Highlight
Two plasma membrane H+-ATPases, encoded by AHA2 and AHA7, enhance primary root elongation and root hair development by mediating H+ efflux in the elongation and root hair zones under low-phosphorus stress.
Keywords: Arabidopsis, low phosphorus, plasma membrane H+-ATPase, proton flux, root growth, root hair.
Abstract
Phosphorus deficiency in soil is one of the major limiting factors for plant growth. Plasma membrane H+-ATPase (PM H+-ATPase) plays an important role in the plant response to low-phosphorus stress (LP). However, few details are known regarding the action of PM H+-ATPase in mediating root proton (H+) flux and root growth under LP. In this study, we investigated the involvement and function of different Arabidopsis PM H+-ATPase genes in root H+ flux in response to LP. First, we examined the expressions of all Arabidopsis PM H+-ATPase gene family members (AHA1–AHA11) under LP. Expression of AHA2 and AHA7 in roots was enhanced under this condition. When the two genes were deficient in their respective Arabidopsis mutant plants, root growth and responses of the mutants to LP were highly inhibited compared with the wild-type plant. AHA2-deficient plants exhibited reduced primary root elongation and lower H+ efflux in the root elongation zone. AHA7-deficient plants exhibited reduced root hair density and lower H+ efflux in the root hair zone. The modulation of H+ efflux by AHA2 or AHA7 was affected by the action of 14-3-3 proteins and/or auxin regulatory pathways in the context of root growth and response to LP. Our results suggest that under LP conditions, AHA2 acts mainly to modulate primary root elongation by mediating H+ efflux in the root elongation zone, whereas AHA7 plays an important role in root hair formation by mediating H+ efflux in the root hair zone.
Introduction
Phosphorus is one of the most important nutrients for plant growth, development, and reproduction. Even when the phosphorus content of soil is high, phosphorus deficiency can arise due to mineralization and fixation processes. Thus, low-phosphorus stress (LP) is one of the major limiting factors for plant growth in many ecosystems and a major constraint for agricultural productivity (Devaiah et al., 2007; Schachtman and Shin, 2007). In order to survive, plants have developed flexible strategies to cope with low-phosphorus soil by improving phosphorus mobilization and uptake from the soil (Vance et al., 2003; Wu et al., 2003; Hoffland et al., 2006; Li et al., 2009). These strategies include the production of enzymes, such as acid phosphatases and nucleases; increased synthesis and secretion of organic acids or protons (H+) in the roots; enhanced expression of high-affinity phosphorus transporters; development of a larger root system capable of exploring a greater soil volume to ensure a sufficient uptake and use of phosphorus; and establishment of differential photosynthate distribution between shoots and roots, resulting in prolific root growth and increased root/shoot ratios. Among these strategies, plant plasma membrane H+-ATPase (PM H+-ATPase) plays the key role in the response to LP. Phosphorus acquisition and utilization are a highly regulated and complex set of processes relying not only on PM H+-ATPase-dependent H+ release, but also on PM H+-ATPase regulatory pathways controlling the abundance and activity of transporters and enzymes involved in phosphorus deficiency responses (Yan et al., 2002).
In plants, PM H+-ATPases constitute a family of proton pumps driven by ATP hydrolysis. Their role is to provide an energy source for the transport of nutrients into the plant cell by generating an electrochemical gradient (Haruta and Sussman, 2012). Most of the membrane-bound transport proteins in plants are driven indirectly by the action of PM H+-ATPase (Palmgren, 2001). PM H+-ATPases form a large family of proteins in plants. For example, in Arabidopsis, there are 11 functional PM H+-ATPase family members, denoted AHA1 to AHA11 (for Arabidopsis H+-ATPase). Of the 11 genes encoding functional PM H+-ATPase proteins in plants, the first to be cloned were AHA1 and AHA3. Using genetic analysis, roles for AHA1 in steroid signaling, AHA2 in iron transport, AHA3 in pollen development, AHA4 in salt stress, and AHA10 in vacuole biosynthesis have been reported (Robertson et al., 2004; Baxter et al., 2005; Gévaudant et al., 2007; Santi and Schmidt, 2009; Caesar et al., 2011). Using RT-PCR, Ueno et al. (2005) determined that eight AHA genes are expressed in green leaves (AHA1, 2, 3, 5, 7, 8, 10, and 11), eight are expressed in roots (AHA1, 2, 3, 4, 7, 8, 10, and 11), and three are expressed in all tissues (AHA1, 2 and 11).
Plant PM H+-ATPases are regulated by a number of factors, including blue light, fusicoccin, protein kinase, 14-3-3 proteins, and auxin (Palmgren, 2001; Hager, 2003; Rodrigues et al., 2014). The C-terminus of plant PM H+-ATPases serves as an autoinhibitory domain to regulate enzymatic activity. The regulatory C-terminal domain of PM H+-ATPases is phosphorylated in vivo. In vitro, the domain becomes phosphorylated within seconds of exposure to blue light and fusicoccin. Phosphorylation of PM H+-ATPase Thr-947 by a protein kinase generates a binding site for 14-3-3 proteins. Activated PM H+-ATPases consist of six phosphorylated PM H+-ATPase molecules, assembled in a hexameric structure, together with six 14-3-3 molecules. Auxin also regulates PM H+-ATPase activity by phosphorylation; auxin-enhanced PM H+-ATPase activity lowers the pH of the cell wall, activates pH-sensitive enzymes and proteins within the cell wall, and initiates cell-wall loosening and extension growth. Auxin can also increase the quantity of certain PM ATPases by influencing protein expression. This results not only in enhanced H+ extrusion, but also eventually in the restoration of the number of PM H+-ATPases in a given membrane area.
Root growth, including primary root elongation and root hair formation, is important for the uptake and use of phosphorus (Shen et al., 2006). Optimal primary root elongation or root hair formation requires fine regulation of root tip H+ secretion by PM H+-ATPase (Palmgren, 2001; Staal et al., 2011; Haruta & Sussmann, 2012). Acidification of the aqueous fraction of the cell wall apoplast by H+ excretion via PM H+-ATPase is a critical growth-promoting event and a key factor in triggering elongation of the primary root (Moloney, 1981). Root hair development in Arabidopsis is associated with alkalinization of the cytoplasm and acidification of the cell wall, and these processes are regulated by PM H+-ATPase-independent H+ influx and efflux (Bibikova et al., 1998). However, little is known regarding the role of PM H+-ATPases in mediating root H+ flux under LP. Thus, in this study, we investigated the involvement and function of different Arabidopsis PM H+-ATPase genes in root H+ flux in response to LP.
Materials and methods
Plant materials, growth conditions, and stress treatment
In this study, the wild-type (WT) Arabidopsis strain used was ecotype Col-0, unless otherwise indicated. The TFT7-overexpressing Arabidopsis plants (denoted OE-TFT7) used in this study was described in a previous publication (Xu et al., 2012). The PM H+-ATPase-deficient mutant Arabidopsis plants (aha1-1: Salk_065288; aha1-2: Salk_118350; aha2-1: Salk_062371; aha2-2: Salk_082786; aha7-1: Salk_042485; aha7-2: Salk_056487) were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University, Columbus, OH, USA); the homozygous Arabidopsis mutant was identified by PCR using primers specific to the insertion T-DNA (see Supplementary Table S1 at JXB online). To generate double-transgenic Arabidopsis plants, knock-out mutants (aha2-2 and aha7-1) were crossed with OE-TFT7 plants. Homozygous double-transgenic plants were selected on the basis of PCR with specific primers and kanamycin resistance.
Before planting, Arabidopsis seeds were surface-sterilized with 70% ethanol for 1 min, then with 1% sodium hypochlorite solution plus sodium dodecyl sulfate for 9 min, and subsequently rinsed with sterile deionized water six times. Seeds were then kept in the dark for 3 days at 4 °C for stratification and grown hydroponically using a sugar-free agar medium-based culture system, as described by Xu and Shi (2008). Sterilized seeds were sown on a sucrose-free medium containing full-strength Murashige and Skoog nutrients and 6 g L–1 agar. This sugar-free agar medium was dispensed into bottom-removed Eppendorf tubes, which were held in a plastic platform. Two to three seeds were sown in each tube. Seedlings were thinned to one plant per tube after 7 days of growth. The nutrient solution consisted of: 5 mM KNO3, 1 mM KH2PO4, 2 mM MgSO4, 2 mM Ca(NO3)2, 0.5 mM Fe-Na-EDTA, 70 μM H3BO3, 14 μM MnCl2, 0.5 μM CuSO4, 1 μM ZnSO4, 0.2 μM Na2MoO4, 10 μM NaCl, and 0.01 μM CoCl2. The pH of the solution was adjusted to 6.0 every day and the solution renewed every 2 days. Low-phosphorous (1 μM) conditions were achieved in the nutrient solution by replacing 1 mM KH2PO4 with 0.999 mM KCl and 0.001 mM KH2PO4. Arabidopsis plants were grown at 22 ± 1 °C, with a light intensity of 120 μmol photons m–2 s–1, a 16 h light/8 h dark photoperiod, and a relative humidity of 70% in the growth chamber (Sanyo Electric Co., Ltd, Kyoto, Japan). Twenty-two-day-old Arabidopsis plants were treated with 1 mM phosphorus (control) or 1 μM phosphorus (LP: low-phosphorus stress) for 8 days within the hydroponic system. Control plants and LP-treated plants were harvested at 0-, 1-, 2-, 4-, 6-, and 8-day time points. After harvesting, Arabidopsis plants were frozen immediately in liquid nitrogen and stored at –80 °C until analysis.
Real-time RT-PCR
Real-time RT-PCR was performed according to the method described by Xu and Shi (2006). Total RNA was extracted from Arabidopsis plants under control and LP conditions. Gene-specific primers for real-time RT-PCR were designed using Primer 5 software (see Supplementary Table S1). At-ACT2 is a strongly and constitutively expressed ‘housekeeping’ gene in Arabidopsis plants; the mRNA expression level of this gene was designated to be 100 relative expression units (REU). Expression levels of the genes of interest are described relative to At-ACT2, specifically, as the ratio of the copy number of the cDNA of interest to the copy number of At-ACT2 multiplied by 100 REU.
Measurement of root growth and phosphorus concentration
Root growth was measured according to the method of Xu et al. (2013b). Root surface area was measured using a root analysis instrument (WinRHIZO; Regent Instruments Inc., Quebec, ON, Canada). The primary root elongation rate of Arabidopsis (μm h–1) was calculated based on the displacement of the primary root apex relative to the primary root length over the duration of the experiment. Root hairs in the region 0–5000 μm from the root cap junction were identified in Arabidopsis plants using confocal laser scanning microscopy (Olympus FV-1000 spectral type SPD mar/G/R IX81 Fluoview laser confocal system). Average root hair densities in 100 × 100 μm2 sections of the 0–5000 μm region were calculated. The phosphorus concentrations of Arabidopsis roots were determined colorimetrically by the phosphovanadate method (Hanson, 1950) after digestion in a mixture of HNO3, HClO4, and H2SO4 (3:1:1, v/v).
Analysis of proton extrusion
The proton extrusion rate of Arabidopsis roots was analysed following the method of Jin et al. (2009). Briefly, 22-day-old Arabidopsis plants were treated with 1 mM phosphorus (control) or 1 μM phosphorus (LP) for 8 days. Arabidopsis plants were transferred to 100-mL pots filled with 1 mM phosphorus solution or 1 μM phosphorus solution. After 12 h, each solution was filtered, and the pH was adjusted to 6.0. The quantity of protons excreted by roots was determined by titrating the solution with 1 mM NaOH.
PM H+-ATPase activity and H+ flux in the root tip of Arabidopsis plants
PM H+-ATPase activity was determined according to the method described by Xu et al. (2012). H+ fluxes were measured non-invasively using the scanning ion-selective electrode technique (SIET; SIET system BIO-003A; Younger USA Science and Technology Corp.; Applicable Electronics Inc.; Science Wares Inc., Falmouth, MA, USA), also according to the method described by Xu et al. (2012). Under normal growth conditions (control) or low-phosphorus stress (LP), proton (H+) flux was measured along the root tip of Arabidopsis plants in the following zones: meristem zone (0–200 μm from the root cap junction), transition zone (200–520 μm from the root cap junction), elongation zone (520–850 μm from the root cap junction), and root hair zone (850–1500 μm from the root cap junction).
Results
Expression of Arabidopsis PM H+-ATPase genes in response to LP
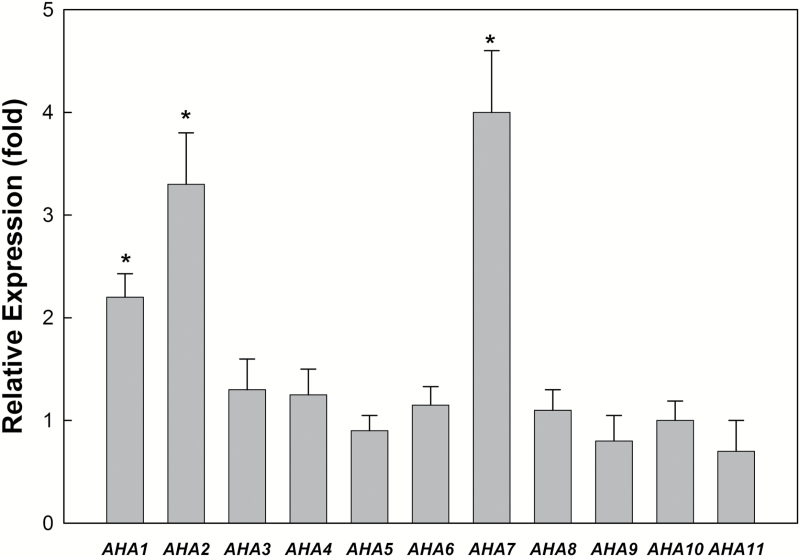
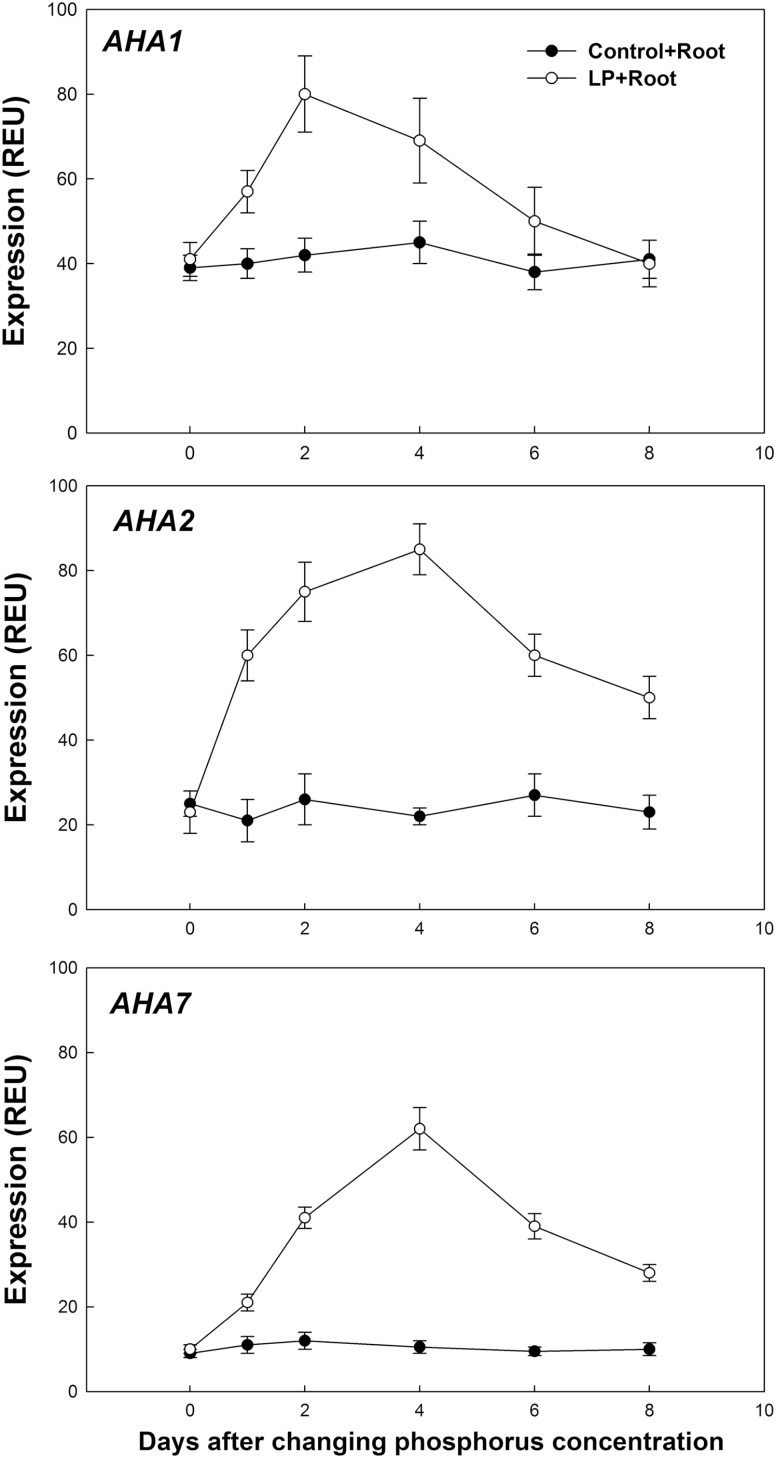
To investigate whether Arabidopsis PM H+-ATPase gene family members are regulated by soil phosphorus levels, we examined the expression of PM H+-ATPase genes (AHA1–AHA11) under LP conditions using real-time RT-PCR. To avoid biased results, real-time RT-PCR data are typically expressed relative to the expression of a housekeeping gene (i.e. At-ACT2) as an internal control. Using real-time RT-PCR, expression patterns of the 11 PM H+-ATPase genes were analysed in whole Arabidopsis plants (leaf and root) under phosphorous-sufficient conditions (control) or low-phosphorus stress (LP) for 4 days (Fig. 1). Under LP, the steady state transcript levels of most PM H+-ATPase gene family members appeared relatively unchanged relative to the control condition, but significant differences in three genes, AHA1, AHA2, and AHA7, were observed. The expression of selected PM H+-ATPase genes (AHA1, AHA2, and AHA7) was further investigated in Arabidopsis roots under control or LP conditions over 0, 1, 2, 4, 6, and 8 days (Fig. 2). During LP, from day 0 to day 4, the expression of AHA1, AHA2, and AHA7 in the roots increased approximately 2-, 4-, and 5-fold, respectively, compared with the control condition. After LP for 8 days, the expression level of AHA2 and AHA7 in the roots was approximately 2.5-fold that of the phosphorus-sufficient plants, but no significant change was found in the expression of AHA1 in roots over this time period.
Fig. 1.
Expression of PM H+-ATPase gene family members (AHA1–AHA11) in Arabidopsis plants in response to low phosphorus stress. Relative expression of various AHA genes in the whole plants of Arabidopsis grown in phosphorus-sufficient conditions (Control; 1 mM) or low phosphorus stress (LP; 1 μM) for 4 days. Values are first normalized to At-ACT2 (housekeeping gene; 100 REU) and then to control (+P) plants. Expression of these studied genes (AHA1–AHA11) under control was taken as 1-fold, and the expression level of these studied genes under LP is shown in this figure. The values are the means and SD of six replicates from two independent experiments. Changes in the relative expression levels of gene were checked for statistical significance according to Student’s t-test (P<0.05).
Fig. 2.
Expression of PM H+-ATPase gene family members (AHA1, AHA2, and AHA7) in the roots of Arabidopsis plants under phosphorus sufficient conditions (Control; 1 mM) or low phosphorus stress (LP; 1 μM) over 0, 1, 2, 4, 6, and 8 days. Relative expression levels were calculated and normalized with respect to At-ACT2 mRNA (=100 REU). Changes in the relative expression levels (REU) of gene mRNA were checked for statistical significance according to Student’s t-test (P<0.05). The values are the means and SD of six replicates from two independent experiments.
Response of PM H+-ATPase-deficient mutant roots to LP
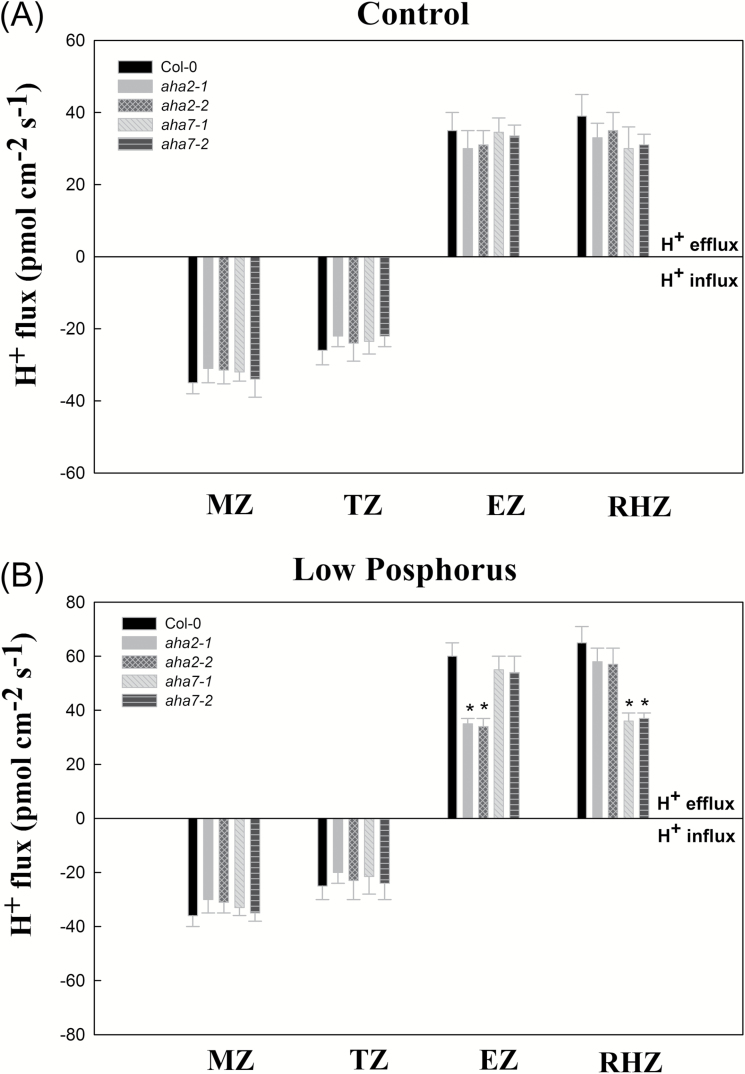
PM H+-ATPases play an important role not only in nutrient uptake by roots, but also in root adaption to environmental stress (Desnos, 2008; Xu et al., 2013a). We therefore analysed root phosphorus content, root surface area, primary root elongation, root hair density, PM H+-ATPase activity, proton extrusion, and proton flux in the roots of Col-0 (WT Arabidopsis plant), aha1 mutant lines (aha1-1 and aha1-2), aha2 mutant lines (aha2-1 and aha2-2), and aha7 mutant lines (aha7-1 and aha7-2) under normal growth conditions (control) and LP (Table 1 and Fig. 3). As shown in Table 1A, there was no significant difference in phosphorus content between the AHA-deficient mutants and Col-0 under control conditions. Under LP, no significant change was found in the phosphorus content of aha1 mutant lines compared with Col-0; however, the phosphorus contents of aha2 and aha7 mutant lines were significantly lower than that of Col-0. As shown in Table 1B, there was no significant difference in the root surface area between AHA-deficient mutants and Col-0 under control conditions. Under LP, no significant change was found in the root surface area of aha1 mutant lines compared with Col-0; however, the root surface areas of aha2 and aha7 mutant lines were significantly lower than that of Col-0. As shown in Table 1C, there was no significant difference in primary root elongation between AHA-deficient mutants and Col-0 under control conditions. Under LP, no significant change was found in the primary root elongation of aha1 or aha7 mutant lines compared with Col-0; however, the primary root elongation of aha2 mutant lines was significantly lower than that of Col-0. As shown in Table 1D, there was no significant difference in root hair density between AHA-deficient mutants and Col-0 under control conditions. Under LP, no significant change was found in root hair density of aha1 or aha2 mutant lines compared with Col-0; however, the root hair density of aha7 mutant lines was significantly lower than that of Col-0. As shown in Table 1E, there was no significant difference in PM H+-ATPase activity between AHA-deficient mutants and Col-0 under control conditions. Under LP, no significant change was found in PM H+-ATPase activity of aha1 mutant lines compared with Col-0; however, the PM H+-ATPase activities of aha2 and aha7 mutant lines were significantly lower than that of Col-0. As shown in Table 1F, there was no significant difference in proton extrusion between AHA-deficient mutants and Col-0 under control condition. Under LP, no significant change was found in proton extrusion in aha1 mutant lines compared with Col-0; however, proton extrusion was significantly lower in aha2 and aha7 mutant lines relative to Col-0. We also investigated proton (H+) flux in the meristem zone (MZ), transition zone (TZ), elongation zone (EZ), and root hair zone (RHZ) in the root tip of WT Arabidopsis (Col-0), aha2 mutant lines (aha2-1 and aha2-2), and aha7 mutant lines (aha7-1 and aha7-2) under normal growth conditions (control) and LP. As shown in Fig. 3, there was no significant difference in proton flux between AHA-deficient mutants and Col-0 under control conditions. Under LP, there was no significant difference in proton flux in the MZ and TZ of AHA-deficient mutants and Col-0. However, under LP, proton efflux in the EZ of aha2 mutant lines was significantly lower than in the EZ of Col-0 and aha7. Finally, under LP, proton efflux in the RHZ of aha7 mutant lines was significantly lower than in the EZ of Col-0 and aha2.
Table 1.
Phosphorus content, root surface area, elongation rate of primary root, root hair density, activity of PM H+-ATPase and proton extrusion in the roots of Col-0 (wild-type Arabidopsis plant), aha1 mutant lines (aha1-1 and aha1-2), aha2 mutant lines (aha2-1 and aha2-2) and aha7 mutant lines (aha7-1 and aha7-2) under normal growth condition (Control) or low phosphorus stress (LP)
Twenty-two-day-old Arabidopsis plants (wild-type and mutant lines) were treated with 1 mM phosphorus (Control) and 1 μM phosphorus (LP) for 8 days under hydroponic system. Then, 30-day-old Arabidopsis plants were used for experimental analysis. The data were subjected to analysis of variance and post hoc comparisons were performed with Duncan’s multiple range test at the P<0.05 level. The statistical software program used was SPSS version 13.0. The values are the means and SD of six replicates from two independent experiments. Values with the same letter (a or b or c) are not significantly different at P<0.05 level under the same treatment (control or LP).
| Col-0 | aha1-1 | aha1-2 | aha2-1 | aha2-2 | aha7-1 | aha7-2 | |
|---|---|---|---|---|---|---|---|
| (A) Root phosphorus content (μg plant–1) | |||||||
| Control | 9.2 ± 0.3 a | 8.9 ± 0.5 a | 9.1 ± 0.2 a | 9.0 ± 0.5 a | 8.8 ± 0.6 a | 9.3 ± 0.2 a | 9.0 ± 0.3 a |
| LP | 6.9 ± 0.5 a | 6.3 ± 0.2 a | 6.5 ± 0.5 a | 5.1 ± 0.3 b | 5.0 ± 0.4 b | 5.2 ± 0.2 b | 4.9 ± 0.6 b |
| (B) Root surface area (cm2 plant–1) | |||||||
| Control | 6.6 ± 0.5 a | 6.0 ± 0.4 a | 6.1 ± 0.3 a | 6.3 ± 0.4 a | 5.9 ± 0.3 a | 6.5 ± 0.6 a | 6.2 ± 0.2 a |
| LP | 4.6 ± 0.3 a | 4.4 ± 0.2 a | 4.5 ± 0.3 a | 3.1 ± 0.2 b | 3.0 ± 0.3 b | 3.3 ± 0.1 b | 3.2 ± 0.2 b |
| (C) Elongation rate of primary root (μm h–1) | |||||||
| Control | 120 ± 8.1 a | 116 ± 9.0 a | 115 ± 7.5 a | 110 ± 9.3 a | 109 ± 8.2 a | 118 ± 8.5 a | 115 ± 9.4 a |
| LP | 90 ± 6.2 a | 88 ± 5.5 a | 91 ± 9.6 a | 61 ± 8.1 b | 66 ± 7.3 b | 85 ± 8.5 a | 80 ± 9.8 a |
| (D) Root hair density (no. in 100 × 100 mm2) | |||||||
| Control | 1.35 ± 0.11 a | 1.30 ± 0.12 a | 1.34 ± 0.09 a | 1.31 ± 0.15 a | 1.32 ± 0.09 a | 1.22 ± 0.09 a | 1.18 ± 0.14 a |
| LP | 1.98 ± 0.15 a | 1.91 ± 0.12 a | 1.89 ± 0.11 a | 1.75 ± 0.20 a | 1.78 ± 0.18 a | 1.32 ± 0.14 b | 1.30 ± 0.17 b |
| (E) Activity of PM H+-ATPase (μmol min–1 mg–1 protein) | |||||||
| Control | 1.28 ± 0.09 a | 1.21 ± 0.13 a | 1.22 ± 0.18 a | 1.10 ± 0.15 a | 1.12 ± 0.19 a | 1.18 ± 0.14 a | 1.19 ± 0.12 a |
| LP | 2.32 ± 0.13 a | 2.06 ± 0.15 a | 2.02 ± 0.11 a | 1.25 ± 0.12 b | 1.18 ± 0.18 b | 1.46 ± 0.13 b | 1.48 ± 0.15 b |
| (F) Rate of root proton extrusion (μmol h–1 10 g–1 FW) | |||||||
| Control | 1.6 ± 0.3 a | 1.4 ± 0.2 a | 1.3 ± 0.2 a | 1.1 ± 0.3 a | 1.2 ± 0.2 a | 1.4 ± 0.1 a | 1.4 ± 0.3 a |
| LP | 2.9 ± 0.3 a | 2.7 ± 0.1 a | 2.6 ± 0.2 a | 1.3 ± 0.2 b | 1.2 ± 0.1 b | 1.7 ± 0.2 b | 1.7 ± 0.1 b |
Fig. 3.
Proton (H+) flux in meristem zone (MZ), transition zone (TZ), elongation zone (EZ), and root hair zone (RHZ) in the root tip of wild-type Arabidopsis (Col-0), aha2 mutant lines (aha2-1 and aha2-2) and aha7 mutant lines (aha7-1 and aha7-2) under normal growth condition (Control; A) or low phosphorus stress (B). Arabidopsis plants were treated as described in Table 1.
Effect of 14-3-3 proteins on the root response of PM H+-ATPase-deficient mutants to LP
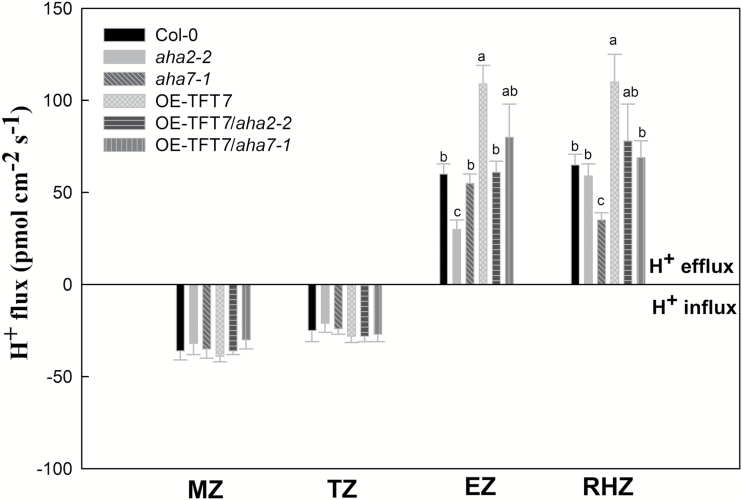
In our previous study (Xu et al., 2012), we demonstrated that the tomato 14-3-3 protein TFT7 directly functions in the root by enhancing Arabidopsis H+ secretion under LP. Subsequently, we set out to establish whether AHA2 or AHA7 is involved in the root response of TFT7-overexpressing Arabidopsis plants (OE-TFT7) to LP. By interbreeding transgenic plant strains, we obtained OE-TFT7 plants that lacked AHA2 or AHA7. Root phosphorus content, root surface area, primary root elongation, root hair density, PM H+-ATPase activity, proton extrusion, and proton flux were analysed in the roots of Col-0 (WT Arabidopsis plant), OE-TFT7 (TFT7-overexpressing Arabidopsis plants), and mutant lines (aha2-2, aha7-1, OE-TFT7/aha2-2, and OE-TFT7/aha7-1) under normal growth conditions (control) and LP (Table 2 and Fig. 4). As shown in Table 2, there was no significant difference in the phosphorus content among these plants (Col-0, OE-TFT7, aha2-2, aha7-1, OE-TFT7/aha2-2, and OE-TFT7/aha7-1) under control conditions. Under LP, the phosphorus contents, root surface areas, primary root elongation, of Col-0, OE-TFT7/aha2-2, and OE-TFT7/aha7-1 (no significance difference was found among these strains) were significantly lower than that of OE-TFT7. Furthermore, the primary root elongation of OE-TFT7/aha7-1 was significantly higher than that of aha2-2, but no significant difference was observed between OE-TFT7/aha7-1 and Col-0, aha7-1, OE-TFT7/aha2-2, and OE-TFT7. Under LP, the root hair densities of Col-0, aha2-2, and OE-TFT7/aha7-1 (no significance difference was found among these strains) were significantly higher than that of aha7-1, but lower than that of OE-TFT7. In addition, the root hair density of OE-TFT7/aha2-2 was significantly higher than that of aha7-1; however, no significant difference was found in the root hair density of OE-TFT7/aha2-2 compared with Col-0, aha2-2, OE-TFT7/aha7-1, and OE-TFT7. As shown in Table 2E, F, there was no significant difference in PM H+-ATPase activity or proton extrusion among Col-0, aha2-2, aha7-1, OE-TFT7/aha2-2, and OE-TFT7/aha7-1 under control conditions; however, H+-ATPase activity and proton extrusion in each of these strains was significantly lower than in OE-TFT7. Under LP, PM H+-ATPase activity and proton extrusion in Col-0, OE-TFT7/aha2-2, and OE-TFT7/aha7-1 (no significance difference was found among these strains) was significantly higher than in aha2-2 or aha7-1, but lower than in OE-TFT7. As shown in Fig. 4, under LP, there was no significant difference in proton flux among these plants (Col-0, OE-TFT7, aha2-2, aha7-1, OE-TFT7/aha2-2, and OE-TFT7/aha7-1) in the MZ or TZ of the root tip. However, in the EZ of the root tip, proton efflux in Col-0, aha7-1, and OE-TFT7/aha2-2 (no significance difference was found among these strains) was significantly higher than in aha2-2, but lower than in OE-TFT7. Proton efflux in OE-TFT7/aha7-1, however, was significantly higher than in aha2-2, but no significant change was found in OE-TFT7/aha7-1 compared with Col-0, aha7-1, OE-TFT7/aha2-2, and OE-TFT7. Further, in the RHZ of the root tip, proton efflux in Col-0, aha2-2, and OE-TFT7/aha7-1 (no significance difference was found among these strains) was significantly higher than in aha7-1, but lower than in OE-TFT7. Proton efflux in OE-TFT7/aha2-2 was significantly higher than in aha7-1, but no significant change was found in OE-TFT7/aha2-2 compared with Col-0, aha2-2, OE-TFT7/aha7-1, and OE-TFT7.
Table 2.
Phosphorus content, root surface area, elongation rate of primary root, root hair density, activity of PM H + -ATPase and proton extrusion in the roots of Col-0 (wild-type Arabidopsis plant), OE-TFT7 (TFT7-overexpressing Arabidopsis plants) and mutant lines (aha2-2, aha7-1, OE-TFT7/aha2-2 and OE-TFT7/aha7-1) under normal growth condition (Control) or low phosphorus stress (LP)
Twenty-two-day-old Arabidopsis plants (wild-type, TFT7-overexpressing and mutant lines) were treated with 1 mM phosphorus (Control) and 1 μM phosphorus (LP) for 8 days under hydroponic system. Then, 30-day-old Arabidopsis plants were used for experimental analysis. The data were subjected to analysis of variance and post hoc comparisons were done with Duncan’s multiple range test at the P<0.05 level. The statistical software program used was SPSS version 13.0. The values are the means and SD of six replicates from two independent experiments. Values with the same letter (a, b or c) are not significantly different at P<0.05 level under the same treatment (control or LP).
| Col-0 | aha2-2 | aha7-1 | OE-TFT7 | OE-TFT7/aha2-2 | OE-TFT7/aha7-1 | |
|---|---|---|---|---|---|---|
| (A) Root phosphorus content (μg plant–1) | ||||||
| Control | 9.1 ± 0.2 a | 9.0 ± 0.4 a | 8.9 ± 0.6 a | 9.5 ± 0.5 a | 9.1 ± 0.2 a | 9.0 ± 0.4 a |
| LP | 6.8 ± 0.2 a | 5.1 ± 0.2 b | 5.0 ± 0.4 b | 8.3 ± 0.2 c | 7.2 ± 0.3 a | 7.0 ± 0.5 a |
| (B) Root surface area (cm2 plant–1) | ||||||
| Control | 6.5 ± 0.4 a | 6.6 ± 0.5 a | 6.4 ± 0.3 a | 6.8 ± 0.6 a | 6.3 ± 0.2 a | 6.2 ± 0.1 a |
| LP | 4.4 ± 0.2 a | 3.1 ± 0.2 b | 3.2 ± 0.3 b | 5.9 ± 0.3 c | 4.6 ± 0.2 a | 4.5 ± 0.3 a |
| (C) Elongation rate of primary root (μm h–1) | ||||||
| Control | 122 ± 7.1 a | 117 ± 8.5 a | 115 ± 9.4 a | 129 ± 9.2 a | 116 ± 9.0 a | 113 ± 9.8 a |
| LP | 91 ± 5.9 a | 63 ± 6.1 b | 83 ± 9.5 a | 118 ± 5.2 c | 93 ± 8.0 a | 101 ± 13.7 ac |
| (D) Root hair density (no. in 100 × 100 mm2) | ||||||
| Control | 1.36 ± 0.15 a | 1.32 ± 0.10 a | 1.26 ± 0.16 a | 1.49 ± 0.25 a | 1.36 ± 0.09 a | 1.30 ± 0.14 a |
| LP | 1.88 ± 0.12 a | 1.80 ± 0.16 a | 1.30 ± 0.12 b | 2.38 ± 0.12 c | 2.12 ± 0.21 ac | 1.98 ± 0.14 a |
| (E) Activity of PM H+-ATPase (μmol min–1 mg–1 protein) | ||||||
| Control | 1.25 ± 0.11 a | 1.15 ± 0.15 a | 1.20 ± 0.16 a | 1.6 ± 0.10 b | 1.28 ± 0.13 a | 1.29 ± 0.17 a |
| LP | 2.30 ± 0.15 a | 1.19 ± 0.11 b | 1.45 ± 0.17 b | 3.17 ± 0.14 c | 2.46 ± 0.22 a | 2.51 ± 0.25 a |
| (F) Rate of root proton extrusion (μmol h–1 10 g–1 FW) | ||||||
| Control | 1.5 ± 0.4 a | 1.3 ± 0.2 a | 1.4 ± 0.1 a | 2.5 ± 0.3 b | 1.7 ± 0.2 a | 1.6 ± 0.3 a |
| LP | 2.8 ± 0.3 a | 1.5 ± 0.1 b | 1.6 ± 0.2 b | 5.0 ± 0.6 c | 3.0 ± 0.3 a | 3.1 ± 0.4 a |
Fig. 4.
Proton (H+) flux in meristem zone (MZ), transition zone (TZ), elongation zone (EZ) and root hair zone (RHZ) in the root tip of wild-type Arabidopsis (Col-0), OE-TFT7 (TFT7-overexpressing Arabidopsis plants) and mutant lines (aha2-2, aha7-1, OE-TFT7/aha2-2 and OE-TFT7/aha7-1) in response to low phosphorus stress. Arabidopsis plants were treated as described in Table 1.
Proton regulation and auxin modulation in the root response of PM H+-ATPase-deficient mutants to LP
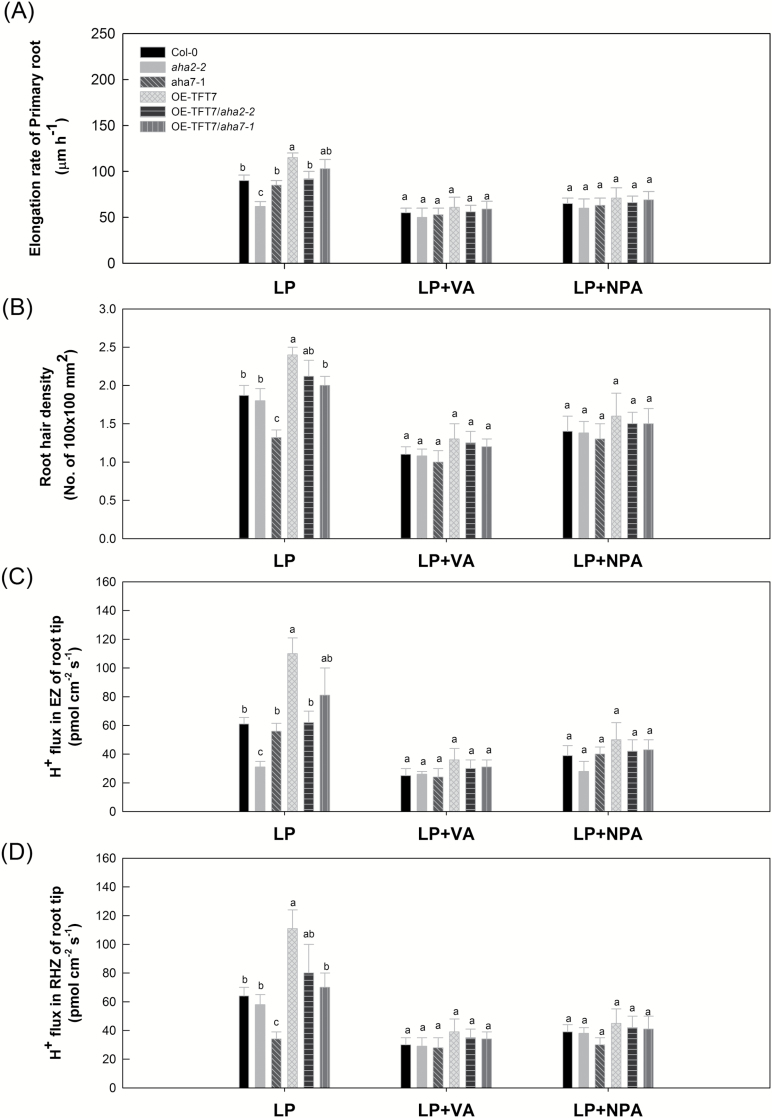
Our previously reported data (Xu et al., 2013b) demonstrate that auxin plays important roles in proton flux in the root tip in response to environmental stress. In the study described here, we investigated the root response of WT Arabidopsis plants (Col-0), OE-TFT7 (TFT7-overexpressing Arabidopsis plants), and mutant lines (aha2-2, aha7-1, OE-TFT7/aha2-2, and OE-TFT7/aha7-1) under LP, and LP with vanadate (VA; a PM H+-ATPase inhibitor) or N-1-naphthylphthalamic acid (NPA; an auxin flux inhibitor). Under LP, primary root elongation in Col-0, aha7-1, OE-TFT7, OE-TFT7/aha2-2, and OE-TFT7/aha7-1 was significantly higher than in aha2-2. However, under LP with VA or NPA, no significant difference in primary root elongation was found among these strains (Fig. 5A). Although the root hair densities of Col-0, aha2-2, OE-TFT7, OE-TFT7/aha2-2, and OE-TFT7/aha7-1 were significantly higher than that of aha7-1 under LP, no significant difference was found among them under LP with VA or NPA (Fig. 5B). H+ flux in the root tip EZ in Col-0, aha7-1, OE-TFT7, OE-TFT7/aha2-2, and OE-TFT7/aha7-1 was significantly higher than in aha2-2 under LP; however, under LP with VA or NPA, no significant differences were found in H+ flux in the root tip EZ among strains (Fig. 5C). Under LP, although H+ flux in the root tip RHZ in Col-0, aha2-2, OE-TFT7, OE-TFT7/aha2-2, and OE-TFT7/aha7-1 was significantly higher than in aha7-1, no significant difference was found among them under LP with VA or NPA (Fig. 5B).
Fig. 5.
Root response of wild-type Arabidopsis plants (Col-0), OE-TFT7 (TFT7-overexpressing Arabidopsis plants) and mutant lines (aha2-2, aha7-1, OE-TFT7/aha2-2 and OE-TFT7/aha7-1) to low phosphorus stress. Twenty-two-day-old Arabidopsis plants were treated with 1 μM phosphorus (LP), LP with 1 mM vanadate (VA; PM H+-ATPase inhibitor) or NPA (auxin flux inhibitor; 10 μM) for 8 days under a hydroponic system. (A) Elongation rate of primary root; (B) root hair density in the root tip of Arabidopsis (0–5000 μm from the root cap junction); (C) H+ flux in elongation zone (EZ); and (D) H+ flux in root hair zone (RHZ) in the root tip of Arabidopsis plants.
Discussion
AHA2 plays a regulatory role in H+ efflux in the root elongation zone in the context of primary root elongation in response to LP
One of the most pronounced adaptations of plants to low-phosphorus soil conditions is the enhancement of root elongation (Richardson, 2009). Primary root elongation, regulated by a sensory zone in the root tip, plays an important role in the plastic acclimation response to fluctuating soil environments (Baluska et al., 2010). PM H+-ATPase-regulated proton (H+) excretion has an important function in this process. The acid growth theory implicates that apoplastic H+ is the major wall-loosening factor that causes root cell extension (Staal et al., 2011). Apoplastic H+ is mainly attributed to H+ efflux mediated by PM H+-ATPase (Palmgren, 2001). Wang et al. (2014) proposed that AHA2 is useful for promoting plant growth by modulating light-induced stomatal opening. AHA2 is also important in maintaining the plasma membrane proton motive force (PMF) during plant seedling growth (Haruta and Sussman, 2012). We found that the expression of AHA2 is up-regulated in roots under LP (Figs 1 and 2). As shown in Table 1, when AHA2 is deficient in Arabidopsis plants, as in the aha2 mutant, root phosphorus content, root surface area, PM H+-ATPase activity, and root proton extrusion are lower than in the Col-0 (WT) and the aha1 mutant. Although there was no significant difference in root hair density between aha2 and Col-0, primary root elongation was lower in aha2 than in Col-0 and aha7. Further, under LP, although no significant difference in H+ efflux of the root hair zone was found between Col-0 and aha2, H+ efflux in the root elongation zone of aha2 was significantly lower than that in Col-0 (Fig. 3). Thus, our results suggest that AHA2 plays a regulatory role in H+ efflux in the root elongation zone in the context of the primary root elongation response to LP.
AHA7-H+-mediated efflux in the root hair zone is important for root hair formation under LP
Plants have developed physiological and morphological adaptations to acquire phosphorus from the soil. These adaptations include not only increased root length for phosphorus uptake over a greater soil volume, but also root hair formation for more efficient acquisition of phosphorus (Raghothama, 1999; Hammond et al., 2004; Lambers et al., 2006). Studies have shown that PM H+-ATPase-regulated H+ flux is important for root hair development (Palmgren, 2001). For instance, PM H+-ATPase protein is highly concentrated in root hairs, and several isoforms of plant PM H+-ATPase localize to root hairs. H+ circuits have also been found in root hairs: H+ enters the root hair at the root tip and leaves the root hair below this tip. AHA7 is the PM H+-ATPase responsible for iron deficiency-induced differentiation of rhizodermic cells. The action of AHA7 is not affected by the modulation of Arabidopsis 14-3-3 protein GRF11 under conditions of iron deficiency (Yang et al., 2013). We also found that the expression of AHA7 is up-regulated in roots under LP (Fig. 1 and Fig. 2). According to Table 1, when AHA7 is deficient in Arabidopsis plants, as in the aha7 mutant, root phosphorus content, root surface area, PM H+-ATPase activity, and root proton extrusion are lower than in the Col-0 (WT) and aha1 mutant. Although there was no significant difference in primary root elongation between aha7 and Col-0, root hair density of aha7 was lower than that of Col-0 and aha2. Further, under LP, although no significant difference was found in H+ efflux in the root elongation zone between Col-0 and aha7, H+ efflux in the root hair zone of aha7 was significantly lower than that of Col-0 (Fig. 3). Thus, our results suggest that AHA7-mediated H+ efflux in the root hair zone is important for root hair formation under LP.
Effect on AHA2 or AHA7 regulation of the action of 14-3-3 proteins or auxin modulation in the root response to LP
In plants, 14-3-3 proteins regulate the activity of PM H+-ATPases (Chevalier et al., 2009). The binding of 14-3-3 proteins to PM H+-ATPases blocks the autoinhibitory action of the PM H+-ATPase C-terminus, thus increasing PM H+-ATPase activity. The 14-3-3 binding sites on Arabidopsis were mapped to the extreme C-terminus, where the penultimate threonine is phosphorylated. The plant 14-3-3 protein family has many members, with high levels of conservation, and most can bind PM H+-ATPase. Environmental factors, such as light, can stimulate 14-3-3 interactions with PM H+-ATPase (Kinoshita and Shimazaki, 1999). This increase in PM H+-ATPase activity causes guard cells to swell, opening the stomata. Our previous work showed that the tomato 14-3-3 protein TFT7 acts mainly in the root and is involved in the local response of plants to LP by activating root plasma membrane H+-ATPase (Xu et al., 2012). By interbreeding strains, we obtained TFT7-overexpressing Arabidopsis plants (OE-TFT7) that lacked AHA2 or AHA7. As shown in Table 2, under LP, root phosphorus content, root surface area, PM H+-ATPase activity, and root proton extrusion of OE-TFT7/aha2 and OE-TFT7/aha7 were significantly lower than that of OE-TFT7. However, primary root elongation in OE-TFT7 was significantly higher than in OE-TFT7/aha2, but not OE-TFT7/aha7. The root hair densities in OE-TFT7 were significantly higher than in OE-TFT7/aha7, but not OE-TFT7/aha2. Further, under LP, H+ efflux in the root elongation zone of OE-TFT7 was significantly higher than in OE-TFT7/aha2, but not OE-TFT7/aha7. Conversely, H+ efflux in the root hair zone of OE-TFT7 was significantly higher than in OE-TFT7/aha7, but not OE-TFT7/aha2 (Fig. 4). It is well known that plants enhance proton extrusion by up-regulating PM H+-ATPases to facilitate phosphorus mobilization and uptake under LP. As shown in Table 2, root proton extrusion of aha2 or aha7 was significantly lower than that of Col-0 (wild-type) under LP. And also, when AHA2 or AHA7 was deficient in TFT7-overexpressing plants (OE-TFT7), root proton extrusion of OE-TFT7 under LP was decreased greatly. Thus, our results suggest that AHA2 or AHA7 is involved in the action of 14-3-3 proteins in the root response to LP.
In plants, auxin also regulates the activity of PM H+-ATPases. The acid growth theory implicates (Hager, 2003) that (i) PM H+-ATPase is responsible for the extrusion of H+ into the cell wall; (ii) auxin enhances the activity of PM H+-ATPase, thus causing a lowering of the cell wall pH; (iii) the reduced pH in the cell wall activates cell wall-loosening enzymes and initiates the enlargement of the cell. Our previous results also suggest that auxin modulation and auxin cell–cell transport are key regulators of acidity-induced cell elongation in the root under alkaline stress (Xu et al., 2013a). In this study, under LP, primary root elongation and H+ efflux in the root elongation zone of OE-TFT7 were significantly higher than in TFT7/aha2 (Fig. 5). In addition, the root hair density and H+ efflux in the root hair zone of OE-TFT7 were significantly higher than in TFT7/aha7. Under PM H+-ATPase inhibition or auxin flux inhibition, however, no significant difference in these indexes was found among these treated plants (Fig. 5). Thus, our results suggest that the H+-mediated regulation of AHA2 and AHA7 is affected by auxin modulation in the root response to LP.
In conclusion, H+ secretion, regulated by PM H+-ATPases, plays an important role in the plant response to LP. On the one hand, H+ extrusion in the rhizosphere is important for the release of phosphorus from the soil. On the other hand, H+ efflux (cell–cell transport) in the root can loosen the cell wall to promote root cell elongation to further increase phosphorus uptake. In this process, AHA2 plays a regulatory role in primary root elongation by modulating H+ efflux in the root elongation zone in response to LP. Similarly, AHA7 is important for root hair formation, because it mediates H+ efflux in the root hair zone under LP. The H+-mediated regulation of AHA2 or AHA7 are affected by 14-3-3 proteins and auxin modulation in the root response to LP. We conclude that AHA2 and AHA7, which encode two Arabidopsis plasma membrane H+-ATPases, differentially modulate H+ efflux in the root tip and thereby have distinct and overlapping roles in response to LP.
Supplementary Material
Acknowledgements
We are grateful for grant support from the National Natural Science Foundation of China (Nos 31422047 and 31272229), National Basic Research Program of China (Nos 2013CB127402 and 2012CB114300), NSFC Major Research Plan project (91317307), Hong Kong Research Grants Council (AoE/M-05/12, CUHK 14122415 and 14160516) and Shenzhen Overseas Talents Innovation & Entrepreneurship Funding Scheme (The Peacock Scheme), Program of New Century Excellent Talent in Universities (NCET-11-0672) and strategic leading special science and technology project of Chinese Academy of Sciences (XDB15030201).
References
- Baluska F, Mancuso S, Volkmann D, Barlow PW. 2010. Root apex transition zone: a signalling-response nexus in the root. Trends in Plant Science 15, 402–408. [DOI] [PubMed] [Google Scholar]
- Baxter IR, Young JC, Armstrong G, et al. 2005. A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 102, 2649–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova TN, Jacob T, Dahse I, Gilroy S. 1998. Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125, 2925–2934. [DOI] [PubMed] [Google Scholar]
- Caesar K, Elgass K, Chen Z, Huppenberger P, Witthöft J, Schleifenbaum F, Blatt MR, Oecking C, Harter K. 2011. A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. The Plant Journal 66, 528–540. [DOI] [PubMed] [Google Scholar]
- Chevalier D, Morris ER, Walker JC. 2009. 14-3-3 and FHA domains mediate phosphoprotein interactions. Annual Review of Plant Biology 60, 67–91. [DOI] [PubMed] [Google Scholar]
- Desnos T. 2008. Root branching responses to phosphate and nitrate. Current Opinion in Plant Biology 11, 82–87. [DOI] [PubMed] [Google Scholar]
- Devaiah BN, Nagarajan VK, Raghothama KG. 2007. Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiology 145, 147–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gévaudant F, Duby G, von Stedingk E, Zhao R, Morsomme P, Boutry M. 2007. Expression of a constitutively activated plasma membrane H+-ATPase alters plant development and increases salt tolerance. Plant Physiology 144, 1763–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager A. 2003. Role of the plasma membrane H+-ATPase in auxin-induced elongation growth: historical and new aspects. Journal of Plant Research 116, 483–505. [DOI] [PubMed] [Google Scholar]
- Hammond JP, Broadley MR, White PJ. 2004. Genetic responses to phosphorus deficiency. Annals of Botany 94, 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson WC. 1950. The photometric determination of phosphorus in fertilizers using the phosphovanado-molybdate complex. Journal of the Science of Food and Agriculture 1, 172–173. [Google Scholar]
- Haruta M, Sussman MR. 2012. The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis. Plant Physiology 158, 1158–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffland E, Wei C, Wissuwa M. 2006. Organic anion exudation by lowland rice (Oryza sativa L.) at zinc and phosphorus deficiency. Plant and Soil 283, 155–162. [Google Scholar]
- Jin CW, Du ST, Chen WW, Li GX, Zhang YS, Zheng SJ. 2009. Elevated carbon dioxide improves plant iron nutrition through enhancing the iron-deficiency-induced responses under iron-limited conditions in tomato. Plant Physiology 150, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Shimazaki Ki. 1999. Blue light activates the plasma membrane H+-ATPase by phosphorylation of the C-terminus in stomatal guard cells. The EMBO Journal 18, 5548–5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers H, Shane MW, Cramer MD, Pearse SJ, Veneklaas EJ. 2006. Root structure and functioning for efficient acquisition of phosphorus: Matching morphological and physiological traits. Annals of Botany 98, 693–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YS, Mao XT, Tian QY, Li HL, Zhang WH. 2009. Phosphorus deficiency-induced reduction in root hydraulic conductivity in Medicago falcate is associated with ethylene production. Environmental and Experimental Botany 67, 172–177. [Google Scholar]
- Moloney MM, Elliott MC, Cleland RE. 1981. Acid growth effects in maize roots: Evidence for a link between auxin-economy and proton extrusion in the control of root growth. Planta 152, 285–291. [DOI] [PubMed] [Google Scholar]
- Palmgren MG. 2001. Plant plasma membrane H+-ATPases: powerhouses for nutrient uptake. Annual Review of Plant Physiology and Plant Molecular Biology 52, 817–845. [DOI] [PubMed] [Google Scholar]
- Raghothama KG. 1999. Phosphate acquisition. Annual Review of Plant Physiology and Plant Molecular Biology 50, 665–693. [DOI] [PubMed] [Google Scholar]
- Richardson AE. 2009. Regulating the phosphorus nutrition of plants: molecular biology meeting agronomic needs. Plant and Soil 322, 17–24. [Google Scholar]
- Robertson WR, Clark K, Young JC, Sussman MR. 2004. An Arabidopsis thaliana plasma membrane proton pump is essential for pollen development. Genetics 168, 1677–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues RB, Sabat G, Minkoff BB, Burch HL, Nguyen TT, Sussman MR. 2014. Expression of a translationally fused TAP-tagged plasma membrane proton pump in Arabidopsis thaliana. Biochemistry 53, 566–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S, Schmidt W. 2009. Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. The New Phytologist 183, 1072–1084. [DOI] [PubMed] [Google Scholar]
- Schachtman DP, Shin R. 2007. Nutrient sensing and signaling: NPKS. Annual Review of Plant Biology 58, 47–69. [DOI] [PubMed] [Google Scholar]
- Shen H, Chen J, Wang Z, Yang C, Sasaki T, Yamamoto Y, Matsumoto H, Yan X. 2006. Root plasma membrane H+-ATPase is involved in the adaptation of soybean to phosphorus starvation. Journal of Experimental Botany 57, 1353–1362. [DOI] [PubMed] [Google Scholar]
- Staal M, De Cnodder T, Simon D, Vandenbussche F, Van der Straeten D, Verbelen JP, Elzenga T, Vissenberg K. 2011. Apoplastic alkalinization is instrumental for the inhibition of cell elongation in the Arabidopsis root by the ethylene precursor 1-aminocyclopropane-1-carboxylic acid. Plant Physiology 155, 2049–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno K, Kinoshita T, Inoue S, Emi T, Shimazaki K. 2005. Biochemical characterization of plasma membrane H+-ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant & Cell Physiology 46, 955–963. [DOI] [PubMed] [Google Scholar]
- Vance CP, Uhde-Stone C, Allan DL. 2003. Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. The New Phytologist 157, 423–447. [DOI] [PubMed] [Google Scholar]
- Wang Y, Noguchi K, Ono N, Inoue S, Terashima I, Kinoshita T. 2014. Overexpression of plasma membrane H+-ATPase in guard cells promotes light-induced stomatal opening and enhances plant growth. Proceedings of the National Academy of Sciences of the United States of America 111, 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Ma L, Hou X, Wang M, Wu Y, Liu F, Deng XW. 2003. Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiology 132, 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Jia L, Shi W, Baluska F, Kronzucker HJ, Liang J, Zhang J. 2013a The Tomato 14-3-3 protein TFT4 modulates H+ efflux, basipetal auxin transport, and the PKS5-J3 pathway in the root growth response to alkaline stress. Plant Physiology 163, 1817–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Jia L, Shi W, Liang J, Zhou F, Li Q, Zhang J. 2013b Abscisic acid accumulation modulates auxin transport in the root tip to enhance proton secretion for maintaining root growth under moderate water stress. The New Phytologist 197, 139–150. [DOI] [PubMed] [Google Scholar]
- Xu WF, Shi WM. 2006. Expression profiling of the 14-3-3 gene family in response to salt stress and potassium and iron deficiencies in young tomato (Solanum lycopersicum) roots: analysis by real-time RT-PCR. Annals of Botany 98, 965–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Shi W. 2008. A “nonsterile” method for selecting and growing Arabidopsis thaliana transformants (T2 Transgenic Lines) resistant to kanamycin. Plant Molecular Biology Reporter 26, 350–357. [Google Scholar]
- Xu W, Shi W, Jia L, Liang J, Zhang J. 2012. TFT6 and TFT7, two different members of tomato 14-3-3 gene family, play distinct roles in plant adaption to low phosphorus stress. Plant, Cell & Environment 35, 1393–1406. [DOI] [PubMed] [Google Scholar]
- Yan F, Zhu Y, Müller C, Zörb C, Schubert S. 2002. Adaptation of H+-pumping and plasma membrane H+ ATPase activity in proteoid roots of white lupin under phosphate deficiency. Plant Physiology 129, 50–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JL, Chen WW, Chen LQ, Qin C, Jin CW, Shi YZ, Zheng SJ. 2013. The 14-3-3 protein GENERAL REGULATORY FACTOR11 (GRF11) acts downstream of nitric oxide to regulate iron acquisition in Arabidopsis thaliana. The New Phytologist 197, 815–824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.