Significance
Understanding how Calmodulin (CaM) is able to recruit more than 300 binding targets is crucial for solving the multispecificity in molecular recognition. To address the molecular mechanism for CaM recognition, we studied the association process that simultaneously involves both the local conformational change of CaM and the global conformational changes of CaM-binding peptide skMLCK with molecular simulations. An energy landscape with a mixture of the atypical “induced fit,” the atypical “conformational selection,” and “simultaneously binding–folding” is uncovered for the CaM–peptide recognition process. The underlying interactions, which lead to this complicated association process, are found to fine-tune the multiple conformational changes during the recognition process, leading to high affinity and specificity. The association characteristic proposed for CaM and skMLCK can provide insights toward multispecific binding of CaM with its target.
Keywords: structure-based model, Calmodulin, mixture binding mechanism, multispecific recognition
Abstract
Calmodulin (CaM) is found to have the capability to bind multiple targets. Investigations on the association mechanism of CaM to its targets are crucial for understanding protein–protein binding and recognition. Here, we developed a structure-based model to explore the binding process between CaM and skMLCK binding peptide. We found the cooperation between nonnative electrostatic interaction and nonnative hydrophobic interaction plays an important role in nonspecific recognition between CaM and its target. We also found that the conserved hydrophobic anchors of skMLCK and binding patches of CaM are crucial for the transition from high affinity to high specificity. Furthermore, this association process involves simultaneously both local conformational change of CaM and global conformational changes of the skMLCK binding peptide. We found a landscape with a mixture of the atypical “induced fit,” the atypical “conformational selection,” and “simultaneously binding–folding,” depending on the synchronization of folding and binding. Finally, we extend our discussions on multispecific binding between CaM and its targets. These association characteristics proposed for CaM and skMLCK can provide insights into multispecific binding of CaM.
Many biological processes are driven by protein–protein binding. The large-scale domain rearrangements in proteins have long been recognized to have a critical role in biological function. This flexibility or conformational dynamics also provide a new viewpoint of binding. In addition to the “lock-and-key” binding mechanism, proposed by Fischer to describe the rigid binding in enzyme catalysis (1), two scenarios, considering flexibility during binding, emerged and are referred as “induced fit” and “conformational selection,” addressing the critical roles of flexibility in protein recognition (2–6).
Calmodulin (CaM) is an ubiquitous binding protein that is involved in a wide range of cellular -dependent signaling pathways. With incorporating ions, -CaM regulates the activity of many kinds of proteins including protein phosphatase, inositol triphosphate kinase, nitric oxide synthase, protein kinases, nicotinamide adenine dinucleotide kinase, pumps, and proteins involved in motility (7–9). The binary complex -CaM is found to have the capability to bind over 300 targets (7–9). Exploring the molecular mechanism of -CaM binding to the different targets is crucial for understanding protein–protein multispecific recognition. X-ray crystallography experiments have been performed to resolve -loaded CaM structure (10). However, complexes of CaM with target enzymes are difficult to study by NMR and the crystallization method, due to the spatial resolution in the experiments. Alternatively, short peptide sequences corresponding to CaM-binding domains are often used to explore CaM–target protein interactions and several studies suggest that these CaM–peptide interactions are excellent models to investigate the interactions between CaM and the fully intact enzyme (11, 12). Multidimensional NMR and X-ray crystallography experimental techniques have been used to resolve numerous structures of peptide–CaM complexes, such as smooth muscle myosin light chain kinase (smMLCK), skeletal myosin light chain kinase (skMLCK), CaM-dependent kinase I (CaMKI), and CaM-dependent kinase II (CaMKII) (13–15). From these experiments, many structural features of the binding between CaM and target peptides can be obtained. The -loaded CaM structure without target peptide binding adopts to a dumbbell conformation, and CaM undergoes a conformational change to form the final compact globular structure with the C domain and the N domain wrapping around the target peptide after binding (SI Appendix, Fig. S1) (10–16). Recently, we performed a fluorescence resonance energy transfer (FRET) experiment on -CaM binding to peptide C28W, showing that CaM possesses an intermediate state with only bound C-terminal domain during its binding (16). On the other hand, the analysis on static structures indicated that the CaM-binding peptides are mostly random coil in the free state and adopt -helical structures in the complex with CaM (17, 18). However, the mechanism and global thermodynamic perspective for the process of coupled folding and binding of -CaM to its targets cannot be fully understood by biochemical and single-molecular methods, due to their limits of spatial or temporal resolution. To meet the challenges, molecular dynamics simulations serve as a powerful tool, which is able to gain more insights and quantitative information toward the underlying mechanism (19–24). Here, we select the skMLCK binding peptide as the target that the -CaM binds to. The binary binding complex skMLCK-CaM has been resolved by NMR techniques [Protein Data Bank (PDB) ID: 2BBM] SI Appendix, Fig. S1B) and provides the structural basis for our simulation (13). The skMLCK peptide, with 26 residues in length, is classified into the typical “1-5-8-14” scenario, which is named by the number of spacings of hydrophobic anchor residues (25).
By developing a coarse-grained structure-based model, we investigated the binding process of CaM to the skMLCK binding peptide. By explicitly taking into account the electrostatic and hydrophobic interactions, we addressed the critical roles of the residues in CaM participating in tuning the binding from high affinity to high specificity. The underlying binding mechanism obtained from the quantified free energy landscape indicated the association process is quite complex with the mixture of induced fit, conformational selection, and simultaneous binding–folding. This unique binding behavior, tuned by multiple conformational changes, is further suggested as the source of multispecificity in CaM recognition. Our results provide a unique way to gain insights into the promiscuity involved in CaM recognition.
Results
Affinity and Flexibility Determine the Landscape of Coupled Folding and Binding of Ca2+-CaM to skMLCK.
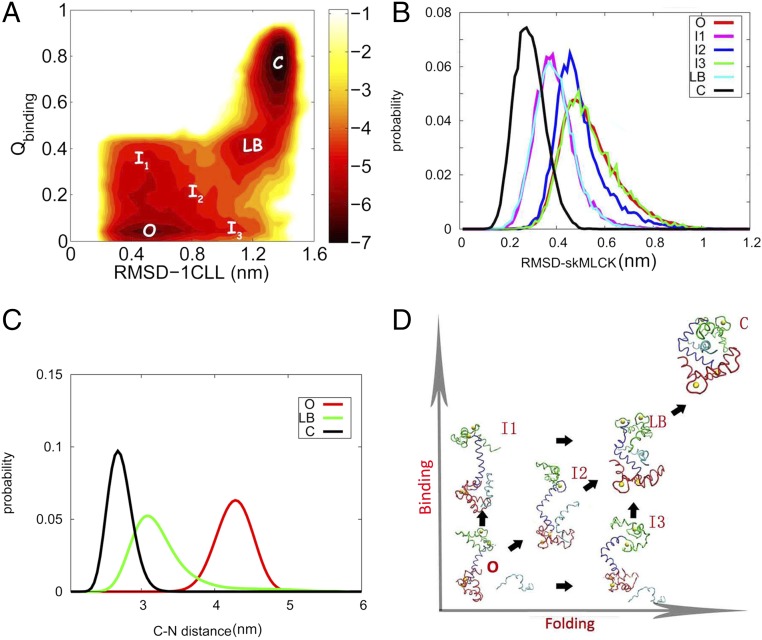
We explored the binding process with replica exchange molecular dynamics (26), which generated the free energy landscape along folding and binding dimensions (Fig. 1A). The reaction coordinates “rmsd-1CLL” and “” were respectively used to monitor the structural change of –CaM and the binding degree between –CaM and skMLCK. To have a clear description of how recognition occurs, we identified three stable states (the O, C, and LB states) and three unstable regions (I1, I2, and I3) on the landscape. The O and C states are open and closed states, corresponding to the target free state of -CaM before binding with both the N and C domains of the CaM open and bound state of -CaM with N and C domains wrapping around skMLCK, respectively, whereas the LB state is an on-pathway binding intermediate state (Fig. 1). We found that there are three parallel pathways going through the intermediate “LB” state from the transition between “O” and “C” states. Each pathway passes through the unstable regions “I1,” “I2,” and “I3,” respectively, before reaching the LB state. We can see the LB state is partly binding with around 0.4 and partly closed with rmsd-1CLL at 1.15 nm (Fig. 1A). Quantitatively, the degree of closeness of the LB is about 3/4 relative to the final completely compact globular state (Fig. 1C). Using the specific native contact probability map to explore the structural features of the LB (SI Appendix, Structural Characteristic for Each State), we found that the long helical structure of the central linker is broken and the bending of the linker contributes to the closeness of -CaM in the LB state. The N terminus of skMLCK mainly binds to the C domain and the linker close to the C domain of CaM (Fig. 1D) (SI Appendix, Structural Characteristic for Each State). Our results provide a dynamical basis for understanding the previous findings that the C-terminal domain of CaM has a higher affinity to target than the N-terminal domain (14, 27, 28) and the high plasticity of the linker is determined by its intrinsic flexibility (13–16). In addition, we used the distributions of rmsd-skMLCK for each state along the pathway to monitor this dynamical process (Fig. 1B). We found that the distributions of rmsd-skMLCK vary in different states and regions. The distribution of rmsd-skMLCK for the C state is narrower and smaller than for the O state and the I3 state. The distributions for the I1 state and the I2 state are in the middle range (Fig. 1B). This indicates that the skMLCK-binding peptide gradually forms -helical structures from a random coil along the binding process to CaM. The distributions of rmsd-skMLCK are almost the same in I1 and LB. The distributions of rmsd-skMLCK for O and I3 are also almost the same. No binding between the -CaM and skMLCK occurs in these two transitions, leading to the fact that skMLCK increases its helicity only by binding to -CaM.
Fig. 1.
(A) The two-dimensional free energy landscape along rmsd-1CLL and . We adopted the root mean square deviation (rmsd-1CLL) relative to the structure 1CLL to monitor the structural change of -CaM and use the fraction of native contacts between -CaM and skMLCK() to monitor binding. We identified three stable states and three unstable regions in the landscape. Besides the O and C states, there is a stable intermediate LB state. We found that all three pathways go through the intermediate LB state in the transition from open to closed states. Each pathway respectively passes through the unstable regions I1, I2, and I3 before getting to the LB state. (B) Conformational distributions of skMLCK at different states and regions. (C) Conformational distributions of CaM in different states. We used the centroid distance between the C domain and N domain of CaM to monitor its degree of closeness. The distributions of “C–N distance” for each state along the pathway are marked by different colors. The red, green, and black lines, whose peaks are located at 4.36 nm, 3.07 nm, and 2.60 nm, stand for the distribution of C–N distance in the O, LB, and C states, respectively. We therefore estimated the degree of closeness of CaM in LB ()relative to C is about 3/4. (D) Structural illustrations for the stable states and unstable regions on the landscape extracted from the simulation. The red, blue, and green regions are respectively the C domain, linker, and N domain of CaM. The O is the target free state. The -CaM adopts a dumbbell conformation before binding of skMLCK. I1 is the conformation extracted from the I1 region; the skMLCK binds only to the C domain of CaM and the conformation of CaM does not change compared with the O state in this region. I2 is the conformation extracted from the I2 region; the skMLCK binds only to the C domain of CaM but the degrees of both binding and closing are less than intermediate LB. I3 is the conformation extracted from the I3 region; the skMLCK does not bind to the CaM and the increasing degree of the closing of CaM opens the door for further skMLCK binding. The intermediate LB is partly binding and partly closed compared to the completely free O state. C is the -CaM–skMLCK complex; CaM adopts the compact conformation of both N- and C-terminal domains binding to skMLCK.
From the free energy landscape, we are able to address a mixture mechanism of coupled folding and binding of -CaM to skMLCK (Fig. 1D). In detail, the pathways O-I1-LB and O-I3-LB correspond to the partial binding between -CaM and skMLCK happening before and after the partial closing of -CaM, respectively. Additionally, in the pathway O-I2-LB, the partial closeness of -CaM is accompanied by the partial binding between -CaM and skMLCK, following the simultaneous binding–closing (folding) mechanism. When the partial binding and partial closeness intermediate LB is formed, all three pathways merge together to form one pathway. LB adopts the simultaneous binding–closing mechanism only to form the completely binding and completely folding state C. At the same time, the skMLCK increases its helicity only when it interacts with CaM.
Nonnative Electrostatic Interactions Act as a “Steering Force” to Facilitate the Binding Preference of CaM–skMLCK Recognition.
Previous structural investigations indicated that most of the -CaM–associated peptides have the propensity to form hydrophobic and electrostatic interactions at the binding interfaces (13, 15, 29–31). Taking these factors into consideration, the residues of both CaM and skMLCK are divided into hydrophobic, electrostatic, and plain ones in our work (SI Appendix, Materials and Methods). Both native interactions and nonnative interactions are supposed to play important roles in the protein–protein binding. The native interactions contribute to the binding affinity and specificity whereas nonnative interactions act on the initial recognition before forming native interactions (32–35). The interchain nonnative electrostatic interactions are sometimes regarded as the steering force to facilitate the protein recognition (32–36). To see the role of native and nonnative interactions in -CaM–skMLCK, we calculated the interaction energies and contact map at each state (Table 1 and SI Appendix, Fig. S6). It is worth noting that the nonnative plain Lennard–Jones interactions are represented only by an exclusive volume repulsive term.
Table 1.
CaM–skMLCK interaction energy in each complex state
| The energy of each state | ||||
| Interaction type | O | LB | LBC | C |
| Nonnative electrostatic | −7.82 | −37.03 | −38.85 | −38.29 |
| Nonnative hydrophobic | −0.04 | −1.90 | −3.61 | −4.56 |
| Native hydrophobic | −0.01 | −13.65 | −21.02 | −30.85 |
| Native electrostatic | −0.00 | −3.97 | −6.17 | −7.63 |
| Plain native LJ | −0.01 | −11.98 | −15.33 | −21.00 |
The unit of energy: kJ · mol−1. LJ, Lennard–Jones.
By exploring interaction energies during binding (Table 1), we found part of nonnative electrostatic interaction is formed and no nonnative hydrophobic interaction is formed in the state O. This implies that the formation of nonnative electrostatic interactions is before the formation of nonnative hydrophobic interactions and native interactions, illustrating again the role of electrostatic interactions as a steering force in -CaM–skMLCK recognition. Regarding the native interactions, we found that they are first formed between the C domain of CaM and the N terminus of the skMLCK and then between the N domain of CaM and the skMLCK. The nonnative electrostatic interactions will contribute to the binding preference between C-domain–CaM and skMLCK from the O to the LB state. This result is consistent with the fact that binding preference is strongly dependent on and adapted through the electrostatic interactions between -CaM and the targets (37). Most nonnative electrostatic interactions are formed in the transition from the O to the LB state (Table 1). E119, E120, and E123 in the C domain of CaM begin to form nonnative electrostatic interactions with K1, R2, R3, and K5 in the N terminus of the skMLCK in the state O. E80 of the linker forms the nonnative electrostatic interaction with R16 of the skMLCK and E47, D50, and E54 of N-domain CaM begin to form nonnative electrostatic interactions with R3 and K5 of skMLCK in the state LB (SI Appendix, Fig. S6A). The native interaction is hardly formed in the N domain of CaM in the LB state (SI Appendix, Fig. S6B). The nonnative electrostatic interactions contribute to the binding preference between N-domain–CaM and skMLCK in the next dynamical binding step from LB to C.
Nonnative Hydrophobic Interactions.
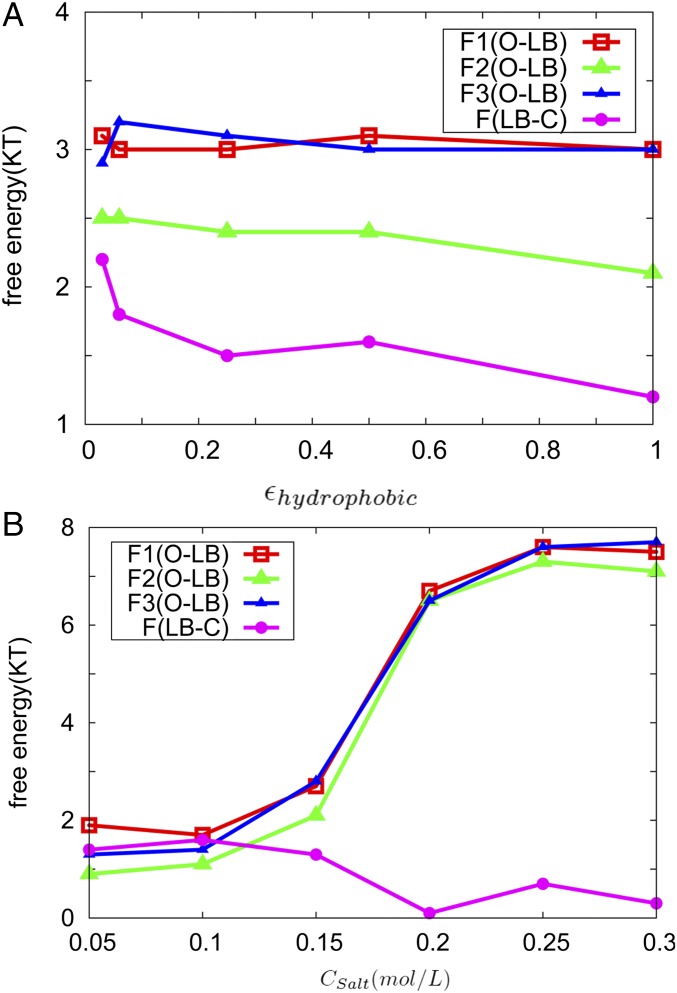
When comparing to the O state, we found a significant amount of nonnative hydrophobic interactions are formed on the C domain of CaM in the LB state (SI Appendix, Fig. S6A). It is due to the fact that hydrophobic interactions are short-ranged, compared with electrostatic interactions, and formed after the initial stage driven by the electrostatic interactions. We found the nonnative hydrophobic interactions are mostly formed around the site forming the native interactions and the distribution of nonnative hydrophobic interactions is wide in CaM (SI Appendix, Fig. S6A). Different from nonnative electrostatic interactions, the nonnative hydrophobic interaction energy and each native interaction change significantly not only in the O–LB transition but also during the LB–C transition (Table 1). To explore the different roles of the nonnative hydrophobic interactions and electrostatic interactions in -CaM–skMLCK recognition, we show the barrier heights along each pathway, respectively, in different electrostatic and nonnative hydrophobic interaction parameters in Fig. 2. Fig. 2A shows that the barrier heights for O–I1–LB and O–I3–LB are almost the same and are higher than that for O–I2–LB in each nonnative hydrophobic interaction strength. The barrier heights of both O–I2–LB and LB–C decrease as the nonnative hydrophobic interactions increase. With the increase of nonnative hydrophobic interactions, the barrier height of LB–C undergoes a significantly larger reduction (4×) than the decrease of barrier height of O–I2–LB. Therefore, the nonnative hydrophobic interactions have more influence on accelerating the transition LB–C than the transition O–LB. The barrier heights for O–I1–LB and O–I3–LB are higher than the barrier height for O–I2–LB in each electrostatic interaction strength and all three barrier heights decrease as the electrostatic interactions increase (Fig. 2B). However, the barrier height for LB–C almost does not change as the electrostatic interaction increases, implying the electrostatic interactions play little role at the last stage of the binding. In addition, we found electrostatic interactions have less significant influence on driving conformational change of the skMLCK compared with the nonnative hydrophobic interactions (SI Appendix, The Nonnative Interactions Drive Conformational Change of the Target).
Fig. 2.
The barrier heights along each pathway in different electrostatic and nonnative hydrophobic interaction strengths. Fi(O–LB) (i = 1,2,3) shows the barrier heights for the pathway O–Ii–LB. F(LB–C) is the barrier height for LB–C. (A) Barrier height changes with different strength of nonnative hydrophobic interactions. is the parameter representing the strength of the LJ potential of the nonnative hydrophobic contacts in the Hamiltonian energy (SI Appendix, Materials and Methods), altering the strength of the nonnative hydrophobic interaction. (B) Barrier height changes with different salt concentrations. is the salt concentration.
Based on the above discussions, the nonnative hydrophobic interactions have a different role compared to nonnative electrostatic interactions in -CaM–skMLCK recognition: At the beginning, nonnative electrostatic interactions steer the two units closer in space; then, nonnative hydrophobic interactions are formed transiently to drive and adapt the interface close to the native bound state.
The Native Interaction Contributes to the Affinity and Specificity of CaM–skMLCK Binding.
Each native interaction changes significantly not only the O–LB transition but also the LB–C transition in our simulation (Table 1). The native interactions contribute to the affinity and specificity of the protein–protein binding (32–34, 36, 37). In the LB state, we found that the residues ranking in the top seven in average contact number for skMLCK and CaM are W4, F8, F17, R2, R3, K5, and V11 and M144, E84, M145, M124, L112, and M109, respectively (SI Appendix, Fig. S6). To investigate the evolution of the native contacts along the routes from LB to C, we show the distribution of native contact for the barrier region (marked by LB–C) between the LB and C states. In the LB–C region, we found that the residues ranking top seven in average contact number for skMLCK and CaM are W4, F8, F17, V11, I9, N7, and R3 and E84, E11, F92, M144, M124, L112, M109, F68, and F19, respectively (SI Appendix, Fig. S6). These results show that the native hydrophobic interactions and native electrostatic interactions play important roles in contributing to the affinity and specificity of the CaM–skMLCK binding. During the recognition, the conserved hydrophobic residues with regular spacing serve as the anchors to form hydrophobic interactions with residues in -CaM. Based on the conserved position of hydrophobic residues, we term the skMLCK corresponding locations in 1-5-8-14 (25, 38). Our simulation shows W4, F8, and F17 of skMLCK rank top three in average contact number in the LB state and W4, F8, F17, and V11 of skMLCK rank top four in average contact number in the LB–C state. It shows the important role of hydrophobic anchors in specific CaM–target binding. Among these four hydrophobic anchors, the W4 anchor contributes the most. Because three electrostatic anchors (R2, R3, and K5) are near W4, the synergistic effect is significant. The number of the hydrophobic residues in MET is high in the skMLCK (SI Appendix, Fig. S6). It again supports that the interactions between hydrophobic anchors in peptides and Met-rich hydrophobic binding patches in -CaM are important for the high-affinity bound complex (38, 39).
The Effects of Flexibility and Ca2+ on the Mixture Binding Mechanism.
To explore how the linker flexibility influences the mixture mechanism of the -CaM binding to the skMLCK, we show the barrier heights along each pathway, respectively, in different strengths of the linker flexibility in Fig. 3. The site-specific constant is the parameter determining local strain energies (SI Appendix, Materials and Methods), which is used to control the strength of the linker flexibility in our simulation. High corresponds to small flexibility of the linker. We found that the increase in the linker flexibility decreases all four barrier heights (Fig. 3A). In addition, we found that the barrier height for O–I3–LB is the lowest in high linker flexibility ( 6) among all of the barrier heights of the O to LB pathway (Fi(O–LB), i = 1,2,3). The barrier height for O–I3–LB also has the biggest variation from low to high linker flexibility. The barrier height for O–I2–LB becomes the lowest for low linker flexibility. This indicates that the pathway O–I3–LB is the easiest to occur in the high linker flexibility. At the same time, we note that the pathway O–I1–LB is the hardest to occur in the high linker flexibility. O–I2–LB becomes the easiest to occur at low linker flexibility. We also see that the barrier height (F(LB–C)) from LB to C is always lower than or comparable to the barrier heights from O to LB in each strength of the linker flexibility.
Fig. 3.
(A) Fi(O–LB)(i = 1,2,3) shows the barrier heights for the pathway O–Ii–LB. F(LB–C) is the barrier height for LB–C. The site-specific constant is the parameter determining local strain energies and modulates linker flexibility in our simulation. Large (small) leads to low (high) flexibility of linker, the smaller the linker flexibility. (B) The red bars are under the condition that the system does not have . The green bars are under the condition that the system has .
Comparing the landscapes (SI Appendix, Fig. S11) of the -CaM binding to skMLCK with and without , we found that C and LB are more stable in the presence of , which is consistent with previous experiments (40). In fact, structural analysis revealed that these hydrophobic residues are buried inside of apo-CaM. Upon binding, the structure of CaM adopts a more extended dumbbell conformation with the hydrophobic interior exposed to solvent to facilitate the subsequent binding with targets (41–43). In addition, although does not interact with the central linker directly, it can control the plasticity of the linker (10). influences both the inherent flexibility and the conformation of each domain of CaM (SI Appendix, Fig. S9) (43–46). In our simulations with , all barrier heights (F1(O–LB), F2(O–LB), F3(O–LB), and F(LB–C)) are decreased (Fig. 3B). The conformation fluctuation caused by the binding of can facilitate the binding process. These results show that plays a positive role in the process of CaM binding to the skMLCK.
Discussion and Conclusion
The Intermediate as a Partially Folded State.
The C-terminal domain of CaM has been previously shown to have a much higher target affinity (about 100 times higher) than the N-terminal domain (16, 27). The final CaM–target complex formation may involve an intermediate state of only the C-terminal domain of CaM bound to its target (27). An intermediate that the targets bind only to the C-terminal domain of CaM without significant change of the conformation in the central linker was proposed by an experimental study (47). From structural analysis based on the modeling results, CaM in the I1 region, although not very stable due to the flexibility of the central linker, provides strong evidence of the intermediate states that the targets bind only to the C-terminal domain of CaM and the conformation of the central linker does not change (Fig. 1D). In addition, a 3/4 closeness intermediate LB is found in our work, which determines a mixture binding mechanism for the binding between CaM and the skMLCK peptide. How did the difference between I1 and LB originate? We have found the key residues in forming the state LB by using the method of averaging native contacts, such as hydrophobic anchors of skMLCK, rich Met residues of the C domain, and electrostatic E84 in the linker. We also used this method to analyze the difference between I1 and LB. We calculated the difference between average native contacts for I1 and LB (ANC) and show it in Fig. 4.
Fig. 4.
(A) The difference between the I1 and LB states in average of native contacts. ANC are the differences between native contact number in the I1 state and the LB state and the values that are larger than 1.0 are marked correspondingly. (B) Key residues in PDB structure. The 1CLL and 2BBM are reference structures for the O and C states, respectively. (C) The structures of I1 and LB states that are extracted from our simulation.
In the I1 state, the residues that have a large difference in average native contacts compared with the LB state are E84, F92, M109, M124, M144, and M145 in CaM and W4, N7, F8, and V11 in skMLCK peptide (Fig. 4A). M109, M124, and F92 have more average contacts in I1 (Fig. 4A). These three residues can be regarded as an “I1 site,” which has a higher tendency to act as binding patches for skMLCK in the I1 state rather than in the LB state (Fig. 4B). In addition, W4 has more average contacts in the I1 state and has a higher tendency to act as an anchor of skMLCK, which interacts with the binding patches in I1 rather than in the LB state (Fig. 4A). For the same reason, E84, M144, and M145 tend to form a binding-patches region (LB site) and F8 tends to act as a binding anchor in the LB state. We note that M109 and M124 in CaM and W4 in skMLCK are among top seven in average contact number in LB, implying the I1 site is also important to the LB state. From I1 to LB, the site near W4 in skMLCK is bound by the I1 site. The site around F8 in the skMLCK approaches and binds with the LB site (Fig. 4C). E84 in the LB site is near the breaking region of the central linker, the binding between the LB site and skMLCK in the LB state led to partial closeness of CaM.
Binding Process Involves Both the Local and Global Conformational Changes.
During the process of coupled folding and binding of -CaM to skMLCK peptide, the peptide undergoes transition from a random coil to -helical structures accompanied by the CaM wrapping itself. The binding process involves both the local conformational change of CaM and the global conformational changes of the skMLCK-binding peptide. Many works have been carried out to study biomolecular folding and binding transitions (19, 20, 48). In addition to the lock-and-key binding mechanism, proposed by Fischer to solve the rigid binding in enzyme catalysis (1), the induced-fit and conformational selection mechanisms (2–6) for flexibility during binding have been proposed for the biological processes driven by protein–protein binding with the local configurational plasticity. For intrinsically random coils, known as “intrinsically disordered proteins” (IDPs), the global conformational changes are always accompanied by their binding (49–51). By investigating the synchronization of binding and folding, the conventional association mechanism can be classified into cooperative “coupled bindingfolding” as well as noncooperative “folding before binding” and “binding before folding” (52, 53). A mechanism of conformational selection followed by induced folding has been also proposed for the binding–folding of IDP (54). Accordingly, What is the mechanism for this binding process simultaneously involving both the local conformational changes of receptor and the global conformational changes of peptide?
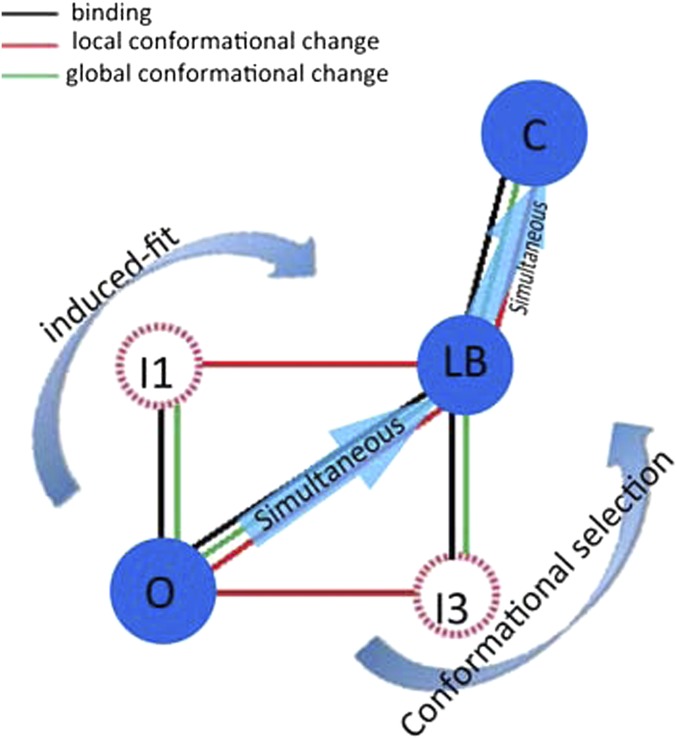
In fact, we found that the global conformational changes are always accompanied by the binding. The global conformational changes of target peptide have better consistency and cooperation with binding than local conformational change. Taking consideration of the recognition between CaM and target peptide as a binding process involved in both local conformational change of CaM and the global conformational changes of the peptide, compared to the simple folding–binding model of induced fit or conformational selection (2–6), -CaM adopts a mixture and complex binding mechanism when it binds to skMLCK. This is because there are three main pathways to the LB state from the O state and each pathway shows a different mechanism (Fig. 5). In fact, O–I1–LB and O–I3–LB take place according to the atypical induced fit and the atypical conformational selection. Different from the classic induced fit and conformational selection, which do not involve the global conformational change of the ligand, during the binding process of both the atypical induced fit and the atypical conformational selection, the ligand skMLCK changes its helicity only when it binds to CaM (Fig. 5). Apart from the atypical induced fit and atypical conformational selection, the pathway O–I2–LB takes place according to a “simultaneous” binding–folding mechanism due to the synchronization of folding (both local conformational change of CaM and the global conformational changes of the skMLCK) and the binding (between the -CaM and skMLCK peptide). The simultaneous binding–folding mechanism is a unique mechanism for the transition from the LB to the C state because of the synchronization of binding and folding (Fig. 5).
Fig. 5.
The mixture-binding mechanism in detail. The red, black, and green lines stand for the local conformational change of CaM, the binding between CaM and skMLCK, and the global conformational change of skMLCK peptide, respectively. O–I1–LB and O–I3 –LB take place according to atypical induced fit and atypical conformational selection, and O–I2–LB takes place according to a simultaneously binding–folding mechanism. The simultaneous mechanism is also the unique mechanism for the transition from LB to C.
The Intrinsically Disordered Properties of Target Peptide.
The distribution of rmsd-skMLCK for the C state is narrower and smaller than that for the O state and the I3 state, and the widths of distributions for the I1 state and the I2 state are in the middle range (Fig. 1B and SI Appendix, Fig. S15). The distributions of rmsd-skMLCK for the states also show the similarity in the conformation of the skMLCK relative to the reference structure 2BBM. In the reference structure 2BBM, the conformation of skMLCK adopts the -helical structures. It indicated that the tightly binding states have higher helicities than the loosely binding states (Fig. 1B). These results indicate that skMLCK has a more disordered conformational ensemble of the loosely binding state than of the tightly binding state, which show the binding nature of the IDP (49–51). Previous work has shown the possibility of first binding and then folding for IDP recognition (55, 56). A recent study addresses how linker flexibility affects the binding mechanism of IDPs (57). The initial binding of an IDP with its target mostly occurs in just a segment instead of the entire IDP and the long-range electrostatic interactions have important biasing effects (55–57). In the present work, we have found that skMLCK and CaM have similar properties in their mixture-binding pathways. In addition, we found the skMLCK binding peptide gradually forms -helical structures binding to CaM, which is in accordance with a “divide-and-conquer” mechanism proposed for the IDP binding–folding (54). In a word, we find many interesting intrinsically disordered properties of skMLCK during its binding process with CaM in our work.
Multispecificity.
-CaM is found to have the capability to bind over 300 targets (7–9). However, the underlying factors to control such multispecific binding remain unclear. Based on our studies, we are able to get some insights on this issue.
-
i)
At the beginning, nonnative electrostatic interactions steer the two units to be closer in space. The nonspecific nonnative electrostatic interactions are responsible for the binding preference between CaM and many kinds of targets due to the dipolar charged distribution of N- and C-terminal domains in CaM. Then nonnative hydrophobic interactions are formed transiently to drive and adapt the interface close to the native bound state, which is beneficial to different targets forming the specific native interaction with CaM. The cooperation between two kinds of nonnative interactions makes the CaM much easier to bind to different targets than mere nonnative electrostatic interaction.
-
ii)
We addressed the importance of conserved hydrophobic anchors in skMLCK contributing to binding to CaM. It again underscores that the native interactions between hydrophobic anchors in the peptide and Met-rich hydrophobic binding patches in CaM are important to the high-affinity bound complex. Although different binding peptides have different locations of hydrophobic anchors based on their biological functions (25, 38), the resulting hydrophobic interactions formed with CaM are able to lead to high-affinity complexes. This multihigh affinity can be regarded as the source of multispecificity.
-
iii)
Arguments i and ii show the speciality of interactions (nonnative, native) between the binding sites of CaM and its peptide provided the precondition for the multispecificity binding. Furthermore, the binding between CaM and its target is essentially a process that their binding sites search for and then bind to each other accompanied by conformational change. Local conformational change of CaM and global conformational change of skMLCK provide the plasticity of their binding sites. The fact that CaM adopts a mixture mechanism binding with the skMLCK indicated that the synchronization of binding and folding may vary from the processes of the CaM binding to its target. Because we found that the barrier heights of the binding pathways are strongly modulated by the strengths of electrostatic and hydrophobic interactions, this implies that the mixture-binding mechanism may depend on the different targets, which are supposed to form different kinds of interactions with CaM. It is expected that different targets are able to select the most suitable ways to bind to CaM, benefiting from the multispecificity binding. There has been increasing evidence recently to support our proposal (40, 47, 58, 59). The experiment has shown that CaM selects the partly rather than entirely compact structure to embrace the peptide, followed by conformational adaption to the bound structure induced by peptide. This leads to a mixture of the two conventional binding scenarios (40). A mutually induced-fit scenario for binding between CaM and CaMKI peptide was proposed for CaM and CaMKI peptide (58, 59), which is essentially a simultaneous binding–folding mechanism and is different from our mixture mechanism for the skMLCK.
-
iv)
It is expected that different targets can select the most suitable ways to bind to CaM, according to their structural features. The intermediate along the binding acts as the decisive factor for the binding mechanism between CaM and target. From our analysis for skMLCK, the distinct conformations of CaM in the I1 state and LB are determined by different binding-patch regions and the binding anchor is selected. To test the validity of our conclusions, we performed simulations of smMLCK peptide binding to CaM with the same simulation procedure used for skMLCK (SI Appendix, The Simulations for CaM and smMLCK). The main difference between smMLCK and skMLCK is that smMLCK lacks the conserved position of hydrophobic “5” (SI Appendix, Fig. S13), which is found to be critical to form the LB state in the simulations of skMLCK. In smMLCK binding, we found that the LB state is hardly populated but the I1 state is more stable compared with that in the skMLCK binding (SI Appendix, Fig. S14). Therefore, smMLCK tends to select a pathway following the atypical induced fit rather than the mixture mechanism for skMLCK.
Materials and Methods
Double-Well Model.
We developed a structure-based model (SBM) to explore the process by which CaM binds to its target skMLCK (32, 60–63). To extend a SBM to systems with two basins, we integrated information of the open (PDB ID: 1CLL) and closed (PDB ID: 2BBM) structures by a mixed-contact map model (10, 13, 32, 60, 61). Because the charged interactions and the hydrophobic interactions play an important role in in CaM recognition (13, 15, 25, 29–31, 54), we added the electrostatic interactions and hydrophobic interactions in our SBM. The electrostatic potential is represented by the Debye–Huckel model (64). The hydrophobic interactions exist only between hydrophobic residues. The CB–CB hydrophobic potential is represented by a 6–12 LJ potential, compared to the 10–12 LJ potential for nonhydrophobic and hydrophobic CA residues. We built a two-bead double-well SBM and the Hamiltonian is given by the expression
The local potential is divided into bond stretching, angle bending, and torsion energy and the repulsive term provides the excluded volume. We integrate a mixed-contact map into our model by in the Hamiltonian, and and are used to introduce the electrostatic interactions and hydrophobic interaction. (Details of the double-well model are in SI Appendix, Materials and Methods.)
Reaction Coordinate.
To describe the process by which the -CaM binds to skMLCK in our simulation, we adopted the rmsd-1CLL relative to structure 1CLL to monitor the structural change of -CaM and used the fraction of native contacts between -CaM and skMLCK() to monitor the binding process. On the other hand, we used the rmsd of skMLCK binding peptide relative to the reference structure 2BBM(rmsd-skMLCK) to monitor the structural change of skMLCK peptide.
Simulation Protocols.
All of the simulations were performed with Gromacs 4.0.5 (65). The time step is 0.0005 ps and the simulation was coupled to a temperature bath via Langevin dynamics with a coupling time of 1.0 ps. To achieve a sufficient sampling, we used replica exchange molecular dynamics (REMD) (26) to explore the thermodynamic energy landscape (Fig. 1A). We used 25 replicas and the neighbor replicas attempted to exchange with each other every 2,000 MD steps. For all replicas, the total simulation time was 1.25 s.
Supplementary Material
Acknowledgments
F.L. and J.W. acknowledge support from National Science Foundation of China Grant 91430217 and Ministry of Science and Technology, China, Grants 2016YFA0203200 and 2013YQ170585. J.W. thanks the support in part from National Science Foundation Grant NSF-PHY-76066. H.P.L. acknowledges support from the National Institutes of Health National Institute of General Medicine Science and the Ohio Eminent Scholar Endowment.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615949114/-/DCSupplemental.
References
- 1.Fischer E. Einfluss der configuration auf die wirkung der enzyme. Ber Dtsch Chem Ges. 1894;27:2985–2993. [Google Scholar]
- 2.Koshland DE. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma BY, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- 4.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: Dynamic landscapes and population shifts. Protein Sci. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsai CJ, Ma BY, Sham YY, Kumar S, Nussinov R. Structured disorder and conformational selection. Proteins. 2001;44:418–427. doi: 10.1002/prot.1107. [DOI] [PubMed] [Google Scholar]
- 6.Bosshard HR. Molecular recognition by induced fit: How fit is the concept? News Physiol Sci. 2001;16:171–173. doi: 10.1152/physiologyonline.2001.16.4.171. [DOI] [PubMed] [Google Scholar]
- 7.Yap KL. Calmodulin target database. J Struct Funct Genomics. 2000;1:8–14. doi: 10.1023/a:1011320027914. [DOI] [PubMed] [Google Scholar]
- 8.Crivici A, Ikura M. Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct. 1995;24:85–116. doi: 10.1146/annurev.bb.24.060195.000505. [DOI] [PubMed] [Google Scholar]
- 9.Chin D, Means AR. Calmodulin: A prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 10.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. Calmodulin structure refined at 1.7 angstrom resolution. J Mol Biol. 1992;228:1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 11.Chin D, Sloan DJ, Quiocho FA, Means AR. Functional consequences of truncating amino acid side chains located at a calmodulin-peptide interface. J Biol Chem. 1997;272:5510–5513. doi: 10.1074/jbc.272.9.5510. [DOI] [PubMed] [Google Scholar]
- 12.Kranz JK, Lee EK, Nairn AC, Wand AJ. A direct test of the reductionist approach to structural studies of calmodulin activity - Relevance of peptide models of target proteins. J Biol Chem. 2002;277:16351–16354. doi: 10.1074/jbc.C200139200. [DOI] [PubMed] [Google Scholar]
- 13.Ikura M, et al. Solution structure of a calmodulin-target peptide complex by multidimensional NMR. Science. 1992;256:632–638. doi: 10.1126/science.1585175. [DOI] [PubMed] [Google Scholar]
- 14.Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin - 2.4-angstrom structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 15.Meador WE, Means AR, Quiocho FA. Modulation of calmodulin plasticity in molecular recognition on the basis of X-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 16.Liu RC, Hu DH, Tan X, Lu HP. Revealing two-state protein-protein interactions of calmodulin by single-molecule spectroscopy. J Am Chem Soc. 2006;128:10034–10042. doi: 10.1021/ja057005m. [DOI] [PubMed] [Google Scholar]
- 17.Kranz JK, Flynn PF, Fuentes EJ, Wand AJ. Dissection of the pathway of molecular recognition by calmodulin. Biochemistry. 2002;41:2599–2608. doi: 10.1021/bi011818f. [DOI] [PubMed] [Google Scholar]
- 18.Ehrhardt MR, Urbauer JL, Wand AJ. The energetics and dynamics of molecular recognition by calmodulin. Biochemistry. 1995;34:2731–2738. doi: 10.1021/bi00009a001. [DOI] [PubMed] [Google Scholar]
- 19.Levy Y, Wolynes PG, Onuchic J. Protein topology determines binding mechanism. Proc Natl Acad Sci USA. 2004;101:511–516. doi: 10.1073/pnas.2534828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levy Y, Cho SS, Onuchic J, Wolynes PG. A survey of flexible protein binding mechanisms and their transition states using native topology based energy landscapes. J Mol Biol. 2005;346:1121–1145. doi: 10.1016/j.jmb.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 21.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 22.Levy Y, Onuchic JN. Mechanisms of protein assembly: Lessons from minimalist models. Acc Chem Res. 2006;39:135–142. doi: 10.1021/ar040204a. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Chu X, Suo Z, Wang E, Wang J. Multidomain protein solves the folding problem by multifunnel combined landscape: Theoretical investigation of a Y-family DNA polymerase. J Am Chem Soc. 2012;134:13755–13764. doi: 10.1021/ja3045663. [DOI] [PubMed] [Google Scholar]
- 24.Okazaki K, Koga N, Takada S, Onuchic JN, Wolynes PG. Multiple-basin energy landscapes for large-amplitude conformational motions of proteins: Structure-based molecular dynamics simulations. Proc Natl Acad Sci USA. 2006;103:11844–11849. doi: 10.1073/pnas.0604375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. Faseb J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 26.Sugita Y, Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 27.Sun HY, Squier TC. Ordered and cooperative binding of opposing globular domains of calmodulin to the plasma membrane Ca-ATPase. J Biol Chem. 2000;275:1731–1738. doi: 10.1074/jbc.275.3.1731. [DOI] [PubMed] [Google Scholar]
- 28.Nandigrami P, Portman JJ. Coarse-grained molecular simulations of allosteric cooperativity. J Chem Phys. 2016;144:105101. doi: 10.1063/1.4943043. [DOI] [PubMed] [Google Scholar]
- 29.Zhang MJ, Vogel HJ. The calmodulin-binding domain of caldesmon binds to calmodulin in an alpha-helical conformation. Biochemistry. 1994;33:1163–1171. doi: 10.1021/bi00171a016. [DOI] [PubMed] [Google Scholar]
- 30.Yuan T, Mietzner TA, Montelaro RC, Vogel HJ. Characterization of the calmodulin-binding domain of SIV transmembrane glycoprotein by NMR and CD spectroscopy. Biochemistry. 1995;34:10690–10696. doi: 10.1021/bi00033a045. [DOI] [PubMed] [Google Scholar]
- 31.Yuan T, Walsh MP, Sutherland C, Fabian H, Vogel HJ. Calcium-dependent and -independent interactions of the calmodulin-binding domain of cyclic nucleotide phosphodiesterase with calmodulin. Biochemistry. 1999;38:1446–1455. doi: 10.1021/bi9816453. [DOI] [PubMed] [Google Scholar]
- 32.Chu X, Liu F, Maxwell BA, Wang Y, Suo Z. Dynamic conformational change regulates the protein-DNA recognition: An investigation on binding of a Y-family polymerase to its target DNA. PLoS Comput Biol. 2014;10:e1003804. doi: 10.1371/journal.pcbi.1003804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: The fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chu X, Wang Y, Gan L, Bai Y, Han W. Importance of electrostatic interactions in the association of intrinsically disordered histone chaperone Chz1 and histone H2A.Z-H2B. PLoS Comput Biol. 2012;8:e1002608. doi: 10.1371/journal.pcbi.1002608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ganguly D, Zhang W, Chen J. Electrostatically accelerated encounter and folding for facile recognition of intrinsically disordered proteins. PLoS Comput Biol. 2013;9:e1003363. doi: 10.1371/journal.pcbi.1003363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, Wang Y, Chu X, Hagen SJ, Han W. Multi-scaled explorations of binding-induced folding of intrinsically disordered protein inhibitor IA3 to its target enzyme. PLoS Comput Biol. 2011;7:e1001118. doi: 10.1371/journal.pcbi.1001118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osawa M, et al. A novel target recognition revealed by calmodulin in complex with Ca2+-calmodulin-dependent kinase kinase. Nat Struct Biol. 1999;6:819–824. doi: 10.1038/12271. [DOI] [PubMed] [Google Scholar]
- 38.Yamniuk AP, Vogel HJ. Calmodulin’s flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol Biotechnol. 2004;27:33–57. doi: 10.1385/MB:27:1:33. [DOI] [PubMed] [Google Scholar]
- 39.Tripathi S, Waxham MN, Cheung MS, Liu Y. Lessons in protein design from combined evolution and conformational dynamics. Sci Rep. 2015;5:14259. doi: 10.1038/srep14259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anthis NJ, Doucleff M, Clore GM. Transient, sparsely populated compact states of apo and calcium-loaded calmodulin probed by paramagnetic relaxation enhancement: Interplay of conformational selection and induced fit. J Am Chem Soc. 2011;133:18966–18974. doi: 10.1021/ja2082813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan T, Ouyang H, Vogel HJ. Surface exposure of the methionine side chains of calmodulin in solution - A nitroxide spin label and two-dimensional NMR study. J Biol Chem. 1999;274:8411–8420. doi: 10.1074/jbc.274.13.8411. [DOI] [PubMed] [Google Scholar]
- 42.Siivari K, Zhang M, Palmer AG, Vogel HJ. NMR studies of the methionine methyl groups in calmodulin. FEBS lett. 1995;366:104–108. doi: 10.1016/0014-5793(95)00504-3. [DOI] [PubMed] [Google Scholar]
- 43.Li WF, Wang W, Takada S. Energy landscape views for interplays among folding, binding, and allostery of calmodulin domains. Proc Natl Acad Sci USA. 2014;111:10550–10555. doi: 10.1073/pnas.1402768111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tripathi S, Portman JJ. Inherent flexibility and protein function: The open/closed conformational transition of the N-terminal domain of calmodulin. J Chem Phys. 2008;128:205104. doi: 10.1063/1.2928634. [DOI] [PubMed] [Google Scholar]
- 45.Tripathi S, Portman JJ. Inherrent flexibility determines the transition mechanisms of the EF-hands of calmodulin. Proc Natl Acad Sci USA. 2009;106:2104–2109. doi: 10.1073/pnas.0806872106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nandigrami P, Portman JJ. Comparing allosteric transitions in the domains of calmodulin through coarse-grained simulations. J Chem Phys. 2016;144:105102. doi: 10.1063/1.4943130. [DOI] [PubMed] [Google Scholar]
- 47.Elshorst B, et al. NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump. Biochemistry. 1999;38:12320–12332. doi: 10.1021/bi9908235. [DOI] [PubMed] [Google Scholar]
- 48.Levy Y, Onuchic JN, Wolynes PG. Fly-casting in protein-DNA binding: Frustration between protein folding and electrostatics facilitates target recognition. J Am Chem Soc. 2007;129:738–739. doi: 10.1021/ja065531n. [DOI] [PubMed] [Google Scholar]
- 49.Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 50.Dunker AK, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 51.Uversky VN. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 53.Chu XK, Gan LF, Wang EK, Wang J. Quantifying the topography of the intrinsic energy landscape of flexible biomolecular recognition. Proc Natl Acad Sci USA. 2013;110:E2342–E2351. doi: 10.1073/pnas.1220699110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, et al. Multiscaled exploration of coupled folding and binding of an intrinsically disordered molecular recognition element in measles virus nucleoprotein. Proc Natl Acad Sci USA. 2013;110:E3743–E3752. doi: 10.1073/pnas.1308381110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Lu Q, Lu HP. Single-molecule dynamics reveals cooperative binding-folding in protein recognition. PLoS Comput Biol. 2006;2:842–852. doi: 10.1371/journal.pcbi.0020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu Q, Lu HP, Wang J. Exploring the mechanism of flexible biomolecular recognition with single molecule dynamics. Phys Rev Lett. 2007;98:128105. doi: 10.1103/PhysRevLett.98.128105. [DOI] [PubMed] [Google Scholar]
- 57.Zhou H-X, Pang X, Lu C. Rate constants and mechanisms of intrinsically disordered proteins binding to structured targets. Phys Chem Chem Phys. 2012;14:10466–10476. doi: 10.1039/c2cp41196b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Q, Zhang PZ. Protein recognition and selection through conformational and mutually induced fit. Proc Natl Acad Sci USA. 2013;110:20545–20550. doi: 10.1073/pnas.1312788110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tripathi S, et al. Conformational frustration in calmodulin-target recognition. J Mol Recognit. 2015;28:74–86. doi: 10.1002/jmr.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y, Tang C, Wang EK, Wang J. Exploration of multi-state conformational dynamics and underlying global functional landscape of maltose binding protein. PLoS Comput Biol. 2012;8:e1002471. doi: 10.1371/journal.pcbi.1002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, Gan L, Wang E, Wang J. Exploring the dynamic functional landscape of adenylate kinase modulated by substrates. J Chem Theory Comput. 2012;9:84–95. doi: 10.1021/ct300720s. [DOI] [PubMed] [Google Scholar]
- 62.Wang J, Verkhivker GM. Energy landscape theory, funnels, specificity, and optimal criterion of biomolecular binding. Phys Rev Lett. 2003;90:188101. doi: 10.1103/PhysRevLett.90.188101. [DOI] [PubMed] [Google Scholar]
- 63.Wang J, Xu L, Wang E. Optimal specificity and function for flexible biomolecular recognition. Biophys J. 2007;92:L109–L111. doi: 10.1529/biophysj.107.105551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Azia A, Levy Y. Nonnative electrostatic interactions can modulate protein folding: Molecular dynamics with a grain of salt. J Mol Biol. 2009;393:527–542. doi: 10.1016/j.jmb.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 65.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.