ABSTRACT
Chronic polymicrobial infections are associated with increased virulence compared to monospecies infections. However, our understanding of microbial dynamics during polymicrobial infection is limited. A recent study by Limoli and colleagues (D. H. Limoli, G. B. Whitfield, T. Kitao, M. L. Ivey, M. R. Davis, Jr., et al., mBio 8:e00186-17, 2017, https://doi.org/10.1128/mBio.00186-17) provides insight into a mechanism that may contribute to the coexistence of Pseudomonas aeruginosa and Staphylococcus aureus in the cystic fibrosis (CF) lung. CF lung infections have frequently been used to investigate microbial interactions due to both the complex polymicrobial community and chronic nature of these infections. The hypothesis of Limoli et al. is that the conversion of P. aeruginosa to its mucoidy phenotype during chronic CF infection promotes coexistence by diminishing its ability to kill S. aureus. Highlighting a new facet of microbial interaction between two species that are traditionally thought of as competitors, this study provides a platform for studying community assembly in a relevant infection setting.
KEYWORDS: Pseudomonas aeruginosa, Staphylococcus aureus, aggregates, biofilms, coinfection, cystic fibrosis, mucoidy, polymicrobial infection
COMMENTARY
A key question underlying investigations into microbial interactions is how can bacteria coexist in the face of competition? As a model system, chronic infection of the cystic fibrosis (CF) lung is excellent for elucidating microbial interactions that shape community composition and function. CF is an inherited condition caused by mutation of the cystic fibrosis transmembrane conductance regulator (CFTR), which results in the accumulation of mucus (sputum) in patient lungs (1). Bacteria use sputum as an energy source to grow to high densities in the CF lung, causing infections that begin in early childhood and persist throughout an individual’s life (2, 3). Despite extensive knowledge of the complex nature of CF lung infections, there is still a lack of understanding regarding how this diversity is maintained. Of particular interest is the interaction of Pseudomonas aeruginosa and Staphylococcus aureus, two of the most prominent bacteria in the CF lung (2, 3). It is well-known that S. aureus is often one of the first bacteria to infect the CF lung during childhood, but it is subsequently outcompeted and displaced by P. aeruginosa, which produces an arsenal of quorum sensing (QS)-regulated virulence factors that are lytic to S. aureus (4). However, the mechanisms that allow S. aureus and P. aeruginosa to coexist during CF lung infection are not well understood.
In a recent article, Limoli and colleagues provide evidence that the conversion of P. aeruginosa to mucoidy during chronic CF infection promotes coexistence with S. aureus by dampening the ability of P. aeruginosa to kill S. aureus (5). Specifically, they found that in standard in vitro culture conditions, survival of S. aureus was limited when it was cultured with wild-type P. aeruginosa. However, when cultured with P. aeruginosa strains that had been isolated alongside S. aureus from CF sputum samples, S. aureus survival improved. Further examination of these isolates showed that many had genetic mutations that led to the overproduction of alginate. The model of mucoid conversion proposed by Limoli et al. is strongly supported by the data presented, despite the fact that mutation as a primary requirement for P. aeruginosa coexistence with S. aureus is not a well-supported idea in the existing literature. While the conversion of P. aeruginosa to a mucoid phenotype in the lung has long been correlated with a poorer clinical outcome (6, 7), populations of P. aeruginosa isolated from CF lungs have been shown to be highly diverse (8), with a mixture of mucoid and nonmucoid phenotypes, despite many months of chronic infection (9). In addition, alternative mutations such as those involved in QS-regulated lytic pathways are frequently observed. Therefore, it is likely that many of these mutations are a result of selective pressures that allow P. aeruginosa to persist within the host, not directly as a result of coexistence with another microbe, despite their impact on microbial community interactions.
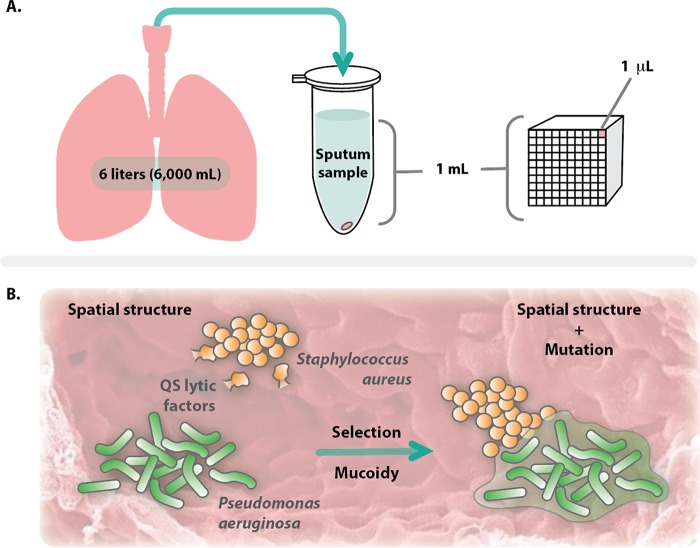
At the macroscale, the CF lung provides a vast landscape in which a bacterial community can develop. A single CF sputum sample often contains ~109 bacteria per ml; however, a bacterial population of this size would volumetrically fill less than 0.1% of 1 ml of sputum (Fig. 1A). In addition, there is evidence that bacteria infecting the CF lung exist as small, high-density clusters of cells (~101 to 104 cells), called aggregates, that form structured and spatially organized communities (10–13). Thus, it may be valuable to consider the CF lung as a vast landscape occupied by sparse bacterial aggregates. Previous studies indicate that aggregates need to be within microns of each other to interact (14, 15), and therefore, it is valuable to consider interactions on the micron scale, rather than as a macroscale clinical sample taken from the lung.
FIG 1 .
Scale and spatial structure impact interactions between microbes in the CF lung. (A) Scale comparison of bacteria residing in the CF lung. The structure of the CF lung provides both a large surface area and volume. From an expectorated 1 ml sputum sample, approximately 109 bacteria occupy ~1 μl of this volume. The observation that bacteria exist as aggregates in the CF lung that must be localized within microns in order to interact suggests either that aggregates are either geographically isolated or in concentrated sites. (B) When aggregates are in concentrated sites, coexistence of S. aureus and P. aeruginosa is likely maintained by spatial structure, preventing lysis of S. aureus by QS-regulated lytic factors produced by P. aeruginosa. Selective pressure applied by the host often causes mucoid conversion of P. aeruginosa, which in turn alters spatial structure and changes how these two bacteria coexist.
It is well-known that when grown in well-mixed communities in vitro, P. aeruginosa actively kills S. aureus, a finding supported by Limoli et al. (5). However, there is evidence that P. aeruginosa and S. aureus can coexist for days in the absence of mutation when grown in spatially structured communities. For example, the Lubbock wound model promotes coexistence of P. aeruginosa and S. aureus for multiple days, even with P. aeruginosa strains that are highly lytic for S. aureus in well-mixed environments (16, 17). The coexistence of these bacteria has been shown to result in their increased tolerance to antimicrobial agents, both in human and murine models of infection, indicating that there can be fitness benefits for both species as a result of their interaction (17, 18). Additionally, work by Fazli et al. has shown that P. aeruginosa and S. aureus occupy distinct spaces during human infection, highlighting a role for spatial structure (19). It would be surprising if biogeography did not play a role in the interaction described here. Similar to alginate, there is increasing support that large polymers such as mucin, DNA, and poly-N-acetylglucosamine play a role in spatially structuring microbial communities during infection (15, 20, 21). For example, our lab has demonstrated that Aggregatibacter actinomycetemcomitans and Streptococcus gordonii interact through cross-feeding, and that this interaction is maintained by precise spatial organization of these organisms through modulation of poly-N-acetylglucosamine levels in the biofilm matrix (15). Additionally, the secreted biofilm matrix of Candida albicans was found to be protective for S. aureus against antimicrobial treatment (22). Such polymers likely play multiple roles in these systems, acting as both a physical barrier and a chemical sink to small molecules. Therefore, an alternate hypothesis for the findings by Limoli et al. (5) is that the alginate produced is not only reducing the production of lytic factors by P. aeruginosa, but may also promote spatial structuring that allows for coexistence (Fig. 1B).
A great strength of this study is the effective use of an in vitro epithelial cell model that supports the coexistence of P. aeruginosa and S. aureus, pushing the biological relevance of this observed interaction beyond well-mixed in vitro batch culture methods that do not provide realistic environments in which to study microbe-microbe interactions (12, 23). The use of the epithelial model in this study raises interesting questions about the spatial structure of the developing P. aeruginosa-S. aureus community. Do the bacteria form aggregates in this system? Is there spatial patterning? Limoli and colleagues (5) have provided an experimentally elegant approach for observing how the host drives selection of phenotypes that potentially alter the ways in which bacteria interact in vivo. Understanding more about these interactions at the micron scale will inform us on how these communities assemble and potentially how to disrupt them. This study provides a platform for some exciting future studies to elucidate the coexistence of two pathogens relevant not only to the CF lung environment, but also to other infection sites such as chronic wounds.
The views expressed in this Commentary do not necessarily reflect the views of this journal or of ASM.
Footnotes
For the article discussed, see https://doi.org/10.1128/mBio.00186-17.
Citation Darch SE, Ibberson CB, Whiteley M. 2017. Evolution of bacterial “frenemies.” mBio 8:e00675-17. https://doi.org/10.1128/mBio.00675-17.
REFERENCES
- 1.Brennan AL, Geddes DM. 2002. Cystic fibrosis. Curr Opin Infect Dis 15:175–182. doi: 10.1097/00001432-200204000-00013. [DOI] [PubMed] [Google Scholar]
- 2.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. 2012. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev 25:193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison F. 2007. Microbial ecology of the cystic fibrosis lung. Microbiology 153:917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- 4.Schuster M, Sexton DJ, Diggle SP, Greenberg EP. 2013. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol 67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 5.Limoli DH, Whitfield GB, Kitao T, Ivey ML, Davis MR Jr, Grahl N, Hogan DA, Rahme LG, Howell PL, O’Toole GA, Goldberg JB. 2017. Pseudomonas aeruginosa alginate overproduction promotes coexistence with Staphylococcus aureus in a model of cystic fibrosis respiratory infection. mBio 8:e00186-17. doi: 10.1128/mBio.00186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryder C, Byrd M, Wozniak DJ. 2007. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldan R, Cigana C, Testa F, Bianconi I, De Simone M, Pellin D, Di Serio C, Bragonzi A, Cirillo DM. 2014. Adaptation of Pseudomonas aeruginosa in cystic fibrosis airways influences virulence of Staphylococcus aureus in vitro and murine models of co-infection. PLoS One 9:e89614. doi: 10.1371/journal.pone.0089614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darch SE, McNally A, Harrison F, Corander J, Barr HL, Paszkiewicz K, Holden S, Fogarty A, Crusz SA, Diggle SP. 2015. Recombination is a key driver of genomic and phenotypic diversity in a Pseudomonas aeruginosa population during cystic fibrosis infection. Sci Rep 5:7649. doi: 10.1038/srep07649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Høiby N, Molin S. 2012. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 10:841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 10.Darch SE, Kragh KN, Abbott EA, Bjarnsholt T, Bull JJ, Whiteley M. 2017. Phage inhibit pathogen dissemination by targeting bacterial migrants in a chronic infection model. mBio 8:e00240-17. doi: 10.1128/mBio.00240-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kragh KN, Alhede M, Jensen PØ, Moser C, Scheike T, Jacobsen CS, Seier Poulsen S, Eickhardt-Sørensen SR, Trøstrup H, Christoffersen L, Hougen HP, Rickelt LF, Kühl M, Høiby N, Bjarnsholt T. 2014. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect Immun 82:4477–4486. doi: 10.1128/IAI.01969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sørensen SR, Moser C, Kühl M, Jensen PØ, Høiby N. 2013. The in vivo biofilm. Trends Microbiol 21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Kragh KN, Hutchison JB, Melaugh G, Rodesney C, Roberts AEL, Irie Y, Jensen PØ, Diggle SP, Allen RJ, Gordon V, Bjarnsholt T. 2016. Role of multicellular aggregates in biofilm formation. mBio 7:e00237-16. doi: 10.1128/mBio.00237-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connell JL, Kim J, Shear JB, Bard AJ, Whiteley M. 2014. Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc Natl Acad Sci U S A 111:18255–18260. doi: 10.1073/pnas.1421211111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. 2014. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci U S A 111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun Y, Dowd SE, Smith E, Rhoads DD, Wolcott RD. 2008. In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen 16:805–813. doi: 10.1111/j.1524-475X.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 17.Trivedi U, Parameswaran S, Armstrong A, Burgueno-Vega D, Griswold J, Dissanaike S, Rumbaugh KP. 2014. Prevalence of multiple antibiotic resistant infections in diabetic versus nondiabetic wounds. J Pathog 2014:173053. doi: 10.1155/2014/173053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalton T, Dowd SE, Wolcott RD, Sun Y, Watters C, Griswold JA, Rumbaugh KP. 2011. An in vivo polymicrobial biofilm wound infection model to study interspecies interactions. PLoS One 6:e27317. doi: 10.1371/journal.pone.0027317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fazli M, Bjarnsholt T, Kirketerp-Møller K, Jørgensen B, Andersen AS, Krogfelt KA, Givskov M, Tolker-Nielsen T. 2009. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J Clin Microbiol 47:4084–4089. doi: 10.1128/JCM.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frenkel ES, Ribbeck K. 2017. Salivary mucins promote the coexistence of competing oral bacterial species. ISME J 11:1286–1290. doi: 10.1038/ismej.2016.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pammi M, Liang R, Hicks J, Mistretta T-A, Versalovic J. 2013. Biofilm extracellular DNA enhances mixed species biofilms of Staphylococcus epidermidis and Candida albicans. BMC Microbiol 13:257. doi: 10.1186/1471-2180-13-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kong EF, Tsui C, Kucharíková S, Andes D, Van Dijck P, Jabra-Rizk MA. 2016. Commensal protection of Staphylococcus aureus against antimicrobials by Candida albicans biofilm matrix. mBio 7:e01365-16. doi: 10.1128/mBio.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts AEL, Kragh KN, Bjarnsholt T, Diggle SP. 2015. The limitations of in vitro experimentation in understanding biofilms and chronic infection. J Mol Biol 427:3646–3661. doi: 10.1016/j.jmb.2015.09.002. [DOI] [PubMed] [Google Scholar]