectric point (pI) of NCRs plays a key role in their function (30). Notable differences were observed in the proportions of anionic, neutral, and cationic peptides in the legumes studied: only G. uralensis lacked cationic NCRs, whereas those species hosting EB bacteroids showed the highest ratio (34–42%) of cationic peptides (Figs. S2D and S5A). The frequencies of NCRs were plotted relative to their predicted pI values, and the data were compared based on those species containing bacteroids with the same morphotype (Fig. 2 A–D). The legumes with EB bacteroids had a highly similar NCR pI profile, with two prominent peaks comprising peptides with pIs of 4–4.9 and 8–9 (Fig. 2D). The other legumes did not have a conserved NCR pI profile associated with the distinct morphotypes; they were, however, characterized by a large set of anionic NCRs, whereas the contribution of neutral and cationic NCRs was highly variable without a clear pattern (Fig. 2 A–C).
Fig. 2.
Isoelectric point profiles of NCR peptides and the relative expression of NCRs with different pIs in the IRLC legumes and nodule zones in M. truncatula. The frequency of nodule-expressed NCRs with given pI values is shown graphically; grouping of the NCRs from different legume species is based on the S, E, SP, and EB morphotypes of bacteroids (A–D, respectively). The contribution [% RPKM or DEseq normalized values (29)] of different isoelectric point categories to whole-nodule NCR expression for seven IRLC legumes is provided (E). The pattern of NCR expression [% DEseq normalized values (29)] in different zones of M. truncatula nodules is shown in F.
The expression of NCRs with different charges was analyzed by calculating the sum of RPKM or normalized DESeq values (29) from the nodule transcriptome data and taking into account the pI of the peptides in those legumes where the RNA-seq information was accessible. The ratio of anionic, neutral, and cationic NCR transcripts expressed in the nodules was similar only between G. orientalis, P. sativum, and M. truncatula—species with EB bacteroids (Fig. S5A and Dataset S3). In these legumes, the expression of cationic NCRs was ∼40% of the NCR transcripts, mainly from genes encoding peptides with a pI of 8–9 (Fig. S5A and Fig. 2E). Conversely, in legumes with S, E, and SP bacteroids, the highest peak in the profile was populated by anionic or neutral NCRs (Fig. 2E). The comparison of the nodule transcriptome among different IRLC legumes shows a positive correlation between expression of cationic NCRs and elongation/branching of bacteroids.
Dominance of Anionic and Cationic NCRs with Particular pI Values Correlates with Early and Late Steps of Bacteroid Differentiation in M. truncatula Nodules.
To investigate whether the activity of any NCR groups of peptides is associated with a distinct step of nodule/bacteroid development, the expression of genes coding for peptides with different pI values was analyzed in the available normalized RNA-seq data obtained from different zones of M. truncatula nodules by laser-capture microdissection (29). The distal (ZIId) and proximal (ZIIp) parts of the infection zone contribute 2% and 16% of the NCR transcripts, respectively, whereas over 50% of the NCR mRNAs are expressed in the interzone (IZ) and 25% in the nitrogen-fixing zone (ZIII) (Fig. S5B). Analyzing the expression of NCRs in each nodule zone revealed that, in ZIId, cationic and anionic NCRs comprise 43% and 40% of the transcripts, respectively (Fig. S5C). However, in ZIIp the profile is different, with the ratio of cationic NCRs dropping to 29% and the anionic NCRs remaining at 40%. In the IZ and ZIII, higher proportions (about 36–38%) of cationic peptides compared with other NCRs were found in both zones. Neutral NCRs made up only 16% of transcripts in zone ZIId that doubled in the ZIII (Fig. S5C). Notably, each nodule zone expressed a prominent set of NCRs with a well-defined pI value. In the distal infection zone, NCRs with a pI of 4–4.4 constituted 19% of the transcripts but made up less than 10% of the NCRs in whole nodules (Fig. 2F and Dataset S3). NCRs with a pI between 8 and 8.4 constitute 36% of the transcripts in ZIId, but only 17% in the nitrogen-fixing zone. As the expression level of these NCR genes decreased in the older regions of the nodules, the proportion of transcripts of cationic peptides (especially in the pI range 8.0–8.9) increased (Fig. 2F and Dataset S3). This analysis supports a correlation between the expression of diverse sets of NCRs and the developmental steps of bacteroids in the different developmental regions of M. truncatula nodules.
NCRs Provoke a Wide Range of Morphological and Physiological Changes in S. meliloti.
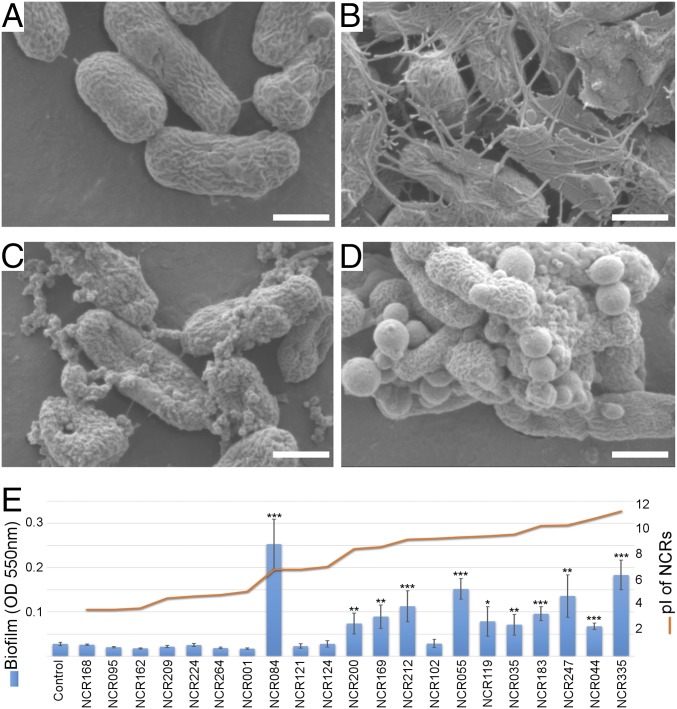
The results described above indicate that the expression and diversification of NCRs are linked to progressive morphological changes of bacteroids. To test this link, free-living S. meliloti cultures were treated individually for 3 h with 23 different (5 anionic, 3 neutral and 15 cationic) synthetic M. truncatula NCR peptides at 8 µM and visualized by scanning electron microscopy (SEM). The responses provoked by these peptides were variable, but could be grouped according to the observed effects. Anionic and neutral NCRs stimulated deposition of extracellular material characterized by a complex set of fibers connecting the bacteria, although the magnitude and complexity of changes were evidently variable depending on the peptides used (Fig. 3 B and C and Fig. S6). Deposition of extracellular material was also induced by cationic NCRs with a pI below 9, whereas cationic peptides with a higher pI provoked protrusions in the cell envelope (Fig. 3D and Fig. S6). These responses were accompanied by alterations in the cell morphology and mimicked occasionally the envelope of nitrogen-fixing bacteroids in G. orientalis and M. truncatula (Fig. S7). In M. truncatula, the most dramatic steps of bacteroid differentiation occur in the IZ. An attempt to reconstruct the effects of IZ NCRs was made with a limited set of synthetic NCRs that were mixed proportionally with the expression of the corresponding pI categories in this region (Dataset S3). Although this treatment did not result in EB-bacteroid morphology, the formation of vesicles and the number of lysed cells provoked by individual cationic NCRs was visibly reduced, indicating that action of neutral and anionic peptides could attenuate the killing effect of cationic peptides (Fig. S8).
Fig. 3.
SEM and biofilm formation of S. meliloti treated with synthetic M. truncatula NCR peptides. SEM images of S. meliloti cells incubated for 3 h in phosphate buffer (A, control) supplemented with anionic (B, NCR095), neutral (C, NCR084), or cationic (D, NCR055) peptides at 8 µM. Biofilm formation was evaluated by crystal violet assays after incubation for 24 h in LSM medium with a range of NCRs with different isoelectric points (E). SEs were calculated with three biological replicates using averages based on three technical repeats. P values of <0.05, 0.01, and <0.001 are marked with one, two, or three asterisks, respectively (Student’s t test). (Scale bars, 1 µm.)
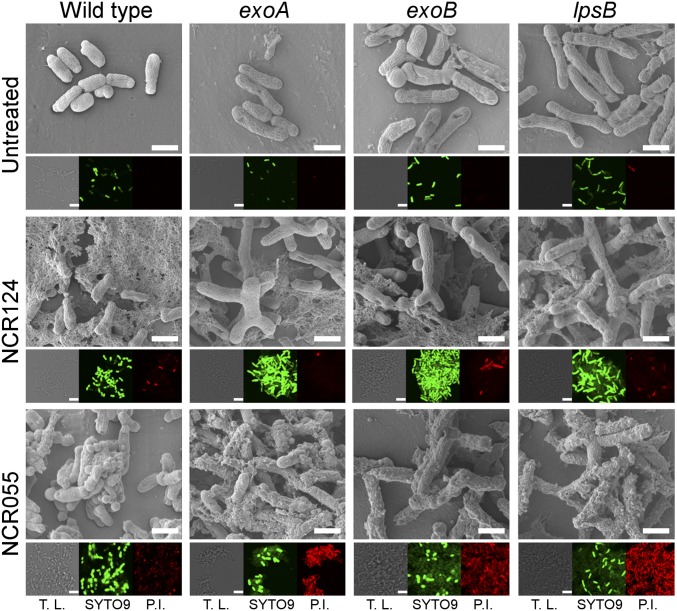
The envelope modifications induced by NCRs prompted us to explore the role of selected genes involved in the synthesis of the cell envelope in the rhizobial partner. S. meliloti mutants affected in the exoA, exoB, or lpsB genes (31, 32) were visualized by SEM after challenging them with the neutral (NCR124) and cationic (NCR055) peptides, which promoted the generation of extracellular material and protrusions in the envelope of the wild-type strain, respectively (Fig. 4 and Fig. S6). Interestingly, the mutant bacteria were larger than the wild-type cells even with mock treatment. In the presence of the neutral NCR124, both the mutants and, to a lesser extent, the wild-type strain produced large multilobed cells. NCR055 stimulated vesicle formation in the mutants, and the production of extracellular material was particularly exacerbated in the exoA and lpsB mutants. The mutants were also more sensitive to the cationic NCR055 and NCR247 peptides than the wild-type strain (Fig. S9). Preincubation with exopolysaccharides (EPS) extracted from the wild-type strain, however, attenuated these responses in both the mutant and the wild-type strains (Fig. S9B). The results collectively indicate that NCRs can influence the bacterial surface, which in turn affects the bacterial responses to NCRs. To gain further insight, biofilm production was assessed by crystal violet staining of S. meliloti cells treated with anionic, neutral, or cationic NCRs. Except for NCR102, significant biofilm formation was induced by the cationic peptides (Fig. 3E). Interestingly, the neutral NCR084 promoted the most pronounced biofilm formation. In contrast, the other neutral and anionic NCRs did not induce biofilm formation (Fig. 3E). The bacterial responses to NCRs with different pI values prompted us to evaluate their effect on bacterial membrane integrity using propidium iodide (PI) and SYTO9. PI can penetrate only cells with permeabilized membranes, whereas SYTO9 stains viable bacteria. This approach showed that integrity of the cell membrane was not affected by anionic NCRs but was greatly compromised by treatment with cationic NCRs and to a lesser extent with neutral NCRs (Fig. S10). Similar effects were provoked by the neutral NCR124 and the cationic NCR055 in the exoA, exoB, and lpsB mutants; however, the PI fluorescence induced by cell-membrane damage was higher in the mutants compared with the wild-type strain following treatment with NCR055 (Fig. 4). As observed previously, the pI charge of NCRs seems to be crucial to their ability to induce different morphological and physiological responses, but these effects apparently do not rely entirely on this characteristic.
Fig. 4.
Morphology and viability of S. meliloti mutants challenged with NCR055 or NCR124. SEM and confocal microscopy images of S. meliloti mutants affected in the exoA, exoB, and lpsB genes after incubation in phosphate buffer only (untreated) or phosphate buffer supplemented with 8 µM NCR124 or NCR055. After incubation, membrane integrity was evaluated by staining with PI (red fluorescence) and SYTO9 (green fluorescence). T.L., transmitted light. (Scale bars, 1 µm.)
Discussion
Terminal differentiation of bacteroids is orchestrated by the plant through the production of symbiotic peptides (9), a process that likely evolved independently in multiple lineages of legumes (7, 8, 31) to increase symbiotic efficiency. Recently, we showed that terminal differentiation in the IRLC can occur with remarkable differences in the morphotype of the bacteroids (6). The current study documents that such variability is correlated with the unique NCR composition of the legume host that evolved by the enrichment and diversification of symbiotic peptides with certain characteristics.
The Bacteroid Morphotypes Reflect the Evolutionary Status of NCR Families in IRLC Legumes.
NCRs were initially identified in IRLC (Medicago, Vicia, Galega) species that accommodate EB bacteroids and all belong to the Vicioid subclade (1, 19–22, 24, 25). However, subsequent studies showed that NCRs are also present in the IRLC species C. arietinum and Astragalus sinicus that accommodate S/SP and E bacteroids, respectively (6, 23). C. arietinum is part of the Vicioid subclade, whereas A. sinicus is placed in the Astragalean subclade, indicating that production of NCRs is not an exclusive feature of the Vicioid subclade (33). This study demonstrates that NCR expression is a common characteristic of IRLC legumes and that the composition and complexity of the NCRs vary substantially among them. Moreover, the complexity does not reflect phylogeny because closely related species can have low and high numbers of NCR genes (Cicer vs. Galega and Ononis spp.). Rather, the morphotype of the endosymbionts reflects the composition and complexity of the NCR families. We infer that the S-bacteroid form is the ancestral morphotype of terminally differentiated bacteroids because few morphological changes occur and the respective host plant species have the lowest diversity of NCRs (seven NCRs in G. uralensis). We propose that, probably along with other nodule-specific proteins, changes in the complexity and composition of NCR families gave rise to SP, E, and EB bacteroids. In fact, transition of morphotypes is a phenomenon observed in several species such as C. arietinum, O. spinosa, and Aeschynomene spp., in which bacteroids become spherical after the formation of an intermediate E morphotype (6, 8).
The evidence collected by SEM and RNA-seq indicates that the expression of cationic NCRs positively correlates with drastic morphological changes of bacteria. The absence of cationic NCRs in G. uralensis is probably a consequence of genome contraction or, more likely, of the fact that the ancestral symbiotic NCR peptides were neutral or anionic, originating from a single evolutionary event in IRLC legumes. Nevertheless, the existing genome and transcript data indicate that NCRs took distinct evolutionary routes in each of the analyzed plant species. It has been noted that NCR genes in M. truncatula are often flanked by transposons (34), which suggests that mobile genetic elements have played an important role in the multiplication and diversification of NCRs in the IRLCs. The fast diversification rate of NCRs in certain species such as G. orientalis, P. sativum, and Medicago spp. led to the emergence of species-specific peptides. However, despite the different evolutionary pathways taken by the legumes analyzed here, it should be noted that legumes with EB bacteroids possess a similar composition of NCRs based on their pI value.
Cationic NCRs Produce a Wide Range of Effects on Rhizobia.
Earlier studies showed that cationic NCR247 and NCR335 provoked modifications in cell size and morphology and affected membrane permeability and cell viability in a concentration-dependent manner (9, 10, 17), general features that might correlate with the charge of the peptides. Similarly, the formation of network-like structures as well as outer membrane vesicles (OMVs) is a phenomenon often caused by the cationic peptides. OMVs are spherical protrusions of the outer membrane in Gram-negative bacteria produced after diverse (a)biotic stimuli that mediate versatile functions. In nonpathogenic bacteria, OMVs promote both survival under stress and nutrient acquisition in bacterial communities (35). The infected plant cells in the nodules may provide a suitable environment for OMV function because a high exchange of chemical signals and nutrients is required. Understanding the functioning, composition, and significance of these NCR-induced vesicles requires further studies. The envelopes of bacteroids in G. orientalis and M. truncatula nodules resemble those of free-living cultured rhizobia treated with cationic peptides. These species accommodate EB bacteroids and showed a high proportion and high expression of cationic peptides. Conversely, such protrusions were absent in the S bacteroids of G. uralensis, which lacks cationic NCRs. The similarities between the free-living cultured rhizobia treated with NCRs and the bacteroids are a good indicator that the in vitro assays simulate to some extent the effect of NCRs in the infected cells. Except for NCR102, all of the cationic NCRs tested enhanced biofilm generation. Similar to OMVs, biofilms are often produced by bacteria in response to adverse conditions (36). The generation of this extracellular matrix is probably a defense mechanism against diverse NCRs in cultured rhizobia.
Insights into the Function of Anionic and Neutral NCRs.
Presently, most knowledge is available for the cationic peptides. However, in bacteroids isolated from M. truncatula nodules, the majority of the 138 NCRs detected by mass spectrometry were anionic (11). Recently, Kim et al. (12) showed that the loss of NCR211, encoding an anionic peptide, produced nonfunctional Fix− nodules in M. truncatula. Differentiation of bacteroids apparently takes place in the nodules of this mutant, but the differentiation is incomplete and the bacteroids are unable to survive in the nodule cells. A protective mechanism is probably one of the putative functions performed by certain anionic and neutral NCRs. The analysis of neutral NCRs has been poorly explored until now. Expression of neutral NCRs is marginal in ZIIp, although their abundance is doubled from the infection zone to the nitrogen-fixing zone. The use of three synthetic neutral NCRs (NCR084, NCR121, and NCR124) on S. meliloti cells showed that these peptides also provoke deposition of extracellular material in the medium, although only NCR084 provoked a remarkable increase in biofilm formation, even greater than that caused by cationic peptides. The fibers induced by these types of NCRs on S. meliloti culture resemble intercellular nanotubes, structures with the capacity to exchange molecules among neighboring bacteria (37). In the majority of the IRLC legumes tested, neutral NCRs represent a small proportion of the NCRs, but five of the seven NCRs in G. uralensis are neutral, raising the possibility that these NCRs, together with the anionic ones, are likely to be the first evolved symbiotic peptides.
Dynamic Interplay Between NCRs and Cell-Wall Components of Rhizobia.
The adequate composition and expression of anionic, neutral, and cationic NCRs within the infected cells seems to be crucial for the differentiation and survival of bacteroids. Both processes might need the modifications in the cell envelope of the bacteria. Indeed, different Rhizobium leguminosarum bv. viciae LPS epitopes (recognized by monoclonal antibodies) were expressed at different stages of nodule and bacteroid development (38). Transcriptome analyses indicated similar changes: Over 150 genes involved in the biogenesis of the cell-envelope constituents were differentially expressed in bacteroids of A. sinicus nodules compared with their free-living state (39), whereas in the bacteroids of M. truncatula nodules, the transcript abundance of several genes associated with LPS synthesis were repressed, but the lpsB gene was induced (40). Investigation of S. meliloti strains defective in surface polysaccharide production (exoA, exoB, lpsB) revealed enlargement of the free-living cells and augmented responses to NCRs such as increased membrane permeabilization caused by the cationic NCR055 and the formation of branched, bloated cells upon treatment with the neutral NCR124. Nodule organogenesis by the S. meliloti lpsB mutant was not affected, but bacteroids were unable to differentiate and showed symptoms of membrane degradation (31). The succinoglycan decoration seems to be crucial for a successful symbiosis between rhizobial strains and M. truncatula ecotypes. The incompatibility of S. meliloti Rm41 with M. truncatula A17 was restored by the introduction of S. meliloti Rm1021 exo genes into S. meliloti Rm41 (41), resulting in alteration of the substitution pattern of EPS. In this context, NCRs may mediate differentiation and survival of rhizobial strains depending on the composition of their cell envelope. This study provides an insight into the NCR evolutionary pathway and the diverse NCR targets, opening a wide range of processes to explore.
Materials and Methods
Sequencing of Nodule Transcriptome and Identification of NCR Genes.
Seedlings of A. canadensis, G. orientalis, G. uralensis, O. viciifolia, O. spinosa, and O. lamberti were inoculated with Mesorhizobium sangaii, Mesorhizobium ciceri, Rhizobium galegae bv. orientalis, Mesorhizobium tianshanense, Rhizobium sp., and M. sangaii, respectively (6). Total RNA was isolated from mature nodules (4–6 wk postinoculation) using the ZR Plant RNA miniprep kit (Zymo Research). Bacterial and plant ribosomal RNA molecules were removed with the Ribo-Zero rRNA Removal Kits for Gram-negative bacteria and Plant Seed/Root (Epicentre). Sequencing libraries were constructed with the help of NEB Next Ultra Directional RNA Library Kit (New England Biolabs) and were sequenced with an Illumina sequencer. Binary short read files (pea RNA-sequencing data) were downloaded from the Sequence Read Archive (SRA) database and were subsequently converted to standard fastq files using the fastq-dump tool from the SRA Toolkit. De novo transcript assembly was carried out with version v2.1.1. of Trinity software (42). NCRs were searched in the assembled nodule transcriptomes and genomic sequences through tBLASTn using M. truncatula NCRs as query in the CLC 9.0.1 workbench (https://www.qiagenbioinformatics.com/). A second round of tBLASTn was performed using the NCRs retrieved from the first tBLASTn as query.
In Silico Analysis of NCR Families of IRLC Legumes.
The presence of signal peptides was predicted by SignalP 4.1 (www.cbs.dtu.dk/services/SignalP/) from the deduced amino acid sequences that were obtained through the tBLASTn search. The amino acid constitution, molecular weight, and isoelectric point were calculated with the Expasy and Sequence Manipulation Site (www.expasy.org/, www.bioinformatics.org/sms2/index.html). For the phylogenetic assembly, the mature peptide sequences were aligned and bootstrapped (1,000 iterations) with ClustalX2 (www.clustal.org/). The data generated were displayed and plotted with Dendroscope 3.5.7 (dendroscope.org/) (43).
SEM, Viability Staining, and Biofilm Formation of NCR-Treated S. meliloti Cells.
Wild-type and mutant strains of S. meliloti 1021 growing in the log phase were diluted to OD600 = 0.1 and incubated for 3 h in phosphate buffer (pH 7) containing a sublethal dose of 8 µM synthetic mature M. truncatula NCRs (www.proteogenix.science/fr/) and later analyzed by SEM. Similarly, treated cells were PI-and SYTO9-stained following the manufacturer’s instructions (Invitrogen) and then observed by confocal microscopy (Olympus Fluoview FV1000). For SEM, cells were fixed with 2.5% (vol/vol) glutaraldehyde in cacodylate buffer (0.05 M, pH 7.2). Later, cells were dehydrated with a graded ethanol series and dried with a critical point dryer, followed by 10 nm of gold coating, and observed by SEM (JEOL JSM-7100F/LV) (17). Biofilm formation was assessed through crystal violet assays in S. meliloti cultures grown in low-salt fungal medium (LSM) supplied with NCRs (8 µM) for 24 h (16). Exopolysaccharides were isolated from supernatants of bacterial culture with ethanol precipitation according to Dusha et al. (44).
Supplementary Material
Acknowledgments
This work was supported by Grant 269067 from the SYM-BIOTICS Advanced Grant of the European Research Council (to É.K.); by Grant NN110979 from the Hungarian National Research Fund (to A.K.); and by a visiting fellowship from the Hungarian Academy of Sciences (to J.A.D.). J.M. benefited from a postdoctoral fellowship from the Consejo Nacional de Ciencia y Tecnología in México (Proposal 231205).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1704217114/-/DCSupplemental.
References
- 1.Mergaert P, et al. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 2003;132:161–173. doi: 10.1104/pp.102.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alunni B, et al. Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula. Mol Plant Microbe Interact. 2007;20:1138–1148. doi: 10.1094/MPMI-20-9-1138. [DOI] [PubMed] [Google Scholar]
- 3.Vasse J, de Billy F, Camut S, Truchet G. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J Bacteriol. 1990;172:4295–4306. doi: 10.1128/jb.172.8.4295-4306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mergaert P, et al. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci USA. 2006;103:5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonaldi K, et al. Nodulation of Aeschynomene afraspera and A. indica by photosynthetic Bradyrhizobium Sp. strain ORS285: The nod-dependent versus the nod-independent symbiotic interaction. Mol Plant Microbe Interact. 2011;24:1359–1371. doi: 10.1094/MPMI-04-11-0093. [DOI] [PubMed] [Google Scholar]
- 6.Montiel J, et al. Terminal bacteroid differentiation is associated with variable morphological changes in legume species belonging to the inverted repeat-lacking clade. Mol Plant Microbe Interact. 2016;29:210–219. doi: 10.1094/MPMI-09-15-0213-R. [DOI] [PubMed] [Google Scholar]
- 7.Oono R, Schmitt I, Sprent JI, Denison RF. Multiple evolutionary origins of legume traits leading to extreme rhizobial differentiation. New Phytol. 2010;187:508–520. doi: 10.1111/j.1469-8137.2010.03261.x. [DOI] [PubMed] [Google Scholar]
- 8.Czernic P, et al. Convergent evolution of endosymbiont differentiation in dalbergioid and inverted repeat-lacking clade legumes mediated by nodule-specific cysteine-rich peptides. Plant Physiol. 2015;169:1254–1265. doi: 10.1104/pp.15.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van de Velde W, et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science. 2010;327:1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- 10.Farkas A, et al. Medicago truncatula symbiotic peptide NCR247 contributes to bacteroid differentiation through multiple mechanisms. Proc Natl Acad Sci USA. 2014;111:5183–5188. doi: 10.1073/pnas.1404169111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durgo H, et al. Identification of nodule-specific cysteine-rich plant peptides in endosymbiotic bacteria. Proteomics. 2015;15:2291–2295. doi: 10.1002/pmic.201400385. [DOI] [PubMed] [Google Scholar]
- 12.Kim M, et al. An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc Natl Acad Sci USA. 2015;112:15238–15243. doi: 10.1073/pnas.1500123112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horváth B, et al. Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc Natl Acad Sci USA. 2015;112:15232–15237. doi: 10.1073/pnas.1500777112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guefrachi I, et al. Extreme specificity of NCR gene expression in Medicago truncatula. BMC Genomics. 2014;15:712. doi: 10.1186/1471-2164-15-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tiricz H, et al. Antimicrobial nodule-specific cysteine-rich peptides induce membrane depolarization-associated changes in the transcriptome of Sinorhizobium meliloti. Appl Environ Microbiol. 2013;79:6737–6746. doi: 10.1128/AEM.01791-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ordögh L, Vörös A, Nagy I, Kondorosi E, Kereszt A. Symbiotic plant peptides eliminate Candida albicans both in vitro and in an epithelial infection model and inhibit the proliferation of immortalized human cells. BioMed Res Int. 2014;2014:320796. doi: 10.1155/2014/320796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mikuláss KR, et al. Antimicrobial nodule-specific cysteine-rich peptides disturb the integrity of bacterial outer and inner membranes and cause loss of membrane potential. Ann Clin Microbiol Antimicrob. 2016;15:43. doi: 10.1186/s12941-016-0159-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penterman J, et al. Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc Natl Acad Sci USA. 2014;111:3561–3566. doi: 10.1073/pnas.1400450111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiménez-Zurdo JI, Frugier F, Crespi MD, Kondorosi A. Expression profiles of 22 novel molecular markers for organogenetic pathways acting in alfalfa nodule development. Mol Plant Microbe Interact. 2000;13:96–106. doi: 10.1094/MPMI.2000.13.1.96. [DOI] [PubMed] [Google Scholar]
- 20.Fruhling M, et al. A small gene family of broad bean codes for late nodulins containing conserved cysteine clusters. Plant Sci. 2000;152:67–77. [Google Scholar]
- 21.Crockard A, Bjourson J, Dazzo B, Cooper JE. A white clover nodulin gene, dd23b, encoding a cysteine cluster protein, is expressed in roots during the very early stages of interaction with Rhizobium leguminosarumbiovar trifolii and after treatment with chitolipooligosaccharide Nod factors. J Plant Res. 2002;115:439–447. doi: 10.1007/s10265-002-0053-7. [DOI] [PubMed] [Google Scholar]
- 22.Kato T, et al. Expression of genes encoding late nodulins characterized by a putative signal peptide and conserved cysteine residues is reduced in ineffective pea nodules. Mol Plant Microbe Interact. 2002;15:129–137. doi: 10.1094/MPMI.2002.15.2.129. [DOI] [PubMed] [Google Scholar]
- 23.Chou MX, Wei XY, Chen DS, Zhou JC. Thirteen nodule-specific or nodule-enhanced genes encoding products homologous to cysteine cluster proteins or plant lipid transfer proteins are identified in Astragalus sinicus L. by suppressive subtractive hybridization. J Exp Bot. 2006;57:2673–2685. doi: 10.1093/jxb/erl030. [DOI] [PubMed] [Google Scholar]
- 24.Kaijalainen S, Schroda M, Lindström K. Cloning of nodule-specific cDNAs of Galega orientalis. Physiol Plant. 2002;114:588–593. doi: 10.1034/j.1399-3054.2002.1140412.x. [DOI] [PubMed] [Google Scholar]
- 25.Scheres B, et al. Sequential induction of nodulin gene expression in the developing pea nodule. Plant Cell. 1990;2:687–700. doi: 10.1105/tpc.2.8.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kant C, Pradhan S, Bhatia S. Dissecting the root nodule transcriptome of chickpea (Cicer arietinum L.) PLoS One. 2016;11:e0157908. doi: 10.1371/journal.pone.0157908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alves-Carvalho S, et al. Full-length de novo assembly of RNA-seq data in pea (Pisum sativum L.) provides a gene expression atlas and gives insights into root nodulation in this species. Plant J. 2015;84:1–19. doi: 10.1111/tpj.12967. [DOI] [PubMed] [Google Scholar]
- 28.O’Rourke JA, et al. The Medicago sativa gene index 1.2: A web-accessible gene expression atlas for investigating expression differences between Medicago sativa subspecies. BMC Genomics. 2015;16:502. doi: 10.1186/s12864-015-1718-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roux B, et al. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 2014;77:817–837. doi: 10.1111/tpj.12442. [DOI] [PubMed] [Google Scholar]
- 30.Kondorosi E, Mergaert P, Kereszt A. A paradigm for endosymbiotic life: Cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu Rev Microbiol. 2013;67:611–628. doi: 10.1146/annurev-micro-092412-155630. [DOI] [PubMed] [Google Scholar]
- 31.Campbell GR, Reuhs BL, Walker GC. Chronic intracellular infection of alfalfa nodules by Sinorhizobium meliloti requires correct lipopolysaccharide core. Proc Natl Acad Sci USA. 2002;99:3938–3943. doi: 10.1073/pnas.062425699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leigh JA, Signer ER, Walker GC. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc Natl Acad Sci USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wojciechowski MF, Lavin M, Sanderson MJ. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot. 2004;91:1846–1862. doi: 10.3732/ajb.91.11.1846. [DOI] [PubMed] [Google Scholar]
- 34.Satge C, et al. Reprogramming of DNA methylation is critical for nodule development in Medicago truncatula. Nat Plants. 2016;2:16166. doi: 10.1038/nplants.2016.166. [DOI] [PubMed] [Google Scholar]
- 35.Schwechheimer C, Kuehn MJ. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat Rev Microbiol. 2015;13:605–619. doi: 10.1038/nrmicro3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinaudi LV, Giordano W. An integrated view of biofilm formation in rhizobia. FEMS Microbiol Lett. 2010;304:1–11. doi: 10.1111/j.1574-6968.2009.01840.x. [DOI] [PubMed] [Google Scholar]
- 37.Dubey GP, Ben-Yehuda S. Intercellular nanotubes mediate bacterial communication. Cell. 2011;144:590–600. doi: 10.1016/j.cell.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 38.Kannenberg EL, Perotto S, Bianciotto V, Rathbun EA, Brewin NJ. Lipopolysaccharide epitope expression of Rhizobium bacteroids as revealed by in situ immunolabelling of pea root nodule sections. J Bacteriol. 1994;176:2021–2032. doi: 10.1128/jb.176.7.2021-2032.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng J, et al. RNA-Seq and microarrays analyses reveal global differential transcriptomes of Mesorhizobium huakuii 7653R between bacteroids and free-living cells. PLoS One. 2014;9:e93626. doi: 10.1371/journal.pone.0093626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker A, et al. Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol Plant Microbe Interact. 2004;17:292–303. doi: 10.1094/MPMI.2004.17.3.292. [DOI] [PubMed] [Google Scholar]
- 41.Simsek S, Ojanen-Reuhs T, Stephens SB, Reuhs BL. Strain-ecotype specificity in Sinorhizobium meliloti-Medicago truncatula symbiosis is correlated to succinoglycan oligosaccharide structure. J Bacteriol. 2007;189:7733–7740. doi: 10.1128/JB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huson DH, Scornavacca C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 44.Dusha I, Olah B, Szegletes Z, Erdei L, Kondorosi A. syrM is involved in the determination of the amount and ratio of the two forms of the acidic exopolysaccharide EPSI in Rhizobium meliloti. Mol Plant Microbe Interact. 1999;12:755–765. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.