Abstract
Based on partial 16S sequences, we previously described a novel group of nonsymbiotic, acetylene reduction activity-positive actinomycetes which were isolated from surface-sterilized roots of Casuarina equisetifolia growing in Mexico. An amplified rRNA restriction analysis confirmed that these actinomycetes are distinct from Frankia, a finding substantiated by a 16S rRNA gene phylogenetic analysis of two of the Mexican isolates. Further support for these actinomycetes being separate from Frankia comes from the very low DNA-DNA homology that was found. Nevertheless, the Mexican isolates may be diazotrophs based not only on their ability to grow in N-free medium and reduce acetylene to ethylene but also on the results from 15N isotope dilution analysis and the finding that a nifH gene was PCR amplified. A comparison of the nifH sequences from the various isolates showed that they are closely related to nifH from Frankia; the similarity was 84 to 98% depending on the host specificity group. An analysis of complete 16S rRNA gene sequences demonstrated that the two strains analyzed in detail are most closely related to actinobacteria in the Thermomonosporaceae and the Micromonosporaceae.
In the course of isolating Frankia from surface-sterilized root nodules of Casuarina equisetifolia growing in Mexico, additional filamentous bacteria were obtained (13). The filaments of these bacteria are smaller in diameter than Frankia hyphae, the filaments do not develop typical nitrogen-fixing diazovesicles ex planta under aerobic conditions, and the strains are unable to reinfect Casuarina or other tested actinorhizal plants upon reinoculation (20). In addition, of 24 fatty acids examined, only 7 were shared by Frankia and the Mexican isolates (17). Based on an analysis of a partial sequence of the 16S rRNA gene, we proposed that these bacteria lie outside of the Frankia clade (20).
In this report, we examined the possibility that these non-Frankia Mexican actinomycetes have nitrogen-fixing genes based on their previously discovered abilities to reduce acetylene to ethylene, an indicator of nitrogenase activity, and to grow in N-free medium (13, 36). Nitrogenase, which catalyzes the reduction of dinitrogen to ammonia, is encoded by the genes of the nifHDK operon and is universally found in diazotrophs. Preliminary results with Southern blot analysis suggested that nifH- and nifD-like sequences were present in some of the Mexican isolates, but the bands weakly hybridized to a Klebsiella pneumoniae nifHD probe (unpublished results). Weak hybridization to DNA isolated from two Mexican strains (L4 and 7702B) was also observed when a Frankia CcI3 nifH probe was used (19).
Because nif genes are conserved among a broad spectrum of bacteria, the use of universal primers has enabled the amplification and analysis of nifH sequences from very different microorganisms and environmental samples (21, 38, 40). However, when we used the universal degenerate primers Zf and Zr (39), the expected nifH gene products were not amplified. Instead, a PCR fragment with sequences encoding a hypothetical reductase with 40 to 43% gene sequence identity and only 19% amino acid similarity to Frankia nifH was amplified (22). Consequently, we developed new primers to determine whether the Mexican isolates have nifH sequences. We also undertook an analysis of the full-length 16S rRNA region to verify that these isolates are outside the Frankia clade and to establish their phylogenetic position in the order Actinobacteria. Our data show that the two isolates examined in detail cluster with the Thermomonosporaceae and the Micromonosporaceae, families that have not previously been described as diazotrophic. The possibility exists, however, that individual taxa fix nitrogen.
MATERIALS AND METHODS
Bacterial strains, cultivation, and microscopy.
We utilized Frankia belonging to different host specificity groups (HSG) as reference strains (Table 1). All the Frankia strains were cultured in DPM (3). The Mexican strains were isolated in DPM and grown in DPM supplemented with glucose instead of propionate as a carbon source (DGM), as described previously (36). Micromonospora aurantiaca (ATCC 27029) was routinely grown in ISP medium 2 (yeast maltose agar) (2).
TABLE 1.
Microbial strains, isolate designation, isolate origin and plant source, and presence or absence of aerial mycelia
| Isolate or strain | Origin or source | Aerial myceliuma | Plant source |
|---|---|---|---|
| Frankia BR | Brazil | − | Casuarina |
| Frankia IPNCe16 | Veracruz, México | − | Casuarina |
| Frankia HFPCcI3 | Florida | − | Casuarina |
| Frankia Mc3 | Veracruz, México | − | Myrica |
| Frankia Mc10 | Veracruz, México | − | Myrica |
| Frankia HFPCeI2 | Harvard Forest, Mass. | − | Elaeagnus |
| L3 | Irapuato, México | + | Casuarina |
| L5 | Irapuato, México | + | Casuarina |
| 7501 | Veracruz, México | + | Casuarina |
| 7702 | Veracruz, México | + | Casuarina |
| 8000 | Veracruz, México | + | Casuarina |
| S. antibioticus | ATCC 11891 | ND | |
| M. aurantiaca | ATCC 27029 | + |
A − indicates the absence of aerial mycelia, and a + indicates the presence of aerial mycelia. ND, not determined.
The strains were grown for 1 month at 30°C, with the spent medium exchanged with fresh medium once a week. The bacteria were examined by using phase or Nomarski optics on a Zeiss Axiophot microscope or by staining aliquots with acridine orange (0.05 mg ml−1 in 50 mM H2PO4, pH 6.8) followed by epifluorescence microscope observation (31). Some strains were also examined by utilizing scanning electron microscopy. The cells were fixed as described previously (19), mounted on stubs, coated with gold-palladium, and examined with an ETEC scanning electron microscope at 10 kV.
Phenotypic characters were assessed by growing the bacteria in modified BAP medium (18) supplemented with phosphatidylcholine (31) or in DPM-DGM broth or medium solidified with 1.5% (wt/vol) agar. The solid medium was supplemented with filter-sterilized propionate or glucose as a C source. Nitrate and ammonia assimilation were assessed by growing the isolates in media containing either NH4NO3 (0.01%) or (NH4)2SO4 (0.1%) or totally lacking combined nitrogen. Other growth responses were assayed by culturing the actinobacteria in ISP medium 2, ISP medium 3 (oatmeal agar), and a modified (L. S. Tisa, personal communication) Czapek agar medium (2). Frankia strain HPFCcI3 (41) and M. aurantiaca were also grown in these media.
Cultures were scored for growth responses after the plates were incubated for 2 to 3 weeks at 28°C.
15N isotope dilution.
Strains L5, 7501, and 7702 were grown in DGM medium with 0.4 mM 15NH4SO4 (enriched to 10% of the total NH4SO4) added as the sole nitrogen source; the natural occurrence of 15N is 0.366%. Frankia IPNCe16 and Burkholderia vietnamiensis TVV75T were grown as positive controls, and Streptomyces antibioticus and Escherichia coli DH5α were used as negative controls. The inoculum size was standardized at 4 μg of protein/ml of culture medium by using the Bradford technique. The flasks were incubated at 28°C for 40 days, after which time the cultures were harvested by centrifugation and washed with deionized water and their dry weights were determined.
Total nitrogen in the dry samples was determined by using the Kjeldahl method. After titration, the solution was brought to an alkaline pH and subsequently evaporated to dryness. The residue was suspended in water to a concentration of 1 μg of NH4Cl/μl.
15N enrichment was determined by using an atomic absorption spectrophotometer (model NOI-6E; Fischer Analysen Instrumente, Leipzig, Germany).
DNA isolation.
Total genomic DNA from the various bacteria was isolated by using a modified cetyltrimethylammonium bromide protocol described previously (23). The concentration and purity of the DNA were evaluated with a spectrophotometer (Gen Quant; Pharmacia, Uppsala, Sweden).
PCR amplification of nifH genes and sequence analysis.
We utilized a two-step PCR strategy to amplify the nifH genes from the Mexican isolates. S. antibioticus (ATCC 11891) (Table 1) was used as a negative control. The first reaction amplified a 1.2-kb fragment comprising the entire nifH gene, the intergenic spacer region, and the 5′ end of nifD. The amplification was done by using the primer IGK (5′-TAC GGY AAR GCB GGY ATC GG-3′) (25) and the reverse primer NDR-1 (5′-TTG GAG CCG GCR TAN GCR CA-3′) (present study). The PCR mix was comprised of 50 ng of genomic DNA, 1.5 mM Mg2+, 200 μM deoxynucleoside triphosphates, 0.1 μmol of each primer and 2.5 U of Taq polymerase. The amplification was done with an initial denaturation step at 95°C followed by 35 cycles at 94°C for 1 min, 56°C for 1 min, and 72°C for 2 min with a final renaturation step for 7 min. The 1.2-kb PCR products were separated on a 1.5% (wt/vol) agarose gel and purified by using the Concert rapid gel extraction system (Gibco, BRL).
The second step amplified an internal nifH fragment of 360 bp and used the primers PolF (5′-TGC GAY CCS ARR GCB GGY ATC GG-3′) and PolR (5′-ATS GCC ATC ATY TCR CCG GA-3′) (25) using 50 ng of the first PCR product as a template. The PCR conditions were the same as those described previously by Poly et al. (25). The reproducibility of the PCR was examined by repeating the reactions at least twice using both the same DNA preparation and DNA from separate extracts of the same isolate or strain.
The 360-bp fragment was purified by using the GeneClean II protocol (Bio 101, Carlsbad, Calif.). The purified PCR fragments were cloned into pBluescript SK(+) by using a TA cloning method that converts the plasmid into linear molecules with T-cohesive ends. We utilized E. coli DH5α for transformation by electroporation (Bio-Rad). Transformants were selected by looking for white colonies, and plasmids were isolated by an alkaline lysis method (30). The insertion of fragments of the correct size was inferred by restriction digestion with PstI and XhoI.
After verifying that a fragment of the correct size was produced, the cloned fragments were sequenced in an ABI PRIM 310 automatic sequencer. Analysis of obtained sequences was done with the BLAST algorithm (1) and DNAman (Lynnon BioSoft, Quebec, Canada). Deduced amino acid sequences were aligned with other nifH sequences from different diazotrophs. The gene sequences were aligned with the Clustal X software (34). Indel-containing regions were excluded from the analysis. Matrix pairwise comparisons were corrected for multiple-base substitutions by the two-parameter method of Kimura. Phylogenetic trees were constructed by the neighbor-joining method (29). A bootstrap confidence analysis was performed on 1,000 replicates to determine the reliability of the distance tree topologies obtained (10). A graphic representation of the resulting tree was obtained by using the NJ plot software (24).
DNA-DNA hybridization analysis.
Total DNA was digested with BamHI, and restriction fragments were subjected to electrophoresis, blotted, and hybridized (5). DNA-DNA homology was based on relative levels of hybridization with two 32P-labeled probes: total DNA from Frankia strain Br and total DNA from the non-Frankia isolate 7702. DNA amounts in gels and radioactivity levels were quantified by densitometry and liquid scintillation, respectively, as described previously (5).
Amplified rRNA gene restriction analysis.
The 16S rRNA gene was amplified by using the universal primers rD1 and fD1 (37) using PCR conditions described previously by Benson et al. (4). These primers target the almost complete 16S rRNA gene (1,500 bp). Aliquots (5 μl) of the 16S PCR amplification product were digested according to the procedures described by the manufacturer. The following enzymes were used: CfoI, NciI, MspI, Sau3A (Boehringer Mannheim, Ottweiler, Germany), RsaI, HinfI, TacI, and HaeIII (Gibco, Los Angeles, Calif.). Restricted DNA was analyzed by electrophoresis in 3% (wt/vol) agarose gels. Sequence divergences between the 16S rRNA genes of pairs of strains were estimated from the proportion of shared restriction fragments by using the Jaccard method (28). A dendrogram was constructed from the distance matrix by using the unweighted-pair group method with arithmetic mean (UPGMA). Both procedures were done with the NTSYSpc 1.8 program (28).
PCR amplification of the 16S gene and sequencing.
The PCR fragments were cloned into pPCRIVTOPO+. Transformants containing inserts were screened by the blue-white phenotype, and plasmids were obtained by the alkaline lysis method. The insertion of fragments of the correct size was inferred by restriction digestion with EcoRI. DNA sequencing of the PCR fragments was performed by Genome Express (Grenoble, France). In order to obtain the whole sequence, internal primers were designed. The primers were 5′-GGTGAAATGCGCAGATAT-3′ (corresponding to coordinate 646 of the Micromonospora 16S sequence) and 5′-GGGCGCTTAATGCGTTAG-3′ (corresponding to coordinate 860 of the Micromonospora 16S sequence). The 16S rRNA gene sequences were analyzed as indicated above for nifH.
Nucleotide sequence accession numbers.
The complete sequence of the PCR fragments is available from GenBank under accession numbers AY534920 for isolate L5 and AY534921 for isolate 7501.
RESULTS
Morphological and physiological characteristics.
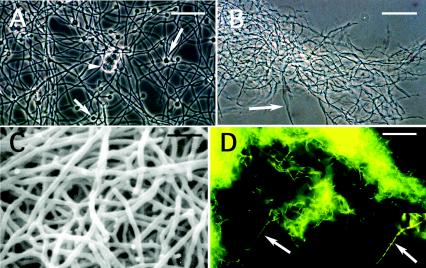
The isolates (7501, L3, L5, 8000, and 7702) examined in this report form aerial hyphae in agar medium and are highly branched and filamentous when grown in liquid media (Fig. 1B to D). The Mexican isolates are significantly smaller than the reference Frankia strain CcI3 (Fig. 1A), measuring 0.25 to 0.4 μm in diameter, compared to 0.5 to 0.6 μm for strain CcI3. Although Frankia filaments are septate, partitions are more obvious in the filaments of the Mexican isolates, especially when stained with acridine orange (Fig. 1D). Under certain growth conditions, the hyphae appeared fragmented.
FIG. 1.
Morphology of some of the actinobacteria analyzed in this report. (A) Casuarina strain CcI3 with multilocular sporangia (arrowhead) and diazovesicles (arrows) grown in liquid BAP minus N medium. Bar, 20 μm. (B) Hyphae (arrow) of L5 from a culture grown in stirred BAP plus N. Bar, 40 μm. (C) Scanning electron micrograph of hyphae from strain 8103 grown in stirred BAP plus N. Bar, 2.5 μm. (D) Hyphae of 7702 stained with acridine orange. Segmented hyphae are indicated with the arrow and in the insert. Bar, 20 μm.
The cells of the Mexican isolates were gram positive and aerobic when grown with combined N. Changes in morphology and growth rate were observed when the bacteria were grown in media containing different C sources (data not shown). They formed numerous spores and produced raised colonies that were yellow-white in color on DGM agar plates. When grown in ISP medium 2, the colonies of the Mexican isolates ranged from light orange to maroon. M. aurantiaca was bright orange in this medium. All the strains, including Frankia CcI3, grew in ISP medium 3 (data not shown).
Unlike Frankia strains Br and CcI3, which produce numerous thick-walled, diazotrophic vesicles when grown under conditions of nitrogen limitation (Fig. 1A), the isolates lacked similar structures. When growing in DGM without N, the Mexican isolates frequently exhibited an altered morphology, consisting of hyphae of different diameters. The thin, highly branched filaments were localized toward the center of the colony, whereas the thicker hyphae were observed on the periphery (data not shown).
15N isotope dilution analysis.
To confirm that the isolates were capable of fixing nitrogen, we performed a 15N isotope dilution analysis. We found that isolates L5, 7501, and 7702 and the positive controls Frankia IPNCe16 and B. vietnamiensis acquired significant mass over the course of the experiment, while the negative controls grew poorly. As evaluated by total N determination using the Kjeldahl method, the negative controls accumulated very low levels of nitrogen. Both positive controls and the isolates L5, 7501, and 7702 exhibited considerable isotopic dilution, indicating active nitrogen fixation. B. vietnamiensis showed the highest level of isotopic dilution, whereas Frankia and isolates L5, 7501, and 7702 were similar to one another, exhibiting a ca. 5% dilution of 15N (Table 2). This result demonstrates that the Mexican isolates possess an active nitrogenase as was previously suggested from our acetylene reduction assay data (36) and from the ability of these strains to grow in N-free media (13).
TABLE 2.
Biomass, total N, and excess of 15N obtained in the isotopic dilution assayb
| Isolate or strain | Biomass (dry weight) (mg) | Total N (Kjeldahl) (mg) | % Excess of 15Na |
|---|---|---|---|
| Frankia IPNCe16 | 6.7 | 0.42 | 4.69 ± 0.05 |
| B. vietnamiensis TVV75T | 14.4 | 0.98 | 1.07 ± 0.002 |
| L5 | 8.6 | 0.29 | 5.43 ± 0.01 |
| 7501 | 14.2 | 0.45 | 5.74 ± 0.003 |
| 7702 | 14 | 0.68 | 5.29 ± 0.01 |
| S. antibioticus | 2.5 | 0.02 | NDb |
| E. coli | 2.1 | ND | ND |
Three replicates ± standard deviations are shown.
ND, not determined.
PCR amplification of nifH genes and sequence analysis.
After the first PCR for the nifH gene amplification using primers IGK and NDR-1, several bands were observed in the lanes containing DNA from the Mexican isolates L3, L5, 7501, and 8000 and from the positive controls, which include Frankia strain IPNCe16, one of several actinobacteria isolated from nodules of C. equisetifolia trees growing in Veracruz, Mexico (23, 35). No bands were observed in the lane containing DNA from S. antibioticus, the negative control (data not shown). Bands with a molecular mass similar to the expected size of the nifH-nifD region (1.2 kb) were purified and used as a template for the second PCR with primers PolR and PolF. All the PCR products were of the expected size of 360 bp (Fig. 2). When the fragments were sequenced, they were found to be very similar to nifH sequences from Frankia (Table 3). The isolates showed 97 to 98% similarity to nifH from Casuarina-nodulating Frankia and 84 to 85% similarity to unculturable Frankia from Ceonothus nodules. This result confirms our earlier findings using Southern blot analysis that these isolates carry at least one gene, nifH, from the nif operon (19).
FIG. 2.
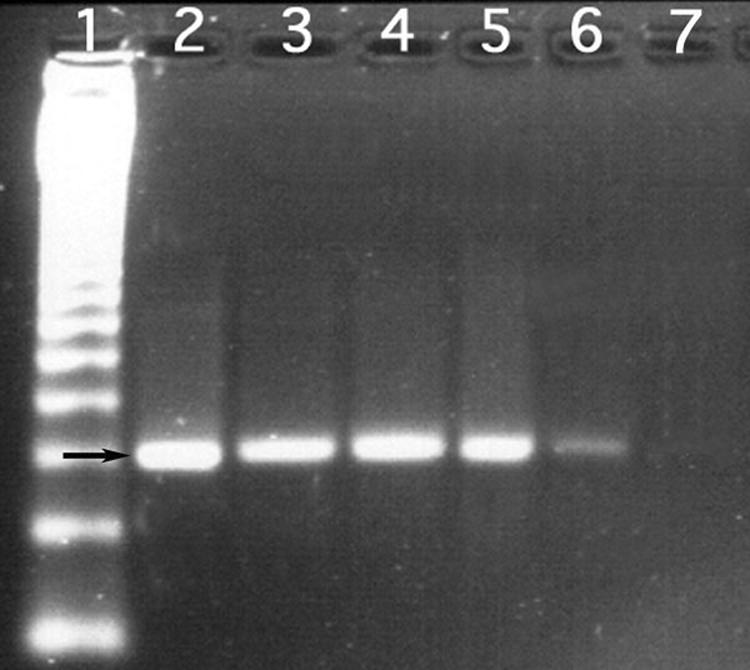
PCR amplification of an internal nifH fragment of 360 bp (indicated by the arrow) using the universal primers PolF and PolR. Lane 1, ladder; lane 2, Frankia IPNCe16 (isolated from C. equisetifolia grown in Mexico); lanes 3 to 6, non-Frankia actinobacteria L3, L5, 7501, and 8000, respectively; lane 7, no-DNA control.
TABLE 3.
Percent similarity between the sequences of a 315-bp fragment of nifH from the Mexican isolates and different diazotrophic actinomycetes
| Reference strain (Genbank accession no.) | % Similarity to Mexican isolate (GenBank accession no.)
|
||
|---|---|---|---|
| L3 (AY714296) | L5 (AY714297) | 7501 (AY714295) | |
| Frankia IPNCe16 (Casuarina) (AY714294) | 97 | 98 | 97 |
| Frankia FaC1 (Alnus) (X73983) | 92 | 93 | 92 |
| Frankia EuIK1 (Elaeagnus) (U53362) | 92 | 93 | 92 |
| Frankia HRN18a (Alnus) (X17522) | 92 | 92 | 91 |
| Frankia ArI3 (Alnus) (L41344) | 91 | 91 | 90 |
| Uncultured Datisca endophyte (X76398) | 87 | 88 | 87 |
| Uncultured Coriaria endophyte (X76399) | 86 | 86 | 85 |
| Uncultured Ceanothus endophyte (U78306) | 84 | 85 | 84 |
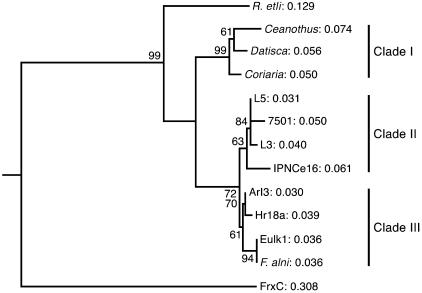
From the partial nifH nucleotide sequences, the deduced amino acid sequences were aligned with homologous regions from various Frankia strains (see the supplemental material), and a phylogenetic tree was constructed. Figure 3 shows the consensus tree based on 100 bootstrap analyses. The nifH gene-containing actinobacteria were grouped into three different clades. Clade I includes unculturable microsymbionts detected in nodules from plants belonging to the Rhamnaceae, Coriariaceae, and Datiscaceae. Clade II includes DNA sequences from the Mexican isolates as well as from Frankia strain IPNCe16 (Casuarina HSG). Clade III encompasses nifH sequences from Frankia strains belonging to the Alnus and Elaeagnus HSG. Another neighbor-joining tree, which included the Casuarina cunninghamiana-isolated strain CcI3 (Table 1), was generated. In this tree, L5, L3, and 7501 clustered with CcI3 and not with the Alnus Frankia strains, further supporting the close relationship between nifH from Casuarina Frankia strains and the Mexican isolates (see the supplemental material).
FIG. 3.
Neighbor-joining tree based on the deduced amino acid sequences derived from partial nifH gene nucleotide sequences of Frankia strains and a new group of putative nitrogen-fixing actinomycetes. Numbers on branches are P values for bootstrap replicates (100). frxC (GenBank accession number X60490) is a Chlamydomonas reinhardtii nifH-like gene, namely the chloroplast chlL gene for light-independent protochlorophyllide reductase. It was used as an outgroup.
Based on nifH gene analysis, the Mexican isolates lie outside the Alnus-Elaeagnus Frankia group but are in the same clade as Frankia strains isolated from nodules of Casuarina.
DNA-DNA hybridization.
DNA relatedness has been used as a genotypic parameter to delineate species (5). Values below 70% are considered to show that the organisms belong to different species (32).
Levels of relatedness were between 43 and 48% among the Mexican isolates when DNA from the non-Frankia strain 7702 was used as a probe (Table 4). The relatively low level of DNA relatedness among the Mexican isolates, ca. 44% relatedness to L5 and 48% relatedness to 7501, suggests that these strains are distinct from isolate 7702. By contrast, DNA from Frankia Br (Casuarina HSG) showed lower levels of DNA relatedness to the isolates, from 27 to 40%. These data give additional support to the conclusion that the Mexican isolates are completely distinct from Frankia.
TABLE 4.
Percent DNA-DNA relatedness between the Mexican actinobacteria and different Frankia species
| Strain or isolate | % DNA-DNA relatedness with:
|
|
|---|---|---|
| Frankia Br | Isolate 7702 | |
| Frankia Br | 100 | 39.21 |
| Frankia IPNCe16 | 98.5 | 39.55 |
| L5 | 39.59 | 43.60 |
| 7501 | 26.06 | 48.43 |
| 7702 | 37.20 | 100 |
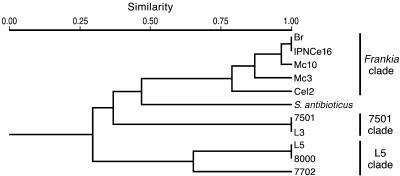
Amplified restriction analysis of 16S rRNA genes.
The 16S rRNA PCR products were digested with nine different restriction enzymes (see Materials and Methods) providing 52 different markers. These markers were treated as binary data and transformed into a distance matrix using the Jaccard coefficient. The constructed dendrogram using UPGMA is shown in Fig. 4. The dendrogram shows that the Frankia clade is clearly separated from the new group of nifH-containing actinobacteria. Moreover, the non-Frankia actinobacteria formed two different clades, one consisting of isolates L5, 8000, and 7702 and the other including isolates L3 and 7501.
FIG. 4.
Dendrogram (UPGMA) of genetic relationships among 16S rRNA genotypes identified by amplified rRNA restriction analysis. A matrix of genetic distances (Jaccard) was used to construct the dendrogram. S. antibioticus was used as the outgroup.
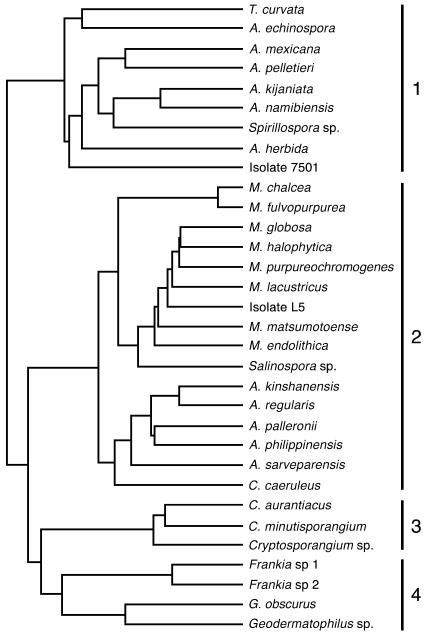
Phylogenetic analysis based on 16S rRNA.
For the phylogenetic analysis, we amplified the complete 16S rRNA regions of two of the isolates, one of each clade determined by amplified rRNA restriction analysis (7501 and L5), and compared them to a broad range of actinomycetes. The DNA amplified from the isolates was a single band of about 1,500 bp, the expected size of the 16S rRNA gene among eubacteria, using the universal 16S rRNA primers. The 16S rRNA nucleotide sequences of the isolates L5 and 7501, representing the two clades, were aligned with homologous regions from various actinomycetes, and a phylogenetic tree was constructed. The dendrogram constructed (UPGMA tree) with the 16S rRNA sequence and compared with the members of the Actinobacteria showed four clades. Besides the Cryptosporangium and Frankia-Geodermatophilus phylogenetic clusters (clades 3 and 4, respectively), clade 1 includes Actinomadura, Actinocorallia, Thermomonospora, and Spirillospora as well as isolate 7501, whereas clade 2 comprises Micromonospora, isolate L5, and Salinospora (Fig. 5).
FIG. 5.
Dendrogram (GCG Program Wisconsin version 10.3) of genetic relationships among 16S rRNA sequences. L5 and 7501 are compared with diverse members of the Actinobacteria. The genus names of the various actinobacteria and the accession numbers (in parentheses) for their 16S RNA DNA sequences are as follows: Thermomonospora curvata (AF002262), Actinomadura echinospora (J420135), Actinomadura mexicana (AF277195), Actinomadura pelletieri (AF163119), Actinomadura kijaniata (X97890), Actinomadura namibiensis (AJ420134), Spirillospora sp. strain JCM3123 (AF163124), Actinocorallia herbida (D85473), Micromonospora chalcea (X92594), Micromonospora fulvopurpurea (X92595), Micromonospora globosa (X92600), Micromonospora halophytica (X92601), Micromonospora purpureochromogenes (X92611), Micromonospora lacustricus (X92622), Micromonospora matsumotoense (AF152109), Micromonospora endolithica (AJ560635), Salinospora sp. strain CNH643 (AY040619), Actinoplanes kinshanensis (AB047496), Actinoplanes regularis (AB047502), Actinoplanes palleronii (AB048216), Actinoplanes philippinensis (AB037007), Actinoplanes sarveparensis (AB047505), Couchioplanes caeruleus subsp. caeruleus (X93202), Cryptosporangium (formerly Actinoplanes [33]) aurantiacus (AB047490), Cryptosporangium (formerly Actinoplanes [33]) minutisporangium (AB037007), Cryptosporangium sp. (AB006168), Frankia sp. 1 (M55343), Frankia sp. 2 (L40622), Geodermatophilus obscurus (L40621), and Geodermatophilus sp. (X92358).
When L5 and 7501 were examined in a neighbor-joining tree using the GCG Program (Wisconsin ver. 10.3) with the complete 16S sequences of selected species in the genus Micromonospora (12), isolate L5 was found to cluster within this genus, with its closest relative being M. aurantiaca (X92604) (see the supplemental material). Isolate 7501 was found to cluster with members of the family Thermomonosporaceae.
DISCUSSION
This study is the first to use molecular data to show that actinobacteria other than Frankia have nifH sequences. Streptomyces thermoautotrophicus UBT1 fixes N2 based on 15N analysis and growth in N-free medium, but it does not reduce acetylene. It has an unusual nitrogen reduction system which is coupled to carbon monoxide reduction and is dependent on oxygen (27). This strain inhabits the soil covering burning charcoal piles and is not associated with plants. In contrast, the Mexican isolates reduce acetylene to ethylene and associate with actinorhizal plants as endophytes (13, 36). They were found in the outer cells of the nitrogen-fixing nodule, a location distinct from the Frankia-containing cells, which are found in the interior of the root nodule. When we tried to isolate Frankia from the nodules of C. equisetifolia, the non-Frankia actinobacteria overgrew the culture medium and masked the presence of the Frankia endosymbiont, which was later designated IPNCe16 (22, 35).
The Mexican isolates were originally obtained from nodules of C. equisetifolia trees growing on the dunes of the Gulf of Mexico near sea level and growing in altitudes of up to 2,300 m. They came from locations that included gardens, agricultural soils, and highly eroded sites (13). Observations of the frequency of endophyte isolation suggest that a medium lacking N was most effective for isolation (data not shown). These actinobacteria, especially members of the L5 clade, utilize several of the C sources recommended for the isolation and growth of Frankia, such as propionate and pyruvate as well as glucose (36). However, isolate 7501 grows better in medium containing glucose as a C source. The Mexican isolates also grew on several media used to support the growth of Streptomyces and other actinomycetes.
Previously, we determined that some of the Mexican isolates could grow on media lacking nitrogen and that they were capable of reducing acetylene to ethylene (13, 36). We also observed weak hybridization of a nifHD probe from K. pneumoniae (unpublished results) and a nifH probe from Frankia CcI3 to the DNA of the Mexican isolates (19). These results led us first to confirm by 15N isotopic dilution assay that the isolates fix nitrogen (Table 2) and second to investigate whether one of the structural genes for nitrogenase, nifH, was present in these strains. Using newly developed primers, we found nifH-homologous sequences in four of the Mexican isolates: L3, L5, 7501, and 8000. The nifH genes of these strains were more fully characterized and found to lie in a clade that includes nifH from IPNCe16, the Frankia strain isolated from C. equisetifolia growing in Mexico.
We expect that the nifDK genes are also present in the Mexican isolates based on our preliminary Southern blots and possibly in the same operon as nifH based on the fact that one of the primers used in this study incorporated the 5′ end of the nifD gene. A PCR-based approach to find the nifDK genes is under way, as is the search for any specialized hyphal structures or physiological adaptations that could potentially permit nitrogenase activity in a microaerophilic environment. If the nifDK genes show robust homology to the cognate genes in Frankia strain IPNCe16, as does nifH, this would strongly suggest that lateral gene transfer from Frankia has occurred. In any case, the combination of features—growth on an N-deficient medium, positive acetylene reduction and 15N isotopic dilution assays, and the presence of the nifH gene—gives strong support to the conclusion that these actinobacteria fix atmospheric N to ammonia.
The relationship of these unusual isolates has been addressed by amplified rRNA restriction analysis, which has been successfully utilized to define phylogenetic relationships among microorganisms belonging to many different bacterial species (16). Recently, by using this approach, several unidentified N2-fixing Burkholderia groups were described (9), and three of these groups have been officially validated as novel Burkholderia species: B. unamae (6), B. tropica (26), and B. xenovorans (12). In this study, the Casuarina nodule-isolated actinobacteria were delineated into two clades by using amplified rRNA restriction analysis, and in addition, DNA-DNA hybridization assays were included to confirm their distinctness from Frankia. Based on the latter, their genetic similarity was 27 to 40%, whereas the DNA-DNA relatedness between the two clades was <50%. Taken together, these findings suggest that there are at least two separate groups of Mexican isolates.
The construction of a dendrogram based on the 16S rRNA sequence of two of the isolates and the published 16S rRNA sequences of a number of actinobacteria confirmed the results of the amplified rRNA restriction analysis and DNA-DNA hybridization assays (Fig. 5). These results showed that isolate 7501 is located in clade 1, forming a distinct phylogenetic cluster among the members of the family Thermomonosporaceae, whereas isolate L5 is included within the genus Micromonospora within clade 2, and may represent a novel species. A neighbor-joining phylogenetic tree of 16S rRNA sequence data of 14 selected species included in the genus Micromonospora (15) confirmed that isolate L5 is closely related to M. aurantiaca and that isolate 7501 clusters with Thermomonosporaceae.
It is noteworthy that isolates 7501 and L3, which have an identical amplified rRNA restriction analysis profile, have been recovered from nodules of Casuarina grown at a high altitude in Irapuato (2,300 m), whereas isolates L5 and 8000, whose amplified rRNA restriction analysis pattern is also identical, have been recovered from Casuarina trees growing in locations widely distant: at sea level, on dunes at Veracruz, 800 km away. This finding suggests that these bacterial species are widely distributed in geographical regions that exhibit very different environmental conditions. The presence of similar actinobacteria in surface-sterilized nodules has been noted by others trying to isolate Frankia from Casuarina nodules in South America (L. Wall, personal communication) and in Thailand (N. Boonkerd and A. Nuntagij, personal communication). Hence, these actinobacteria may be widespread.
Micromonospora has been isolated from soil and plants (14) and as an endophyte of healthy wheat plants (7). Several species of Micromonospora have the potential for biological control because they are able to parasitize the fungal phytopathogen Pythium (8). The isolation of a selected group of actinobacteria from within the tissue of nitrogen-fixing root nodules suggests that the host derives some benefit from harboring the endophyte. Whether these actinobacteria are involved in improving the life of the Casuarina plants by fixing nitrogen, by producing compounds that facilitate plant growth, or by competing with pathogenic bacteria or fungi in the rhizosphere is not known at this time. Actinobacteria are well known for their ability to produce a broad range of antibacterial, antifungal, and plant growth-regulatory metabolites (11).The ability to fix nitrogen by a select group of actinobacteria in addition to Frankia may be added to this list.
Supplementary Material
Acknowledgments
This research was partially supported by the Mexican Council for Science and Technology (CONACYT) grant 1370/N9206 and by the Research and Graduate Studies Section of the Mexican Polytechnic Institute.
This work represents part of the Ph.D. thesis of N.-O.P., who received a fellowship from CONACYT.
We thank Margaret Kowalczyk (UCLA) for her help with the figures and Charles R. Marshall (Harvard University) for useful discussions.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atlas, R. M. 1993. Alphabetical listing of media. In L. C. Parks (ed.), Handbook of microbiological media. CRC Press, Inc., Boca Raton, Fla.
- 3.Baker, D. D., and D. O'Keefe. 1984. A modified sucrose fractionation procedure for the isolation of frankiae from actinorhizal root nodules and soil samples. Plant Soil 78:23-28. [Google Scholar]
- 4.Benson, D. R., D. W. Stephens, M. L. Clawson, and W. B. Silvester. 1996. Amplification of 16S rRNA genes from Frankia strains in root nodules of Ceanothus griseus, Coriaria arborea, Coriaria plumose, Discaria toumatou, and Purshia tridentata. Appl. Environ. Microbiol. 62:2904-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caballero-Mellado, J., L. E. Fuentes-Ramírez, V. M. Reis, and E. Martínez-Romero. 1995. Genetic structure of Acetobacter diazotrophicus populations and identification of a new genetically distant group. Appl. Environ. Microbiol. 61:3008-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caballero-Mellado, J., L. Martínez-Aguilar, G. Paredes-Valdez, and P. Estrada-de los Santos. 2004. Burkholderia unamae sp nov., an N2-fixing rhizospheric and endophytic species. Int. J. Syst. Evol. Microbiol. 54:1165-1172. [DOI] [PubMed] [Google Scholar]
- 7.Coombs, J. T., and C. M. M. Franco. 2003. Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 69:5603-5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Tarabily, K. A., G. E. S. J. Hardy, K. Sivasithamparam, A. M. Hussein, and D. L. Kurtboke. 1997. The potential for biological control of cavity-spot disease of carrots, caused by Pythium coloratum, by streptomycete and non-streptomycete actinobacteria. New Phytol. 137:495-507. [DOI] [PubMed] [Google Scholar]
- 9.Estrada-de los Santos, P., M. R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 11.Franco, C. M. M., and L. E. L. Coutinho. 1991. Detection of novel secondary metabolites. Crit. Rev. Biotechnol. 11:193-276. [DOI] [PubMed] [Google Scholar]
- 12.Goris, J., P. De Vos, J. Caballero-Mellado, J.-H. Park, E. Falsen, J. F. Quensen III, J. M. Tiedje, and P. Vandamme. 2004. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400T and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:1677-1681. [DOI] [PubMed] [Google Scholar]
- 13.Guillén, G. M., M. Valdés, J. Liao, and A. M. Hirsch. 1993. Identificación de actinobacterias aisladas de nódulos de Casuarina, por técnicas tradicionales y moleculares. Rev. Lat. Amer. Microbiol. 35:195-200. [Google Scholar]
- 14.Hayakawa, M. T., T. Sadakata, T. Kijura, and H. Nonomura. 1991. New methods for the highly selective isolation of Micromonospora and Microbispora from soil. J. Ferment. Bioeng. 72:320-326. [Google Scholar]
- 15.Kasai, H., T. Tamura, and S. Harayama. 2000. Intragenic relationships among Micromonospora species deduced from gyrB-based phylogeny and DNA relatedness. Int. J. Syst. Evol. Microbiol. 50:127-134. [DOI] [PubMed] [Google Scholar]
- 16.Laguerre, G., M. R. Allard, F. Revoy, and N. Amarger. 1994. Rapid identification of rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl. Environ. Microbiol. 60:56-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munive, J.-A. 1997. Características fisiológicas y quimiotaxonómicas de un nuevo grupo de actinomicetos diazótrofos. Tesis de Maestro en Ciencias. ENCB, IPN, México, D.F., México.
- 18.Murry, M. A., M. S. Fontaine, and J. G. Torrey. 1984. Growth kinetics and nitrogenase reduction in Frankia sp. HFP ArI3 grown in batch culture. Plant Soil 78:61-78. [Google Scholar]
- 19.Niner, B. M. 1995. Studies of luteolin-regulated and ribosomal genes in symbiosis. Ph.D. thesis. University of California, Los Angeles.
- 20.Niner, B. M., J. P. Brandt, M. C. Villegas, C. R. Marshall, A. M. Hirsch, and M. Valdés. 1996. Analysis of partial sequences of genes coding for 16S rRNA of actinomycetes isolated from Casuarina equisetifolia nodules in Mexico. Appl. Environ. Microbiol. 62:3034-3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohkuma, M., S. Koda, R. Usami, K. Horikoshi, and T. Kudo. 1996. Diversity of nitrogen fixation genes in the symbiotic intestinal microflora of the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:2747-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérez, N. O. 2001. Filogenia molecular entre actinomicetos fijadores de nitrógeno. Tesis de Doctorado. ENCB, IPN, México, D.F., México.
- 23.Pérez, N. O., L. Vásquez, H. Olivera, and M. Valdés. 1999. Genetic characterization of Mexican Frankia strains isolated from Casuarina equisetifolia. Can. J. Bot. 77:1214-1219. [Google Scholar]
- 24.Perrière, G., and M. Gouy. 1996. WWW-Query: an on-line retrieval system for biological sequence banks. Biochimie 78:364-369. [DOI] [PubMed] [Google Scholar]
- 25.Poly, F., L. Jocteur-Monrozier, and R. Bally. 2001. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 152:95-103. [DOI] [PubMed] [Google Scholar]
- 26.Reis, V. M., P. Estrada-de los Santos, S. Tenorio-Salgado, J. Vogel, M. Stoffels, S. Guyon, P. Mavingui, V. L. D. Baldani, M. Schmid, J. I. Baldani, J. Balandreau, A. Hartmann, and J. Caballero-Mellado. 2004. Burkholderia tropica sp. nov., a novel nitrogen-fixing plant-associated bacterium. Int. J. Syst. Evol. Microbiol. 54:2155-2162. [DOI] [PubMed] [Google Scholar]
- 27.Ribbe, M., D. Gadkari, and O. Meyer. 1997. N2 fixation by Streptomyces thermoautotrophicus involves a molybdenum-dinitrogenase and a manganese-superoxide oxidoreductase that couple N2 reduction to the oxidation of superoxide produced from O2 by a molybdenum-CO dehydrogenase. J. Biol. Chem. 272:26627-26633. [DOI] [PubMed] [Google Scholar]
- 28.Rohlf, F. J. 1994. NTSYS-pc. Numerical taxonomy and multivariate analysis system. Exter Software, Setauket, New York.
- 29.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 31.Schwencke, J. 1991. Rapid exponential growth and increased biomass yield of some Frankia strains in buffered and stirred mineral medium (BAP) with phosphatidyl choline. Plant Soil 136:37-41. [Google Scholar]
- 32.Stackebrant, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 33.Tamura, T., and K. Hatano. 2001. Phylogenetic analysis of the genus Actinoplanes and transfer of Actinoplanes minutisporangius Ruan et al. 1986 and ‘Actinoplanes aurantiacus’ to Cryptosporangium minutisporangium comb. nov. and Cryptosporangium aurantiacum sp. nov. Int. J. Syst. Evol. Microbiol. 51:2119-2125. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTALX windows interphase: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vásquez, L., N. O. Pérez, and M. Valdés. 2000. Isolation and symbiotic characteristics of Mexican Frankia strains associated with Casuarina. Appl. Soil Ecol. 14:249-255. [Google Scholar]
- 36.Villegas, M., L. Vásquez, and M. Valdés. 1997. Growth and nitrogenase activity conditions of a new diazotrophic group of filamentous bacteria isolated from Casuarina root nodules. Rev. Lat. Amer. Microbiol. 39:65-72. [Google Scholar]
- 37.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Widmer, F., B. T. Shaffer, L. A. Porteous, and R. J. Seidler. 1999. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade Mountain Range. Appl. Environ. Microbiol. 65:374-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zehr, J., and L. McReynolds. 1989. Use of degenerate oligonucleotide primers for amplification of the nifH gene from the marine cyanobacterium Tricodesmium thiebautii. Appl. Environ. Microbiol. 55:2522-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zehr, J., M. T. Mellon, and S. Zani. 1998. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Z., and J. G. Torrey. 1985. Studies of an effective strain of Frankia from Allocasuarina lehmanniana of the Casuarinaceae. Plant Soil 87:1-16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.