ABSTRACT
Streptococcus pyogenes (group A Streptococcus [GAS]) is an obligate human pathogen responsible for a broad spectrum of human disease. GAS has a requirement for metal homeostasis within the human host and, as such, tightly modulates metal uptake and efflux during infection. Metal acquisition systems are required to combat metal sequestration by the host, while metal efflux systems are essential to protect against metal overload poisoning. Here, we investigated the function of PmtA (PerR-regulated metal transporter A), a P1B-4-type ATPase efflux pump, in invasive GAS M1T1 strain 5448. We reveal that PmtA functions as a ferrous iron [Fe(II)] efflux system. In the presence of high Fe(II) concentrations, the 5448ΔpmtA deletion mutant exhibited diminished growth and accumulated 5-fold-higher levels of intracellular Fe(II) than did the wild type and the complemented mutant. The 5448ΔpmtA deletion mutant also showed enhanced susceptibility to killing by the Fe-dependent antibiotic streptonigrin as well as increased sensitivity to hydrogen peroxide and superoxide. We suggest that the PerR-mediated control of Fe(II) efflux by PmtA is important for bacterial defense against oxidative stress. PmtA represents an exemplar for an Fe(II) efflux system in a host-adapted Gram-positive bacterial pathogen.
KEYWORDS: iron efflux, PmtA, oxidative stress response, PerR, group A Streptococcus, Streptococcus pyogenes
INTRODUCTION
Defense against peroxide is recognized as one of the most widespread stress responses in prokaryotes. A characteristic of the peroxide stress response is the increased expression of peroxidases that can directly remove hydrogen peroxide as well as the induction of systems that can repair damaged proteins and nucleic acids (1, 2). Streptococcus pyogenes (group A Streptococcus [GAS]) coordinates oxidative stress responses through the action of the regulator PerR, which functions as a repressor of oxidative stress defense genes under normal conditions through binding to DNA at per boxes upstream of these genes (3, 4). This stable complex typically exists with Fe as a prosthetic group; in the presence of hydrogen peroxide, Fe causes metal-catalyzed oxidation and damage of the complex, leading to its dissociation and the subsequent transcription of peroxide response genes (3–6). In GAS, the PerR-regulated oxidative stress response encompasses both direct mechanisms, involving the detoxification of reactive oxygen species (ROS), and indirect mechanisms, which involve the repair of biomolecules damaged by oxidative stress. Direct mechanisms involving the enzymatic detoxification of reactive oxygen species are achieved through alkyl hydroperoxidases and glutathione peroxidases (1, 2, 5) as well as a single superoxide dismutase, SodA, which is Mn dependent (7). An example of an indirect response mechanism is PolAI, a DNA polymerase that repairs oxidatively damaged DNA (8).
Another strategy employed by bacteria to ameliorate the effects of peroxide stress is to control the intracellular concentration of prooxidant metal ions so as to minimize the Fenton reaction (10, 11). Cu(I) and Fe(II) are biologically available metal elements capable of potentiating Fenton chemistry. In the case of Fe(II), one aspect of the bacterial peroxide stress response involves the decreased expression of Fe uptake systems and the increased expression of Fe storage proteins that effectively sequester Fe and thus prevent prooxidant activity (12–14). In contrast, copper concentrations within the bacterial cell are held very low as a consequence of the action of a Cu efflux system (CopYAZ) that is controlled by a Cu-sensing regulator (CopY) (15–17). The possibility that Fe ions might also be effluxed from the bacterial cell during peroxide stress has not been widely investigated. Recently, however, two P-type ATPases that mediate bacterial Fe efflux, PfeT of Bacillus subtilis, which is regulated by Fur and PerR (18), and FrvA of Listeria monocytogenes, which is regulated by Fur (19), have been described.
GAS is an obligate human pathogen responsible for approximately 600,000 deaths annually (20). Disease states range from mild infections of the skin (impetigo) and pharynx (“strep throat”) to severe invasive disease such as necrotizing fasciitis and streptococcal toxic shock syndrome. Repeated infection can also lead to postinfectious immune sequelae such as poststreptococcal glomerulonephritis and rheumatic heart disease (21). As a host-adapted human pathogen, GAS deploys multiple systems for resisting innate and adaptive immunity during colonization and infection (21, 22). These systems include mechanisms for defense against oxidative stress (23). To date, the capacity of GAS to efflux Fe as part of the oxidative stress response has not been investigated.
In this work, we describe the role of the cation transport protein denoted PmtA (PerR-regulated metal transporter A) (SPy1167), a P-type ATPase, as a PerR-regulated efflux protein of GAS with specificity for Fe(II). Additionally, we demonstrate that Fe(II) efflux provides increased resistance to oxidative stress through the efflux of prooxidant Fe(II) under conditions of oxidative stress.
RESULTS
PmtA is a P1B-4-type ATPase.
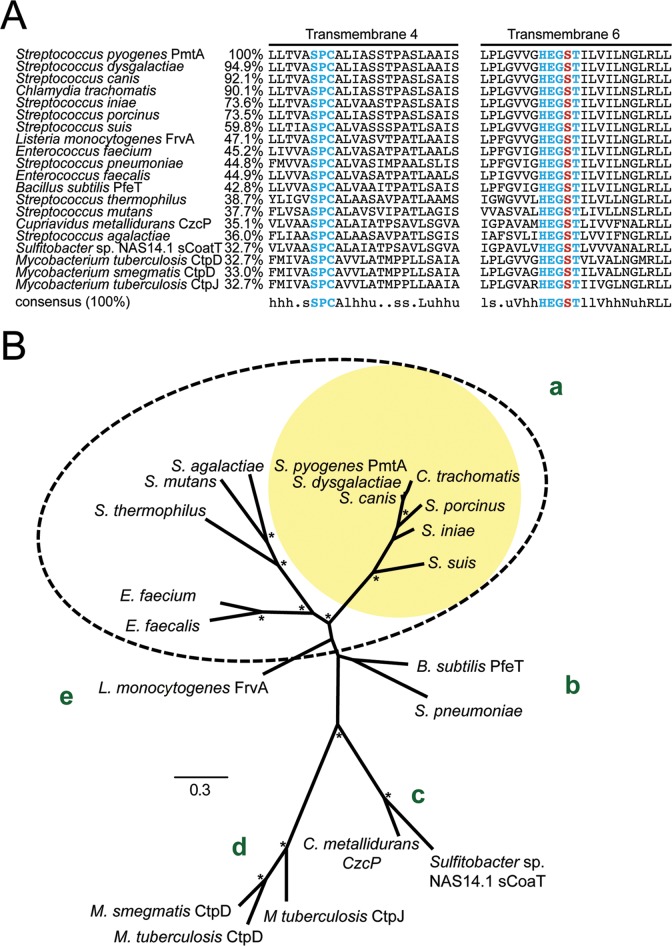
PmtA (SPy1167) is annotated in the M1T1 reference genome (MGAS5005) (24) as a P-type ATPase (25). However, there are multiple subclasses of class P1B-type ATPases in bacteria, each with various metal specificities. Metal ion specificity has previously been shown to depend on specific amino acid sequences within transmembrane (TM) regions of the protein (26). In order to classify PmtA within P-type ATPase class designations, an amino acid alignment of GAS PmtA with other annotated P1B-4-type ATPases from different bacteria (NCBI nonredundant GenBank database) was performed using MUSCLE (27). Following amino acid alignment and hydrophobicity analysis using TMHMM (28), the transmembrane region was analyzed in order to determine the P1B subclass (29, 30). P1B-4 ATPases feature a characteristic SPC amino acid metal binding motif in transmembrane region 4 and an HEG(G/S)T metal binding motif in transmembrane region 6 (29). This analysis confirmed that PmtA is a P1B-4 P-type ATPase, a classification identical to that of PfeT and FrvA (Fig. 1A; see also Fig. S1 in the supplemental material for the full alignment).
FIG 1.
Phylogenetic clustering and high-level conservation of key motifs within transmembrane helices of S. pyogenes PmtA and homologous P1B-4-type ATPase family transporters. Amino acid sequences of P1B-4-type ATPases from 20 bacterial species with close homology to GAS PmtA as well as previously characterized P1B-4-type ATPases were used for analysis. (A) Multiple-sequence alignment of TM helices 4 and 6 from the PmtA and P1B-4-type ATPase family protein alignment (see Fig. S1 in the supplemental material). Key conserved motifs within TM helices 4 and 6 are highlighted in blue, and the essential serine residue within TM helix 6 is highlighted in red. Consensus patterns based on discriminating equivalence class at a 100% threshold are indicated below the alignment. Percent amino acid identities relative to S. pyogenes PmtA (TM helices 1 to 6) are indicated next to the species name. (B) Unrooted maximum-likelihood phylogenetic tree of S. pyogenes PmtA-related sequences based on protein sequences derived from the six core transmembrane helices common to the P1B-4-type ATPase family. The corresponding multiple-sequence alignment is presented in Fig. S1 in the supplemental material. The bar indicates the number of amino acid substitutions per site under the WAG substitution model. Asterisks indicate bootstrap values greater than 90 (out of 100 replicates). The dashed oval denotes clade a, and yellow shading highlights subclade a containing PmtA of GAS.
Phylogenetic analysis indicates that GAS PmtA is closely related to the P1B-4-type ATPases from other lactic acid bacteria (clade a) (Fig. 1B). Within this clade, the P1B-4-type ATPase lineage is differentiated into two clusters. GAS PmtA is closely related to PmtA homologs found in other genetically related streptococcal species, including S. dysgalactiae and S. canis, and a Chlamydia trachomatis PmtA homolog, which share >90% amino acid identity (Fig. 1B, yellow shading, and Fig. S1). Furthermore, S. pyogenes PmtA is genetically distinct from the lineages made up of functionally characterized P1B-4-type ATPase family members B. subtilis PfeT (43% identity) (clade b) and L. monocytogenes FrvA (47% identity) (clade e) (Fig. 1B).
A 5448ΔpmtA mutant is sensitive to high extracellular Fe(II) concentrations.
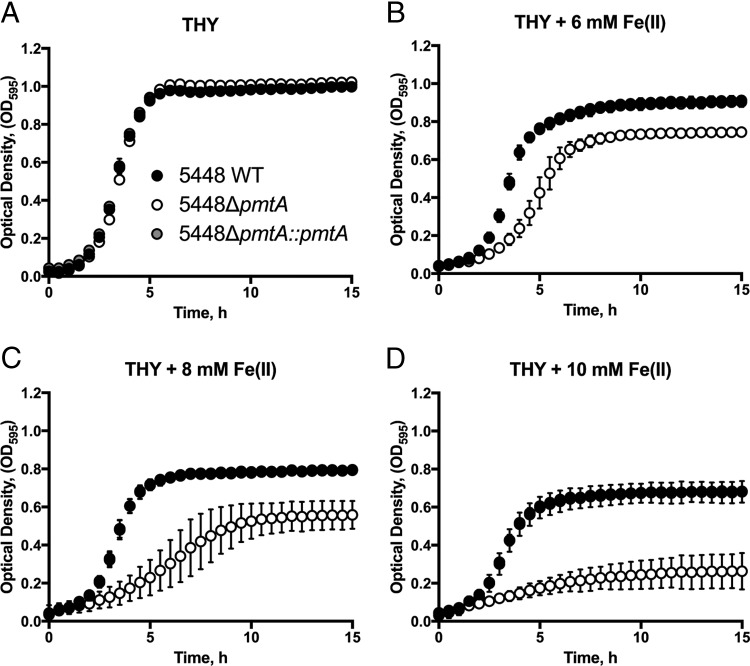
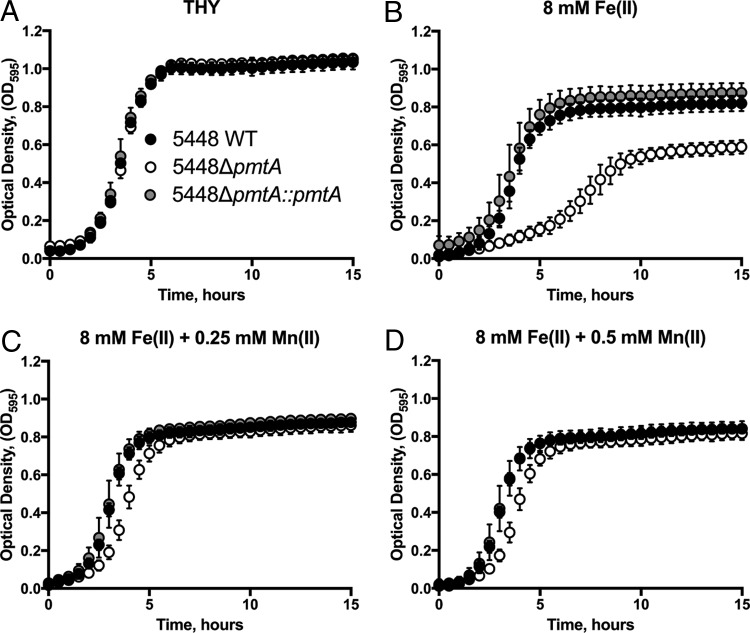
To investigate the role of pmtA in GAS physiology, we created an isogenic gene deletion mutant in invasive GAS M1T1 isolate 5448 (31). Briefly, the 5448ΔpmtA strain was constructed by allelic exchange of the ahpA3 cassette with pmtA (SPy1167). The 5448ΔpmtA mutant was complemented through the exchange of ahpA3 with the wild-type (WT) pmtA allele. We examined the growth characteristics of wild-type strain 5448 (5448 WT), 5448ΔpmtA, and 5448ΔpmtA::pmtA with various metal concentrations. In Todd-Hewitt broth supplemented with 1% (wt/vol) yeast extract (THY broth) alone, all strains grew equivalently (Fig. 2A). However, in the presence of 8 mM Fe(II), 5448ΔpmtA exhibited a prolonged doubling time (3.4 ± 0.2 h [standard deviation {SD}] versus 1.3 ± 0.2 h [SD] for the WT and 1.4 ± 0.1 h [SD] for 5448ΔpmtA::pmtA; P < 0.0001) and reached a lower final cell yield than did 5448 WT and 5448ΔpmtA::pmtA with identical treatments (Fig. 2C). This reduction in the growth rate and endpoint density in the 5448ΔpmtA mutant became more pronounced with increasing concentrations of Fe(II) (Fig. 2B to D). A similar effect was noted with Co(II), where 5448ΔpmtA showed a diminished growth rate and reached a lower stationary-phase optical density (OD) than did 5448 WT and 5448ΔpmtA::pmtA under the same conditions (see Fig. S2 in the supplemental material). This growth defect of the 5448ΔpmtA mutant was restored in the complemented mutant. Growth analysis in the presence of Mn(II), Ni(II), Cu(II), or Zn(II) showed no difference in the growth of the 5448ΔpmtA mutant relative to 5448 WT or 5448ΔpmtA::pmtA (Fig. S3 to S6).
FIG 2.
Growth curve analysis in the presence of Fe(II). Cultures of strains 5448 WT, 5448ΔpmtA, and 5448ΔpmtA::pmtA grown overnight were diluted to a starting OD600 of 0.05 in THY broth (A) or THY broth supplemented with 6 mM (B), 8 mM (C), or 10 mM (D) Fe(II). Growth at 37°C was monitored by recording the OD595. Graphs represent means ± standard deviations of data from 3 independent biological replicates.
Intracellular metal accumulation by 5448ΔpmtA.
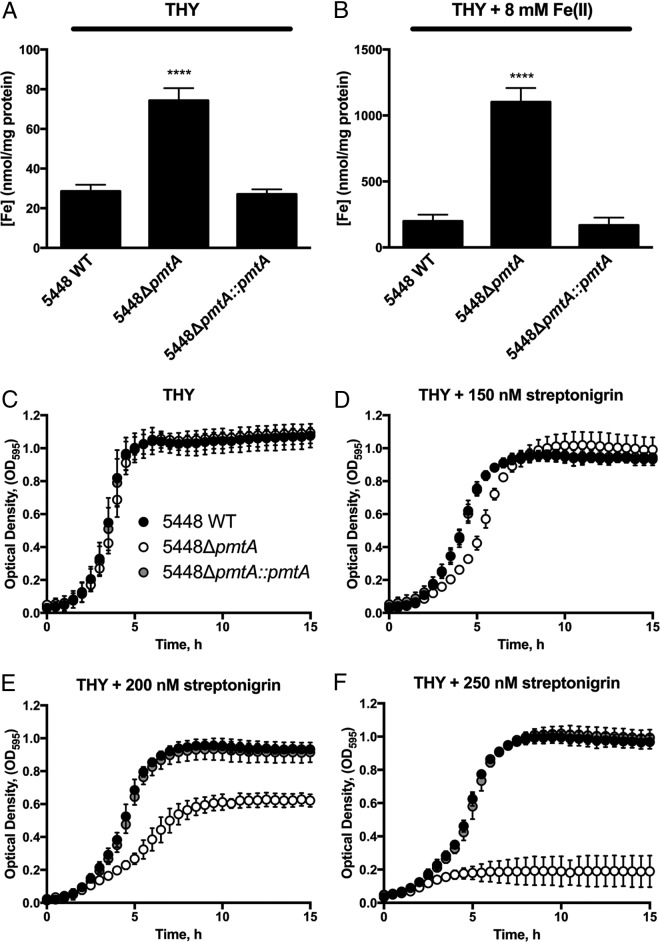
Based on the finding that 5448ΔpmtA is sensitive to high concentrations of Fe(II), we sought to determine whether this sensitivity arose through an inability to efflux this metal ion. Inductively coupled plasma mass spectrometry (ICP-MS) was used to analyze the intracellular metal accumulation of 5448 WT, 5448ΔpmtA, and 5448ΔpmtA::pmtA grown in THY broth after challenge with Fe(II). In the absence of additional Fe(II) in the medium, 5448ΔpmtA accumulated approximately 2-fold-higher levels of intracellular Fe than did 5448 WT and the complemented mutant (Fig. 3A) (P < 0.001). Intracellular Fe accumulation was increased in all strains following the addition of 8 mM Fe(II). However, the 5448ΔpmtA mutant accumulated approximately 5-fold more Fe than did 5448 WT and the complemented mutant (Fig. 3B) (P < 0.0001). ICP-MS was also used to analyze intracellular metal accumulation after the addition of Co(II) and Zn(II). Only Co significantly accumulated in the 5448ΔpmtA background (see Fig. S7 in the supplemental material). In addition to the lack of growth inhibition of the 5448ΔpmtA mutant by Zn(II) (Fig. S6), the lack of significantly higher Zn accumulation in the 5448ΔpmtA strain when challenged by Zn(II) suggests that PmtA does not function as a Zn exporter, as previously hypothesized (25).
FIG 3.
Intracellular Fe accumulation and streptonigrin sensitivity. (A and B) Mid-exponential-growth-phase cultures (OD600 = 0.6 to 0.8) of strains were challenged with sterile water (A) or 8 mM Fe(II) (B). Cells were analyzed by ICP-MS, with metal accumulation normalized to the total protein content. Graphs represent means and standard deviations of data from 3 independent experiments (1-way ANOVA was used for all comparisons to 5448 WT under each condition [**, P < 0.01; ***, P < 0.001; ****, P < 0.0001]). (C to F) Growth curve analysis of strains 5448 WT, 5448ΔpmtA, and 5448ΔpmtA::pmtA. Cultures grown overnight were diluted to an OD600 of 0.05 in THY broth alone (C) or THY broth with 150 nM (D), 200 nM (E), or 250 nM (F) streptonigrin. Growth at 37°C was monitored by recording the OD595. Graphs represent means ± standard deviations of data from 3 independent biological replicates.
Streptonigrin exerts a toxic effect against 5448ΔpmtA.
Based on the observation of increased intracellular Fe levels within the 5448ΔpmtA mutant, we hypothesized that this mutant would show greater susceptibility to streptonigrin. Streptonigrin is an aminoquinone antibiotic that exerts Fe-dependent toxicity against bacteria (32). The addition of 200 nM streptonigrin to THY broth resulted in an increased doubling time (2.4 ± 0.3 h [SD] versus 1.3 ± 0.2 h [SD] for the WT and 1.4 ± 0.4 h [SD] for 5448ΔpmtA::pmtA; P < 0.01) and a reduced stationary-phase optical density of the 5448ΔpmtA mutant (Fig. 3D and E). Furthermore, 250 nM streptonigrin completely inhibited the growth of the 5448ΔpmtA mutant compared to 5448 WT and the complemented mutant, which grew normally (Fig. 3F). This result further supports the view that PmtA is involved in Fe efflux and that elevated intracellular Fe levels in the 5448ΔpmtA mutant (due to the inability to efflux the metal) result in toxicity to the cell.
Changes in pmtA gene expression profiles under different conditions.
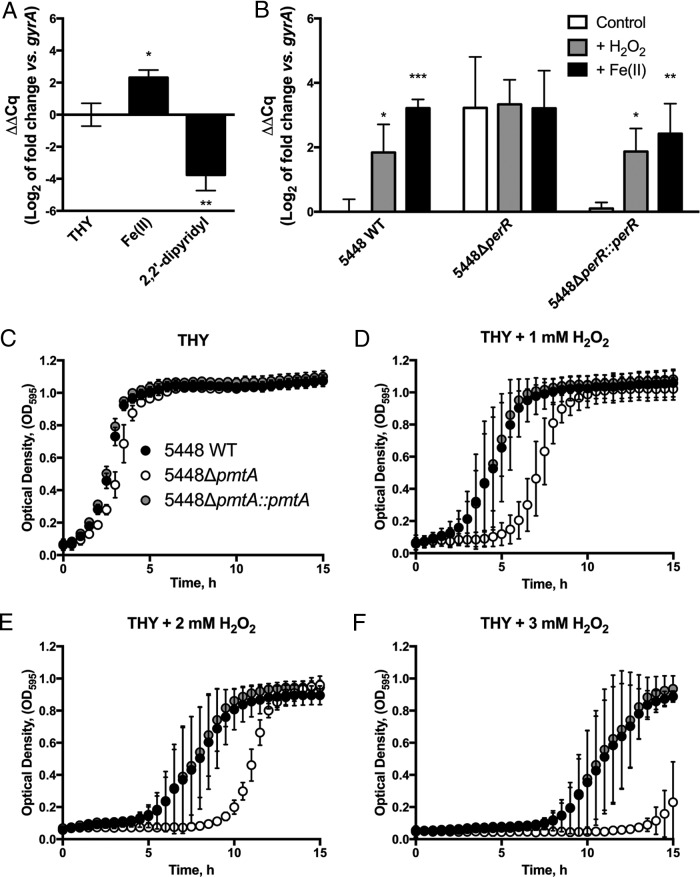
Given that the 5448ΔpmtA mutant was found to be sensitive to Fe(II), we sought to assess the nature of pmtA regulation. Thus, 5448 WT was grown in the presence of subinhibitory concentrations of Fe(II) and the Fe chelator 2,2′-dipyridyl, and the gene expression of pmtA was examined by reverse transcriptase quantitative real-time PCR (RT-qPCR). Consistent with data from our growth analysis, we observed a 4-fold upregulation of pmtA in the presence of Fe(II) (Fig. 4A), while the addition of the Fe chelator 2,2′-dipyridyl resulted in an approximately 16-fold downregulation of pmtA (Fig. 4A).
FIG 4.
Gene expression analysis of pmtA and H2O2 stress sensitivity. (A) 5448 WT was grown in the presence of Fe(II) or 2,2′-dipyridyl, and gene expression of pmtA was analyzed. (B) 5448 WT, 5448ΔperR, and 5448ΔperR::perR cells grown at 37°C to mid-exponential phase (OD600 = 0.6 to 0.8) were challenged with water (control), 5 mM H2O2, or 8 mM Fe(II) for 30 min. pmtA gene expression was analyzed by qPCR using gyrA as the reference gene. Data represent the mean ΔΔCq (log2) values ± standard deviations from 3 independent biological replicates (2-way ANOVA was used for all comparisons to the control for each strain [*, P < 0.05; ** P < 0.01; ***, P < 0.001]). (C to F) Growth curve analysis of 5448 WT, 5448ΔpmtA, and 5448ΔpmtA::pmtA in hydrogen peroxide. Strains were grown in THY broth plus 2 mM Fe(II) to mid-exponential phase (OD = 0.6 to 0.8) and diluted to an OD600 of 0.05 in THY broth with 0.00 mM (C), 1 mM (D), 2 mM (E), or 3 mM (F) H2O2. Growth at 37°C was monitored by recording the OD595. Graphs represent means ± standard deviations of data from 3 independent biological replicates.
A consensus per box, the regulatory element bound by PerR, is found immediately upstream of the pmtA gene (6, 25, 33). We therefore sought to determine whether pmtA is upregulated by hydrogen peroxide, consistent with PerR regulation. Quantitative gene expression data for 5448 WT indicated that pmtA is upregulated approximately 4-fold when exposed to 5 mM H2O2 and approximately 8-fold when exposed to 8 mM Fe(II) (Fig. 4B). This upregulation was PerR dependent, as pmtA gene expression was constitutive in 5448ΔperR. Gene regulation was restored to wild-type levels in the 5448ΔperR complemented mutant, suggesting that hydrogen peroxide and Fe(II) induce pmtA in a PerR-dependent manner.
Growth of 5448ΔpmtA is inhibited by reactive oxygen species.
As excess intracellular Fe is capable of potentiating oxidative stress in biological systems, we hypothesized that Fe accumulation in the 5448ΔpmtA mutant may result in increased susceptibility to oxidative stress. Thus, growth analysis of strains 5448 WT, 5448ΔpmtA, and 5448ΔpmtA::pmtA was performed in the presence of hydrogen peroxide. In the presence of up to 3 mM hydrogen peroxide alone, the 5448ΔpmtA mutant grew at a rate equivalent to those of 5448 WT and the complemented mutant (see Fig. S8 in the supplemental material). However, when cells were pretreated with a subinhibitory concentration of Fe(II) (2 mM), the 5448ΔpmtA mutant exhibited a prolonged lag phase and impaired growth in the presence of increasing H2O2 concentrations (Fig. 4D to F). These data suggest that PmtA plays a role in the GAS response to hydrogen peroxide stress through the efflux of intracellular Fe(II). We additionally examined the sensitivity of the 5448ΔpmtA mutant using the superoxide generator methyl viologen (paraquat). In contrast to its growth in THY broth (Fig. S9A), the 5448ΔpmtA mutant showed an increased doubling time relative to those of 5448 WT and the complemented mutant with 1 mM paraquat (4.1 ± 0.8 h [SD] versus 2.5 ± 0.3 h [SD] for the WT and 2.5 ± 0.6 h [SD] for 5448ΔpmtA::pmtA; P < 0.01) and 1.25 mM paraquat (Fig. S9B and S9C). In a humanized plasminogen mouse model of invasive disease, the 5448ΔpmtA mutant was equally as virulent as 5448 WT and the complemented mutant (Fig. S10).
Growth of 5448ΔpmtA in Fe(II) can be restored by Mn(II) supplementation.
Based on previously reported findings that the growth of a B. subtilis pfeT mutant in Fe(II) could be restored by the addition of Mn(II) (18), we sought to determine if Fe(II) toxicity in the 5448ΔpmtA mutant could be ameliorated by the addition of Mn(II). Growth of the 5448ΔpmtA mutant was inhibited by 8 mM Fe(II); however, growth could be restored to near-wild-type levels by the addition of 0.25 mM Mn(II) (Fig. 5). These data confirm that, similar to a B. subtilis pfeT mutant, Fe(II)-mediated toxicity in the 5448ΔpmtA mutant could be abrogated by the addition of Mn(II). Similarly, the growth defect of the 5448ΔpmtA mutant with 250 nM streptonigrin could also be restored by the addition of 0.25 mM Mn(II) (see Fig. S11 in the supplemental material). The ability of Mn(II) to restore the growth of the 5448ΔpmtA mutant was also observed for Co(II) toxicity (Fig. S12).
FIG 5.
Growth curve analysis of 5448 WT, 5448ΔpmtA, and 5448ΔpmtA::pmtA in Fe(II) and rescue of the 5448ΔpmtA mutant by Mn(II). Cultures of strains 5448 WT, 5448ΔpmtA, and 5448ΔpmtA::pmtA grown overnight were diluted to an OD600 of 0.05 in THY broth (A) or THY broth with 8 mM Fe(II) (B), 8 mM Fe(II) plus 0.25 mM Mn(II) (C), or 8 mM Fe(II) plus 0.5 mM Mn(II) (D). Growth at 37°C was monitored by recording the OD595. Graphs represent means ± standard deviations of data from 3 independent biological replicates.
DISCUSSION
During infection, GAS encounters reactive oxygen species. GAS encodes the regulator PerR, which acts to sense and coordinate the bacterial response to oxidative stress. Previous research examining the PerR regulon has shown that this regulator controls the direct detoxification of reactive oxygen species, mechanisms to repair ROS-mediated damage, and a number of genes involved in metal homeostasis (6, 33, 34). Here, we provided evidence to suggest that the PerR-regulated GAS gene pmtA encodes an Fe(II) efflux pump that contributes to defense against reactive oxygen species.
The acquisition of transition metal ions, such as Zn, Fe, and Mn, has emerged as an important factor in bacterial pathogenesis. Although bacterial pathogens are starved for Fe by the human host, they produce a variety of high-affinity uptake systems that can acquire this metal ion (35). Fe uptake is important for GAS pathogenesis (36); the bacterium can acquire Fe from hemoglobin (37) but not transferrin or lactoferrin (38). GAS utilizes the ABC transporters Siu and Sia in conjunction with Shr and Shp for heme uptake (36, 39, 40). Given the importance of Fe as a difficult nutrient to acquire, it had previously been assumed that Fe export is not required, but instead, excess Fe would be sequestered in ferritin or ferritin-like proteins (41), and the import of Fe would be downregulated (42). Here, we demonstrate that PmtA is a PerR-regulated GAS P1B-4 P-type ATPase that contributes to resistance to oxidative stress through the removal of prooxidant Fe(II) from the intracellular environment. Our observations coincide with the P1B-4 P-type ATPases PfeT of B. subtilis (18) and FrvA of L. monocytogenes (19), both of which are also Fe(II) efflux pumps regulated by the Fur family of regulators. The likely role of PmtA as an Fe(II) exporter is further supported by the observation that the pmtA deletion mutant accumulated higher levels of Fe(II) than did the WT strain under both normal and Fe(II) stress conditions.
The first described bacterial Fe efflux system was the cation diffusion facilitator FieF of Cupriavidus metallidurans, which provided resistance to high-Fe(II) stress and also had metal specificities for Cd(II), Co(II), Ni(II), and Zn(II) (43). GAS PmtA also demonstrated specificity for Co(II). While we observed that PmtA is able to efflux Fe(II) from GAS, it can also efflux Co(II), which is consistent with the function of P1B-4-type ATPases (18, 29). PmtA was previously described to be involved in Zn(II) transport (25). Those authors' conclusions were based on microarray data demonstrating that PerR differentially regulated pmtA alongside genes involved in Zn transport. Furthermore, those authors reported that PmtA had high homology to ZosA of B. subtilis [before this protein was redefined as the Fe(II)-transporting P1B-4-type ATPase PfeT] and that growth of the pmtA perR double mutant restored the Zn(II) hyperresistance phenotype observed in the perR mutant to near-WT levels (25). However, the accumulation of Zn in the pmtA mutant was not demonstrated. Here, we have shown that Zn does not accumulate in the 5448ΔpmtA mutant (see Fig. S7 in the supplemental material).
Under oxidative stress conditions, excess Fe(II) is capable of reacting with superoxide and H2O2 (10, 44). Fe-bound proteins may be vulnerable to damage by these reactive oxygen species, leading to the liberation of Fe as labile ions. If allowed to accumulate, this increased amount of Fe could further potentiate oxidative stress; thus, GAS possesses PmtA to efflux Fe(II) from the cell under oxidative stress conditions. As such, the 5448ΔpmtA deletion mutant, which contained higher levels of intracellular Fe(II) than did the WT strain, grew less efficiently in the presence of hydrogen peroxide and superoxide. Similarly, the Salmonella enterica serovar Typhimurium Fe efflux pump STM3944 mutant was shown to be more sensitive to hydrogen peroxide (45). The PerR regulon in GAS controls a variety of genes involved in the response to hydrogen peroxide stress (5). Here, we confirm that pmtA expression is upregulated by both Fe(II) and hydrogen peroxide and is regulated via PerR. The GAS ferritin-like protein Dpr (formerly MrgA) is also regulated by PerR (14) and has been shown to chelate Fe and protect GAS under conditions of oxidative stress (46). We hypothesize that GAS uses Dpr and PmtA to sequester and export Fe(II) under oxidative stress conditions, respectively. Upon initial entry into the host, GAS undoubtedly encounters oxidative stress (23, 47). However, the lack of a virulence defect of the 5448ΔpmtA mutant in the mouse invasive disease model may reflect a niche-specific effect where this mutant is still able to survive and cause disease. Dpr may compensate for the loss of PmtA in the mouse model of invasive disease.
The necessity of maintaining the Fe/Mn ratio in Gram-positive bacteria such as Streptococcus pneumoniae and GAS is particularly important for defense against oxidative stress and pathogenesis (48–50). During oxidative stress, mismetallation of proteins can occur when the Fe/Mn ratio is disrupted (44). THY broth is a metal-rich microbiological medium; we have determined that THY broth as used in our laboratory contains approximately 15 μM Fe, which may explain the level of Fe accumulation that we observed in the 5448ΔpmtA mutant. Thus, when media and cells were treated with 2,2′-dipyridyl, the downregulation of pmtA may have been caused by a decreased amount of available Fe. Furthermore, the Mn rescue of the 5448ΔpmtA mutant when stressed with streptonigrin, Fe(II), and Co(II) is consistent with results obtained by examining PfeT of B. subtilis (18). We speculate that Fe might exert a toxic effect by mismetallating into Mn-containing metalloproteins. Thus, the addition of Mn relieves the toxic effect of excess Fe(II). In summary, PmtA represents an essential system for ensuring Fe(II) homeostasis in GAS, thus providing an important additional mechanism for resistance to oxidative stress.
MATERIALS AND METHODS
Phylogenetic and protein sequence analysis.
Sequences that share homology with S. pyogenes PmtA (see Table S1 in the supplemental material) were identified on the basis of a BLASTP search of the nonredundant GenBank database (NCBI) and included functionally characterized P1B-4 ATPase amino acid sequences from B. subtilis PfeT (GenBank accession number CUB17201.1), L. monocytogenes FrvA (accession number NP_464168), and GAS PmtA (accession number AAZ51785.1). Determination of the P-type ATPase subclass was performed by using previously described methods (26). Protein sequences were aligned by using MUSCLE v3.8.31 (27) and trimmed to the common transmembrane domains 1 and 6 relative to the well-characterized PfeT protein from Bacillus subtilis (18). A best-fit phylogenetic tree was estimated by using RAxML v8.2.8 (51) based on the WAG substitution model with gamma correction of among-site rate variation. One hundred nonparametric bootstraps were applied.
Bacterial strains and growth conditions.
The characterized invasive GAS M1T1 clinical strain 5448 was used in this study (31). 5448 WT and isogenic mutant strains were routinely cultured at 37°C in Todd-Hewitt broth (Difco) supplemented with 1% (wt/vol) yeast extract (Merck) (THY broth). For growth on solid medium, strains were cultured on 5% horse blood agar or THY agar (1.5%, wt/vol). Escherichia coli strains were grown in lysogeny broth (LB) (52). E. coli strain MC1061 was used for the maintenance of pHY304-derived plasmids, and the E. coli Top10 strain was used for the maintenance of pJRS233-derived plasmids (see Table S2 in the supplemental material). For the selection of GAS, the antibiotics kanamycin (300 μg/ml), spectinomycin (100 μg/ml), and erythromycin (2 μg/ml) were used. To select for E. coli plasmids, kanamycin (50 μg/ml), spectinomycin (100 μg/ml), and erythromycin (500 μg/ml) were used.
DNA manipulations and mutant construction.
All GAS mutants were constructed according to standard operating procedures (50). Briefly, the construction of 5448ΔpmtA was achieved by the amplification of 5448 WT genomic DNA with primers pmtA-KO-1 (incorporating an XhoI site at the 5′ end) and pmtA-KO-2 (with homology to Kmr at the 5′ end) to create the 5′-flanking region of pmtA. Primers pmtA-KO-3 (with homology to Kmr at the 5′ end) and pmtA-KO-4 (incorporating a BamHI site at the 5′ end) were used to amplify the 3′-flanking region of pmtA (see Table S3 in the supplemental material). The Kmr cassette was amplified from pUC4ΩKm2 by using primers km-F and km-R. The resultant fragments were amplified via 3-way PCR to form the construct pmtA-KO, which was ligated into the temperature-sensitive shuttle vector pHY304 to yield pHY304-pmtA-KO. The plasmid was electroporated into electrocompetent GAS cells as previously described (53). A double-crossover event was selected for, resulting in 5448ΔpmtA, which contains Kmr (marker) and has lost Eryr (shuttle vector). Complementation of mutants was achieved by PCR amplification of the original pmtA allele using primers pmtA-KO-1 and pmtA-KO-4 and ligation into vector pJRS233, yielding pJRS233-pmtA, which was electroporated into 5448ΔpmtA. The plasmid was integrated via double crossover at 37°C for the replacement of Kmr with pmtA at the original locus to yield 5448ΔpmtA::pmtA. All strains were confirmed by DNA sequencing conducted at the Australian Equine Genetics Research Centre at the University of Queensland.
Growth analysis.
Growth analysis of 5448 WT, 5448ΔpmtA, and 5448ΔpmtA::pmtA was performed by using various amounts of CoSO4·4H2O, CuSO4·5H2O, FeSO4·7H2O, MnSO4·4H2O, NiSO4·6H2O, ZnSO4·7H2O, streptonigrin (Sigma), and methyl viologen (paraquat) (Sigma). All salts were analytical grade (Sigma). Metal solutions were prepared in deionized distilled water and filter sterilized. Streptonigrin was prepared with 100% ethanol and paraquat in deionized water. Cultures of GAS grown overnight in THY broth were diluted in fresh THY broth to an OD at 600 nm (OD600) of 0.05 and supplemented with various concentrations of the compound. The optical density was measured at 595 nm every 30 min while cells were grown statically at 37°C in a FLUOstar Optima plate reader (BMG Labtech) in flat-bottom 96-well plates (Greiner) (final volume of 200 μl/well). For growth in the presence of hydrogen peroxide, GAS strains 5448 WT, 5448ΔpmtA, and 5448ΔpmtA::pmtA were cultured at 37°C in THY broth or THY broth supplemented with 2 mM Fe(II). Bacteria were harvested at the mid-log growth phase and diluted to an OD600 of 0.05 in THY broth with various concentrations of hydrogen peroxide, and the optical density at 595 nm was recorded. The doubling time was calculated by using the reciprocal of the gradient of natural-log-transformed OD595 values during the exponential growth phase.
Quantitative intracellular metal accumulation analysis.
Strains 5448 WT, 5448ΔpmtA, and 5448ΔpmtA::pmtA were grown to the mid-exponential growth phase (OD600 = 0.6 to 0.8) in THY broth and challenged with an equal volume of either sterile deionized H2O or Fe(II) in H2O to a final concentration of 8 mM Fe(II), 1 mM Co(II), or 1 mM Zn(II) for 1 h at 37°C. Cells were harvested by centrifugation at 3,200 × g and washed 3 times with 1× phosphate-buffered saline (PBS) plus 250 mM EDTA and 3 times with 1× PBS, and a sample was taken to determine the total protein content by a bicinchoninic acid (BCA) assay (Thermo Scientific). Samples were digested in 70% nitric acid at 70°C for 48 h, diluted to 2% nitric acid, and analyzed by ICP-MS for Fe, Zn, and Co at the School of Geosciences at the University of Queensland. The metal content was normalized to the total protein content obtained from the BCA assay.
RNA extraction.
Cultures of GAS grown overnight were diluted to an OD600 of 0.025 in THY broth or THY medium supplemented with 8 mM Fe(II) or 50 μM 2,2′-dipyridyl. At the mid-exponential growth phase (OD600 = 0.6 to 0.8), 5 ml of the culture was harvested by centrifugation at 8,000 × g for 10 min, and pellets were resuspended in TRIzol (Invitrogen), transferred to lysing matrix B tubes (MP Biomedicals), and mechanically lysed by using the FastPrep 120 instrument (Thermo Scientific). Alternatively, in experiments with 5448 WT, 5448ΔperR, and 5448ΔperR::perR, strains were grown to the mid-exponential phase and exposed to 5 mM H2O2 or 8 mM Fe(II) for 30 min, and the resultant bacterial pellets were processed identically. RNA was extracted from lysates by the addition of 0.2 volumes of chloroform, and phase separation of RNA was undertaken by centrifugation at 12,000 × g for 30 min at 4°C. The aqueous phase was removed, combined with 70% ethanol, and loaded onto RNeasy columns (Qiagen) for purification according to the manufacturer's recommendations. Following RNA elution in ultrapure water, RNA was DNase digested by using RNase-free DNase (Turbo; Ambion) according to the manufacturer's directions. RNA integrity was examined by gel electrophoresis and quantified by spectrophotometric analysis on the NanoDrop instrument (Thermo Scientific). PCR amplification of the RNA samples was performed to confirm the absence of genomic DNA contamination.
Quantitative gene expression analysis.
A total of 500 ng of RNA was converted to cDNA by using Superscript III (Invitrogen) with random hexamers according to the manufacturer's directions. Quantitative PCR (qPCR) was performed by using SYBR green master mix (Applied Biosystems) with primer mixes at 100 nM and cDNA at 2 ng per reaction mixture on the Viia7 instrument (Applied Biosystems). qPCR primers used in this study are described in Table S3 in the supplemental material. Data were analyzed by using LinRegPCR (54) to obtain quantification cycle (Cq) values with gyrA as the reference gene, and data were plotted as ΔΔCq values (55), using either THY broth alone or 5448 WT plus H2O as a control.
Invasive model of GAS infection.
Transgenic humanized plasminogen mice heterozygous for the human plasminogen transgene (AlbPLG1+/−) were infected with a dose of 2 × 108 to 4 × 108 CFU of 5448, 5448ΔpmtA, or 5448ΔpmtA::pmtA. Mice (n = 10) were subcutaneously infected with freshly prepared GAS strains in 100 μl of 1× PBS, and virulence was assessed as previously described (56). All animal experiments were conducted according to the Guidelines for the Care and Use of Laboratory Animals (National Health and Medical Research Council, Australia) and were approved by the University of Queensland Animal Ethics Committee.
Data analysis.
All data were analyzed by using GraphPad Prism 7. Analyses of ICP-MS and gene expression data were performed by using either 1-way analysis of variance (ANOVA) (for WT-only and single-condition experiments) or 2-way ANOVA (for comparison of strains under mixed conditions). Murine survival curves were analyzed by using the Mantel-Cox log rank test. Growth curve data represent means ± standard deviations of results from 3 independent biological replicates.
Supplementary Material
ACKNOWLEDGMENTS
A.G.T. is a recipient of the Australian Postgraduate Award. C.Y.O. is a recipient of a Garnett Passe & Rodney Williams Memorial Foundation research fellowship, and M.J.W. is an NHMRC Principal Research Fellow. This work was supported by the National Health and Medical Research Council (Australia).
We thank Matt Sweet for critically reading the manuscript.
A.G.T., C.Y.O., K.Y.D., A.G.M., and M.J.W. designed experiments. A.G.T. performed experimentation. M.R.D. performed phylogenetic analyses. N.P.W. provided reagents and critiqued experimental design. A.G.T., C.Y.O., K.Y.D., M.R.D., A.G.M., and M.J.W. wrote the manuscript, and all authors edited the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00140-17.
REFERENCES
- 1.Brenot A, King KY, Janowiak B, Griffith O, Caparon MG. 2004. Contribution of glutathione peroxidase to the virulence of Streptococcus pyogenes. Infect Immun 72:408–413. doi: 10.1128/IAI.72.1.408-413.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.King KY, Horenstein JA, Caparon MG. 2000. Aerotolerance and peroxide resistance in peroxidase and PerR mutants of Streptococcus pyogenes. J Bacteriol 182:5290–5299. doi: 10.1128/JB.182.19.5290-5299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JW, Helmann JD. 2006. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 4.Makthal N, Rastegari S, Sanson M, Ma Z, Olsen RJ, Helmann JD, Musser JM, Kumaraswami M. 2013. Crystal structure of peroxide stress regulator from Streptococcus pyogenes provides functional insights into the mechanism of oxidative stress sensing. J Biol Chem 288:18311–18324. doi: 10.1074/jbc.M113.456590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenot A, King KY, Caparon MG. 2005. The PerR regulon in peroxide resistance and virulence of Streptococcus pyogenes. Mol Microbiol 55:221–234. doi: 10.1111/j.1365-2958.2004.04370.x. [DOI] [PubMed] [Google Scholar]
- 6.Grifantini R, Toukoki C, Colaprico A, Gryllos I. 2011. Peroxide stimulon and role of PerR in group A Streptococcus. J Bacteriol 193:6539–6551. doi: 10.1128/JB.05924-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerlach D, Reichardt W, Vettermann S. 1998. Extracellular superoxide dismutase from Streptococcus pyogenes type 12 strain is manganese-dependent. FEMS Microbiol Lett 160:217–224. doi: 10.1111/j.1574-6968.1998.tb12914.x. [DOI] [PubMed] [Google Scholar]
- 8.Toukoki C, Gryllos I. 2013. PolA1, a putative DNA polymerase I, is coexpressed with PerR and contributes to peroxide stress defenses of group A Streptococcus. J Bacteriol 195:717–725. doi: 10.1128/JB.01847-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reference deleted.
- 10.Anjem A, Imlay JA. 2012. Mononuclear iron enzymes are primary targets of hydrogen peroxide stress. J Biol Chem 287:15544–15556. doi: 10.1074/jbc.M111.330365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imlay JA. 2003. Pathways of oxidative damage. Annu Rev Microbiol 57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 12.Cornelis P, Wei Q, Andrews SC, Vinckx T. 2011. Iron homeostasis and management of oxidative stress response in bacteria. Metallomics 3:540–549. doi: 10.1039/c1mt00022e. [DOI] [PubMed] [Google Scholar]
- 13.Zhang TF, Ding Y, Li TT, Wan Y, Li W, Chen HC, Zhou R. 2012. A Fur-like protein PerR regulates two oxidative stress response related operons dpr and metQIN in Streptococcus suis. BMC Microbiol 12:85. doi: 10.1186/1471-2180-12-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsou CC, Chiang-Ni C, Lin YS, Chuang WJ, Lin MT, Liu CC, Wu JJ. 2008. An iron-binding protein, Dpr, decreases hydrogen peroxide stress and protects Streptococcus pyogenes against multiple stresses. Infect Immun 76:4038–4045. doi: 10.1128/IAI.00477-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson MD, Kehl-Fie TE, Klein R, Kelly J, Burnham C, Mann B, Rosch JW. 2015. Role of copper efflux in pneumococcal pathogenesis and resistance to macrophage-mediated immune clearance. Infect Immun 83:1684–1694. doi: 10.1128/IAI.03015-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young CA, Gordon LD, Fang Z, Holder RC, Reid SD. 2015. Copper tolerance and the characterization of a copper-responsive operon, copYAZ, in an M1T1 clinical strain of Streptococcus pyogenes. J Bacteriol 197:2580–2592. doi: 10.1128/JB.00127-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu Y, Tsui HC, Bruce KE, Sham LT, Higgins KA, Lisher JP, Kazmierczak KM, Maroney MJ, Dann CE III, Winkler ME, Giedroc DP. 2013. A new structural paradigm in copper resistance in Streptococcus pneumoniae. Nat Chem Biol 9:177–183. doi: 10.1038/nchembio.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guan G, Pinochet-Barros A, Gaballa A, Patel SJ, Arguello JM, Helmann JD. 2015. PfeT, a P1B4-type ATPase, effluxes ferrous iron and protects Bacillus subtilis against iron intoxication. Mol Microbiol 98:787–803. doi: 10.1111/mmi.13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pi H, Patel SJ, Arguello JM, Helmann JD. 2016. The Listeria monocytogenes Fur-regulated virulence protein FrvA is an Fe(II) efflux P-type ATPase. Mol Microbiol 100:1066–1079. doi: 10.1111/mmi.13368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carapetis JR, Steer AC, Mulholland EK, Weber M. 2005. The global burden of group A streptococcal diseases. Lancet Infect Dis 5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- 21.Walker MJ, Barnett TC, McArthur JD, Cole JN, Gillen CM, Henningham A, Sriprakash KS, Sanderson-Smith ML, Nizet V. 2014. Disease manifestations and pathogenic mechanisms of group A Streptococcus. Clin Microbiol Rev 27:264–301. doi: 10.1128/CMR.00101-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cole JN, Barnett TC, Nizet V, Walker MJ. 2011. Molecular insight into invasive group A streptococcal disease. Nat Rev Microbiol 9:724–736. doi: 10.1038/nrmicro2648. [DOI] [PubMed] [Google Scholar]
- 23.Henningham A, Dohrmann S, Nizet V, Cole JN. 2015. Mechanisms of group A Streptococcus resistance to reactive oxygen species. FEMS Microbiol Rev 39:488–508. doi: 10.1093/femsre/fuu009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. 2005. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis 192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- 25.Brenot A, Weston BF, Caparon MG. 2007. A PerR-regulated metal transporter (PmtA) is an interface between oxidative stress and metal homeostasis in Streptococcus pyogenes. Mol Microbiol 63:1185–1196. doi: 10.1111/j.1365-2958.2006.05577.x. [DOI] [PubMed] [Google Scholar]
- 26.Smith AT, Smith KP, Rosenzweig AC. 2014. Diversity of the metal-transporting P1B-type ATPases. J Biol Inorg Chem 19:947–960. doi: 10.1007/s00775-014-1129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 29.Argüello JM. 2003. Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J Membr Biol 195:93–108. doi: 10.1007/s00232-003-2048-2. [DOI] [PubMed] [Google Scholar]
- 30.Argüello JM, González-Guerrero M, Raimunda D. 2011. Bacterial transition metal P1B-ATPases: transport mechanism and roles in virulence. Biochemistry 50:9940–9949. doi: 10.1021/bi201418k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chatellier S, Ihendyane N, Kansal RG, Khambaty F, Basma H, Norrby-Teglund A, Low DE, McGeer A, Kotb M. 2000. Genetic relatedness and superantigen expression in group A Streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect Immun 68:3523–3534. doi: 10.1128/IAI.68.6.3523-3534.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yeowell HN, White JR. 1982. Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob Agents Chemother 22:961–968. doi: 10.1128/AAC.22.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gryllos I, Grifantini R, Colaprico A, Cary ME, Hakansson A, Carey DW, Suarez-Chavez M, Kalish LA, Mitchell PD, White GL, Wessels MR. 2008. PerR confers phagocytic killing resistance and allows pharyngeal colonization by group A Streptococcus. PLoS Pathog 4:e1000145. doi: 10.1371/journal.ppat.1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ricci S, Janulczyk R, Bjorck L. 2002. The regulator PerR is involved in oxidative stress response and iron homeostasis and is necessary for full virulence of Streptococcus pyogenes. Infect Immun 70:4968–4976. doi: 10.1128/IAI.70.9.4968-4976.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skaar EP. 2010. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog 6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montanez GE, Neely MN, Eichenbaum Z. 2005. The streptococcal iron uptake (Siu) transporter is required for iron uptake and virulence in a zebrafish infection model. Microbiology 151:3749–3757. doi: 10.1099/mic.0.28075-0. [DOI] [PubMed] [Google Scholar]
- 37.Francis RT Jr, Booth JW, Becker RR. 1985. Uptake of iron from hemoglobin and the haptoglobin-hemoglobin complex by hemolytic bacteria. Int J Biochem 17:767–773. doi: 10.1016/0020-711X(85)90262-9. [DOI] [PubMed] [Google Scholar]
- 38.Eichenbaum Z, Muller E, Morse SA, Scott JR. 1996. Acquisition of iron from host proteins by the group A Streptococcus. Infect Immun 64:5428–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nygaard TK, Blouin GC, Liu M, Fukumura M, Olson JS, Fabian M, Dooley DM, Lei B. 2006. The mechanism of direct heme transfer from the streptococcal cell surface protein Shp to HtsA of the HtsABC transporter. J Biol Chem 281:20761–20771. doi: 10.1074/jbc.M601832200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanks TS, Liu M, McClure MJ, Lei B. 2005. ABC transporter FtsABCD of Streptococcus pyogenes mediates uptake of ferric ferrichrome. BMC Microbiol 5:62. doi: 10.1186/1471-2180-5-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith JL. 2004. The physiological role of ferritin-like compounds in bacteria. Crit Rev Microbiol 30:173–185. doi: 10.1080/10408410490435151. [DOI] [PubMed] [Google Scholar]
- 42.Frawley ER, Fang FC. 2014. The ins and outs of bacterial iron metabolism. Mol Microbiol 93:609–616. doi: 10.1111/mmi.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Munkelt D, Grass G, Nies DH. 2004. The chromosomally encoded cation diffusion facilitator proteins DmeF and FieF from Wautersia metallidurans CH34 are transporters of broad metal specificity. J Bacteriol 186:8036–8043. doi: 10.1128/JB.186.23.8036-8043.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sobota JM, Imlay JA. 2011. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proc Natl Acad Sci U S A 108:5402–5407. doi: 10.1073/pnas.1100410108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frawley ER, Crouch ML, Bingham-Ramos LK, Robbins HF, Wang W, Wright GD, Fang FC. 2013. Iron and citrate export by a major facilitator superfamily pump regulates metabolism and stress resistance in Salmonella Typhimurium. Proc Natl Acad Sci U S A 110:12054–12059. doi: 10.1073/pnas.1218274110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsou CC, Chiang-Ni C, Lin YS, Chuang WJ, Lin MT, Liu CC, Wu JJ. 2010. Oxidative stress and metal ions regulate a ferritin-like gene, dpr, in Streptococcus pyogenes. Int J Med Microbiol 300:259–264. doi: 10.1016/j.ijmm.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 47.Babior BM. 2000. Phagocytes and oxidative stress. Am J Med 109:33–44. doi: 10.1016/S0002-9343(00)00481-2. [DOI] [PubMed] [Google Scholar]
- 48.Ong CLY, Potter AJ, Trappetti C, Walker MJ, Jennings MP, Paton JC, McEwan AG. 2013. Interplay between manganese and iron in pneumococcal pathogenesis: role of the orphan response regulator RitR. Infect Immun 81:421–429. doi: 10.1128/IAI.00805-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosch JW, Gao G, Ridout G, Wang YD, Tuomanen EI. 2009. Role of the manganese efflux system MntE for signalling and pathogenesis in Streptococcus pneumoniae. Mol Microbiol 72:12–25. doi: 10.1111/j.1365-2958.2009.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turner AG, Ong CL, Gillen CM, Davies MR, West NP, McEwan AG, Walker MJ. 2015. Manganese homeostasis in group A Streptococcus is critical for resistance to oxidative stress and virulence. mBio 6:e00278–. doi: 10.1128/mBio.00278-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Venturini C, Ong CY, Gillen CM, Ben-Zakour NL, Maamary PG, Nizet V, Beatson SA, Walker MJ. 2013. Acquisition of the Sda1-encoding bacteriophage does not enhance virulence of the serotype M1 Streptococcus pyogenes strain SF370. Infect Immun 81:2062–2069. doi: 10.1128/IAI.00192-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 55.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔ method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, McArthur JD, Dinkla K, Aziz RK, Kansal RG, Simpson AJ, Buchanan JT, Chhatwal GS, Kotb M, Nizet V. 2007. DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med 13:981–985. doi: 10.1038/nm1612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.