Significance
The principal plant model species, Arabidopsis thaliana, is central to our understanding of how molecular variants lead to phenotypic change. In this genome-sequencing effort focused on accessions from Africa, we show that African populations represent the most ancient lineages and provide new clues about the origin of selfing and the species itself. Population history in Africa contrasts sharply with the pattern in Eurasia, where the vast majority of samples result from the recent expansion of a single clade. This previously unexplored reservoir of variation is remarkable given the large number of genomic studies conducted previously in this well-studied species and implies that assaying variation in Africa may often be necessary for understanding population history in diverse species.
Keywords: evolution, population history, self-compatibility, climate, migration
Abstract
Over the past 20 y, many studies have examined the history of the plant ecological and molecular model, Arabidopsis thaliana, in Europe and North America. Although these studies informed us about the recent history of the species, the early history has remained elusive. In a large-scale genomic analysis of African A. thaliana, we sequenced the genomes of 78 modern and herbarium samples from Africa and analyzed these together with over 1,000 previously sequenced Eurasian samples. In striking contrast to expectations, we find that all African individuals sampled are native to this continent, including those from sub-Saharan Africa. Moreover, we show that Africa harbors the greatest variation and represents the deepest history in the A. thaliana lineage. Our results also reveal evidence that selfing, a major defining characteristic of the species, evolved in a single geographic region, best represented today within Africa. Demographic inference supports a model in which the ancestral A. thaliana population began to split by 120–90 kya, during the last interglacial and Abbassia pluvial, and Eurasian populations subsequently separated from one another at around 40 kya. This bears striking similarities to the patterns observed for diverse species, including humans, implying a key role for climatic events during interglacial and pluvial periods in shaping the histories and current distributions of a wide range of species.
The plant Arabidopsis thaliana is the principal plant model species, and as such has been useful not only to examine basic biological mechanisms but also to elucidate evolutionary processes. The exceptional resources available in this species, including seed stocks collected from throughout Eurasia for over 75 y, have been a valuable tool for learning about the natural history of A. thaliana on this continent (1, 2). Previous studies have shown that current variation in Eurasia is mainly a result of expansions and mixing from refugia in Iberia, Central Asia, and Italy/Balkans after the end of the last glacial period ∼10 kya (3–8). The main finding of the recent analysis of 1,135 sequenced genomes was that a few Eurasian samples represent divergent relict lineages, whereas the vast majority derived from the recent expansion of a single clade (4). Given the large number of studies that examine the natural history of A. thaliana, one would expect that this history would by now be described rather completely and there would be no major surprises left to uncover. However, there are still many open questions about the ancient history of the species.
Several features differentiate A. thaliana from its closest relatives. Although most members of the Arabidopsis genus are obligate out-crossing perennials with large flowers and genome sizes of over 230 Mb and 8 chromosomes, A. thaliana is a predominantly selfing annual with reduced floral morphology and a reduced genome size of ∼150 Mb and 5 chromosomes. The transition to predominant selfing in A. thaliana was likely the catalyst for these derived morphological and genomic features (9–13). These changes, in particular the rearranged and shrunken genome, created a strong reproductive barrier between A. thaliana and its closest relatives (14).
Although the genetic basis of self-compatibility in A. thaliana is known, the specific events that occurred during the transition to predominant selfing are still unclear. In obligate out-crossing Arabidopsis species, many highly divergent S-locus haplogroups (S-haplogroups) are maintained by balancing selection, providing a mechanism for inbreeding avoidance. In A. thaliana, three S-haplogroups are found, and each contains mutations that obliterate function of the S-locus genes (15–17). Loss-of-function occurred independently in each S-haplogroup (18–21), but because these three S-haplogroups were never found together in the same geographic region, self-compatibility is inferred to have evolved separately in multiple locations (16, 21, 22). However, the hypothesis of geographically distinct origins is difficult to reconcile with the major genomic and phenotypic changes that render A. thaliana incompatible with its out-crossing congeners (9–13). Shifts from out-crossing to predominant selfing are common and have been considered the most prevalent evolutionary transitions in flowering plants (23). Reconstructing the evolutionary history of the transition to selfing in A. thaliana could provide general insights into this common evolutionary transition. However, because substantial time has passed since this transition (24–26) and no intermediate forms have been found between A. thaliana and its obligate out-crossing relatives, this reconstruction is challenging.
We sequenced the genomes of 78 African samples and analyze these in combination with 1,135 previously sequenced samples (4) (Fig. 1 and SI Appendix, Table S1). We find that African variation reveals the ancient history of the species and clarifies details concerning the transition to selfing. Congruence of A. thaliana population history with major climatic events and paleontological observations illustrates the relevance of population genetic studies for understanding climate-mediated demography more generally.
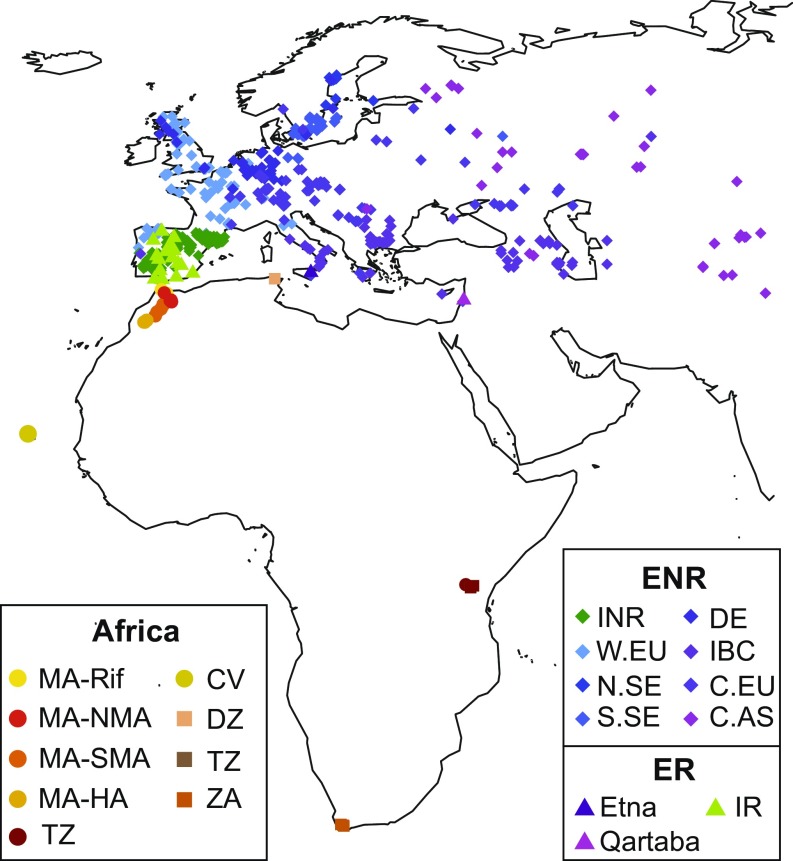
Fig. 1.
Sample map of accessions included in this study. Herbarium samples are shown as squares. Abbreviations are as follows: Algeria (DZ), Cape Verde (CV), Central Asia (C.AS), Central Europe (C.EU), Eurasian nonrelicts (ENR), Eurasian relicts (ER), Germany (DE), Italy, Balkans, and Caucasus (IBC), Iberian nonrelicts (INR), Iberian relicts (IR), Morocco (MA), North Sweden (N.SE), South Africa (ZA), South Sweden (S.SE), Tanzania (TZ), Western Europe (W.EU).
Results
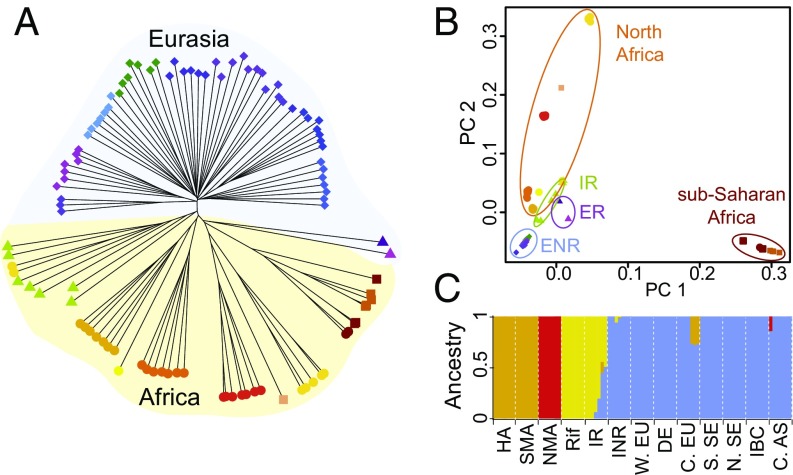
To examine the relationship between African individuals and other worldwide samples, we used three complementary clustering approaches. Distance-based clustering by neighbor-joining reveals a clear split between Eurasian and African samples, indicating deep divergence between the continents (Fig. 2A and SI Appendix, Fig. S1). The majority of Eurasian samples form a nearly star-shaped phylogeny, consistent with recent expansion of these lineages. Conversely, longer more bifurcated branches separate African subclusters and previously identified Eurasian relicts from each other and from the nonrelict clade. In general, the Eurasian clades cluster consistently with the nine groups defined previously (4). Exceptions are the Central Europe clade, which separates into two clusters, and the Iberian relicts, which cluster with the Moroccan Rif-Zin population. Moroccan samples separate into four clades, reflecting their geographic distribution and South Africa and Tanzania cluster together in a single clade. The results for South Africa and Tanzania are striking because A. thaliana populations outside of Eurasia and North Africa were previously thought to be recently introduced by humans (27).
Fig. 2.
Global population structure. (A) Unrooted neighbor-joining tree, (B) PCA, (C) ADMIXTURE results for K = 4.
Similarly, principal component analysis (PCA) separates African populations from each other and from Eurasian populations (Fig. 1B). The first PC distinguishes sub-Saharan Africa, and the second separates the four Moroccan clusters from Eurasians. Subsequent PCs mainly discriminate populations within Africa, whereas Eurasian populations remain tightly clustered (SI Appendix, Fig. S3). Results from ADMIXTURE (28) reinforce this finding. Moroccan populations separate into three clusters and are distinguished from a single cluster of Eurasian samples (Fig. 1C, and SI Appendix, Figs. S4 and S5, and Table S2). PCA and ADMIXTURE results also suggest a Moroccan origin of the relicts in Iberia, which are spread between Rif-Zin and Iberian nonrelicts in PCA, and sizable portions of the Iberian relict genomes match the Moroccan clusters in ADMIXTURE. This finding is consistent with previous work (29) and with the accepted phylogeographical history of Mediterranean and North African flora characterized by a complex history of expansions and contractions driven by important climatic changes experienced in this vast region, particularly since the Pliocene (30, 31).
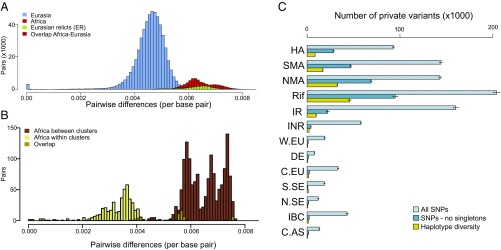
Furthermore, from pairwise differences, we recover the previously reported difference between Eurasian relicts and nonrelicts (4) and find that all African accessions are at least as divergent as samples previously classified as relicts (Fig. 3A and SI Appendix, Fig. S6). Therefore, in contrast to Eurasia, where most samples represent a single recently spread clade, all African individuals represent relict samples. The distribution of pairwise differences within Africa (Fig. 3B) further demonstrates the high diversity in these samples.
Fig. 3.
Patterns of diversity across geographic regions. Distributions of genome-wide pairwise differences per base pair in: (A) worldwide comparison and (B) within and between African populations, where overlap between distributions is shown as described in legends. (C) Numbers of private SNPs and haplotypes found in each cluster. Error bars denote 95% confidence intervals.
If populations in Africa truly are more ancient than the Eurasian clusters, we should also expect higher numbers of private variants in Africa. Indeed, we find that the Moroccan clusters and Iberian relicts, which appear likely derived from Morocco, harbor the highest numbers of private SNPs (Fig. 3C and SI Appendix, Fig. S8 and Table S3). This signal intensifies when we exclude recently arisen variants, which we find constitute the majority of the private variation in Eurasia. First, we excluded singletons, the class of SNPs most influenced by recent population growth, and found a 7.0- to 23.2-fold enrichment in Morocco compared with the top nonrelict cluster (Fig. 3C). Next, we considered the spatial distribution of private variants across the genome. Because novel private variants are unlikely to be tightly linked, clustering of several, contiguous private SNPs indicates old haplotypes. At the haplotype level, we again found extremely high enrichment (3.9- to 20.9-fold) for Moroccan clusters and Iberian relicts (4.5-fold) relative to the top Eurasian cluster (Fig. 3C). Notably, the Eastern Mediterranean and Caucasus regions, which were previously favored as points of origin and major centers of diversity of the species (3), do not exhibit a striking pattern for any of the metrics examined (see IBC in Fig. 3C). These findings evoke a model in which polymorphism in Africa is because of ancient variation and Eurasian polymorphism is mainly because of recent expansion.
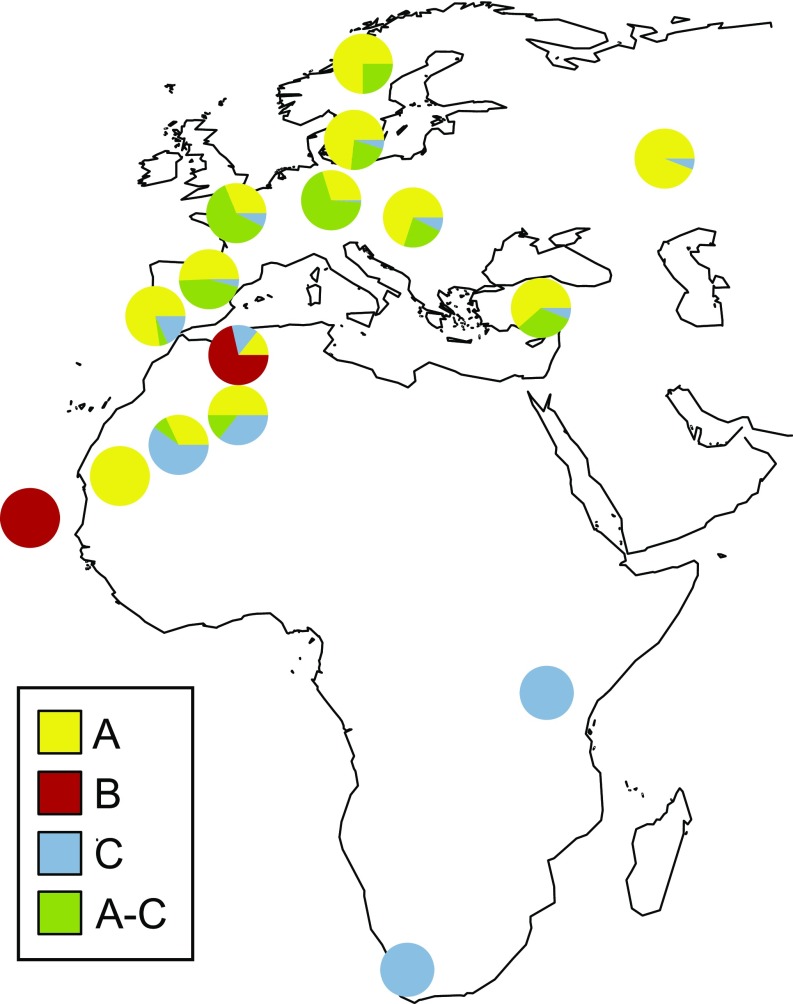
To better understand the relevance of variation in Africa for the early history of the A. thaliana lineage, we examined variation at the locus that confers self-compatibility. S-locus variation in Africa differs in several ways from what is found in Eurasia (Fig. 4 and SI Appendix, Table S4). We found all three S-haplogroups together in a single geographic region, with S-haplogroup B private to Africa. In addition, the A-C recombinant is also present at low frequency in Morocco, in contrast to what is found in Eurasia (32). Finally, we discovered deletion haplotypes in haplogroups A and C in Morocco (SI Appendix, Fig. S9). The finding that all S-locus haplogroups are present together implies that selfing evolved in a single geographic region.
Fig. 4.
Map of S-locus haplogroup diversity.
Taken together, the patterns in population structure, and levels of variation across the genome and at the S-locus, specifically, imply a deep history in Africa. To clarify the details of past demographic events, we inferred historical effective population sizes (Ne) and split times among populations based on cross-coalescent rates (CCR) using a multiple sequentially Markovian coalescent approach (MSMC) (33).
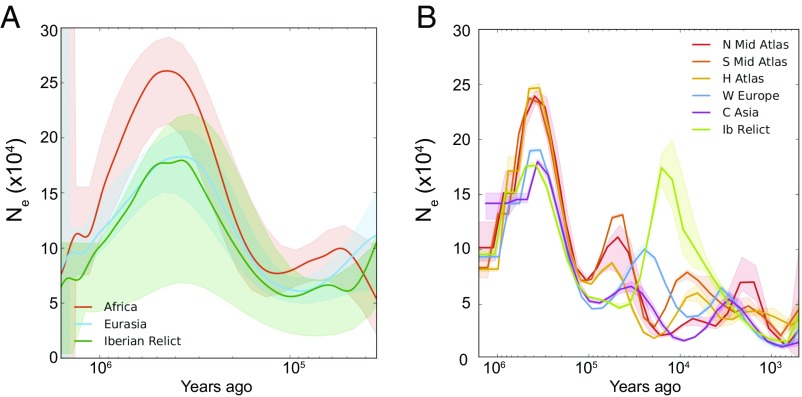
In ancient times, we find the highest Ne is in Africa, peaking at around 500–400 kya (Figs. 5 and SI Appendix, Fig. S10). All Eurasian populations (including Iberian relicts) show the same trajectories as the Africans, but with lower amplitudes. Given that these curves are in phase with one another and we do not see evidence for a population split until 120 kya (Fig. 6A), we interpret this as population structure in the ancestral population combined with bottlenecks as more derived populations migrated away from this ancestral population. This finding is consistent with our finding that variation in Eurasia is often a subset of variation present within Africa and is similar to the situation in humans (34). Notably, the IBC cluster, which includes previously hypothesized A. thaliana origins and refugia (3, 5, 7), exhibits a much lower ancient population size than Africa (SI Appendix, Fig. S10).
Fig. 5.
Historical effective population size of A. thaliana inferred using MSMC. Although two-haplotype analysis provides more resolution in the distant past, eight-haplotype analysis provides better resolution in the recent past. (A) Inference using pairs of haplotypes, with lines representing medians and shading representing ±1 SD calculated across pairs. This analysis is expected to produce unbiased estimates between 40 kya and 1.6 Mya (SI Appendix). (B) Inference based on sets of eight haplotypes with lines representing medians. This analysis is expected to produce unbiased estimates as recently as 1.6 kya.
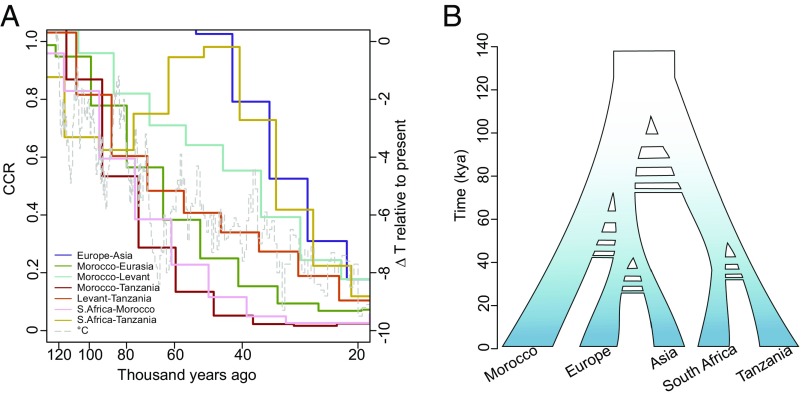
Fig. 6.
Inferred timing of population splits. (A) Relative CCR between populations. Decreasing values from 1.0 indicate population separation. The dashed line represents historical temperature (63). (B) A schematic model for the demographic history of A. thaliana based on CCR results, with hashes to represent uncertainty regarding possible timing of gene flow events.
At 120–90 kya, there are bottlenecks in all populations (Fig. 5) and a split among the major clades (Moroccan, Tanzanian, and Levant) (Fig. 6 and SI Appendix, Fig. S11). This roughly corresponds to the Abbassia Pluvial, a period when migration corridors were open because of high precipitation and humidity in Africa (35) and also marks the last interglacial at Marine Isotope Stage 5e (130–116 kya), when temperatures were 1–2° warmer than present-day conditions, providing favorable conditions in Eurasia. As this interglacial period came to a close, there was a worldwide shift toward cooler, drier conditions as the most recent and severe Pleistocene glaciation phase began (36). Beginning at this time, CCR implies a progressive decline in population connectivity, consistent with decreasing temperature and increasing aridity (Fig. 6A). We checked for consistency using a complementary method that relies on the joint site frequency spectrum between populations (δaδi) (37) and found a slightly older estimate for the split and overlapping confidence intervals (141–116 kya) (SI Appendix, Table S5). We propose that the most likely scenario is that A. thaliana was colonizing broadly within Africa as well as in the Levant during the last interglacial (130–116 kya), but that connections between populations began to break down as populations spread and as climate became cooler and drier at the end of this period.
MSMC based on eight haplotypes detects several changes in Ne in more recent times (Fig. 5B). Since ∼120 kya, the population size changes in Europe and Asia are often out of phase with those in Africa, consistent with geographical separation and exposure to different climatic regimes. Maxima in African populations occur at around 60–40 kya and 11–5 kya, corresponding to orbital-scale climate shifts from arid to moist conditions (i.e., pluvial periods) that occurred at 11–5 kya and 59–47 kya in North Africa (38, 39). We find relatively recent split times between sub-Saharan African clades (South Africa and Tanzania) and between Western Europe and Central Asia, at around 40 kya (Fig. 6).
There are a few caveats to consider regarding the demographic inference. Because MSMC can spread instantaneous population size changes over time, maxima and minima are informative but the slope should be interpreted with caution (33). In addition, the precise timing associated with inferred population size changes and splits is dependent on parameters that are difficult to measure and may vary over space and time, including mutation rate, degree of purifying selection, and the possible input from a seed bank. We used the best available data for mutation rate [based on mutation accumulation experiments (40)] and made the usual simplifying assumptions for other parameters (one generation per year), but the timing we infer would need to be revised if these assumptions turned out to be incorrect.
Discussion
Genomic studies thus far have amassed data for nearly 2,000 Eurasian A. thaliana accessions but were unable to provide insight into the early history of the species. Here, in a genome-scale sequencing effort focused on African accessions, we find clear evidence for a deep history of African A. thaliana populations, which harbor variation that was either lost or never present in Eurasia. Several specific results were unexpected based on current knowledge in this well-studied species. First, we discovered surprising and clear evidence that A. thaliana is native not only to North Africa but also to Afro-alpine regions of sub-Saharan Africa. Second, our results revealed that the deepest splits species-wide separate the African lineages from one another and that in ancient times, the effective population size was largest in Africa. Finally, we learned that variation at the S-locus is highest in Africa and that all three S-haplogroups are present there.
Based on our results, we can outline a model for the early history and transition to selfing in A. thaliana (detailed in SI Appendix, Fig. S12). In the first step, we infer that the population ancestral to A. thaliana became geographically separated from its parental out-crossing population. Our results suggest that this separation involved migration of the ancestral subpopulation into Africa by 1.2–0.8 Mya. This timing corresponds to the Middle Pleistocene Transition, a shift to drier more variable climate and more open habitats in Africa (i.e., grasslands versus woodlands), as evidenced by soil carbon analysis showing an increase in the ratio of C4 to C3 plants (41, 42).
Although the estimated divergence times between A. thaliana and Arabidopsis lyrata center around 5–7 Mya (9, 43), the origin of A. thaliana itself appears to be much younger. Our model predicts that there was an initial bottleneck as the subpopulation that led to A. thaliana split from a A. lyrata-like ancestral population [similar to that observed in Mimulus nasutus (44) and Capsella rubella (45–47)], followed by an expansion in Ne as the selfing population began to spread. In this case, we could interpret the MSMC results to suggest that the transition to selfing occurred between 1 Mya and 500 kya, before the most ancient maximum in Ne. This finding is in line with an estimate based on the depth of the A. thaliana genealogy (0.84% maximum divergence among individuals sampled here) under a simple model (T ∼ D/2 μ ∼ 598 kya). Our estimated timing is also consistent with previous estimates for the loss of self-incompatibility and origin of selfing (24–26).
Once selfing was established, traits associated with the “selfing syndrome” would have been favored, including reduced pollen number and petal size (48). Such phenotypic shifts are common in predominantly selfing species and have occurred in A. thaliana compared with its closest relatives (26). At the genomic level, A. thaliana exhibits major chromosomal rearrangements and a reduction in genome size and number of chromosomes (49). This genomic reduction is also likely a by-product of the shift to predominant selfing in A. thaliana (9–11), consistent with an observed link between reduced genome size and selfing in other plant species (11–13). These changes introduce a strong reproductive barrier as found in hybrids of A. thaliana and A. lyrata, which are infertile because of the chromosomal rearrangements that occurred in A. thaliana (14).
Given that all three S-haplogroups co-occur in Morocco, we hypothesize that the transition to predominant selfing occurred in a single region, best represented today in Morocco. This finding differs from previous assertions that these events likely happened separately in geographically distinct populations (15, 16). Moreover, it allows for the possibility that the transition to selfing was aided by a shared precursor mutation, a shared climate, and the bottleneck that occurred during the migration away from the ancestral population (22). Our proposed model parallels observations in partially selfing populations of A. lyrata (50). Here, self-compatibility is associated with two different S-haplogroups in Great Lakes populations and self-compatibility may have been favored because of the bottleneck that initially limited S-haplogroup diversity and thus mate availability.
After the origin and initial population size increase of A. thaliana, we infer several demographic changes that are congruent with known climatic shifts. At 120–90 kya, we find evidence from MSMC and δaδi for splitting among the major clades: Morocco, Levant and sub-Saharan Africa. This split corresponds to the Abbassia pluvial, which produced migration corridors within Africa (120–90 kya) (35, 39) as well as Marine Isotope Stage 5e (130–116 kya), the last interglacial period, when worldwide temperatures were 1–2° warmer than they are currently (51, 52). This is consistent with a model in which A. thaliana spread widely throughout Africa and into Eurasia when conditions were favorable (∼120 kya), with isolation as gene flow was reduced (SI Appendix, Fig. S12). More recent major demographic events include the split between European and Asian populations at around 40 kya and the increase in Ne within Africa during the most recent pluvial.
The patterns we observe and their concordance with climatic events suggest that the transition to selfing and speciation occurred within Africa, with subsequent migration out of Africa into Eurasia. However, it is also possible that the initial transition to selfing occurred within Eurasia followed by migration into Africa and concomitant loss of variation in Eurasia. This alternative would require that the ancient variation in the A. thaliana lineage was either lost or has not been sampled in Eurasia and the bottleneck into Africa was mild enough to preserve high levels of genetic variation.
Overall, the patterns in A. thaliana bear striking similarities to those observed for human populations, particularly in the larger effective population size in Africa (34), the exodus from Africa approximately 120 kya (39, 53–55), and the splitting of major human populations in Europe and Asia (approximately 45–35 kya) (53, 54). Analogous to what we propose here, demographic events in human populations have been attributed to major climate transitions (35, 39, 56).
Moreover, the timing and types of demographic events we infer during the history of A. thaliana are consistent with previous observations in a broad range of other plant species. Specifically, the shift to predominance of C4 plants across Africa at 1.2–0.8 Mya and the intensification of glacial cycles worldwide (57) correspond with our estimated timing of the evolution of selfing in A. thaliana and a clustering of speciation events more generally (58). The geographic expansion approximately 120 kya corresponds to an African pluvial and worldwide interglacial, which resulted in expansion of forests across Africa (59) and Eurasia (51, 52). Finally, we see evidence of an increase in effective population size overlapping with the most recent and well-described African pluvial at 11–5 kya, when the Sahara was heavily vegetated and filled with lakes (38, 60, 61). The concordance between inferred population size changes, climate, and reports for other species implies that the patterns we observe in A. thaliana may be representative of climate-mediated population dynamics across diverse taxa.
Materials and Methods
For full materials and methods, please see SI Appendix, Supplementary Text.
We sequenced the genomes of 79 A. thaliana individuals, including 70 fresh samples and 9 herbarium samples (SI Appendix, Table S1). For fresh leaf samples, sequencing libraries were prepared using Illumina TruSeq DNA sample prep kits (Illumina) and sequenced on Illumina Hi-Seq instruments. DNA from herbarium specimens was extracted, authenticated, and treated with uracil glycosylase to remove damaged nucleotides in a clean room facility at the University of Tübingen. To align the sequences to the TAIR10 reference genome and to call variants, we used two different pipelines: the MPI-SHORE pipeline (62) and a more conservative pipeline designed to reduce false positives resulting from indels.
For population structure analyses, we subsampled the complete dataset to match sample sizes across clusters as some Eurasian geographic regions are heavily oversampled, which could cause biases in some analyses, and we pruned SNPs based on LD to select a representative set. For ADMIXTURE, the number of clusters (K) was determined based on the outcome of cross-validation analyses.
To infer patterns of effective population size and population separations over historical time, we used a MSMC v2 (33). Because A. thaliana accessions are inbred, we created pseudodiploids by combining chromosomes from pairs of individuals from the same populations and ran MSMC in the two- and eight-haplotype configurations (Fig. 5). We assumed a mutation rate of 7.1 × 10−9 based on results of mutation accumulation experiments (40) and a generation time of 1 y. We confirmed inferences using δa δi (37) on joint site frequency spectra from pairs of populations.
Supplementary Material
Acknowledgments
We thank the South African National Biodiversity Institute, the University of Vienna, and Naturalis for access to herbarium samples; and V. Castric and M. Horton for helpful comments on the manuscript. Support for sequencing and plant growth were provided by the Vienna Biocenter Core Campus Science Support Facilities and the Max Planck Gesellschaft (MPG). Computational resources were from the Vienna Scientific Cluster, Center for Integrative Bioinformatics Vienna, and MPG were used. The project was supported by European Union CIG 304301, European Research Council Grant CVI_ADAPT 638810, and MPG (to A.M.H.); Austrian Science Funds FWF W1225; Grants BIO2016-75754-P (to C.A.-B.) and CGL2016-77720-P (to F.X.P.) from the Agencia Estatal de Investigación of Spain and the Fondo Europeo de Desarrollo Regional; Japan Society for the Promotion of Science KAKENHI 15K18583 (to T.T.); and the Presidential Innovation Fund of the MPG (to H.A.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper has been deposited in the European Nucleotide Archive/Sequence Read Archive database, study PRJEB19780 (accession nos. ERS1575066–ERS1575147). Analysis scripts are available at https://github.com/HancockLab/African_A.thaliana.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616736114/-/DCSupplemental.
References
- 1.Provart NJ, et al. 50 years of Arabidopsis research: Highlights and future directions. New Phytol. 2016;209:921–944. doi: 10.1111/nph.13687. [DOI] [PubMed] [Google Scholar]
- 2.Somerville C, Koornneef M. A fortunate choice: The history of Arabidopsis as a model plant. Nat Rev Genet. 2002;3:883–889. doi: 10.1038/nrg927. [DOI] [PubMed] [Google Scholar]
- 3.Beck JB, Schmuths H, Schaal BA. Native range genetic variation in Arabidopsis thaliana is strongly geographically structured and reflects Pleistocene glacial dynamics. Mol Ecol. 2008;17:902–915. doi: 10.1111/j.1365-294X.2007.03615.x. [DOI] [PubMed] [Google Scholar]
- 4.Consortium G. 1001 Genomes Consortium. Electronic address: magnus.nordborg@gmi.oeaw.ac.at 1001 Genomes Consortium 1135 genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell. 2016;166:481–491. doi: 10.1016/j.cell.2016.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.François O, Blum MG, Jakobsson M, Rosenberg NA. Demographic history of European populations of Arabidopsis thaliana. PLoS Genet. 2008;4:e1000075. doi: 10.1371/journal.pgen.1000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picó FX, Méndez-Vigo B, Martínez-Zapater JM, Alonso-Blanco C. Natural genetic variation of Arabidopsis thaliana is geographically structured in the Iberian peninsula. Genetics. 2008;180:1009–1021. doi: 10.1534/genetics.108.089581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharbel TF, Haubold B, Mitchell-Olds T. Genetic isolation by distance in Arabidopsis thaliana: Biogeography and postglacial colonization of Europe. Mol Ecol. 2000;9:2109–2118. doi: 10.1046/j.1365-294x.2000.01122.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee C-R, et al. On the post-glacial spread of human commensal Arabidopsis thaliana. Nat Commun. 2017;8:14458. doi: 10.1038/ncomms14458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu TT, et al. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oyama RK, et al. The shrunken genome of Arabidopsis thaliana. Plant Syst Evol. 2008;273:257–271. [Google Scholar]
- 11.Wright SI, Ness RW, Foxe JP, Barrett SCH. Genomic consequences of outcrossing and selfing in plants. Int J Plant Sci. 2008;169:105–118. [Google Scholar]
- 12.Albach DC, Greilhuber J. Genome size variation and evolution in Veronica. Ann Bot (Lond) 2004;94:897–911. doi: 10.1093/aob/mch219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trivers R, Burt A, Palestis BG. B chromosomes and genome size in flowering plants. Genome. 2004;47:1–8. doi: 10.1139/g03-088. [DOI] [PubMed] [Google Scholar]
- 14.Nasrallah ME, Yogeeswaran K, Snyder S, Nasrallah JB. Arabidopsis species hybrids in the study of species differences and evolution of amphiploidy in plants. Plant Physiol. 2000;124:1605–1614. doi: 10.1104/pp.124.4.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman-Broyles S, et al. S locus genes and the evolution of self-fertility in Arabidopsis thaliana. Plant Cell. 2007;19:94–106. doi: 10.1105/tpc.106.048199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu KK, Shimizu-Inatsugi R, Tsuchimatsu T, Purugganan MD. Independent origins of self-compatibility in Arabidopsis thaliana. Mol Ecol. 2008;17:704–714. doi: 10.1111/j.1365-294X.2007.03605.x. [DOI] [PubMed] [Google Scholar]
- 17.Tsuchimatsu T, et al. Evolution of self-compatibility in Arabidopsis by a mutation in the male specificity gene. Nature. 2010;464:1342–1346. doi: 10.1038/nature08927. [DOI] [PubMed] [Google Scholar]
- 18.Dwyer KG, et al. Molecular characterization and evolution of self-incompatibility genes in Arabidopsis thaliana: The case of the Sc haplotype. Genetics. 2013;193:985–994. doi: 10.1534/genetics.112.146787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu P, Sherman-Broyles S, Nasrallah ME, Nasrallah JB. A cryptic modifier causing transient self-incompatibility in Arabidopsis thaliana. Curr Biol. 2007;17:734–740. doi: 10.1016/j.cub.2007.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasrallah JB. Recognition and rejection of self in plant reproduction. Science. 2002;296:305–308. doi: 10.1126/science.296.5566.305. [DOI] [PubMed] [Google Scholar]
- 21.Boggs NA, Nasrallah JB, Nasrallah ME. Independent S-locus mutations caused self-fertility in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000426. doi: 10.1371/journal.pgen.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vekemans X, Poux C, Goubet PM, Castric V. The evolution of selfing from outcrossing ancestors in Brassicaceae: What have we learned from variation at the S-locus? J Evol Biol. 2014;27:1372–1385. doi: 10.1111/jeb.12372. [DOI] [PubMed] [Google Scholar]
- 23.Stebbins GL. Flowering Plants: Evolution Above the Species Level. Harvard Univ Press; Cambridge, MA: 1974. [Google Scholar]
- 24.Bechsgaard JS, Castric V, Charlesworth D, Vekemans X, Schierup MH. The transition to self-compatibility in Arabidopsis thaliana and evolution within S-haplotypes over 10 Myr. Mol Biol Evol. 2006;23:1741–1750. doi: 10.1093/molbev/msl042. [DOI] [PubMed] [Google Scholar]
- 25.Tang C, et al. The evolution of selfing in Arabidopsis thaliana. Science. 2007;317:1070–1072. doi: 10.1126/science.1143153. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu KK, Tsuchimatsu T. Evolution of selfing: Recurrent patterns in molecular adaptation. Annu Rev Ecol Evol Syst. 2015;46:593–622. [Google Scholar]
- 27.Hoffmann MH. Biogeography of Arabidopsis thaliana (L.) Heynh. (Brassicaceae) J Biogeogr. 2002;29:125–134. [Google Scholar]
- 28.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brennan AC, et al. The genetic structure of Arabidopsis thaliana in the south-western Mediterranean range reveals a shared history between North Africa and southern Europe. BMC Plant Biol. 2014;14:17. doi: 10.1186/1471-2229-14-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Désamoré A, et al. Out of Africa: Northwestwards Pleistocene expansions of the heather Erica arborea. J Biogeogr. 2011;38:164–176. [Google Scholar]
- 31.Quézel P. Analysis of the flora of Mediterranean and Saharan Africa. Ann Mo Bot Gard. 1978;65:479–534. [Google Scholar]
- 32.Tsuchimatsu T, et al. Patterns of polymorphism at the self-incompatibility locus in 1,083 Arabidopsis thaliana genomes. Mol Biol Evol. April 4, 2017 doi: 10.1093/molbev/msx122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Auton A, et al. 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osborne AH, et al. A humid corridor across the Sahara for the migration of early modern humans out of Africa 120,000 years ago. Proc Natl Acad Sci USA. 2008;105:16444–16447. doi: 10.1073/pnas.0804472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quante M. The changing climate: Past, present, future. In: Habel JC, Assmann T, editors. Relict Species: Phylogeography and Conservation Biology. Springer; Berlin: 2010. [Google Scholar]
- 37.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tierney JE, deMenocal PB. Abrupt shifts in Horn of Africa hydroclimate since the Last Glacial Maximum. Science. 2013;342:843–846. doi: 10.1126/science.1240411. [DOI] [PubMed] [Google Scholar]
- 39.Timmermann A, Friedrich T. Late Pleistocene climate drivers of early human migration. Nature. 2016;538:92–95. doi: 10.1038/nature19365. [DOI] [PubMed] [Google Scholar]
- 40.Ossowski S, et al. The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science. 2010;327:92–94. doi: 10.1126/science.1180677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerling TE, Hay RL. An isotopic study of paleosol carbonates from Olduvai Gorge. Quat Res. 1986;25:63–78. [Google Scholar]
- 42.deMenocal PB. African climate change and faunal evolution during the Pliocene-Pleistocene. Earth Planet Sci Lett. 2004;220:3–24. [Google Scholar]
- 43.Hohmann N, Wolf EM, Lysak MA, Koch MA. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell. 2015;27:2770–2784. doi: 10.1105/tpc.15.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brandvain Y, Kenney AM, Flagel L, Coop G, Sweigart AL. Speciation and introgression between Mimulus nasutus and Mimulus guttatus. PLoS Genet. 2014;10:e1004410. doi: 10.1371/journal.pgen.1004410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brandvain Y, Slotte T, Hazzouri KM, Wright SI, Coop G. Genomic identification of founding haplotypes reveals the history of the selfing species Capsella rubella. PLoS Genet. 2013;9:e1003754. doi: 10.1371/journal.pgen.1003754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foxe JP, et al. Recent speciation associated with the evolution of selfing in Capsella. Proc Natl Acad Sci USA. 2009;106:5241–5245. doi: 10.1073/pnas.0807679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guo YL, et al. Recent speciation of Capsella rubella from Capsella grandiflora, associated with loss of self-incompatibility and an extreme bottleneck. Proc Natl Acad Sci USA. 2009;106:5246–5251. doi: 10.1073/pnas.0808012106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sicard A, Lenhard M. The selfing syndrome: A model for studying the genetic and evolutionary basis of morphological adaptation in plants. Ann Bot (Lond) 2011;107:1433–1443. doi: 10.1093/aob/mcr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnston JS, et al. Evolution of genome size in Brassicaceae. Ann Bot (Lond) 2005;95:229–235. doi: 10.1093/aob/mci016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mable BK, et al. What causes mating system shifts in plants? Arabidopsis lyrata as a case study. Heredity (Edinb) 2017;118:110. doi: 10.1038/hdy.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaspar F, Kühl N, Cubasch U, Litt T. A model-data comparison of European temperatures in the Eemian interglacial. Geophys Res Lett. 2005;32:L11703. [Google Scholar]
- 52.Kukla GJ, et al. Last interglacial climates. Quat Res. 2002;58:2–13. [Google Scholar]
- 53.Henn BM, Cavalli-Sforza LL, Feldman MW. The great human expansion. Proc Natl Acad Sci USA. 2012;109:17758–17764. doi: 10.1073/pnas.1212380109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mallick S, et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pagani L, et al. Genomic analyses inform on migration events during the peopling of Eurasia. Nature. 2016;538:238–242. doi: 10.1038/nature19792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.deMenocal PB, Stringer C. Human migration: Climate and the peopling of the world. Nature. 2016;538:49–50. doi: 10.1038/nature19471. [DOI] [PubMed] [Google Scholar]
- 57.Tzedakis PC, Crucifix M, Mitsui T, Wolff EW. A simple rule to determine which insolation cycles lead to interglacials. Nature. 2017;542:427–432. doi: 10.1038/nature21364. [DOI] [PubMed] [Google Scholar]
- 58.deMenocal PB. Plio-Pleistocene African climate. Science. 1995;270:53–59. doi: 10.1126/science.270.5233.53. [DOI] [PubMed] [Google Scholar]
- 59.Dupont L. Orbital scale vegetation change in Africa. Quat Sci Rev. 2011;30:3589–3602. [Google Scholar]
- 60.Cohmap M. COHMAP MEMBERS Climatic changes of the last 18,000 years: Observations and model simulations. Science. 1988;241:1043–1052. doi: 10.1126/science.241.4869.1043. [DOI] [PubMed] [Google Scholar]
- 61.Kuper R, Kröpelin S. Climate-controlled Holocene occupation in the Sahara: Motor of Africa’s evolution. Science. 2006;313:803–807. doi: 10.1126/science.1130989. [DOI] [PubMed] [Google Scholar]
- 62.Ossowski S, et al. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kawamura K, et al. Northern Hemisphere forcing of climatic cycles in Antarctica over the past 360,000 years. Nature. 2007;448:912–916. doi: 10.1038/nature06015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.