Highlight
The low molecular weight LOW SULPHUR UPREGULATED (LSU) network hubs mediate stomatal closure during several abiotic stress conditions by stimulating reactive oxygen species production; bacterial virulence effectors interfere with this function.
Keywords: Abiotic stress, Arabidopsis thaliana, biotic stress, combinatorial stress, LOW SULPHUR UPREGULATED, LSU1
Abstract
In natural environments, plants often experience different stresses simultaneously, and adverse abiotic conditions can weaken the plant immune system. Interactome mapping revealed that the LOW SULPHUR UPREGULATED (LSU) proteins are hubs in an Arabidopsis protein interaction network that are targeted by virulence effectors from evolutionarily diverse pathogens. Here we show that LSU proteins are up-regulated in several abiotic and biotic stress conditions, such as nutrient depletion or salt stress, by both transcriptional and post-translational mechanisms. Interference with LSU expression prevents chloroplastic reactive oxygen species (ROS) production and proper stomatal closure during sulphur stress. We demonstrate that LSU1 interacts with the chloroplastic superoxide dismutase FSD2 and stimulates its enzymatic activity in vivo and in vitro. Pseudomonas syringae virulence effectors interfere with this interaction and preclude re-localization of LSU1 to chloroplasts. We demonstrate that reduced LSU levels cause a moderately enhanced disease susceptibility in plants exposed to abiotic stresses such as nutrient deficiency, high salinity, or heavy metal toxicity, whereas LSU1 overexpression confers significant disease resistance in several of these conditions. Our data suggest that the network hub LSU1 plays an important role in co-ordinating plant immune responses across a spectrum of abiotic stress conditions.
Introduction
In nature and in farm fields, plants commonly encounter different forms of abiotic and biotic stress simultaneously. Due to global warming and the increasing need to farm on suboptimal soil, the incidence of combinatorial stress conditions is likely to increase. In molecular studies, stress responses are usually investigated in isolation. Although these approaches proved powerful for elucidating homeostatic response mechanisms for a variety of isolated stress conditions, it is becoming increasingly clear that the molecular responses to multiple simultaneous stressors differ from those to individual stressors. On a systems level, transcriptional profiling studies have demonstrated that the response to combinatorial stress qualitatively differs from the additive combination of single-stress responses (Atkinson et al., 2013; Prasch and Sonnewald, 2013; Rasmussen et al., 2013; Suzuki et al., 2014). In particular, lack of nutrients, water shortage, or high salinity can weaken the plant immune system (Triky-Dotan et al., 2005; Amtmann et al., 2008; Al-Sadi et al., 2010; You et al., 2011; Kissoudis et al., 2015).
Bacterial pathogens such as Pseudomonas syringae pv. tomato (Pst) can enter the apoplast through stomata, which mediate water and gas exchange and play a central role in many abiotic stress responses. During the first steps of infection, plants recognize conserved bacterial protein patterns such as flagellin and activate a primary immune response known as pattern-triggered immunity (PTI), which includes increased reactive oxygen species (ROS) production, callus deposition, and other defence mechanisms (Jones and Dangl, 2006). Adapted pathogens can overcome initial immune responses by delivering virulence effector proteins into host cells that reach different subcellular compartments and interfere with PTI. Plants conversely possess intracellular receptors to detect the presence of virulence effectors by direct or indirect recognition, and trigger enhanced immune responses resulting in effector-triggered immunity (ETI). Beyond the idealized zig-zag model of the plant immune system (Jones and Dangl, 2006), it is clear that the perpetual arms race between pathogens and their hosts has resulted in a highly complex quantitative interplay of defence and counter-defence mechanisms.
To identify host interaction partners of virulence effectors and thus potentially novel components of the plant immune system, we recently conducted large-scale protein interaction mapping experiments between Arabidopsis proteins and virulence effectors from three evolutionarily distant pathogens. We discovered that effectors from Pst, the oomycete Hyaloperonospora arabidopsidis, and the fungus Golovinomyces orontii converge onto common host proteins, several of which are hubs in the plant protein network (Arabidopsis Interactome Mapping Consortium, 2011; Mukhtar et al., 2011; Wessling et al., 2014). Hubs in binary interaction networks were previously shown to mediate and integrate diverse cellular processes (Han et al., 2004; Yu et al., 2008). Moreover, we observed a correlation between the extent of effector convergence onto host targets and the manifestation of immune phenotypes in the corresponding genetic nulls (Mukhtar et al., 2011; Wessling et al., 2014). Incorporating population genetic data, we further discovered that protein products of Arabidopsis genes that experience the strongest balancing and positive selection preferentially interact with effector-targeted proteins (Wessling et al., 2014). Thus, despite the fact that no immune function had previously been described for most of the new effector targets, our collective data suggest that these are important in host–pathogen interactions.
Among the highly targeted host proteins are Arabidopsis LOW SULPHUR (S) UPREGULATED (LSU) proteins. Arabidopsis thaliana has four members of the LSU protein family (LSU1–LSU4), which can be found in all higher land plants. LSU genes were named for their strong transcriptional induction in response to S deficiency (–S) (Maruyama-Nakashita et al., 2003), and a general function for the Nicotiana benthamiana LSU orthologue UP9C during –S stress was demonstrated (Lewandowska et al., 2010). However, the molecular and precise physiological functions of LSU proteins remain to be clarified. The observation that LSU family members are intensely targeted by evolutionarily diverse pathogens (Mukhtar et al., 2011; Wessling et al., 2014) suggests a hitherto unknown function in plant immunity.
Here we report that LSU1 and LSU2 levels are increased by transcriptional and post-translational mechanisms in a variety of abiotic stress conditions. In response to –S or salt stress, LSU gene expression is required for ROS production in guard cell chloroplasts and subsequent stomatal closure. Using genetic and biochemical approaches we demonstrate that LSU1, which is expressed in guard cells, physically interacts with the iron (Fe)-dependent superoxide dismutase (SOD) FSD2 and can activate its enzymatic activity. Virulence effectors from P. syringae interfere with the function and subcellular localization of LSU1. Correspondingly, in conditions of abiotic stress, we observe a moderate enhanced disease susceptibility (EDS) phenotype in seedlings with reduced LSU levels and a corresponding enhanced disease resistance (EDR) phenotype in LSU1 overexpressors. Together we uncover a physiological function for LSU proteins during combinatorial biotic and abiotic stress and propose a working mechanism of action for LSU1.
Materials and methods
Plant cultivation and manipulation
Arabidopsis thaliana Col-0 was cultivated on half-strength Murashige and Skoog (1/2 MS) medium or with S compounds substituted by equivalent chloride salts and supplemented with 0.5% (w/v) MES and Plant Preservative Mixture at 0.04% (v/v; Plant Cell Technology) under long-day conditions (16 h light–21 °C/8 h dark–16 °C). For stress treatments 7-day-old wild-type (WT) seedlings grown on 1/2 MS plates were transferred to liquid 1/2 MS medium without S or Fe or with 10 µM Cu, 2 mM DTT, 150 mM NaCl, or pH 8 for 1 d. For ROS detection and P. syringae assays, seedlings were grown on standard medium, without S, or in the presence of 25 µM Cu or 50 mM NaCl. Stable transgenic lines were generated with the Agrobacterium tumefaciens GV3101 (pMP90) strain by floral dipping (Koncz and Schell, 1986; Clough and Bent, 1998). The fsd2-2 mutant corresponds to the SALK line SALK_080457C (ID NASC: N663088) (Myouga et al., 2008).
Gene expression analysis
RNA was prepared with the NucleoSpin RNA kit (Macherey-Nagel) following the manufacturer’s instructions. RNA quantity and quality was evaluated by spectrometry and in agarose gels prior to reverse transcription to cDNA using M-MLV Reverse Transcriptase, RNase H Minus, Point Mutant (Promega). Quantitative PCRs (qPCRs) were conducted using the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) in a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad) using an initial cycle at 95 °C for 3 min and 40 cycles consisting of 95 °C for 10 s, 58 °C for 20 s, and 72 °C for 20 s. To assess expression changes of LSU genes, the measured levels were first normalized to ACTIN2 (ACT2) and ELONGATION FACTOR1 (EF1) levels, except for salt and pH 8 treatments where these controls showed strong regulation and where ACT8 and UBIQUITIN10 (UBQ10) were used instead. The normalized expression levels are represented as fold change in expression relative to control conditions.
Protein analysis and biochemical fractionation
Total protein extracts were prepared in 100 mM NaCl; 50 mM Tris–HCl pH 7.5; 0.5% (v/v) Triton X-100; 1 mM DTT; 1× Complete Protease Inhibitor Cocktail Tablet (Roche), and spectrophotometrically quantified using ROTIQUANT (Carl Roth). A 10–20 µg aliquot of protein extract was loaded onto an SDS-polyacrylamide gel, blotted on nitrocellulose membranes, probed with antibodies listed in Supplementary Table S3 at JXB online, and developed with the SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) in a Luminescent Image Analyzer LAS4000 System (Fujifilm). Biochemical fractionation was conducted as previously described (Garcia-Molina et al., 2014) from 7-day-old CaMV35S:HA-LSU1/2 seedlings in a Sorvall™ MTX 150 Micro-Ultracentrifuge (Thermo Scientific) with a S55A2 rotor. Fractions were assayed by western blots with antibodies anti-H3, anti-UGPase, anti-V-ATPase, and anti-haemagglutinin (HA) (Supplementary Table S3).
Subcellular localization experiments
CaMV35S:GFP-LSU1/2 and CaMV35S:GFP (Cutler et al., 2000) transgenic seedlings were examined in a FLUOVIEW FV1000 confocal laser microscope (Olympus) using specific green fluorescent protein (GFP) and chlorophyll filters.
Generation of constructs and amiRNA lines
Full-length ORFs were available as GATEWAY™ entry clones (Arabidopsis Interactome Mapping Consortium, 2011). To generate constructs for GFP and HA fusion proteins, entry clones were transferred into pMDC43 or pALLIGATOR2, respectively, using GATEWAY recombination reactions (Life Technologies) (Curtis and Grossniklaus, 2003; Bensmihen et al., 2004). For bimolecular fluorescence complementation (BiFC) assays, pYFN43 and pYFC43 were used (Belda-Palazón et al., 2012). Translational fusions to glutathione S-transferase (GST) and maltose-binding protein (MBP) were generated with pGEX6 and pMAL-DEST plasmids (Braun et al., 2002), respectively. Artificial miRNAs (amiRNAs) targeting LSU1–LSU4 were designed as in Schwab et al. (2006) with the oligonucleotides listed in Supplementary Table S2, GATEWAY-recombined into pDONR207 entry vector (Life Technologies), and subsequently into pALLIGATOR3 (Bernaudat et al., 2011).
Stomatal aperture and water loss determination
To determine stomatal aperture, 12-day-old Arabidopsis seedlings grown on S-sufficient or -deficient media were transferred to the indicated media supplemented with 10 µM abscisic acid (ABA; Sigma-Aldrich), or the corresponding mock treatment, for 3 h. First leaflets were abaxially observed by light microscopy, and the ratio between the width and length of ostiols (Rwl) was measured. Water loss of 12-day-old seedlings was inferred from the change in fresh weight over 60 min after removing seedlings from plates. For each biological replicate, at least 30 stomata were analysed.
ROS production in guard cells
ROS production was traced as previously described (Miao et al., 2006; Zou et al., 2015) with modifications. Briefly, detached first leaflets of 14-day-old seedlings were incubated for 10 min on 50 µM H2DCFDA (Life Technologies), washed twice with water, and observed either under laser confocal (Olympus FV1000) or BX61 epifluorescence microscopy (Olympus) with a GFP band-pass filter. 3,3’-Diaminobenzidine (DAB) staining was used to detect H2O2 specifically. To this end, 14-day-old seedlings were incubated with 1 ml of DAB solution [1 mg ml–1 (w/v) DAB; 0.05% (v/v) Tween-20; 10 mM Na2HPO4] and vacuum-infiltrated for 5 min in the dark. Samples were left at room temperature for 5 h protected from the light, and de-stained with two washes with ethanol:acetic acid:glycerol (3:1:1) solution prior to microscopy.
Production of recombinant proteins
Escherichia coli Rosetta™ (Novagen) transformants were grown to log phase (OD600 0.4–0.8) in LB medium supplemented with antibiotics and with 0.2% glucose for MBP constructs; protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 28 °C. Pelleted cells were resuspended in 1× phospahte-buffered saline (PBS) supplemented with 1× Complete Protease Inhibitor Cocktail Tablet (Roche), sonicated, and lysates were rotated with either Amylose Resin (New England Biolabs) or Protino Glutathione Agarose 4B (Macherey-Nagel) for 2 h at 4 °C. The matrix was washed twice with 3 vols of PBS or MBP wash buffer (200 mM NaCl; 20 mM Tris–HCl pH 7.4; 1 mM EDTA; 1 mM DTT), respectively, and eluted with 2 vols of 10 mM Tris–HCl pH 8, 10 mM glutathione, or MBP wash buffer supplemented with 10 mM maltose.
Protein–protein interaction assays
Yeast two-hybrid (Y2H) experiments were conducted with the indicated ORFs cloned into pDEST-AD or pDEST-DB as described (Dreze et al., 2010). BiFC assays were carried out by agro-injection of pYFN43-LSU1/2 and pYFC43-FSD2, pYFC43-PTEN1, or pYFC43-LSU1/2 in N. benthamiana epidermal cells at OD600 0.1 and observed after 1–2 d under epifluorescence microscopy (Olympus BX61) with a yellow fluorescent protein (YFP) band-pass filter. For in vitro pull-down assays, Protino Glutathione Agarose 4B (Macherey-Nagel) alone or coated with GST–FSD2 was incubated with equimolar combinations of purified MBP–LSU1 and/or MBP–NIMIN1, MBP–AvrXccC, MBP–AvrB2, or MBP–HopR1 in a final volume of 100 µl of PBS and incubated with gentle rotation for 3 h at 4 °C. Beads were washed four times with 3 vols of PBS each and eluted with 10 mM Tris–HCl pH 8, 10 mM glutathione. MBP–LSU1 pull-downs were analysed by western blot using anti-MBP antibodies (Supplementary Table S3).
Determination of superoxide dismutase enzymatic activity
SOD activity was determined as described by Kuo et al. (2013) with modifications. Briefly, samples were incubated with 1 ml of reaction buffer [5 mM Nitro Blue Tetrazolium (NBT); 50 mM PBS pH 7.8; 10 mM methionine; 0.5 mM EDTA; and 20 µM riboflavin] for 30 min in the dark, and then illuminated for 10–15 min. SOD activity was monitored by measuring inhibition of NBT–diformazan formation at A560. Nicotiana benthamiana samples were prepared in extraction buffer [50 mM PBS pH 7.8; 0.05% (v/v) NP-40; 2% (w/v) polyvinylpolypyrrolidone (PVPP); 1 mM EDTA]. For in vitro assays, purified recombinant GST–FSD2 (3 µg) or GST–FSD1 (1.5 µg) were incubated with the indicated recombinant proteins in 50 µl of PBS at room temperature for 30 min prior to enzymatic activity determination.
Pseudomonas syringae infection assays
Infection assays were conducted as previously described in Ishiga et al. (2011). Pseudomonas syringae pv. tomato DC3000, COR–, or hrcC was cultured overnight at 28 °C in NYGA medium [0.5% (w/v) bactopeptone, 0.3% (w/v) yeast extract; 2% (v/v) glycerol] supplemented with antibiotics. Infection assays were performed on 12-day-old seedlings grown in vitro by flooding with bacterial suspension at OD600 0.01 (5 × 106 CFU ml–1) for 3 min. Pseudomonas syringae density was determined after 3 d. For this, shoots were harvested, disinfected with 5% (w/w) H2O2, and rinsed twice with water. Samples were ground and spotted in serial dilutions on selective medium (LB supplemented with 50 mg l–1 rifampicin and 30 mg l–1 kanamycin for all strains and also 30 mg l–1 spectinomycin for COR–) to obtain colony counts after 1 d of incubation at 28 °C. To ensure equal infection rates, WT and amiR-LSU lines were cultivated together.
Miscellaneous methods
Student’s t-test was used to determine significant differences from control lines or treatments as indicated. Fluorescence and band intensities were quantified with the image-processing package Fiji. Sequence analysis and multiple alignments were performed with the CLC Sequence Viewer 7 (Qiagen) software. Protein distance measure is indicated according to Jukes and Cantor (1969).
Results
Transcriptional and post-translational regulation of LSU levels in abiotic stress
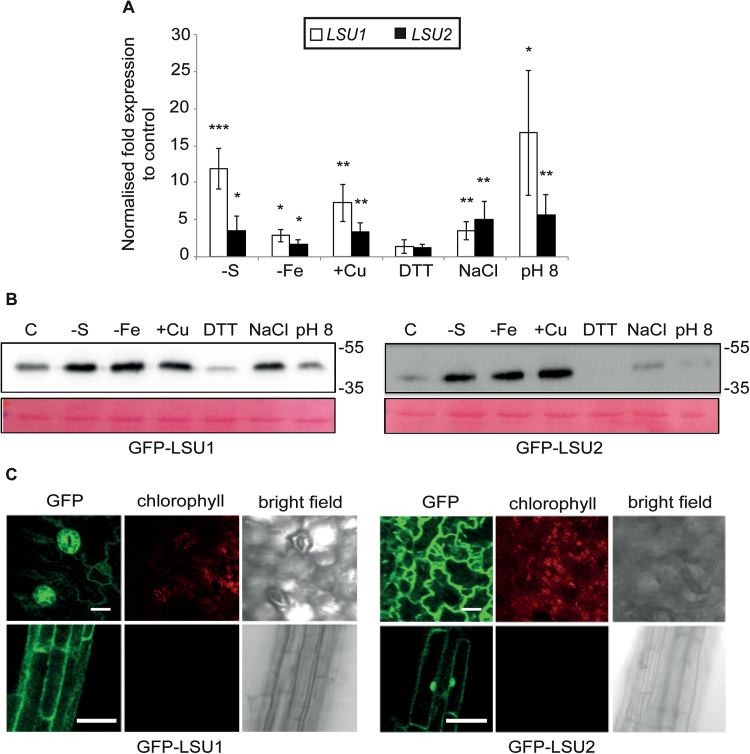
Hubs in binary interaction networks were shown to co-ordinate different cellular processes (Han et al., 2004; Yu et al., 2008). Therefore, we wondered whether LSU genes might also function in abiotic stress conditions other than –S conditions (Nikiforova et al., 2003; Arabidopsis Interactome Mapping Consortium, 2011; Hawkesford et al., 2012. We investigated transcript and protein abundance in several abiotic stress conditions selecting LSU1 and LSU2 (LSU1/2) as representative members of the family (Supplementary Fig. S1A–C). LSU1/2 transcript levels were determined by qPCR in 7-day-old WT seedlings transferred to liquid media representing different abiotic stress conditions. In addition to the induction by –S, LSU transcript levels increased between 2- and 15-fold in WT seedlings exposed to increased salinity, iron (Fe) deficiency, copper (Cu) excess, or basic pH; no change was observed in response to DTT (Fig. 1A). To investigate potential post-translational regulation in the absence of transcriptional changes, we used transgenic lines expressing LSU1/2 fusion proteins from ectopic promoters. CaMV35S:GFP-LSU1/2 transgenic seedlings accumulated more GFP–LSU1 upon –S, Fe deprivation, Cu excess, and salt excess in comparison with control treatments, whereas increased GFP–LSU2 amounts were found on –S, Fe deficiency, and high Cu, but not salt excess. For the rest of the treatments, no differences or even reduced levels were observed (Fig. 1B). The LSU1 and LSU2 responsiveness towards several abiotic stress conditions suggests that LSU proteins could mediate plant responses to a variety of abiotic stress conditions.
Fig. 1.
Characterization of LSU1 and LSU2. (A and B) LSU1 and LSU2 expression is induced under certain abiotic stress conditions. (A) Expression of LSU1 and LSU2 transcripts in different abiotic stress conditions was determined by qPCR and expressed as the ratio between experimental and control treatments. Error bars correspond to the SD of three biologically independent measurements, and significant differences were assessed by the Student’s t-test relative to control treatments (*P<0.05; **P<0.01; ***P<0.001). (B) α-GFP western blots on total lysates of GFP–LSU1- and GFP–LSU2-expressing seedlings grown as indicated. Ponceau staining shows the large RuBisCO subunit and indicates equal loading; molecular weight marker sizes are indicated in kDa. (C) Representative confocal images of cotyledons and roots of GFP–LSU1 and GFP–LSU2 seedlings grown on –S media showing the GFP signal, chlorophyll autofluorescence, and bright field are provided (scale bar=50 µm).
LSU1 and LSU2 display differential subcellular and tissue localizations
Despite the high similarity of LSU proteins (Supplementary Fig. S1A–C), the interaction partners of LSU1/2 proteins previously identified in the Arabidopsis interactome only partially overlap (Arabidopsis Interactome Mapping Consortium, 2011). We wondered whether the localizations of LSU proteins further support a functional specialization and interrogated the tissue and subcellular localization of LSU1/2 in planta. A biochemical fractionation following heterologous expression in N. benthamiana showed that LSU1/2 can be found in nuclear, cytosolic, and microsmal fractions (Supplementary Fig S2A, B). We therefore investigated the tissue and subcellular localization of GFP–LSU1/2 proteins in Arabidopsis. Microscopically, GFP–LSU1 lines display a diffuse fluorescent signal in the root cytoplasm/membrane (Fig. 1C, lower panel, left). In leaflets of 5-day-old seedlings, GFP–LSU1 was prominently detected in guard cells, suggesting a function in stomatal regulation (Fig. 1C, upper panel, left). GFP–LSU2 exhibited a diffuse but ubiquitous cytosolic signal in leaves and roots, and also illuminated nuclei in root cells (Fig. 1C, right). Soluble GFP alone showed an overall similar pattern, which is expected given the small size of LSU proteins and GFP. Nonetheless, clear differences can be seen especially in the strong stomatal localization of GFP–LSU1 compared with pavement cells and more prominent nuclear localization of LSU2 in root cells (Fig. 1C; Supplementary Fig. S2C). The results support the conclusion that LSU proteins can localize to multiple cell compartments and suggest at least partially specialized and tissue-specific roles for LSU1 and LSU2. Importantly, the data point towards a likely function of LSU1 in guard cells. This conclusion is supported by markedly reduced LSU1 transcript levels in cotyledons of the stomataless mutants spch-3 and mute-3 (de Marcos et al., 2015).
Abrogation of LSU expression constrains stomatal movements upon sulphur deficiency
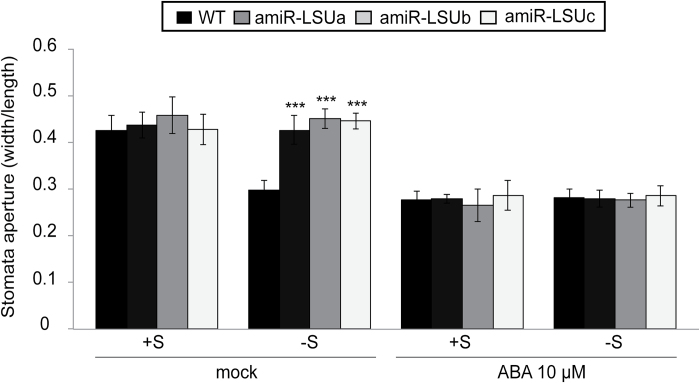
No T-DNA mutants were available for LSU1 and LSU3, and the paired duplications of the LSU1/3 and LSU2/4 paralogues further challenge generation of higher order mutants. Hence, to characterize the LSU genes phenotypically and avoid problems due to potential redundancy, three different amiRNAs targeting all LSU family members at different positions were designed and used to generate three amiR-LSU lines (amiR-LSUa–c; Supplementary Fig. S3A). Transcript levels of the four LSU genes were determined by qPCR in seedlings grown on –S. Compared with WT lines, LSU1–LSU3 transcripts were reduced by >80% and LSU4 levels by 50% in the amiR-LSUa–c (Supplementary Fig. S3B). Based on these results, the amiR-LSU lines were considered knock-down lines and used for subsequent phenotypic studies.
The amiR-LSU lines did not exhibit obvious phenotypes on soil or in vitro culture in comparison with the WT (data not shown). Since GFP–LSU1 localized to guard cells and stomatal closure is a physiological response to –S (Terry, 1976; Karmoker et al., 1991), guard cell movements in the amiR-LSU lines were characterized. The apertures of abaxial stomata of first leaflets were estimated as the ratio between the width and length of the ostiols (Rwl). Stomata were mostly open (Rwl ~0.45) in both WT and amiR-LSUa–c lines grown on S-sufficient (+S) media. S deficiency, however, led to partial stomatal closure in WT lines indicated by an ~30% decrease in Rwl. In contrast, stomata remained open in amiR-LSU lines in –S media (Fig. 2A; Supplementary Fig. S4A), indicating that stomatal closure during –S conditions is an LSU-dependent process. In agreement with these observations, the amiR-LSUa–c lines exhibited a more severe water loss on –S media compared with the WT (Supplementary Fig. S4B; Supplementary Table S1). Notably, stomatal closure in response to exogenous ABA was not impaired in either condition, indicating that ABA-dependent stomatal closure is not affected in the amiR-LSU lines (Fig. 2). Thus, LSU proteins, probably the prominently guard cell-localized LSU1, participate in the reg4ulation of stomatal closure in response to –S.
Fig. 2.
Abrogation of LSU constrains stomatal closure during abiotic stress conditions. Twelve-day-old WT and amiR-LSUa–c seedlings grown in +S or –S conditions were transferred to the same media supplemented with ABA or mock treated for 3 h. Stomatal aperture was quantified as the ratio of ostiol width to length (Rwl). Error bars correspond to the SD of ≥3 biologically independent measurements (n ≥30 stomata), and significant differences were assessed by the Student’s t-test relative to the WT in the same conditions (***P<0.001).
LSU down-regulation compromises ROS production in guard cells upon certain abiotic stress conditions
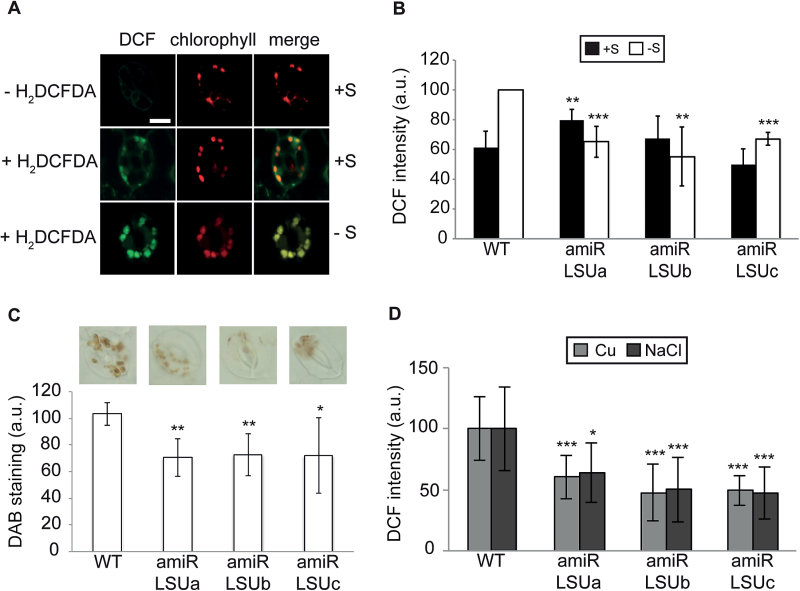
Guard cell movements are controlled by a complex interplay of hormones and secondary signals (Daszkowska-Golec and Szarejko, 2013; Arnaud and Hwang, 2014), which culminate in an increase of H2O2 that triggers ion channel opening and a change in guard cell turgor (Pei et al., 2000; Zhang et al., 2001; Desikan et al., 2004; Wang et al., 2012). To address whether ROS production during –S stress is impaired in guard cells, we pharmacologically monitored ROS production using the ROS-mediated conversion of 2’,7’-dichlorodihydro-fluorescein diacetate (H2DCFDA) into fluorescent 2’,7’-dichlorofluorescein (DCF) (Brandt and Keston, 1965). In +S conditions, DCF fluorescence in guard cells exhibited a diffuse pattern partly co-localizing with chloroplast autofluorescence (Fig. 3A). In contrast, in –S conditions, DCF fluorescence intensity nearly doubled (~40%) compared with +S and exclusively localized to speckles overlapping the chlorophyll autofluorescence (Fig. 3A, B). In the amiR-LSU lines, the –S-induced stimulation of ROS production was either dramatically reduced (amiR-LSUc) or completely abrogated (amiR-LSUa, b; Fig. 3B; Supplementary Fig. S4C). Among the various ROS species, H2O2 is of particular interest due to its varied roles as a defence and signalling molecule. To test if the observed differences in guard cell ROS production could be attributed to an H2O2 accumulation, DAB staining on S-deprived seedlings was conducted and the signal intensity per guard cell pair quantified. As depicted in Fig. 3C, the amiR-LSUa–c lines showed a reduced DAB staining pattern (~30%) compared with the WT. In addition, we tested whether ROS production in the amiR-LSU lines is affected in conditions other than –S. Consistent with the induction of LSU transcripts in response to high salt or Cu, the amiR-LSU lines exhibited 40–50% lower DCF signal intensity compared with the WT (Fig. 3D). These data demonstrate that LSU proteins stimulate ROS production, among them H2O2, in guard cell chloroplasts in response to different abiotic stress conditions.
Fig. 3.
Abrogation of LSU impairs ROS production during abiotic stress conditions. (A) Reactive oxygen species (ROS) in chloroplast guard cells increase during sulphur starvation. Representative confocal pictures of the abaxial guard cell stomata of WT seedlings grown under +S or –S conditions showing DCF and chlorophyll fluorescence and a merged signal (scale bar=10 µm). (B) Quantification of relative DCF fluorescence intensity of stomata (n ≥30) of WT and amiR-LSUa–c seedlings grown in +S and –S conditions. (C) Hydrogen peroxide production in guard cells of amiR-LSU lines is compromised during sulphur starvation. Representative pictures of 3,3’-diaminobenzidine (DAB) staining in guard cells of seedlings grown on –S and quantification of signal intensity per guard cell pair. (D) ROS production in chloroplast guard cells is compromised during copper and salt treatments. Quantification of DCF signal intensity in guard cells of seedlings grown on 25 µM CuCl2 and 50 mM NaCl, respectively. In all cases, error bars correspond to the SD of ≥3 biologically independent measurements, and significant differences were assessed by the Student’s t-test relative to the WT in the same conditions (*P<0.05; **P<0.01; ***P<0.001).
LSU1 physically interacts with and activates the iron superoxide dismutase FSD2
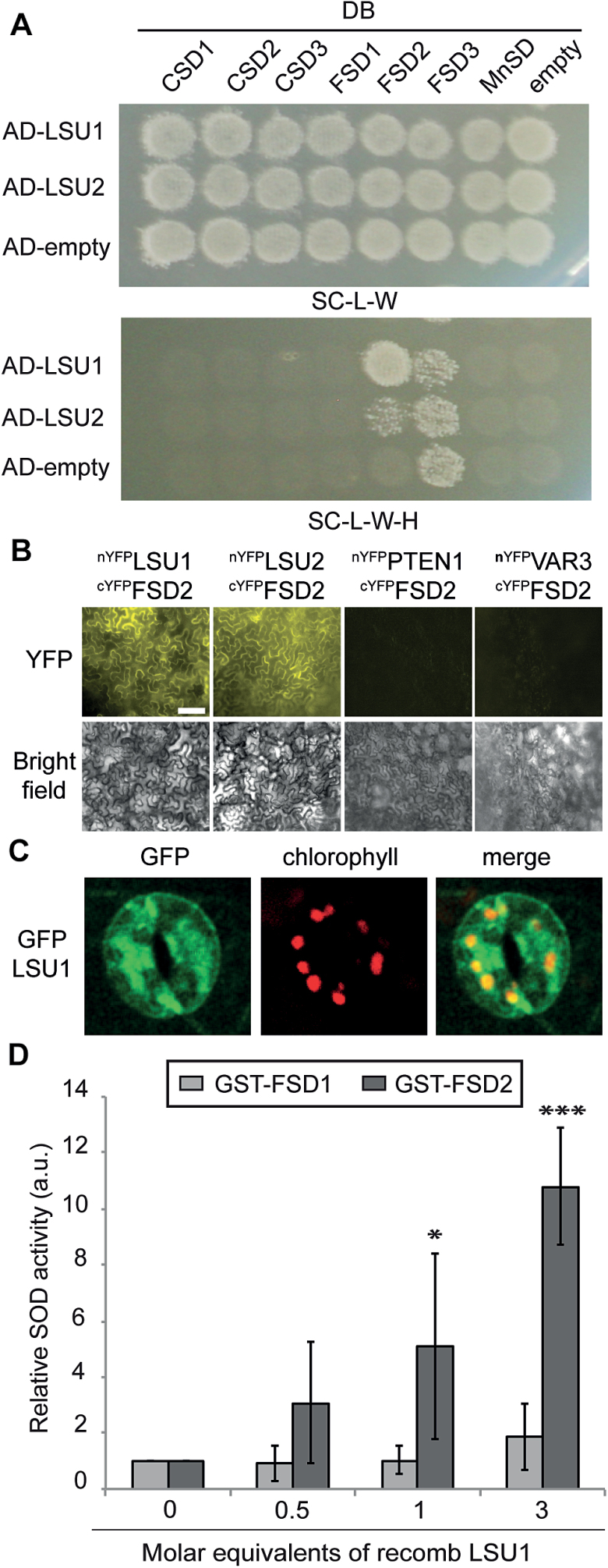
H2O2 mainly occurs in cells as a result of the enzymatic conversion of the superoxide radical (O2–) by SODs. Arabidopsis possesses three different SOD isoforms classified according to their respective metal cofactors: manganese (MnSOD), Cu/zinc (CSD1/2), and Fe (FSD1–FSD3) (Kliebenstein et al., 1998). Of these, FSD2, FSD3, and CSD2 localize to plastids and may therefore mediate the H2O2 production during –S stress, whereas the others function in different cell compartments. We previously identified FSD2 as an interaction partner of LSU1 and LSU2 (Arabidopsis Interactome Mapping Consortium, 2011) and aimed to verify this interaction; to ascertain its specificity, we also included all other Arabidopsis SOD proteins in this verification experiment. Y2H experiments using GAL4 activation domain (AD)–LSU1/2 and all Arabidopsis SODs fused to the GAL4 DNA-binding domain (DB) confirmed the specific interaction of LSU1/2 with the chloroplast-localized FSD2 (Fig. 4A). Due to self-activation of FSD3, a possible association with LSU1/2 could not be tested (Fig. 4A). BiFC in N. benthamiana demonstrates that LSU1 and LSU2 can interact with FSD2 in planta (Fig. 4B). Additional support for the interactions was obtained in biochemical pull-down experiments (see below). Moreover, upon closer inspection of confocal images, we found that in –S conditions GFP–LSU1 partly co-localized with chlorophyll autofluorescence in guard cells, suggesting plastidial localization of LSU1 in these conditions [Pearson correlation coefficient (PCC) 0.52; Fig. 4C]. Thus, LSU1 can physically interact with FSD2 in vitro and in vivo.
Fig. 4.
LSU1 and LSU2 physically and functionally interact with FSD2. (A) Specific interaction of AD–LSU1 and AD–LSU2 with DB–FSD2 by Y2H. The top panel shows diploid yeasts and successful mating; the bottom panel shows growth on selective media indicating interactions of AD–LSU1/2 with DB–FSD2 and autoactivation of DB–FSD3. (B) Bimolecular fluorescence complementation (BiFC). Nicotiana benthamiana epidermal leaves transiently co-expressing cYFP–FSD2 and nYFP–LSU1/2 restore YFP fluorescence, whereas co-infiltration of cYFP–FSD2 and nYFP fusions with the cytosolic Arabidopsis PHOSPHATASE AND TENSIN HOMOLOG DELETED ON CHROMOSOME TEN 1 (PTEN1, AT5G39400) and with the chloroplast-localized nYFP–VARIEGATED 3 (VAR3, AT5G17790) do not (scale bar=100 µm). (C) LSU1 partly co-localizes with guard cell chloroplasts. Representative confocal images of guard cells in first leaflets of –S-grown GFP–LSU1 seedlings. Shown are GFP signal (left), chlorophyll autofluorescence (middle), and merged channels (right) (scale bar=25 µm). (D) Relative in vitro SOD activity of recombinant GST–FSD1 and GST–FSD2, respectively, in the presence of increasing concentrations of MBP–LSU1. Error bars correspond to the SD of three independent experiments, and significant differences from controls were assessed using Student’s t-test (*P<0.05; ***P<0.001).
Given that reduced LSU levels lead to decreased ROS production in guard cells under abiotic stress conditions, we wondered if LSU proteins could directly stimulate FSD2 enzyme activity. In vivo activity measurements using N. benthamiana leaves transiently expressing GFP–LSU1 and GFP–FSD2 together or alone showed only incremental stimulation of FSD2 activity by LSU1 (Supplementary Fig. S5A, B). To eliminate confounding effects by endogenous proteins, we purified recombinant proteins and investigated the effect of LSU1 and LSU2 on FSD2 activity in vitro, using the non-interacting FSD1 to assess specificity (Fig. 5A). The functionality of GST-tagged FSD1 and FSD2 purified from Escherichia coli was confirmed by plotting SOD activity against the enzyme concentration, which yielded classic saturation curves (Supplementary Fig. S5C). Non-saturating enzyme concentrations were then incubated with increasing amounts of MBP–LSU1, and SOD activity was determined. Recombinant LSU1 specifically induced FSD2 enzyme activity, but not the activity of the non-interacting FSD1 (Fig. 4D). A similar trend was observed upon addition of GST–LSU2 but not GST alone or MBP fused to the unrelated NIM1-INTERACTING 1 (NIMIN1) (Supplementary Fig. S5D). Thus, mechanistically LSU1 and LSU2 can stimulate H2O2 production by direct interaction with and activation of FSD2.
Fig. 5.
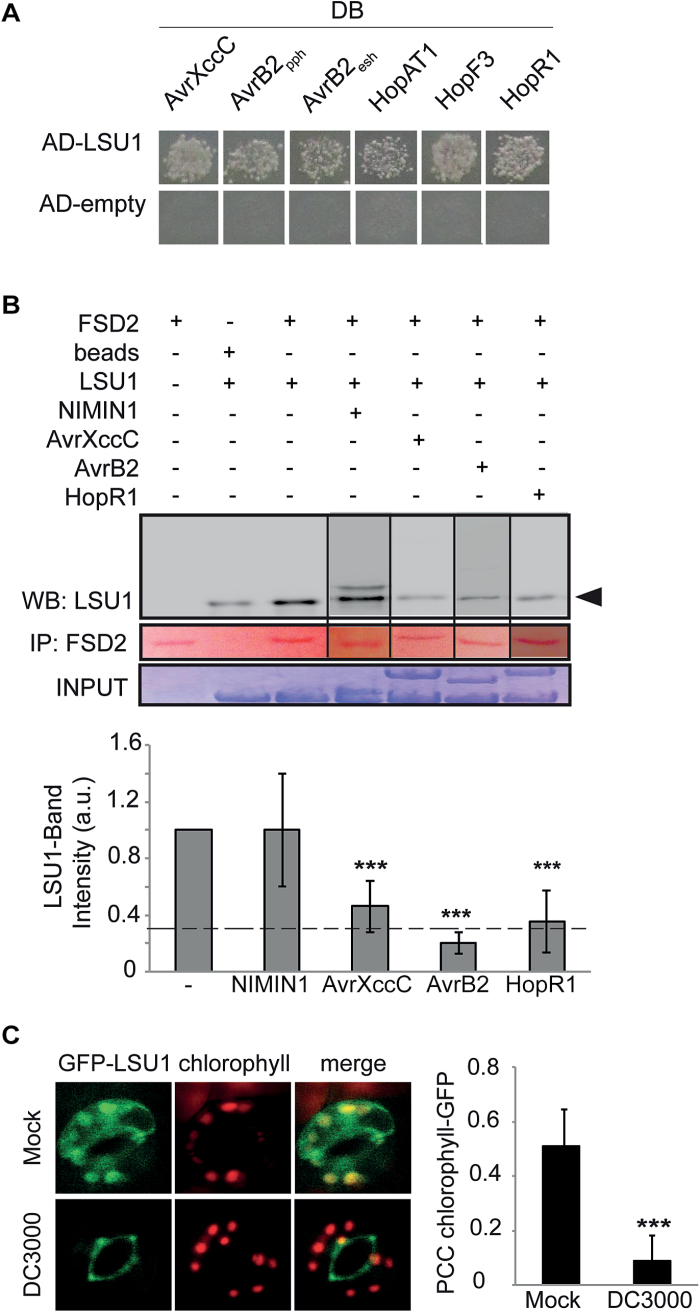
Pseudomonas virulence effectors interact with LSU1 and interfere with its functions. (A) Interactions in the Y2H system between AD–LSU1 and DB fusions with bacterial virulence effectors: Xcc AvrXccC (G9BA82_XANCA), Psp AvrB2 (PSPPH_A0120, AvrB2pph and A2BCU4_PSESH, AvrB2esh), Pst HopAT1 (PSPTO_5618), HopF3 (PSPTO_0502), and HopR1 (PSPTO_0883, C-terminal region). (B) Representative α-MBP western blot analysis of pull-down experiments of recombinant MBP–LSU1 by GST–FSD2 in the presence of the negative control protein MBP–NIMIN1 or the MBP-tagged virulence effectors AvrXccC, AvrB2 (AvrB2pph), and HopR1 (C-term). Ponceau staining shows precipitated GST–FSD2. Coomassie brilliant blue (CBB) shows input material. Quantification of precipitated MBP–LSU1 relative to no competitor summarizing ≥3 independent experiments is shown in the lower panel. For all quantifications, the no-competitor positive control was loaded on the same gel. The dashed line indicates background MBP–LSU1 in the beads-only control. (C) Representative confocal images of GFP–LSU1 seedlings grown on –S after Pst or mock infection; shown are GFP fluorescence, chlorophyll autofluorescence, and merged channels. The bar graph shows the PCC of the GFP and chlorophyll fluorescent patterns in Pst- and mock-infected leaves (n >10). In (B, lower panel) and (C) the significance of differences was assessed using the Student’s t-test (*P<0.05; ***P<0.001).
Pseudomonas syringae virulence effectors interfere with LSU1 functions
Stomata are a major entry point for bacterial pathogens, and of all LSU family members the guard cell-localized LSU1 preferentially interacted with bacterial virulence effectors in our Y2H-based network mapping experiments (Melotto et al., 2006; Mukhtar et al., 2011; Wessling et al., 2014). H2O2 can trigger stomatal closure, but may also have signalling and defence functions (Desikan et al., 2001; Van Aken and Whelan, 2012; de Torres Zabala et al., 2015). Lastly, chloroplastic H2O2 production was recently shown to be an important element of PTI with which Pst virulence effectors can interfere (de Torres Zabala et al., 2015). We therefore wondered if the LSU1–FSD2 interaction may be relevant in defence responses and if LSU-interacting bacterial effectors can interfere with it. First, we confirmed by Y2H the interaction of LSU1 with several bacterial virulence effectors (Fig. 5A), including the putatively chloroplast-localized Pst HopAT1, HopF3, and the C-terminal fragment of HopR1 (Mukhtar et al., 2011; Wessling et al., 2014; de Torres Zabala et al., 2015). Using in vitro GST pull-down experiment, we tested if Xanthomonas campestris pv. campestris (Xcc) AvrXccC, P. syringae pv. phaseolica (Psp) AvrB2, and Pst HopR1 interfere with the LSU1–FSD2 interaction; the others were not tested for technical reasons. After pull-down of GST–FSD2, co-precipitation of LSU1 was assessed by western blot with antibodies to MBP, and band intensity was quantified (Fig. 5B). All three effectors effectively and specifically interfered with the interaction such that only 20–40% of MBP–LSU1 was recovered in the GST–FSD2 pull-down compared with controls (Fig. 5B, lower panel). Furthermore, microscopic analysis revealed that Pst infection during –S stress interfered with the chloroplastic localization of GFP–LSU1 such that the proportion of chloroplasts overlapping GFP fluorescence decreased from 73% to 41% and the PCC of the two fluorescence patterns decreased from 0.52 and 0.51 in untreated and mock-treated leaves, respectively, and to 0.09 in Pst-infected seedlings (Fig. 5C). Thus, virulence effectors interfere with several aspects of LSU1 function in vitro and in vivo.
LSU proteins mediate resistance to Pseudomonas syringae infection under certain abiotic stress conditions
These results led us to investigate genetically the immune function of LSU proteins, especially during abiotic stress. First we asked if LSU down-regulation alters plant susceptibility towards Pst. To perform infection assays under nutrient deficiency conditions, 12-day-old WT and amiR-LSUa–c seedlings grown in vitro on +S or –S were flooded with Pst suspensions and colony density was counted 3 days post-infection (dpi). As depicted on Fig. 6A, consistent with the lack of detectable LSU expression, no significant differences in pathogen susceptibility were found among lines under standard conditions (+S). However, growth of seedlings on –S resulted in mild but significant EDS in the amiR-LSU lines, as the colony count increased ~2-fold compared with the WT (Fig. 6A; Supplementary Table S1). Consistent with this phenotype and our findings above, the amiR-LSUa–c lines showed reduced ROS production in guard cell chloroplasts following Pst challenge 1 dpi (Fig. 6B).
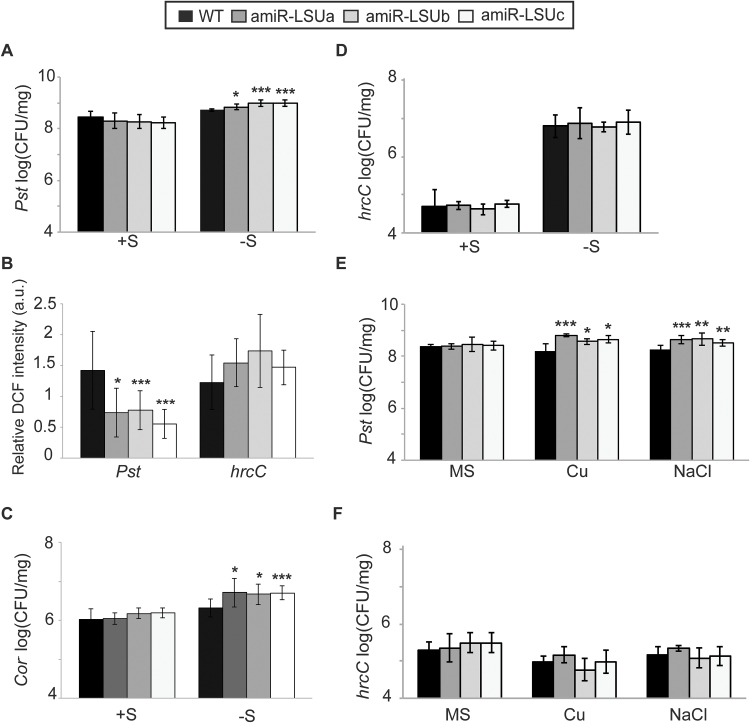
Fig. 6.
LSU proteins mediate defence responses during abiotic stress. WT and amiR-LSUa–c seedlings were flooded with bacterial suspension at 5 × 106 CFU ml–1 of the P. syringae strains: Pst DC3000 (Pst), COR–, and hrcC. In (A) and (C–F), colony-forming units were determined 3 days post-infection (dpi) and normalized to fresh weight. Seedlings were grown in normal (+S) and –S conditions, or 1/2 MS conditions, or submitted to high Cu or NaCl, as indicated. (B) DCF fluorescence in guard cells of –S-grown lines at 1 dpi with the indicated bacterial strain normalized to mock treatment. Error bars correspond to the SD of at least three biologically independent experiments, and statistical differences from the WT were assessed using Student’s t-test and are indicated with asterisks (*P<0.05; **P<0.01; ***P<0.001).
Part of the Pst infectivity relies on the production of coronatine (COR), a jasmonic acid analogue that mediates stomatal re-opening and promotes bacterial virulence by suppressing salicylic acid-mediated defence responses (Cui et al., 2005; Melotto et al., 2006). Upon infection by the COR-defective mutant COR–, the amiR-LSUa–c lines still exhibit moderate EDS on –S (on average ~2.5 times higher than in the WT) (Fig. 6C). Thus, the EDS phenotype in the amiR-LSU lines is not part of the COR-induced responses. To test if this phenotype would be a component of PTI, the same assay was repeated using the Type III Secretion System (T3SS)-defective hrcC mutant, which elicits PTI but is unable to deliver effectors into the plant cytosol (Cunnac et al., 2011) and therefore does not elicit ETI. The amiR-LSU lines did not exhibit an altered susceptibility towards hrcC in either growth condition (Fig. 6D), suggesting that the LSU-dependent immune function requires effector delivery or, formally, the T3SS itself. Consistently, infection of –S-stressed plants by hrcC, in contrast to WT Pst, did not cause an additionally increased ROS production in guard cell chloroplasts (Fig. 6B). Analysis of guard cell dynamics during initial phases of Pst infection further excluded impairments of Pst-triggered stomatal closure in amiR-LSU lines and revealed an altered pattern of stomatal dynamics in –S conditions. The WT and the amiR-LSUa–c lines effectively closed stomata 1 h post-infection (hpi) in +S and –S conditions, indicating that LSU proteins do not function during this stage of infection (Supplementary Fig. S6). However, in WT plants grown on –S media, stomata remained closed even at 4 hpi (Supplementary Fig. S6B). In amiR-LSUa and amiR-LSUc, stomata were ~20% and 30%, respectively, more open at 4 hpi compared with the WT (Supplementary Fig. S6B; Supplementary Table S3). Together, these observations reinforce the notion that in –S stress conditions, LSU proteins have a defence function that involves stimulating ROS production, but which is not mediated by COR and is not part of the hrcC-activated PTI. Consistent with this idea, the knock-out line fsd2-2 also displayed a weak EDS phenotype under our S-deficient conditions (~1.5-fold more colonies than the WT; Supplementary Fig. S7).
Since LSU proteins are up-regulated in several stress conditions, we wondered if they might also have an immune function in abiotic stresses other than S deprivation. Indeed, amiR-LSU plants grown in the presence of high salt or Cu concentrations also exhibited moderate but significant EDS towards Pst, but not hrcC, demonstrating that LSU proteins support immune function in several abiotic stress conditions (Fig. 6D, E).
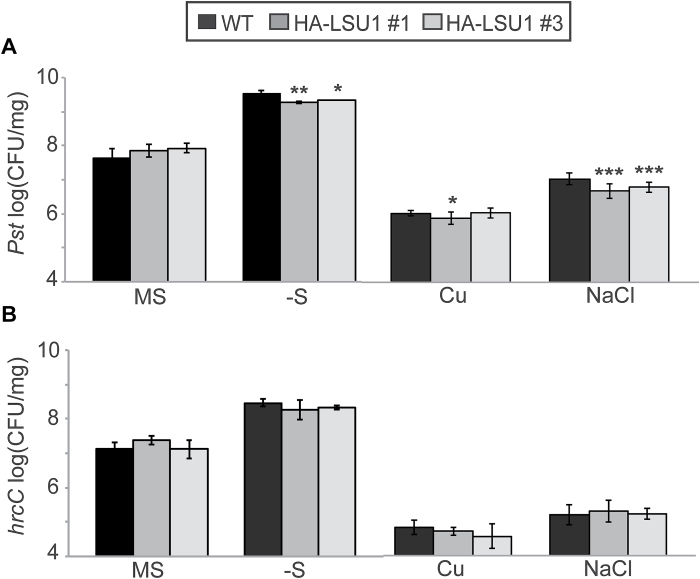
To test if LSU overexpression results in complementary EDR, and to gather support for our suggestion that LSU1 might be responsible for the observed amiR-LSU phenotypes, we phenotypically characterized two independent 35S:HA-LSU1 overexpression lines using Pst and hrcC. A phenotypic pattern complementary to the amiR-LSUa–c lines was observed such that LSU1 overexpression conferred significant EDR towards Pst during –S and high salt stress but had no effect under normal conditions or towards hrcC (Fig. 7; Supplementary Table S1).
Fig. 7.
Ectopic expression of LSU1 results in a moderate resistance to P. syringae during abiotic stress. The WT and two independent HA–LSU1 lines were grown and treated as in Fig. 5. Error bars correspond to the SD of at least three biologically independent experiments, and statistical differences from the WT were assessed using Student’s t-test and are indicated with asterisks (*P<0.05; **P<0.01; ***P<0.001).
Discussion
In addition to understanding molecular plant response mechanisms to isolated abiotic and biotic stress, it is also important to characterize the integrated responses to combinatorial stress conditions. LSU proteins, which had only been implicated in –S stress, are highly connected hubs in a plant protein interactome network and intensely targeted by virulence effectors from evolutionarily distant pathogens (Arabidopsis Interactome Mapping Consortium, 2011; Mukhtar et al., 2011; Wessling et al., 2014). These observations motivated us to investigate the function of the encoded proteins to illuminate their normal physiological role and find out why effectors may have evolved to interfere with this function.
Our primary characterization of LSU1 and LSU2, as representatives of this protein family, suggests a general function of these genes in abiotic stress responses. Both LSU1 and LSU2 transcripts are up-regulated in several abiotic stress conditions, and apparent post-translational regulation leads to protein accumulation in seedlings and, for LSU1, to stabilization of GFP–LSU1 in guard cells. Since all LSU family members have a serine-rich domain that includes putative phosphorylation sites (Supplementary Fig. S1A) and phosphorylation of LSU3 has been detected in vivo (Phosphat; http://phosphat.uni-hohenheim.de/), the impact of this post-translational modification on protein stability warrants further investigation.
The prominent localization of GFP–LSU1 in guard cells, reinforced by the LSU1 down-regulation in cotyledons of the stomataless mutants spch-3 and mute-3 (de Marcos et al., 2015), prompted us to investigate potential functions of LSU in stomatal dynamics. Indeed, transgenic lines with reduced LSU levels display impaired stomatal closure under –S. Moreover, the amiR-LSU lines exhibited profoundly reduced ROS production in guard cells when exposed to stress conditions that induce LSU transcription, such as high salinity and –S. This suggests that LSU proteins, most proobably LSU1, promote stomatal closure in response to abiotic stress by stimulating chloroplastic ROS production. In guard cells, ROS levels depend on a complex interplay between ABA and ethylene whereby ethylene negatively regulates ABA-dependent stomatal closure by alleviating ROS levels, while, conversely, ABA represses ethylene production (Tanaka et al., 2005, 2006; Watkins et al., 2014). However, upon exogenous ABA treatment, stomata of amiR-LSU lines closed normally, indicating that the ABA-triggered stomatal closure is not impaired. It is possible that, similarly to ethylene, LSUs function upstream of ABA. This appears unlikely given that S limitation negatively impacts ethylene biosynthesis due to reduced methionine availability (Bürstenbinder et al., 2007) and also leads to drastically reduced ABA levels (Cao et al., 2014). We therefore hypothesize that LSU proteins may participate in an alternative mechanism to trigger stomatal closure in some stress conditions. The exclusive presence of LSU proteins among gymno- and angiosperms provides evolutionary support for the proposed role of LSU proteins in guard cells.
A hint towards a possible mechanism by which LSUs induce ROS production came from our Arabidopsis interactome map, where LSU proteins were found to interact with FSD2 (Arabidopsis Interactome Mapping Consortium, 2011). We demonstrate here that LSU1 and LSU2 specifically interact with FSD2 in vitro and in planta, and stimulate its enzyme activity. Consistent with the idea that LSU1 activates FSD2 in planta, GFP–LSU1, but not GFP–LSU2, was found in guard cell patterns that largely overlap with chloroplast autofluorescence, and upon S deprivation amiR-LSU lines exhibit reduced H2O2 production in guard cells. The increased H2O2 levels can trigger stomatal closure and potentially other effects, such as transcriptional regulation (Vandenabeele et al., 2003; Van Aken and Whelan, 2012) (Supplementary Fig. S8). A heterodimeric complex of FSD2/FSD3 was previously shown to protect chloroplast nucleoids from ROS (Myouga et al., 2008). This study found that FSD3 localized exclusively to speckles in the nucleoids, while, consistent with our data, FSD2 exhibited a wider chloroplastic localization suggesting additional functions (Myouga et al., 2008). We propose that abiotic stress conditions lead to stabilization of LSU1, especially in guard cells, where it enters chloroplasts and stimulates ROS production by activating FSD2. The question of how LSU1, despite the lack of transit peptide, is imported to the chloroplast remains to be answered, though examples of chloroplast- and nuclear-localized proteins using non-canonical importing signals or interacting partners have been reported (Miras et al., 2002, 2007; Withers et al., 2012). Biochemically, it is likely that FSD2 activation by LSU1 represents another example of factor-associated SOD activation, which has been previously reported for all SOD isoforms (Luk et al., 2003; Chu et al., 2005; Kuo et al., 2013). The capacity of LSU to stimulate FSD2, the small size, and lack of clearly defined globular domains moreover beg the question of whether LSU proteins might also have chaperone-like activity for other clients.
Besides their function for gas exchange, stomata are natural orifices that constitute entry points for bacterial pathogens into the plant apoplast (Melotto et al., 2006). Although plants reinforce stomatal immunity upon bacterial recognition during PTI, bacterial pathogens such as Pst can counteract this effect by producing COR and virulence effectors to re-open stomata or prevent stomatal closure, respectively (Melotto et al., 2006; Gudesblat et al., 2009). Given that we identified LSU proteins as targets of virulence effectors, we hypothesized that LSU-mediated stomatal closure could also have a defence function with which pathogens may aim to interfere. In support of this, we found that amiR-LSU lines do exhibit an EDS phenotype in conditions of abiotic stress whereas LSU1 overexpression lines display complementary EDR phenotypes. Upon infection by hrcC, this phenotype was not detectable, whereas it was unaltered upon infection by a COR– strain, indicating that the LSU-dependent immune function is not due to a general LSU function in these conditions and not due to PTI or COR-triggered pathways. Abrogation of LSU expression did not directly impair stomatal closure in response to Pst infection during S scarcity; however, it caused a moderate and faster stomatal re-opening in comparison with the WT at 4 hpi. The weak immune effects may be due to the inability of the amiR-LSU lines to maintain closed stomata during the early phase of the infection, but independent mechanisms cannot be excluded and should be further investigated.
Chloroplastic ROS were recently shown to be an important defence signal that Pst can suppress by only partly known mechanisms (de Torres Zabala et al., 2015). Here, we demonstrated that Pst effectors can interfere with the activating interaction of LSU1 with FSD2. In addition, Pst infection prevents GFP–LSU1 from entering guard cell chloroplasts, and our genetic data confirm the immune function of LSU proteins during high salinity or –S stress. In these conditions, artificially reduced LSU levels abolish the normally increased ROS production in response to Pst infection and cause a modest EDS phenotype. Lack of FSD2 activity in the fsd2-2 line resulted in a similar degree of Pst susceptibility, whereas ectopic expression of LSU1 results in modest EDR phenotypes.
Recent studies already implicated virulence effectors in modulating stomatal function by demonstrating that Pst HopF2 and HopM1 can suppress stomatal immune responses and ROS burst in planta during bacterial infection (Hurley et al., 2014; Lozano-Duran et al., 2014). Thus, we suggest that upon infection with virulent Pst, the antagonistic interplay of effectors triggering weak ETI (Gassmann, 2005; Fabro et al., 2011) and effectors interfering with LSU results in an overall small, but significant immune phenotype in amiR-LSU lines, demonstrating the immune function of LSU proteins in plants experiencing –S or salt stress (Supplementary Fig. S8).
Taken together, we demonstrate that LSU proteins stimulate production of H2O2 and potentially other ROS, and effect stomatal closure in response to several types of abiotic stress, including high salinity or S depletion. Mechanistically, this stimulation is at least in part achieved via LSU up-regulation and subsequent interaction with and activation of the SOD FSD2. Moreover, we show that LSU proteins are elements of the plant immune response when plants experience simultaneous abiotic stress.
Author contributions
experiments and figures: AG-M. Assistance with Y2H experiments: MA and AA. Design of experiments: AG-M, PE, JLD, and PB. Manuscript writing: AG-M and PB. Critical manuscript reading and editing: PE and JLD. All authors discussed and commented on the article.
Supplementary Material
Acknowledgements
We thank Andreas Keymer (LMU; München) for help and assistance in confocal microscopy imaging. The 35SCaMV-GFP line was kindly provided by Kay Schneitz (TUM; München). We thank François Parcy (CEA-CNRS; Grenoble) and Alejandro Ferrando (IBMCP-CSIC; València) for pALLIGATOR2 and pYFN43 and pYFC43 vectors, respectively. We thank Jens Steinbrenner (University Giessen) for fruitful discussions, and we thank Freddy Monteiro, Sarah R. Grant, and Nora Marín de la Rosa for discussions and critical reading of the manuscript. This work was funded by grants to PB by the Human Frontiers Science Organisation (grant RGY0080/2013) and by the DFG collaborative research grant (CRC) 924 (CRC924/SFB924), and to JLD from the National Science Foundation (IOS-1257373), the National Institutes of Health (1RO1 GM107444), the Gordon and Betty Moore Foundation (GBMF3030), the HHMI, and by a Distinguished Guest Professorship, Eberhard-Karls-Universität, Tübingen, Germany. JLD is an Investigator of the Howard Hughes Medical Institute. The authors declare that there are no conflicts of interest.
References
- Al-Sadi AM, Al-Masoudi RS, Al-Habsi N, Al-Said FA, Al-Rawahy SA, Ahmed M, Deadman ML. 2010. Effect of salinity on pythium damping-off of cucumber and on the tolerance of Pythium aphanidermatum. Plant Pathology 59, 112–120. [Google Scholar]
- Amtmann A, Troufflard S, Armengaud P. 2008. The effect of potassium nutrition on pest and disease resistance in plants. Physiologia Plantarum 133, 682–691. [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium. 2011. Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaud D, Hwang I. 2015. A sophisticated network of signaling pathways regulates stomatal defenses to bacterial pathogens. Molecular Plant 8, 566–581. [DOI] [PubMed] [Google Scholar]
- Atkinson NJ, Lilley CJ, Urwin PE. 2013. Identification of genes involved in the response of Arabidopsis to simultaneous biotic and abiotic stresses. Plant Physiology 162, 2028–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belda-Palazón B, Ruiz L, Martí E, Tárraga S, Tiburcio AF, Culiáñez F, Farràs R, Carrasco P, Ferrando A. 2012. Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS One 7, e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensmihen S, To A, Lambert G, Kroj T, Giraudat J, Parcy F. 2004. Analysis of an activated ABI5 allele using a new selection method for transgenic Arabidopsis seeds. FEBS Letters 561, 127–131. [DOI] [PubMed] [Google Scholar]
- Bernaudat F, Frelet-Barrand A, Pochon N, et al. 2011. Heterologous expression of membrane proteins: choosing the appropriate host. PLoS One 6, e29191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt R, Keston AS. 1965. Synthesis of diacetyldichlorofluorescin: a stable reagent for fluorometric analysis. Analytical Biochemistry 11, 6–9. [DOI] [PubMed] [Google Scholar]
- Braun P, Hu Y, Shen B, Halleck A, Koundinya M, Harlow E, LaBaer J. 2002. Proteome-scale purification of human proteins from bacteria. Proceedings of the National Academy of Sciences, USA 99, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürstenbinder K, Rzewuski G, Wirtz M, Hell R, Sauter M. 2007. The role of methionine recycling for ethylene synthesis in Arabidopsis. The Plant Journal 49, 238–249. [DOI] [PubMed] [Google Scholar]
- Cao MJ, Wang Z, Zhao Q, Mao JL, Speiser A, Wirtz M, Hell R, Zhu JK, Xiang CB. 2014. Sulfate availability affects ABA levels and germination response to ABA and salt stress in Arabidopsis thaliana. The Plant Journal 77, 604–615. [DOI] [PubMed] [Google Scholar]
- Chu CC, Lee WC, Guo WY, Pan SM, Chen LJ, Li HM, Jinn TL. 2005. A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiology 139, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Cui J, Bahrami AK, Pringle EG, Hernandez-Guzman G, Bender CL, Pierce NE, Ausubel FM. 2005. Pseudomonas syringae manipulates systemic plant defenses against pathogens and herbivores. Proceedings of the National Academy of Sciences, USA 102, 1791–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunnac S, Chakravarthy S, Kvitko BH, Russell AB, Martin GB, Collmer A. 2011. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proceedings of the National Academy of Sciences, USA 108, 2975–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. 2000. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proceedings of the National Academy of Sciences, USA 97, 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daszkowska-Golec A, Szarejko I. 2013. Open or close the gate—stomata action under the control of phytohormones in drought stress conditions. Frontiers in Plant Science 4, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Marcos A, Triviño M, Pérez-Bueno ML, Ballesteros I, Barón M, Mena M, Fenoll C. 2015. Transcriptional profiles of Arabidopsis stomataless mutants reveal developmental and physiological features of life in the absence of stomata. Frontiers in Plant Science 6, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, A-H-Mackerness S, Hancock JT, Neill SJ. 2001. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiology 127, 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan R, Cheung MK, Bright J, Henson D, Hancock JT, Neill SJ. 2004. ABA, hydrogen peroxide and nitric oxide signalling in stomatal guard cells. Journal of Experimental Botany 55, 205–212. [DOI] [PubMed] [Google Scholar]
- de Torres Zabala M, Littlejohn G, Jayaraman S, et al. 2015. Chloroplasts play a central role in plant defence and are targeted by pathogen effectors. Nature Plants 1, 15074. [DOI] [PubMed] [Google Scholar]
- Dreze M, Monachello D, Lurin C, Cusick ME, Hill DE, Vidal M, Braun P. 2010. High-quality binary interactome mapping. Methods in Enzymology 470, 281–315. [DOI] [PubMed] [Google Scholar]
- Fabro G, Steinbrenner J, Coates M, et al. 2011. Multiple candidate effectors from the oomycete pathogen Hyaloperonospora arabidopsidis suppress host plant immunity. PLoS Pathogens 7, e1002348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Molina A, Xing S, Huijser P. 2014. Functional characterisation of Arabidopsis SPL7 conserved protein domains suggests novel regulatory mechanisms in the Cu deficiency response. BMC Plant Biology 14, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann W. 2005. Natural variation in the Arabidopsis response to the avirulence gene hopPsyA uncouples the hypersensitive response from disease resistance. Molecular Plant-Microbe Interactions 18, 1054–1060. [DOI] [PubMed] [Google Scholar]
- Gudesblat GE, Torres PS, Vojnov AA. 2009. Xanthomonas campestris overcomes Arabidopsis stomatal innate immunity through a DSF cell-to-cell signal-regulated virulence factor. Plant Physiology 149, 1017–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han JD, Bertin N, Hao T, et al. 2004. Evidence for dynamically organized modularity in the yeast protein–protein interaction network. Nature 430, 88–93. [DOI] [PubMed] [Google Scholar]
- Hawkesford M, Horst W, Kichey T, Lambers H, Schjoerring J, Møller IS, White P. 2012. Functions of macronutrients. In: Marschner P, ed. Marschner’s mineral nutrition of higher plants, 3rd edn San Diego: Academic Press, 135–189. [Google Scholar]
- Hurley B, Lee D, Mott A, Wilton M, Liu J, Liu YC, Angers S, Coaker G, Guttman DS, Desveaux D. 2014. The Pseudomonas syringae type III effector HopF2 suppresses Arabidopsis stomatal immunity. PLoS One 9, e114921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y, Ishiga T, Uppalapati SR, Mysore KS. 2011. Arabidopsis seedling flood-inoculation technique: a rapid and reliable assay for studying plant–bacterial interactions. Plant Methods 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jukes TH, Cantor CR. 1969. Evolution of protein molecules. New York: Academic Press, 21–132. [Google Scholar]
- Karmoker JL, Clarkson DT, Saker LR, Rooney JM, Purves JV. 1991. Sulphate deprivation depresses the transport of nitrogen to the xylem and the hydraulic conductivity of barley (Hordeum vulgare L.) roots. Planta 185, 269–278. [DOI] [PubMed] [Google Scholar]
- Kissoudis C, Chowdhury R, Heusden S, Wiel C, Finkers R, Visser RGF, Bai Y, Linden G. 2015. Combined biotic and abiotic stress resistance in tomato. Euphytica 202, 317–332. [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL. 1998. Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiology 118, 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. 1986. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Molecular and General Genetics 204, 383–396. [Google Scholar]
- Kuo WY, Huang CH, Liu AC, Cheng CP, Li SH, Chang WC, Weiss C, Azem A, Jinn TL. 2013. CHAPERONIN 20 mediates iron superoxide dismutase (FeSOD) activity independent of its co-chaperonin role in Arabidopsis chloroplasts. New Phytologist 197, 99–110. [DOI] [PubMed] [Google Scholar]
- Lewandowska M, Wawrzynska A, Moniuszko G, et al. 2010. A contribution to identification of novel regulators of plant response to sulfur deficiency: characteristics of a tobacco gene UP9C, its protein product and the effects of UP9C silencing. Molecular Plant 3, 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Bourdais G, He SY, Robatzek S. 2014. The bacterial effector HopM1 suppresses PAMP-triggered oxidative burst and stomatal immunity. New Phytologist 202, 259–269. [DOI] [PubMed] [Google Scholar]
- Luk E, Carroll M, Baker M, Culotta VC. 2003. Manganese activation of superoxide dismutase 2 in Saccharomyces cerevisiae requires MTM1, a member of the mitochondrial carrier family. Proceedings of the National Academy of Sciences, USA 100, 10353–10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A, Inoue E, Watanabe-Takahashi A, Yamaya T, Takahashi H. 2003. Transcriptome profiling of sulfur-responsive genes in Arabidopsis reveals global effects of sulfur nutrition on multiple metabolic pathways. Plant Physiology 132, 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. 2006. Plant stomata function in innate immunity against bacterial invasion. Cell 126, 969–980. [DOI] [PubMed] [Google Scholar]
- Miao Y, Lv D, Wang P, Wang XC, Chen J, Miao C, Song CP. 2006. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. The Plant Cell 18, 2749–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras S, Salvi D, Ferro M, Grunwald D, Garin J, Joyard J, Rolland N. 2002. Non-canonical transit peptide for import into the chloroplast. Journal of Biological Chemistry 277, 47770–47778. [DOI] [PubMed] [Google Scholar]
- Miras S, Salvi D, Piette L, Seigneurin-Berny D, Grunwald D, Reinbothe C, Joyard J, Reinbothe S, Rolland N. 2007. Toc159- and Toc75-independent import of a transit sequence-less precursor into the inner envelope of chloroplasts. Journal of Biological Chemistry 282, 29482–29492. [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M, et al. 2011. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, Shono Y, Nagata N, Ikeuchi M, Shinozaki K. 2008. A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. The Plant Cell 20, 3148–3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R. 2003. Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. The Plant Journal 33, 633–650. [DOI] [PubMed] [Google Scholar]
- Pei ZM, Murata Y, Benning G, Thomine S, Klüsener B, Allen GJ, Grill E, Schroeder JI. 2000. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734. [DOI] [PubMed] [Google Scholar]
- Prasch CM, Sonnewald U. 2013. Simultaneous application of heat, drought, and virus to Arabidopsis plants reveals significant shifts in signaling networks. Plant Physiology 162, 1849–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen S, Barah P, Suarez-Rodriguez MC, Bressendorff S, Friis P, Costantino P, Bones AM, Nielsen HB, Mundy J. 2013. Transcriptome responses to combinations of stresses in Arabidopsis. Plant Physiology 161, 1783–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. 2006. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell 18, 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R. 2014. Abiotic and biotic stress combinations. New Phytologist 203, 32–43. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Sano T, Tamaoki M, Nakajima N, Kondo N, Hasezawa S. 2005. Ethylene inhibits abscisic acid-induced stomatal closure in Arabidopsis. Plant Physiology 138, 2337–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Sano T, Tamaoki M, Nakajima N, Kondo N, Hasezawa S. 2006. Cytokinin and auxin inhibit abscisic acid-induced stomatal closure by enhancing ethylene production in Arabidopsis. Journal of Experimental Botany 57, 2259–2266. [DOI] [PubMed] [Google Scholar]
- Terry N. 1976. Effects of sulfur on the photosynthesis of intact leaves and isolated chloroplasts of sugar beets. Plant Physiology 57, 477–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triky-Dotan S, Yermiyahu U, Katan J, Gamliel A. 2005. Development of crown and root rot disease of tomato under irrigation with saline water. Phytopathology 95, 1438–1444. [DOI] [PubMed] [Google Scholar]
- Van Aken O, Whelan J. 2012. Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsis. Frontiers in Plant Science 3, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenabeele S, Van Der Kelen K, Dat J, et al. 2003. A comprehensive analysis of hydrogen peroxide-induced gene expression in tobacco. Proceedings of the National Academy of Sciences, USA 100, 16113–16118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WH, Yi XQ, Han AD, Liu TW, Chen J, Wu FH, Dong XJ, He JX, Pei ZM, Zheng HL. 2012. Calcium-sensing receptor regulates stomatal closure through hydrogen peroxide and nitric oxide in response to extracellular calcium in Arabidopsis. Journal of Experimental Botany 63, 177–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JM, Hechler PJ, Muday GK. 2014. Ethylene-induced flavonol accumulation in guard cells suppresses reactive oxygen species and moderates stomatal aperture. Plant Physiology 164, 1707–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weßling R, Epple P, Altmann S, et al. 2014. Convergent targeting of a common host protein-network by pathogen effectors from three kingdoms of life. Cell Host and Microbe 16, 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withers J, Yao J, Mecey C, Howe GA, Melotto M, He SY. 2012. Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proceedings of the National Academy of Sciences, USA 109, 20148–20153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You MP, Colmer TD, Barbetti MJ. 2011. Salinity drives host reaction in Phaseolus vulgaris (common bean) to Macrophomina phaseolina. Functional Plant Biology 38, 984–992. [DOI] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, et al. 2008. High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang L, Dong F, Gao J, Galbraith DW, Song CP. 2001. Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiology 126, 1438–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou JJ, Li XD, Ratnasekera D, Wang C, Liu WX, Song LF, Zhang WZ, Wu WH. 2015. Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. The Plant Cell 27, 1445–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.