HIGHLIGHT
The chromatin-modifying complex Paf1 is required for shoot and root thigmomorphogenesis and touch-inducible gene expression.
Keywords: Histone methylation, Paf1 complex, TCH genes, thigmomorphogenesis, touch response, VIP3.
Abstract
Thigmomorphogenesis is a stereotypical developmental alteration in the plant body plan that can be induced by repeatedly touching plant organs. To unravel how plants sense and record multiple touch stimuli we performed a novel forward genetic screen based on the development of a shorter stem in response to repetitive touch. The touch insensitive (ths1) mutant identified in this screen is defective in some aspects of shoot and root thigmomorphogenesis. The ths1 mutant is an intermediate loss-of-function allele of VERNALIZATION INDEPENDENCE 3 (VIP3), a previously characterized gene whose product is part of the RNA polymerase II-associated factor 1 (Paf1) complex. The Paf1 complex is found in yeast, plants and animals, and has been implicated in histone modification and RNA processing. Several components of the Paf1 complex are required for reduced stem height in response to touch and normal root slanting and coiling responses. Global levels of histone H3K36 trimethylation are reduced in VIP3 mutants. In addition, THS1/VIP3 is required for wild type histone H3K36 trimethylation at the TOUCH3 (TCH3) and TOUCH4 (TCH4) loci and for rapid touch-induced upregulation of TCH3 and TCH4 transcripts. Thus, an evolutionarily conserved chromatin-modifying complex is required for both short- and long-term responses to mechanical stimulation, providing insight into how plants record mechanical signals for thigmomorphogenesis.
Introduction
It is firmly established that development in multicellular organisms relies on the local concentration of key biochemical signals such as hormones or growth factors (Wolpert, 1969). However, it is becoming increasingly clear that the temporal pattern of changes in the concentration of these biochemical signals is equally important for the final phenotypic output (Raj and van Oudenaarden, 2008; Lander, 2011; Oates, 2011; Webb et al., 2016). For instance, the intrinsic delays associated with transcription and translation have been proposed to play an instructive role in patterning (Monk, 2003; Gaffney and Monk, 2006). Understanding how the intensity as well as the frequency of a stimulus are measured and recorded is a key challenge for plant biologists, as post-embryonic development is continuously impacted by the environmental conditions in which the plant grows.
Mechanical stimuli are coded both in intensity and in frequency [for examples see (Chehab et al., 2009; Leblanc-Fournier et al., 2014)] but how this is accomplished at the molecular and genetic level is unknown. Here we begin to address the genetic pathways by which a repeated mechanical stimulus leads to a developmental change during plant thigmomorphogenesis, a stereotypical developmental alteration in the plant body plan that can be induced by shaking, rubbing, bending, or brushing leaves or stems (Chehab et al., 2009; Coutand, 2010). Thigmomorphogenesis is classically defined as producing a shorter and thicker plant due to decreased stem height and increased stem and/or petiole diameter relative to control plants (Jaffe, 1980; Biddington, 1986; Chehab et al., 2009; Coutand, 2010). Repeated mechanical stimulation can also result in a more flexible stem and an increase in root mass relative to the shoot. Taken together, these morphological changes are thought to produce a body plan that is more resistant to damage upon future mechanical challenges. This presents an intriguing conversion of energy from an elastic deformation (i.e. bending) into a developmental response (i.e. growth).
Three hallmarks of thigmomorphogenesis are: a dose responsiveness that prevents the plant from responding to isolated events, a systemic nature such that a stimulus applied to one organ leads to changes in overall plant morphology, and a characteristic delay between the initial stimulus and the developmental response (Chehab et al., 2009; Leblanc-Fournier et al., 2014; Moulia et al., 2015). The mechanical stimulus received by cells in the rosette leaves of plants that have been touched, sprayed, blown or bent must be transmitted to the cells of the shoot apical meristem in order to control elongation of the future shoot or stem (Moulia et al., 2015).
Indeed, there is evidence that plants are able to record the number and frequency of mechanical stimuli. The Venus flytrap uses action potentials to measure the number of times an insect touches the trigger hairs of its trap over the course of an hour (Böhm et al., 2016). Plants are also able to sense the number and frequency of stem bending events. When several bends are imposed on poplar stems, the impact on the transcriptome becomes less pronounced from the second bending event onwards. It also takes no less than a week without bending for a stem to recover full molecular sensitivity to such mechanical deformation (Martin et al., 2010). Thus, plants are indeed capable of recording successive mechanical deformations.
Plants also respond to touch in the short-term, though how these events are connected to long-term changes in body plan remains unclear. Following a single touch event, plants rapidly upregulate a large number of genes; in Arabidopsis as much as 2.5% of its genome (Braam and Davis, 1990; Lee et al., 2005). The best characterized of these in Arabidopsis are the TOUCH (TCH) genes, which encode calmodulins and calmodulin-like proteins, cell wall-modifying enzymes, and wound- and defense-inducible genes (Braam and Davis, 1990; Ling et al., 1991; Perera and Zielinski, 1992; Kagaya and Hattori, 2009; Lee et al., 2005). The timing of the induction of most TCH genes is between 5 and 30 minutes after the application of stimuli, suggesting a role in the early response to touch stimulation. The cml24 (tch2) mutation affects root morphology on hard agar and is implicated in microtubule structure and starvation-induced autophagy (Wang et al., 2011; Tsai et al., 2013) but a functional role for TCH genes in thigmomorphogenesis has yet to be established. While these results demonstrate that touch is perceived at the molecular level after a single event, no link between short-term and long-term responses to touch has been identified. In trees, the transcription factor gene ZFP2 is rapidly induced in response to bending (Leblanc-Fournier et al., 2008; Coutand et al., 2009). Interestingly, this induction can be attenuated after multiple bending events, suggesting that plants are able to record the number of mechanical perturbations, albeit through an unknown mechanism (Martin et al., 2010).
The growth response is likely to involve the phytohormone jasmonic acid (JA). JA accumulates in the stems of plants after touching of their rosette leaves, and JA biosynthesis and signaling are required for thigmomorphogenesis (Chehab et al., 2012). Conversely, application of the growth regulatory hormone gibberellin or a loss-of-function mutation in the gibberellin-catabolizing enzyme AtGA2ox7 can prevent thigmomorphogenesis (Lange and Lange, 2015). However, these effects are potentially far downstream in the pathway. No real known regulator of thigmomorphogenesis capable of sensing repetition has been identified yet.
We therefore performed a novel forward genetic screen designed to identify the key molecular and genetic components that link successive elastic deformations to thigmomorphogenesis. Arabidopsis exhibits several stereotypical thigmomorphogenic responses that require multiple days of touch, including the inhibition of stem elongation, shortened petioles, reduced rosette diameter and delayed transition to flowering (Braam and Davis, 1990; Chehab et al., 2012; Cazzonelli et al., 2014; Lange and Lange, 2015). Here we concentrated on stem thigmomorphogenesis to identify new regulators of the touch response. We screened for plants that did not exhibit shortened stems after repeated touch. The first mutant isolated in this screen identified a known regulator of gene expression, the RNA Polymerase II-associated factor 1 (Paf1) complex, as a key element of thigmomorphogenesis that serves to integrate touch stimuli over time.
Materials and methods
Touch response assays
For Fig. 1 and Fig. 2, plants were grown for one week in long day conditions of 16 hours of light and 8 hours of darkness at 23°C, then transferred to 24 hours of light at 25°C for one more week. These conditions were established during the screen and were replicated in later experiments to reduce variability due to growth conditions. Seedlings were allowed to germinate and grow in a large 16-hour chamber, then moved to a smaller 24-hour chamber that was set aside only for touch assay experiments. Two-week-old seedlings were then subjected to touch stimulus for 8–10 days, stopping when bolts were visible. For hand touching, a gloved hand was held parallel to the tray and moved across the pots, touching the tops of all plants within the tray with the palm once and then repeated for a total of ten passes each day. For paintbrushing, seedling leaves were brushed each day with ten passes of a 3-inch standard paintbrush held at a 45 degree angle to the soil surface. Touched and untouched plants of the same genotype were grown side-by-side in the same tray and trays were rotated 180 degrees every day to reduce variability in temperature and airflow within the growth chambers. Stem height was measured 10–14 days later, whenever the stem height of wild type untouched plants reached over 15 cm high or approximately 30 days after germination. The number of days required for stems to reach 15 cm was dependent on the light and temperature conditions of the particular experiment and chamber used; similar results were obtained when wild type untouched stems were measured anytime beyond this point. In Fig. 3, plants were grown in 16 hours of light in order to allow the null vernalization independence (vip) mutants to grow as tall as possible. In Fig. 4, 12-day-old seedlings were grown in long day conditions of 16 hours light and 8 hours darkness at 21°C and were either untouched or touched with a large, soft paintbrush every 5–7 seconds for 2 minutes, with on average 18 brushes per treatment. Material was collected 30 minutes after the first touch treatment or 30 minutes after plants were subjected to a second touch treatment 2 hours following the first one.
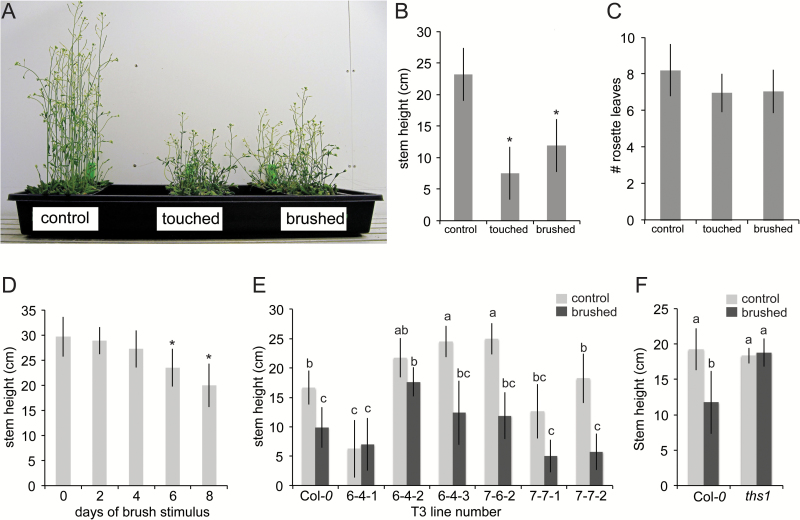
Fig. 1.
A forward genetic screen identifies an Arabidopsis mutant insensitive to repeated touch. (A-C) Characterization of hand touch-induced and paintbrush-induced thigmomorphogenesis. Two-week-old Col-0 seedlings grown on soil were either untreated, touched by a gloved hand or brushed with a paintbrush (10 passes per day). Plants were then grown for an additional 10 days without stimulus. (A) Image of control, hand-touched, and paintbrushed plants at the time point of stem height measurement. (B) Average stem height of plants in (A). At least 12 plants were assessed per treatment. (C) Flowering time, measured as the number of rosette leaves at the time of bolting, of plants shown in (A). (D) Brush dose response curve. At least 22 plants were assessed per treatment. This experiment was repeated once. (E) Secondary screen of T-DNA insertion lines for insensitivity to paintbrushing. Eight plants were assessed per line for each treatment. Line 6-4-2 was selected for further analysis and renamed touch-insensitive 1 (ths1). This experiment was not replicated but seeds were collected to assess in the next generation. (F) ths1 mutants are heritably insensitive to touch. Twenty plants were assessed per genotype for each treatment. Error bars indicate standard deviation. This experiment was repeated twice with similar results. In (B) and (D), asterisks mark difference from untouched controls, P<0.01 (Student’s t-test). In (E) and (F), statistical groups represented with letters were determined by ANOVA followed by Tukey’s HSD test, P<0.05.
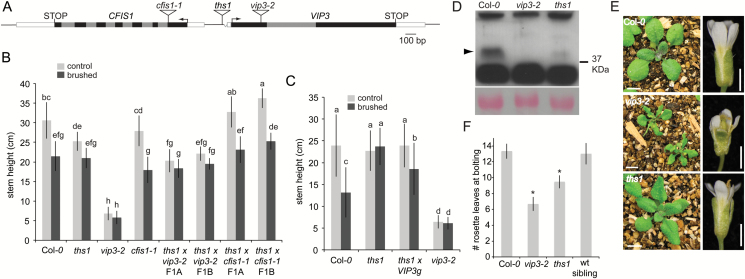
Fig. 2.
Touch insensitivity in the ths1 mutant is due to partial loss of function of the VIP3 gene. (A) Schematic of a region of chromosome 4 containing VIP3 and CFIS1 in ths1, vip3-2 (SALK_083364) and cfis1-1 (SAIL_367_F03) mutants. Black and grey boxes indicate exons and introns, respectively. Note that the deletion in the 5’UTR of VIP3 is present only in the ths1 allele. Arrows indicate transcriptional start sites. White boxes are 5’ and 3’ UTRs. (B) Complementation test, comparing stem height in parental and F1 lines in response to 8 days of paintbrushing as described in the legend to Fig. 1. At least 15 plants were used per treatment for each genotype. Error bars indicate standard deviation. This experiment was repeated once with similar results. Statistical groups represented with letters were determined by ANOVA followed by Scheffé’s test, P<0.05. (C) Partial rescue of touch insensitivity in the ths1 background with VIP3g. n=50 plants per treatment for each genotype. Error bars indicate standard deviation. Statistical groups represented with letters were determined by ANOVA followed by Scheffé’s test, P<0.05. This experiment was repeated twice with similar results. (D) Top, anti-VIP3 immunoblot. Arrow indicates the band corresponding to VIP3. Bottom, Ponceau S staining of the large subunit of Rubisco. (E) Three-week-old seedlings (left) and flowers (right) from the indicated genotypes grown in soil under 24 hours of light. (F) Number of rosette leaves prior to bolting in the indicated genotypes grown in 16 hours of light. n=30 plants per genotype. Asterisks mark significant difference from the untouched control P<0.01 (Student’s t-test). Error bars indicate standard deviation.
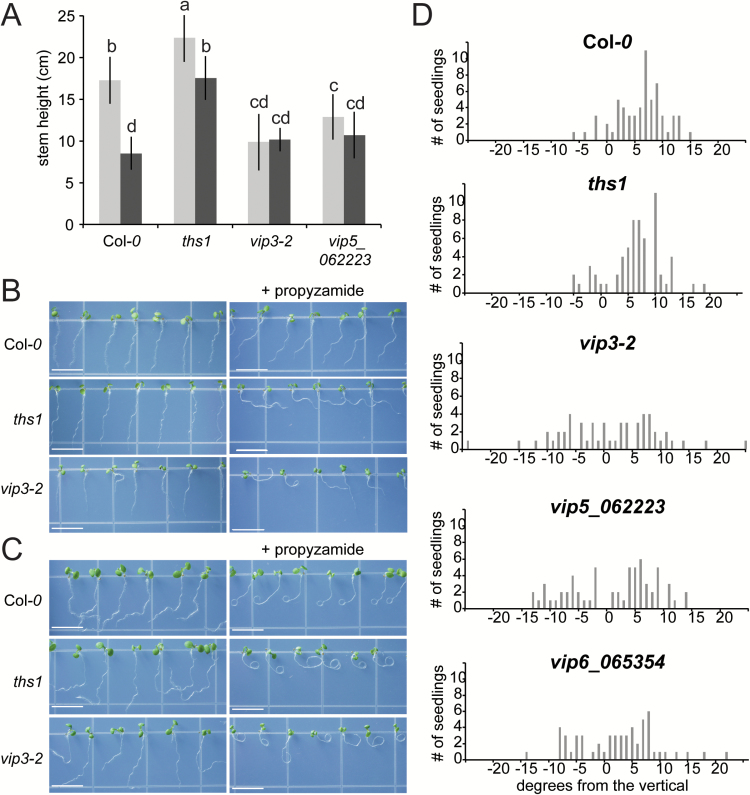
Fig. 3.
The Paf1 complex is required for stem and root thigmomorphogenesis. (A) Comparison of Col-0, ths1, vip3-2 and vip5_062223 plants after no touch (light bars) and after 8 days of paintbrushing (dark bars). Plants were grown under long day conditions of 16 hours of light in order to produce taller plants. At least seven plants were used for each genotype and treatment. Statistical groups represented with letters were determined by ANOVA followed by Scheffé’s test, P<0.05. Error bars indicate standard deviation. This experiment was also conducted in 24 hours of light with similar results. (B) In the slanting assay, wild type and mutant seedlings were grown horizontally on normal media or media supplemented with 3 μM propyzamide for 2 days, then tilted 30 degrees back from the vertical for 3 days. Size bar, 7 mm. (C) In the coiling assay, wild type and mutant seedlings were grown horizontally on normal media or media supplemented with 3 μM propyzamide for 2 days, then placed vertically for 3 days. (D) Distribution of root slanting angle in Col-0 and vip mutant seedlings in the absence of propyzamide. Data from three independent experiments are included in each chart, providing a total of n=55 seedlings for each genotype.
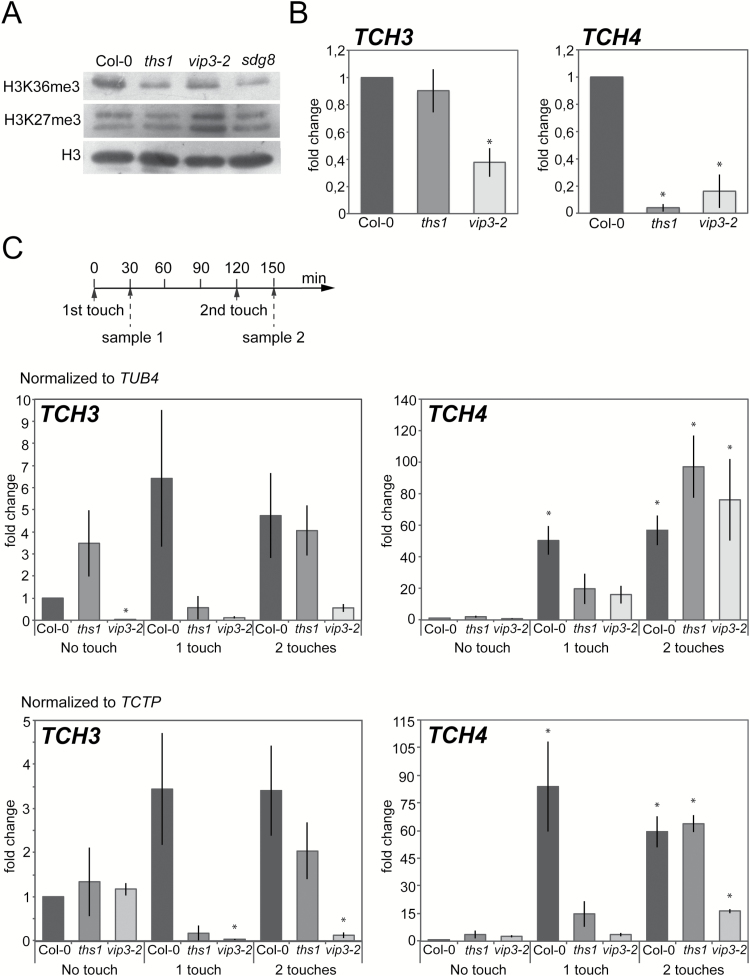
Fig. 4.
ths1 and vip3-2 mutants have altered global and genic histone H3 methylation patterns and fail to induce TCH3 and TCH4 in response to touch. (A) Chromatin extracts from cauline leaves of the indicated genotypes were separated by SDS-PAGE and detected with antibodies specific to H3K36me3 or H3K27me3. (B) Relative enrichment for H3K36me3 at the TCH3 and TCH4 loci in cauline leaf chromatin. Chromatin was isolated as in (A), then immunoprecipitated with the H3K36me3 antibody or without antibody and precipitated DNA amplified by qRT-PCR. Data presented is a percentage of the input, normalized to the S-ADENOSYLMETHIONINE SYNTHASE (SAM) gene and represented as a fold difference with the wild type non-treated sample. Error bars indicate the SEM for three biological replicates with two technical replicates each. (C) Quantitative reverse-transcriptase PCR analysis of TCH3 and TCH4 expression in response to touch in wild type and mutant seedlings. Seedlings were brushed for 2 minutes with a paintbrush according to the scheme shown in the top panel. RNA was prepared from the aerial tissues of treated and untreated plants and cDNA amplified with gene specific primers. The data was normalized to TUB4 (upper panels) or TCTP (lower panels) as reference genes and is presented as fold change compared to wild type non-treated samples. The error bars indicate SEM from three replicates. Asterisks mark significant difference from the untouched control P<0 05.
Identifying T-DNA
The left border of the T-DNA in ths1 plants was amplified with left border primers and AD1 by TAIL PCR as described (Liu et al., 1995). The third round TAIL PCR product was cloned using the pGEM-T Easy vector kit (Promega) and sequenced. The location of the T-DNA was verified by PCR genotyping. Oligos 29830.F and 29830.R were used to amplify the left border region from genomic DNA for sequencing and independent verification of the junction.
Characterizing ths1 T-DNA
Templates in PCR reactions were either 1 μl genomic DNA or 1 μl cDNA made from 5 mg leaf tissue from wild type or ths1 mutant plants. PCR reactions were carried out using Hot Star Taq (Qiagen) and the following primer pairs: 29830-QPCR.F2/ 29830.F3, 29830-QPCR.F2/LBb1, 29830-QPCR.F2/Lba1, or ACTF2/ACTR2. The 29830-QPCR.F2/29830.F3 product includes an intron of approximately 450 bp from the VIP3 gene and the ACTF2/ACTR2 product includes an intron approximately 100 bp from the ACT genes. 5–10 μl of each PCR reaction was separated on an ethidium gel and photographed.
Mutant lines
The cfis1-1 (SAIL_367_F03), vip3-2 (SALK_083364), vip5-062223 (SALK_062223) and vip6-065364/elf8-2 (SALK_065364) lines were obtained from the Arabidopsis Biological Resource Center. For genotyping, plant genomic DNA was isolated as described (Edwards et al., 1991) and wild type or mutant alleles amplified using the primer combinations listed in Supplementary Table S1 at JXB online.
VIP3 immunoblotting
50 mg of leaf tissue from 3–4 week old wild type, vip3-2, or ths1 mutant plants was ground in liquid nitrogen and immediately resuspended in 200 μl 2X SDS PAGE sample buffer. Equal volumes were run on an 8% SDS PAGE gel, blotted to polyvinylidene difluoride (PVDF), blocked in 5% milk/Tris-buffered saline with Tween 20 (TBST), and probed with a 1:1000 dilution of anti-VIP3 antibody overnight at 4°C. The blot was washed three times for 30 minutes each with TBST and incubated with anti-Rabbit HRP (Sigma) at 1:5000 for 60 minutes at room temperature, then washed again with TBST for 30 minutes. Peroxidase activity was detected with the SuperSignal West Femto kit (Thermo Scientific) and imaged on X-ray film. Equal loading was confirmed by Ponceau S staining and by comparison of non-specific bands.
Root slanting and coiling
Seeds were sown on square 100 x 15 mm plates (Falcon) containing 0.5X Murashige and Skoog medium, 1% sucrose and 1.6% phytagar, and in some cases with 3 μM propyzamide (Sigma). Plates were sealed with porous tape. After incubation at 4°C for 2 days in the dark, plates were transferred to a growth chamber and grown vertically for 2 days and then tilted 30° back from the vertical for 2–3 days. All plates were grown in 16 hours of light at 23°C. For curling assays, seedlings were grown on the same media for 2 days vertically and then 3 days horizontally.
Chromatin immunoprecipitation
All experiments were performed in triplicate with two technical repeats for each of the three biological repeats. For ChIP, 0.5 g of cauline leaf material was harvested from plants grown for three weeks under short day conditions of 8 hours light and 16 hours dark at 21°C followed by 2 weeks under continuous light at 16°C (as shown previously (Zhang et al., 2003). These conditions attenuate the phenotypic defects of the vip3 mutant, allowing us to collect sufficient tissue. Harvested tissue was placed on ice and crosslinked in 40 ml of infiltration buffer (13.69 g sucrose, 1 ml 100 mM PMSF, 1 ml 1M Tris/HCl pH 8, 200 μl 0.5M EDTA, 2.7 ml 37% formaldehyde in 100 ml distilled water). Vacuum was slowly introduced to the samples on ice, maintained for 10 minutes and slowly released. This was repeated three times. To stop the reaction 2 ml of 2M glycine was added to each sample. The samples were then infiltrated again for 5 minutes. Samples were washed with 500 ml of distilled water, dried on filter paper and frozen in liquid nitrogen.
Extraction of nuclei was performed as described (Jaskiewicz et al., 2011) in extraction buffer containing 0.44 M sucrose, 1.25% Ficoll, 2.5% Dextran T40, 20 mM Hepes KOH pH 7.4, 10 mM MgCl2, 0.5% Triton X-100, 5 mM DTT, 1 mM PMSF and 1X complete protease inhibitor cocktail (Roche, Product No 04693116001). After spinning down, the pellet was diluted in 500 µl of lysis buffer (50 mM Tris-HCl pH 8, 10 mM EDTA, 1% SDS and 1X complete protease inhibitor cocktail) and sonicated with a Bioruptor UCD-200 to fragments of 0.2–0.5 kilobases.
8 µg of chromatin diluted to a total volume of 500 µl with dilution buffer [1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 8, 167 mM NaCl, 0.2% complete protease inhibitor cocktail (Roche, EDTA-Free, Product No 04693116001)] was used for immunoprecipitation with specific antibodies on Protein G Plus-Agarose beads (Santa Cruz Technology, sc2002L) overnight on a spinning wheel at 4°C. Two independent immunoprecipitations with the H3K36me3 antibody (Abcam: ab9050, 3 µl per sample) and a no-antibody control were performed using each chromatin sample. Three independent chromatin samples were prepared for each genotype. Following the incubation, all samples were washed with wash buffer (150 mM NaCl, 20 mM Tris HCl pH 8, 2 mM EDTA, 1% Triton X-100, 0.1% SDS, 1 mM PMSF and 1X complete protease inhibitor cocktail). Precipitated DNA was extracted with the NucleoSpin PCR purification kit (Machery-Nagel REF-740809.250).
The quantity of DNA in each sample was determined using quantitative reverse-transcriptase PCR with the LightCycler 480 SYBR Green I Master Mix (Roche, Product No 04887352001) and the primers listed in Supplementary Table S1 to amplify a portion of exon 2 of TCH3, or the junction between exon 1 and exon 2 of TCH4. Enrichment for H3K36me3 at each region of interest was initially calculated as a percentage of the DNA recovered in the corresponding input samples. All samples were normalized against the enrichment for H3K36me3 at the S-ADENOSYL METHIONINE SYNTHASE (SAM; At4g01850) locus (Cazzonelli et al., 2014). The signal intensity after immunoprecipitation with specific antibodies was at least 10 times higher than with the no-antibody control for the corresponding samples. The final result is represented as a fold difference over the wild type sample.
Chromatin immunoblotting
2.5 µg of chromatin extract from 0.5 g of cauline leaf tissue, prepared as described above, from each sample were run on a 10% SDS PAGE gel, blotted to PVDF, blocked in 3% BSA/phosphate-buffered saline (PBS) overnight at 4°C and probed with a 1:5000 dilution of each primary antibody. The antibodies Abcam ab9050 and mAbcam ab6002 were used to detect H3K36me3 and H3K27me3, respectively. Anti-Histone H3 antibody (Agrisera, AS10710) was applied as a loading control. After washing the membranes with 0.5% BSA, 0.1% Tween-20 in PBS, they were probed with a secondary anti-Rabbit HRP (Sigma) antibody at 1:10 000 for 90 minutes at room temperature. The signal was visualized with the Pierce™ ECL Western Blotting Substrate (ThermoFisher Scientific, No32106)
Quantitative RT-PCR (qPCR)
All experiments were performed in triplicate, with three technical repeats for each of the three biological repeats. For Fig. 4, RNA extraction from the aerial portion of 12-day-old plants grown under long day conditions of 16 hours of light and 8 hours of dark at 21°C was performed with the Spectrum™ Plant Total RNA Kit (STRN250 SIGMA). 800 ng of total RNA was used for cDNA synthesis with the oligo(dT)20 primer and RevertAid Reverse Transcriptase (Thermo Scientific #EPO441). Expression of target genes was measured using LightCycler 480 SYBR Green I Master Mix (Roche, Product No 04887352001) in a StepOne Cycler (Applied Biosciences) in a reaction volume of 15 µl. The sequences of primers designed to amplify the coding regions of TUB4 (AT5G44340), TCTP (AT3G16640), TCH3 (AT2G41100), and TCH4 (AT5G57560) are listed in Supplementary Table S1. TUB4 and TCTP are classically used as reference genes for normalization because of their relatively invariant expression (Lee et al., 2005; Szecsi et al., 2006). Data were analyzed by the ΔΔCT approach with TCTP or TUB4 as reference genes.
For Supplementary Fig. S1, cDNAs were generated using an oligo(dT)20 primer and M-MLV Reverse Transcriptase (Promega) from template RNA extracted from rosette leaves of 4-week-old plants with TRIzol Reagent (Invitrogen). Primer mixes designed to amplify CFIS1 (29820-QPCR.F/29820-QPCR.R), exon 1 of VIP3 (29830-QPCR.F/29830-QPCR.R), exon 2 of VIP3 (29830-QPCR.F3/29830-QPCR.R3), or ACTIN2/7/8 (ACTF-QPCR/equal volumes of Actin2.R-QPCR, Actin7.R-QPCR and Actin8.R-QPCR) were added to a cocktail containing 1X SYBR Green PCR Master Mix (Applied Biosciences) and 0.5 μl cDNA to make a final 25 μl reaction. After amplification, the data were analyzed using StepOne software (Applied Biosciences).
Statistical analyses
ANOVAs and regressions were performed in R with the agricolae and car packages. Type II sum of squares was used for two-way ANOVAs and Tukey’s or Scheffé’s methods used as post-hoc means separation tests for balanced and unbalanced datasets, respectively. Student’s t-tests were performed in Excel.
Results
A forward genetic screen identifies a mutant defective in stem thigmomorphogenesis
The leaves of two-week-old soil-grown Arabidopsis thaliana seedlings, ecotype Columbia-0 (Col-0), were mechanically stimulated either by touching or brushing with a paintbrush. Stimulus was only administered to rosette leaves prior to bolting. We found that paintbrushing was almost as effective as hand touching at reducing subsequent stem height (Fig. 1A and 1B) and had the advantage of avoiding contact between experimenter and plant. The difference in stem heights between touched or paintbrushed and control plants was maintained until senescence. Repeated touch can lead to a delay in flowering (Chehab et al., 2012; Lange and Lange, 2015) but in our hands flowering time was not significantly different between hand-touched or paintbrushed plants and untouched plants (Fig. 1C). The reason for this discrepancy is not clear, though it is possible that it can be attributed to differences in growth conditions or in the severity of the mechanical stimulus. As expected, the decrease in stem height was proportional to the number of days of stimulus (Fig. 1D).
Twenty-four pools of 100 pROK2 T-DNA insertion lines (Alonso et al., 2003) were sown in flats and all seedlings subjected to 10 days of paintbrushing prior to bolting as described above. Thirty-three plants with stems taller than the wild type after touch were selected for further analysis. A representative example of a secondary screen in the next generation is shown in Fig. 1E. Line 6-4-2 showed insensitivity to touch as well as stem height similar to Col-0 and was named touch insensitive 1 (ths1). Twenty ths1 siblings were tested in the next generation and heritable insensitivity to touch was confirmed (Fig. 1F).
Touch insensitivity is caused by a T-DNA insertion located in the 5’UTR of the VERNALIZATION INDEPENDENCE 3 (VIP3) gene.
Using thermal asymmetric interlaced PCR (Liu et al., 1995), we located a T-DNA insertion in the ths1 genome on chromosome 4, in the presumptive promoter of VERNALIZATION INDEPENDENCE 3 (VIP3) (At4g29830) (Fig. 2A). The first 23 bp of the VIP3 5’UTR were deleted, placing the left border of the T-DNA 57 bp upstream of the ATG of VIP3 and 332 bp upstream of the ATG of At4g29820 (CFIS1), which is expressed from the other strand of chromosome 4. We were unable to amplify the right border of the T-DNA. This T-DNA insertion upstream of VIP3 segregated with the observed touch insensitivity in ths1 mutants. When ths1 mutant plants were backcrossed to wild type Col-0 plants and the F2 progeny screened for inheritance of the T-DNA, the seven F2 lines homozygous for the T-DNA did not significantly respond to touch by reducing stem height, while F2 lines lacking the T-DNA did (see Supplementary Fig. S2). VIP3 encodes a WD-repeat protein that is implicated in chromatin remodeling, mRNA turnover, flowering time and vernalization (Zhang et al., 2003; Oh et al., 2004; Dorcey et al., 2012; Takagi and Ueguchi, 2012). CFIS1 encodes a protein proposed to function as a cleavage factor during mRNA polyadenylation (Hunt et al., 2008).
To determine which gene was responsible for the touch insensitive phenotype of the ths1 mutant, homozygous vip3-2 (Jolivet et al., 2006) and cfis1-1 (Fig. 2A and Supplementary Fig. S1) mutants were crossed to homozygous ths1 mutants and the F1 generation analyzed for stem touch responsiveness as in Fig. 1F. The offspring of two independent crosses between cfis1-1 and ths1 mutants were touch sensitive, ruling out CFIS1 as responsible for touch sensitivity in the simplest genetic scenario and establishing that ths1 is a recessive mutation (Fig. 2B). However, the offspring of two independent crosses between vip3-2 and ths1 mutants were touch insensitive, implicating VIP3 as responsible for touch insensitivity in ths1 mutants (Fig. 2B). The introduction of a transgene encoding a wild type copy of the VIP3 gene partially rescued touch sensitivity in the ths1 mutant background (Fig. 2C). Taken together, these data demonstrate that a defect in the VIP3 locus is responsible for the touch insensitive phenotype of the ths1 mutant. We therefore renamed ths1 as vip3-6, with other existing alleles named vip3-1, vip3-2, vip3-3, vip3zwgand bouquet-1 (Zhang et al., 2003; Jolivet et al., 2006; Dorcey et al., 2012; Takagi and Ueguchi, 2012).
Immunoblotting with an anti-VIP3 antibody (Zhang et al., 2003) was performed on whole cell extracts from the leaves of wild type, ths1 and vip3-2 plants. A VIP3-specific band of approximately 34 kDa present in extracts from wild type plants and absent from extracts from vip3-2 plants was only faintly detectable after long exposures in extracts from ths1 plants (Fig. 2D). We used quantitative RT-PCR to compare transcript levels from the first or second exon of VIP3 and exons 1–3 of CF1S1 in ths1, vip3-2, and wild type plants. The ths1 mutant showed a modest 1.5 to 3-fold increase in VIP3 and CFIS1 transcript levels compared to wild type (see Supplementary Fig. S3A). We note that the data shown in Fig. 2B show that the touch insensitive phenotype of ths1 is not due to a recessive defect in CFIS1 and that the ths1 phenotype is not dominant; thus this modest overexpression of CFIS1 due to the T-DNA insertion is unlikely to be responsible for the observed phenotype.
Semi-quantitative RT-PCR detected an abundant transcript stretching from the left border of the T-DNA to the second exon of VIP3, along with several larger, less abundant transcripts (see Supplementary Fig. S3B). These transcripts may be derived from the NOS or CaMV 35S promoters present inside the pROK2 vector. Though present at levels similar to or higher than the wild type transcript, this transcript may lack proper start site or splicing information due to its aberrant start site. Thus, while ths1 plants produce mRNA from the VIP3 locus, it is unstable and/or not properly translated.
vip3-2 mutants are small with narrow, crinkled leaves and short, misshapen floral organs, as shown previously (Jolivet et al., 2006). ths1 mutants grown side-by-side were smaller and had narrower leaves than wild type plants and exhibited mild floral organ defects (Fig. 2E). We also assessed the flowering time of wild type and ths1 homozygous mutant siblings derived from a ths1 x Col-0 backcross, Col-0, and vip3-2 mutants. ths1 mutant siblings showed slightly but significantly earlier flowering, while wild type siblings were indistinguishable from Col-0, and vip3-2 mutants had a clear early flowering phenotype (Fig. 2F). Taken together, the data presented in Fig. 2 show that an intermediate loss-of-function allele of VIP3 is responsible for touch insensitivity in ths1 mutants. From these results we infer that the low amount of VIP3 protein produced in the ths1 background (Fig. 2D) is sufficient for most developmental functions of VIP3.
Two members of the Paf1 complex are required for stem thigmomorphogenesis
The VIP3 protein has been detected both in the Paf1 complex and in the Superkiller (SKI) complex (Dorcey et al., 2012). Several other proteins (VIP4, VIP5, VIP6/ELF8 and VIP2/ELF7) are known to be part of the Paf1 complex but not the SKI complex (Zhang and van Nocker, 2002; Zhang et al., 2003; He et al., 2004; Oh et al., 2004; Oh et al., 2008; Shiraya et al., 2008; Park et al., 2010). To determine if the touch insensitivity of ths1 can be attributed to the action of the Paf1 complex, we tested null vip5-062223 mutants (Oh et al., 2004) in the paintbrush assay. When grown in 16 hours of light to produce taller plants, vip3-2 and vip5-062223 mutants were similarly touch insensitive, while wild type plants still showed an approximately 40% reduction of stem height when touched (Fig. 3A). Early flowering is not sufficient to confer touch insensitivity as two early flowering lines showed a normal touch response (Supplementary Fig. S4).
Root skewing and coiling is abnormal in ths1 and vip mutants
Assays for root touch response that involve growing seedlings on agar plates include waving (Okada and Shimura, 1990), skewing or slanting (Simmons et al., 1995; Rutherford and Masson, 1996), and curling or looping (Mirza, 1987; Simmons et al., 1995; Buer et al., 2000). These assays are thought to reveal the interplay between the response to touching of the agar surface and the response to gravity (Okada and Shimura, 1990; Thompson and Holbrook, 2004). Forward and reverse genetic approaches have revealed that slanting is affected by a diverse array of pathways, including the cortical microtubule array, hormones such as auxin and ethylene and environmental factors such as sucrose and salt stress (Buer et al., 2000; Shoji et al., 2006 ; Oliva and Dunand, 2007). In addition, lateral root initiation can be modulated by physical bending with forceps or by growth into barriers (Ditengou et al., 2008; Laskowski et al., 2008; Richter et al., 2009). This touch perception pathway appears to modulate the endogenous auxin-regulated pathway for lateral root initiation (Ditengou et al., 2008; Laskowski et al., 2008; Richter et al., 2009).
According to publically available gene expression databases, components of the Paf1 complex are widely expressed in most plant tissues, including the root (Winter et al., 2007). We therefore analyzed root thigmomorphogenetic responses using root slanting and coiling assays (Bidzinski et al., 2014; Swanson et al., 2015). As previously observed, the roots of wild type plants slant to the left when grown on hard agar plates tilted 30 degrees from the vertical, and this phenomenon was enhanced in the presence of the microtubule-destabilizing drug propyzamide (Furutani et al., 2000; Nakamura et al., 2004), while ths1 and vip3-2 mutant roots slanted strongly to the right under these conditions (Fig. 3B). Similar results were obtained for root coiling assays, where seedlings were grown horizontally on hard agar plates (Fig. 3C). Plotting the distribution of slanting angle within a population of seedlings grown in the absence of propyzamide revealed that wild type plants slanted on average 6 degrees to the left when viewed from above the agar surface (Fig. 3D). While the ths1 mutant did not show a clear difference from wild type, vip3-2, vip5-062223 and null vip6-065364 (elf8-2) (He et al., 2004; Oh et al., 2004; Shiraya et al., 2008) mutants showed a dramatic difference in how their roots responded to growth on hard agar, with a wider range of slanting angles and no preference for slanting direction. In summary, ths1 mutants have defects in root skewing and coiling assays when propyzamide is added, and vip null mutants have defects in root skewing and coiling assays both with and without propyzamide.
ths1 and vip3-2 mutants exhibit a histone H3 trimethylation pattern associated with inactive chromatin globally and at TCH3 and TCH4 loci
The Paf1 complex is known to promote histone methylation patterns associated with active chromatin. At Flowering Locus C (FLC), the Paf1 complex promotes lysine methylation of histone H3 associated with transcriptionally active regions, lysine 4 (H3K4) and lysine 36 (H3K36), while reducing lysine methylation associated with transcriptional repression at lysine 27 (H3K27) (Zhao et al., 2005; Ko et al., 2010). Global levels of trimethylation at H3K36 and H3K27 in cauline leaves were assessed by western blotting with specific antibodies. Under these conditions, a reduction in the level of H3K36 trimethylation (H3K36me3) was detected in ths1 and vip3-2 mutants (Fig. 4A). As a positive control, we also observed reduced H3K36me3 levels in the sdg8 background under the same conditions (Fig. 4A); SDG8 encodes an H3K36-directed methyltransferase and is required for global and genic H3K36me3 (Zhao et al., 2005; Xu et al., 2008). H3K27me3 levels were higher than the wild type in vip3 mutants but indistinguishable from the wild type in ths1 and sdg8 mutants.
H3K36me3 levels were also assessed by chromatin immunoprecipitation at TCH3 and TCH4 loci. TCH3 is one of the most studied TCH genes and encodes a calmodulin-like calcium-binding protein (Sistrunk et al., 1994). TCH4 encodes a xyloglucan endotransglycosylase (Xu et al., 1995). We found that levels of H3K36me3 were not significantly changed at the TCH3 locus in vip3-6/ths1. However, in the null vip3-2 mutant they were reduced two-fold compared to the wild type (Fig. 4B). Levels of H3K36me3 were significantly reduced at the TCH4 locus in both the vip3-6/ths1 and vip3-2 mutants when compared to wild type.
To determine if these chromatin marks were related to TCH gene expression, we tested the rapid touch induction of TCH4. Thirty minutes after one touch treatment with a paintbrush, a significant induction of TCH4 expression was observed in the wild type (Fig. 4C). The same trend, albeit not statistically significant, was observed for TCH3. Two touch treatments also led to high levels of expression of both TCH genes in the wild type (Fig. 4C). These responses were reduced and/or delayed in both vip3 mutants (Fig. 4C). Note that the induction was not totally absent in both vip3 mutants and could even reach wild type levels after two subsequent touches for TCH4 expression in the vip3-6/ths1 mutant. Nonetheless, the induction was never detected after a single touch event in the vip3 mutants (Fig. 4C). VIP3 transcript levels were not significantly altered in response to touch (Supplementary Fig. S5). Thus, the Paf1 complex is not only required for long-term thigmomorphogenesis responses but is also involved in short-term touch-induced transcription.
Discussion
We show here that the Arabidopsis Paf1 complex is a critical factor for stem and root thigmomorphogenesis, stereotypical alterations in plant form that are made in response to repeated mechanical disturbance. We also show that THS1/VIP3 is required for both global and genic levels of H3K36 methylation and for upregulation of two canonical touch-inducible loci in response to touch stimulation. VIP3 was identified in a genetic screen with no a priori expectations and our findings are consistent with previous observations that sgd8 mutants exhibit defects in wind-induced alterations in leaf morphology and in H3K4 trimethylation at the TCH3 gene (Cazzonelli et al., 2014). It has been proposed that mitotically stable chromatin marks serve to establish and stabilize certain transcriptional states (Iwasaki and Paszkowski, 2014; Alabert et al., 2015; Avramova, 2015; Crisp et al., 2016). The data presented here are consistent with this model, demonstrating that the state of chromatin is modulated by the Paf1 complex and facilitates both the spatial and temporal aspects of thigmomorphogenesis, namely its systemic nature and need for repetitive touch.
Linking VIP3 and the Paf1 complex to stem and root thigmorphogenesis.
We present four lines of evidence to support the conclusion that a lesion in VIP3 is responsible for the thigmomorphic defects seen in ths1 mutants: 1) The T-DNA inserted into the VIP3 3’ UTR in ths1 mutants segregates with its touch insensitive phenotype, ruling out a causal mutation at a second site (Supplementary Fig. S2); 2) While the T-DNA insertion is associated with a modest increase in expression of both VIP3 and CFIS1 (Supplementary Fig. S3A), the complementation test shown in Fig. 2B, where the cfis1 mutant complements the ths1 mutant but a vip3 null mutant does not, indicates that it is a recessive lesion in VIP3 that is responsible for the touch insensitivity of ths1 mutants; 3) A VIP3g genomic construct partially restores touch responsiveness to the ths1 mutant. This result is difficult to explain if a gene other than VIP3 were involved given that no CFIS1 sequences were included in the VIP3g construct; and 4) vip3-2 null mutants do not respond to touch.
These lines of evidence link VIP3 to stem thigmomorphogenesis. We also present evidence that other components of the Paf1 complex are required for thigmomorphic responses in both the stem and the root. The stems of vip3-2 and vip5 null mutants do not shorten in response to paintbrushing (Fig. 3A) and ths1, vip3-2, vip5, and vip6 mutants show defects in root coiling and slanting assays (Fig. 4). However, we cannot rule out the possibility that vip null mutants are simply too short, even when grown in 16 hours of light, to get any shorter. Furthermore, we acknowledge that the root coiling and slanting assays used here do not solely assess touch responses but also integrate the perception of gravity (Thompson and Holbrook, 2004). Nonetheless, these additional data help support our hypothesis that the plant Paf1 complex is required for normal thigmomorphogenesis in the stem and in the root.
One challenging aspect of the paintbrush assay used here is the variability in stem height; both in the average untouched ths1 stem heights compared to average untouched wild type stem heights and in the extent to which stem height in the ths1 mutant is insensitive to touch (compare Fig. 1F, 2B, 2C and 3A). We found that the average stem height of any genotype depends on a number of growth conditions including, but not limited to, light and temperature. We controlled for these variables as much as possible by using the same shelf and chamber, rotating trays and growing touched and untouched pots side-by-side. We were nonetheless unable to completely prevent stem height variability. However, we note that the key comparison in this study is between touched and untouched individuals of a single genotype, and that ths1 mutants were consistently less sensitive to paintbrushing than the wild type under all conditions tested.
The Paf1 complex as a conserved and multitasking regulator of gene expression
First identified in yeast as a transcriptional regulator, the Paf1 complex was later shown to be present and functionally conserved in all kingdoms (Koch et al., 1999; Penheiter et al., 2005; Sheldon et al., 2005; Nordick et al., 2008; Oh et al., 2008; Dermody and Buratowski, 2010; Chu et al., 2013; Sadeghi et al., 2015). The Paf1 complex plays a role in a number of transcription-related processes, such as facilitating elongation, recruitment and regulation of chromatin-modifying factors like histone-methylation factors, transcription termination and polyadenylation (Krogan et al., 2003; Ng et al., 2003; Nordick et al., 2008; Penheiter et al., 2005; Rozenblatt-Rosen et al., 2009; Zhang et al., 2009; Tomson and Arndt, 2013; Sadeghi et al., 2015). In yeast, it was also shown to participate in small nucleolar RNA formation (Sheldon et al., 2005; Tomson et al., 2013). Paf1 complex activity has been reported to be necessary for the maintenance of the active chromatin marks histone H3K4me3 and H3K36me3 in yeast, plant, human and mouse cells (Jaehning, 2010; Zhang et al., 2013), providing further evidence for its functional conservation among kingdoms.
Impaired Paf1 complex function leads to developmental defects in animals. The activity of the Paf1 complex is required for heart and neural crest development in zebrafish (Nguyen et al., 2010), embryonic epidermal morphogenesis in C. elegans (Kubota et al., 2014), Notch and Wnt signaling in Drosophila (Bray et al., 2005; Mosimann et al., 2009; Bahrampour and Thor, 2016), key lineage specific factor expression in mice embryos (Zhang et al., 2013), oligodendrocyte differentiation (Kim et al., 2012) and cardiomyocyte specification (Langenbacher et al., 2011). It also affects cancer progression in human cell lines (Dey et al., 2011; Tomson and Arndt, 2013) and anti-viral immune responses (Liu et al., 2011). In plants, the Paf1 complex functions in the regulation of flowering time through establishment of histone marks at the FLC locus (Zhao et al., 2005; Oh et al., 2008; Xu et al., 2008).
Chromatin, a new frontier in plant mechanotransduction?
Given that we find that TCH gene induction in response to touch is delayed in vip3 mutants, the function of the Paf1 complex does not seem absolutely required for TCH induction, but instead may prime a quick response to mechanical perturbations through histone modifications at TCH loci. The role of the Paf1 complex in thigmomorphogenesis might therefore reside in providing chromatin with the ability to respond to mechanical perturbations rapidly.
This scenario echoes a number of recent reports in animal systems suggesting that mechanotransduction incorporates a chromatin component. In fact, because chromatin modifications change the shape of the nucleus, notably through chromatin condensation and decondensation states, it has been proposed that mechanical forces may in turn be the primary modifiers of chromatin (Pagliara et al., 2014). Global chromatin modifications have consistently been associated with nuclear stiffness [for example (Schreiner et al., 2015)] and nuclear stiffness has been shown to increase with differentiation (Hampoelz and Lecuit, 2011). Given that chromatin is associated with the nuclear envelope through lamins, its modification may result from deformations originating within the extracellular matrix that are propagated through the cytoskeleton and the LINC (linker of the nucleoskeleton and cytoskeleton) complex (Isermann and Lammerding, 2013). Applying mechanical strains to mesenchymal stem cells in culture leads to a lamin-dependent decrease in histone deacetylase activity and an increase in histone acetylation (Li et al., 2011). The finding that the Paf1 complex is involved in thigmomorphogenesis in plants may thus provide an opportunity to unravel potential connections between cytoskeletal and gene expression responses to mechanical perturbation.
Our results build a foundation for future investigations into the possible function of the Paf1 complex and chromatin marks in priming TCH genes for rapid induction upon mechanical stimulation and in translating discrete touch events into a continuous growth response. Given that the Paf1 complex is conserved across kingdoms and because animals also alter their body plan in response to repeated mechanical stresses (Kahn et al., 2009; Heckel et al., 2015; Steed et al., 2016), these findings also raise the possibility of a common mechanism for recording multiple mechanical deformations and translating them into developmental changes.
Supplementary Material
Acknowledgements
This research was supported by the Washington University/Monsanto Plant Biology Program Fund (to ESH), NIH R01GM084211 (to ESH), and the European Research Council (ERC-2013-CoG-615739 ‘‘MechanoDevo’’ to OH). Steve Van Nocker provided anti-VIP3 antibodies. vip mutant lines and the pROK T-DNA pools were obtained from the Arabidopsis Biological Resource Center. We thank Hao Yu (National University of Singapore) for providing p35S:AGL24 transgenic and svp-41 mutant lines.
Glossary
Abbreviations:
- Col-0
Columbia-0;
- FLC
Flowering Locus C;
- JA
jasmonic acid;
- LINC
linker of the nucleoskeleton and cytoskeleton;
- me3
trimethylation;
- Paf1
RNA polymerase II-associated factor 1;
- PBS
phosphate-buffered saline;
- PVDF
polyvinylidene difluoride;
- qRT-PCR
quantitative reverse-transcriptase PCR;
- SAM
S-ADENOSYLMETHIONINE SYNTHASE;
- TBST
Tris-buffered saline with Tween 20;
- TCH
TOUCH
- ths1
touch insensitive 1;
- VIP3
VERNALIZATION INDEPENDENCE 3
References
- Alabert C, Barth TK, Reverón-Gómez N, Sidoli S, Schmidt A, Jensen ON, Imhof A, Groth A. 2015. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes & development 29, 585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science (New York, N.Y.) 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Avramova Z. 2015. Transcriptional ‘memory’ of a stress: transient chromatin and memory (epigenetic) marks at stress-response genes. The Plant journal: for cell and molecular biology 83, 149–159. [DOI] [PubMed] [Google Scholar]
- Bahrampour S, Thor S. 2016. Ctr9, a Key Component of the Paf1 Complex Affects Proliferation and Terminal Differentiation in the Developing Drosophila Nervous System. G3 (Bethesda). Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddington NL. 1986. The effects of mechanically-induced stress in plants--a review. Plant Growth Regulation 4, 103–123. [Google Scholar]
- Bidzinski P, Noir S, Shahi S, Reinstädler A, Gratkowska DM, Panstruga R. 2014. Physiological characterization and genetic modifiers of aberrant root thigmomorphogenesis in mutants of Arabidopsis thaliana MILDEW LOCUS O genes. Plant, cell & environment 37, 2738–2753. [DOI] [PubMed] [Google Scholar]
- Böhm J, Scherzer S, Krol E, et al. 2016. The Venus Flytrap Dionaea muscipula Counts Prey-Induced Action Potentials to Induce Sodium Uptake. Current Biology: CB 26, 286–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J, Davis RW. 1990. Rain-, wind-, and touch-induced expression of calmodulin and calmodulin-related genes in Arabidopsis. Cell 60, 357–364. [DOI] [PubMed] [Google Scholar]
- Bray S, Musisi H, Bienz M. 2005. Bre1 is required for Notch signaling and histone modification. Developmental cell 8, 279–286. [DOI] [PubMed] [Google Scholar]
- Buer CS, Masle J, Wasteneys GO. 2000. Growth conditions modulate root-wave phenotypes in Arabidopsis. Plant & cell physiology 41, 1164–1170. [DOI] [PubMed] [Google Scholar]
- Cazzonelli CI, Nisar N, Roberts AC, Murray KD, Borevitz JO, Pogson BJ. 2014. A chromatin modifying enzyme, SDG8, is involved in morphological, gene expression, and epigenetic responses to mechanical stimulation. Frontiers in Plant Science 5, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab EW, Eich E, Braam J. 2009. Thigmomorphogenesis: a complex plant response to mechano-stimulation. Journal of Experimental Botany 60, 43–56. [DOI] [PubMed] [Google Scholar]
- Chehab EW, Yao C, Henderson Z, Kim S, Braam J. 2012. Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests. Current Biology: CB 22, 701–706. [DOI] [PubMed] [Google Scholar]
- Chu X, Qin X, Xu H, Li L, Wang Z, Li F, Xie X, Zhou H, Shen Y, Long J. 2013. Structural insights into Paf1 complex assembly and histone binding. Nucleic Acids Research 41, 10619–10629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutand C. 2010. Mechanosensing and thigmomorphogenesis, a physiological and biomechanical point of view. Plant Science 179, 168–182. [Google Scholar]
- Coutand C, Martin L, Leblanc-Fournier N, Decourteix M, Julien JL, Moulia B. 2009. Strain mechanosensing quantitatively controls diameter growth and PtaZFP2 gene expression in poplar. Plant Physiology 151, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp PA, Ganguly D, Eichten SR, Borevitz JO, Pogson BJ. 2016. Reconsidering plant memory: Intersections between stress recovery, RNA turnover, and epigenetics. Science Advances 2, e1501340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermody JL, Buratowski S. 2010. Leo1 subunit of the yeast paf1 complex binds RNA and contributes to complex recruitment. The Journal of biological chemistry 285, 33671–33679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P, Ponnusamy MP, Deb S, Batra SK. 2011. Human RNA polymerase II-association factor 1 (hPaf1/PD2) regulates histone methylation and chromatin remodeling in pancreatic cancer. PLoS One 6, e26926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, et al. 2008. Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 105, 18818–18823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorcey E, Rodriguez-Villalon A, Salinas P, Santuari L, Pradervand S, Harshman K, Hardtke CS. 2012. Context-dependent dual role of SKI8 homologs in mRNA synthesis and turnover. PLoS Genetics 8, e1002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. 1991. Nucleic Acids Research 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani I, Watanabe Y, Prieto R, Masukawa M, Suzuki K, Naoi K, Thitamadee S, Shikanai T, Hashimoto T. 2000. The SPIRAL genes are required for directional control of cell elongation in Aarabidopsis thaliana. Development (Cambridge, England) 127, 4443–4453. [DOI] [PubMed] [Google Scholar]
- Gaffney EA, Monk NA. 2006. Gene expression time delays and Turing pattern formation systems. Bulletin of Mathematical Biology 68, 99–130. [DOI] [PubMed] [Google Scholar]
- Hampoelz B, Lecuit T. 2011. Nuclear mechanics in differentiation and development. Current Opinion in Cell Biology 23, 668–675. [DOI] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM. 2004. PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes & development 18, 2774–2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel E, Boselli F, Roth S, Krudewig A, Belting HG, Charvin G, Vermot J. 2015. Oscillatory Flow Modulates Mechanosensitive klf2a Expression through trpv4 and trpp2 during Heart Valve Development. Current Biology: CB 25, 1354–1361. [DOI] [PubMed] [Google Scholar]
- Hedtke B, Grimm B. 2009. Silencing of a plant gene by transcriptional interference. Nucleic Acids Research 37, 3739–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt AG, Xu R, Addepalli B, et al. 2008. Arabidopsis mRNA polyadenylation machinery: comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genomics 9, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isermann P, Lammerding J. 2013. Nuclear mechanics and mechanotransduction in health and disease. Current biology: CB 23, R1113–R1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki M, Paszkowski J. 2014. Epigenetic memory in plants. The EMBO journal 33, 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaehning JA. 2010. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochimica et Biophysica Acta 1799, 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe MJ. 1980. Morphogenetic Responses of Plants to Mechanical Stimuli or Stress. Bioscience 30, 239–243. [Google Scholar]
- Jaskiewicz M, Peterhansel C, Conrath U. 2011. Detection of histone modifications in plant leaves. Journal of Visual Experiments 3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolivet S, Vezon D, Froger N, Mercier R. 2006. Non conservation of the meiotic function of the Ski8/Rec103 homolog in Arabidopsis. Genes to Cells: devoted to molecular & cellular mechanisms 11, 615–622. [DOI] [PubMed] [Google Scholar]
- Kagaya Y, Hattori T. 2009. Arabidopsis transcription factors, RAV1 and RAV2, are regulated by touch-related stimuli in a dose-dependent and biphasic manner. Genes & genetic systems 84, 95–99. [DOI] [PubMed] [Google Scholar]
- Kahn J, Shwartz Y, Blitz E, et al. 2009. Muscle contraction is necessary to maintain joint progenitor cell fate. Developmental Cell 16, 734–743. [DOI] [PubMed] [Google Scholar]
- Kim S, Kim JD, Chung AY, et al. 2012. Antagonistic regulation of PAF1C and p-TEFb is required for oligodendrocyte differentiation. The Journal of neuroscience: the official journal of the Society for Neuroscience 32, 8201–8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JH, Mitina I, Tamada Y, Hyun Y, Choi Y, Amasino RM, Noh B, Noh YS. 2010. Growth habit determination by the balance of histone methylation activities in Arabidopsis. The EMBO Journal 29, 3208–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Wollmann P, Dahl M, Lottspeich F. 1999. A role for Ctr9p and Paf1p in the regulation G1 cyclin expression in yeast. Nucleic Acids Research 27, 2126–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Molecular Cell 11, 721–729. [DOI] [PubMed] [Google Scholar]
- Kubota Y, Tsuyama K, Takabayashi Y, Haruta N, Maruyama R, Iida N, Sugimoto A. 2014. The PAF1 complex is involved in embryonic epidermal morphogenesis in Caenorhabditis elegans. Developmental Biology 391, 43–53. [DOI] [PubMed] [Google Scholar]
- Lander AD. 2011. Pattern, growth, and control. Cell 144, 955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange MJ, Lange T. 2015. Touch-induced changes in Arabidopsis morphology dependent on gibberellin breakdown. Nature Plants 1, 14025. [DOI] [PubMed] [Google Scholar]
- Langenbacher AD, Nguyen CT, Cavanaugh AM, Huang J, Lu F, Chen JN. 2011. The PAF1 complex differentially regulates cardiomyocyte specification. Developmental Biology 353, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Marée AF, Scheres B. 2008. Root system architecture from coupling cell shape to auxin transport. PLoS Biology 6, e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc-Fournier N, Coutand C, Crouzet J, Brunel N, Lenne C, Moulia B, Julien JL. 2008. Jr-ZFP2, encoding a Cys2/His2-type transcription factor, is involved in the early stages of the mechano-perception pathway and specifically expressed in mechanically stimulated tissues in woody plants. Plant, cell & environment 31, 715–726. [DOI] [PubMed] [Google Scholar]
- Leblanc-Fournier N, Martin L, Lenne C, Decourteix M. 2014. To respond or not to respond, the recurring question in plant mechanosensitivity. Frontiers in Plant Science 5, 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Polisensky DH, Braam J. 2005. Genome-wide identification of touch- and darkness-regulated Arabidopsis genes: a focus on calmodulin-like and XTH genes. The New phytologist 165, 429–444. [DOI] [PubMed] [Google Scholar]
- Li Y, Chu JS, Kurpinski K, Li X, Bautista DM, Yang L, Sung KL, Li S. 2011. Biophysical regulation of histone acetylation in mesenchymal stem cells. Biophysical Journal 100, 1902–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling V, Perera I, Zielinski RE. 1991. Primary structures of Arabidopsis calmodulin isoforms deduced from the sequences of cDNA clones. Plant Physiology 96, 1196–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Oliveira NM, Cheney KM, et al. 2011. A whole genome screen for HIV restriction factors. Retrovirology 8, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YG, Mitsukawa N, Oosumi T, Whittier RF. 1995. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. The Plant journal: for cell and molecular biology 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Martin L, Leblanc-Fournier N, Julien JL, Moulia B, Coutand C. 2010. Acclimation kinetics of physiological and molecular responses of plants to multiple mechanical loadings. Journal of Experimental Botany 61, 2403–2412. [DOI] [PubMed] [Google Scholar]
- Mirza JI. 1987. The Effects of Light and Gravity on the Horizontal Curvature of Roots of Gravitropic and Agravitropic Arabidopsis thaliana L. Plant Physiology 83, 118–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk NA. 2003. Oscillatory expression of Hes1, p53, and NF-kappaB driven by transcriptional time delays. Current Biology: CB 13, 1409–1413. [DOI] [PubMed] [Google Scholar]
- Mosimann C, Hausmann G, Basler K. 2009. The role of Parafibromin/Hyrax as a nuclear Gli/Ci-interacting protein in Hedgehog target gene control. Mechanisms of Development 126, 394–405. [DOI] [PubMed] [Google Scholar]
- Moulia B, Coutand C, Julien JL. 2015. Mechanosensitive control of plant growth: bearing the load, sensing, transducing, and responding. Frontiers in Plant Science 6, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Naoi K, Shoji T, Hashimoto T. 2004. Low concentrations of propyzamide and oryzalin alter microtubule dynamics in Arabidopsis epidermal cells. Plant & cell physiology 45, 1330–1334. [DOI] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. The Journal of biological chemistry 278, 33625–33628. [DOI] [PubMed] [Google Scholar]
- Nguyen CT, Langenbacher A, Hsieh M, Chen JN. 2010. The PAF1 complex component Leo1 is essential for cardiac and neural crest development in zebrafish. Developmental Biology 341, 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordick K, Hoffman MG, Betz JL, Jaehning JA. 2008. Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryotic Cell 7, 1158–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oates AC. 2011. What’s all the noise about developmental stochasticity? Development (Cambridge, England) 138, 601–607. [DOI] [PubMed] [Google Scholar]
- Oh S, Park S, van Nocker S. 2008. Genic and global functions for Paf1C in chromatin modification and gene expression in Arabidopsis. PLoS Genetics 4, e1000077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh S, Zhang H, Ludwig P, van Nocker S. 2004. A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. The Plant cell 16, 2940–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Shimura Y. 1990. Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science (New York, N.Y.) 250, 274–276. [DOI] [PubMed] [Google Scholar]
- Oliva M, Dunand C. 2007. Waving and skewing: how gravity and the surface of growth media affect root development in Arabidopsis. The New phytologist 176, 37–43. [DOI] [PubMed] [Google Scholar]
- Pagliara S, Franze K, McClain CR, Wylde GW, Fisher CL, Franklin RJ, Kabla AJ, Keyser UF, Chalut KJ. 2014. Auxetic nuclei in embryonic stem cells exiting pluripotency. Nature Materials 13, 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Oh S, Ek-Ramos J, van Nocker S. 2010. PLANT HOMOLOGOUS TO PARAFIBROMIN is a component of the PAF1 complex and assists in regulating expression of genes within H3K27ME3-enriched chromatin. Plant Physiology 153, 821–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penheiter KL, Washburn TM, Porter SE, Hoffman MG, Jaehning JA. 2005. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Molecular Cell 20, 213–223. [DOI] [PubMed] [Google Scholar]
- Perera IY, Zielinski RE. 1992. Structure and expression of the Arabidopsis CaM-3 calmodulin gene. Plant Molecular Biology 19, 649–664. [DOI] [PubMed] [Google Scholar]
- Raj A, van Oudenaarden A. 2008. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter GL, Monshausen GB, Krol A, Gilroy S. 2009. Mechanical stimuli modulate lateral root organogenesis. Plant Physiology 151, 1855–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Nagaike T, Francis JM, Kaneko S, Glatt KA, Hughes CM, LaFramboise T, Manley JL, Meyerson M. 2009. The tumor suppressor Cdc73 functionally associates with CPSF and CstF 3’ mRNA processing factors. Proceedings of the National Academy of Sciences of the United States of America 106, 755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford R, Masson PH. 1996. Arabidopsis thaliana sku mutant seedlings show exaggerated surface-dependent alteration in root growth vector. Plant Physiology 111, 987–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghi L, Prasad P, Ekwall K, Cohen A, Svensson JP. 2015. The Paf1 complex factors Leo1 and Paf1 promote local histone turnover to modulate chromatin states in fission yeast. EMBO Reports 16, 1673–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner SM, Koo PK, Zhao Y, Mochrie SG, King MC. 2015. The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nature Communications 6, 7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon KE, Mauger DM, Arndt KM. 2005. A Requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3’ end formation. Molecular Cell 20, 225–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraya T, Sato S, Kato T, Tabata S, Iwasaki T. 2008. Arabidopsis VIP6/ELF8, the homolog of CTR9 component of the transcriptional complex PAF1, is essential for plant development. Plant Biotechnology 25, 447–455. [Google Scholar]
- Shoji T, Suzuki K, Abe T, Kaneko Y, Shi H, Zhu JK, Rus A, Hasegawa PM, Hashimoto T. 2006. Salt stress affects cortical microtubule organization and helical growth in Arabidopsis. Plant & cell physiology 47, 1158–1168. [DOI] [PubMed] [Google Scholar]
- Simmons C, Soll D, Migliaccio F. 1995. Circumnutation and gravitropism cause root waving in Arabidopsis thaliana. Journal of Experimental Botany 46, 143–150. [Google Scholar]
- Sistrunk ML, Antosiewicz DM, Purugganan MM, Braam J. 1994. Arabidopsis TCH3 encodes a novel Ca2+ binding protein and shows environmentally induced and tissue-specific regulation. The Plant cell 6, 1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed E, Boselli F, Vermot J. 2016. Hemodynamics driven cardiac valve morphogenesis. Biochimica et Biophysica Acta 1863, 1760–1766. [DOI] [PubMed] [Google Scholar]
- Swanson SJ, Barker R, Ye Y, Gilroy S. 2015. Evaluating mechano-transduction and touch responses in plant roots. Methods in molecular biology (Clifton, N.J.) 1309, 143–150. [DOI] [PubMed] [Google Scholar]
- Szécsi J, Joly C, Bordji K, Varaud E, Cock JM, Dumas C, Bendahmane M. 2006. BIGPETALp, a bHLH transcription factor is involved in the control of Arabidopsis petal size. The EMBO Journal 25, 3912–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi N, Ueguchi C. 2012. Enhancement of meristem formation by bouquet-1, a mis-sense allele of the vernalization independence 3 gene encoding a WD40 repeat protein in Arabidopsis thaliana. Genes to Cells: devoted to molecular & cellular mechanisms 17, 982–993. [DOI] [PubMed] [Google Scholar]
- Thompson MV, Holbrook NM. 2004. Root-gel interactions and the root waving behavior of Arabidopsis. Plant Physiology 135, 1822–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomson BN, Arndt KM. 2013. The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochimica et Biophysica Acta 1829, 116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomson BN, Crisucci EM, Heisler LE, Gebbia M, Nislow C, Arndt KM. 2013. Effects of the Paf1 complex and histone modifications on snoRNA 3’-end formation reveal broad and locus-specific regulation. Molecular and Cellular Biology 33, 170–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YC, Koo Y, Delk NA, Gehl B, Braam J. 2013. Calmodulin-related CML24 interacts with ATG4b and affects autophagy progression in Arabidopsis. The Plant journal: for cell and molecular biology 73, 325–335. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang B, Gilroy S, Chehab EW, Braam J. 2011. CML24 is Involved in Root Mechanoresponses and Cortical Microtubule Orientation in Arabidopsis. Journal of Plant Growth Regulation, 467–479. [Google Scholar]
- Webb AB, Lengyel IM, Jorg DJ, Valentin G, Julicher F, Morelli LG, Oates AC. 2016. Persistence, period and precision of autonomous cellular oscillators from the zebrafish segmentation clock. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L. 1969. Positional information and the spatial pattern of cellular differentiation. Journal of Theoretical Biology 25, 1–47. [DOI] [PubMed] [Google Scholar]
- Xu L, Zhao Z, Dong A, Soubigou-Taconnat L, Renou JP, Steinmetz A, Shen WH. 2008. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Molecular and Cellular Biology 28, 1348–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Braam J. 1995. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. The Plant cell 7, 1555–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ransom C, Ludwig P, van Nocker S. 2003. Genetic analysis of early flowering mutants in Arabidopsis defines a class of pleiotropic developmental regulator required for expression of the flowering-time switch flowering locus C. Genetics 164, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, van Nocker S. 2002. The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. The Plant journal: for cell and molecular biology 31, 663–673. [DOI] [PubMed] [Google Scholar]
- Zhang K, Haversat JM, Mager J. 2013. CTR9/PAF1c regulates molecular lineage identity, histone H3K36 trimethylation and genomic imprinting during preimplantation development. Developmental Biology 383, 15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Sikes ML, Beyer AL, Schneider DA. 2009. The Paf1 complex is required for efficient transcription elongation by RNA polymerase I. Proceedings of the National Academy of Sciences of the United States of America 106, 2153–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Yu Y, Meyer D, Wu C, Shen WH. 2005. Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nature Cell Biology 7, 1256–1260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.