Abstract
Granulocyte–macrophage colony-stimulating factor (GM-CSF) is a major regulator of monocyte to macrophage differentiation. In both humans and mice, the main phenotype of decreased GM-CSF function is pulmonary proteinosis due to aberrant function of alveolar macrophages. Recently, this cytokine has been shown to up-regulate a cyclic nucleotide phosphodiesterase, PDE1B. Two PDE1B variants with unique N-terminal sequences, PDE1B1 and PDE1B2, have been identified. Here, we report that the previously uncharacterized PDE1B2 is selectively increased by GM-CSF by stimulation of transcription at a previously unknown transcriptional start site. Analysis of the exon and intron organization of the PDE1B gene reveals that PDE1B2 has a different N-terminal sequence because of a separate first exon that is located 11.5 kb downstream from the PDE1B1 first exon. By using 5′-RACE, alignment of EST sequences, and a luciferase-reporter system, we provide evidence that PDE1B2 has a separate transcriptional start site from PDE1B1 that can be activated by monocyte differentiation. Furthermore, IL-4 treatment in the presence of GM-CSF, which shifts the differentiation from a macrophage to a dendritic cell phenotype, suppresses the up-regulation of PDE1B2. Induction of PDE1B2 is also found in T cells upon activation by PHA. Therefore, PDE1B2 may have a regulatory role in multiple immune cell types. Last, characterization of the catalytic properties of recombinant PDE1B2 shows that it prefers cGMP over cAMP as a substrate and, thus, is likely to regulate cGMP in macrophages. Also, PDE1B2 has a nearly 3-fold lower EC50 for activation by calmodulin than PDE1B1.
Keywords: cAMP, cGMP, phosphodiesterase
Monocytes are circulating peripheral blood cells that can be differentiated by cytokines into macrophages of different phenotypes as well as into dendritic cells. The resulting phenotype of differentiated monocytes is determined by the cytokine or combination of cytokines and other local stimuli to which the cell is exposed. One of the principal promoters of monocyte differentiation is the cytokine granulocyte–macrophage colony-stimulating factor (GM-CSF) (1, 2). The phenotype of monocytes differentiated in culture with GM-CSF alone resembles alveolar macrophages (1, 3). Moreover, GM-CSF deficiency in both mice (4–6) and humans (7, 8) is manifest by pulmonary alveolar proteinosis that is linked to alveolar macrophage dysfunction. Alternatively, monocytes treated with GM-CSF in the presence of IL-4 differentiate into dendritic cells (9, 10) instead of macrophages. Dendritic cells are professional antigen-presenting cells that have some of the same functional capabilities as alveolar macrophages but are especially adept at activating naive T cells. Hence, the combination of cytokines and other local stimuli can have selective effects on expression of genes that, in turn, determine the resulting phenotype of the differentiated monocyte.
Cyclic nucleotide phosphodiesterases (PDEs) regulate the amplitude and duration of cAMP and cGMP signals by controlling their rates of degradation. The expression patterns of PDEs change with monocyte differentiation (11–13) and the resulting PDE-expression profiles vary with the cytokine used for differentiation (14). So far, 11 families of PDEs have been identified that have different tissue and cellular expression, enzymatic characteristics, specificities for cAMP or cGMP, cellular localization, and regulation by allosteric inhibitors and activators, as well as different profiles of pharmacological inhibition (15–17). Most PDE families contain several different isoforms that are products of separate genes. Also, many of these genes encode several variants that are the products of different mRNAs formed by use of alternative start sites or alternative splicing.
Previous characterization of the changes in PDE expression with GM-CSF treatment of monocytes has shown that PDE1B expression is increased significantly by differentiation (14). However, the identity of the PDE1B variant that was induced was not determined. This issue is significant because different PDE variants can have disparate properties. For example, variation in the first 18–34 aa of PDE1A determines the affinity of the isozyme for Ca2+/calmodulin (18–20), whereas the first 17–25 aa of PDE2A determine whether the enzyme will be localized to the cytosol (PDE2A1) (21) or membrane [PDE2A2 (22) and PDE2A3 (23)]. Thus, expression of variants of different PDE isoforms can provide a mechanism for imparting highly specific properties to individual PDEs.
Two PDE1B variants (PDE1B1 and PDE1B2) that possess different N-terminal sequences have been identified. PDE1B1 has a unique 38-aa N-terminal sequence, whereas PDE1B2 has 18 aa that are unique to its N terminus. PDE1B1 was identified >10 years ago (24, 25), and it has been characterized extensively (26, 27). However, only one article concerning PDE1B2 has been published (28). This article showed strong expression of PDE1B2 mRNA in the spinal cord and weak expression in thyroid, thymus, uterus, small intestine, putamen, and caudate nucleus (28). The tissue expression of PDE1B2 protein has not yet been demonstrated, nor have the kinetic properties of PDE1B2 been reported.
Here, we report that the PDE1B2 mRNA, protein, and activity are up-regulated upon monocyte differentiation. Furthermore, we elucidate a mechanism for the differential up-regulation of PDE1B2 expression and also present kinetic constants for enzymatic activity. Our results suggest that PDE1B2 is important for regulation of macrophages by GM-CSF and possibly also for the function of other immune cells. This work is significant because it characterizes a previously undefined enzyme and suggests that targeting of PDE1B2 activity may have clinical usefulness for treatment of inflammatory diseases.
Experimental Procedures
Cell Culture and Differentiation. Monocytes and T cells were purified from buffy coats obtained from human donors by the Red Cross (Portland, OR) as described (14). Monocytes were purified by using density-gradient centrifugation with Ficoll-Paque Plus (Amersham Pharmacia Biotech), followed by positive selection with magnetic CD 14 microbeads (Miltenyi Biotec, Auburn, CA). T cells were purified by negative selection using a pan T cell isolation kit (Miltenyi Biotec). Monocytes were differentiated in RPMI medium 1640 supplemented with penicillin, streptomycin, and 10% FBS and buffered with 10 mM Hepes. Monocytes were differentiated at a concentration of 5 × 105 cells per ml with 100 ng/ml GM-CSF in the absence or presence of various concentrations of IL-4. T cells were activated in RPMI medium 1640 supplemented with penicillin, streptomycin, and 10% FBS at a concentration of 1 × 106 cells per ml with 10 μg/ml PHA. The HL-60 cell line was purchased from the American Type Culture Collection and maintained in the same media as primary monocytes. For HL-60 cell differentiation, cells were incubated at 7 × 105 cells per ml with 50 nM phorbol-12-myristate-13-acetate (PMA). HEK 293T cells were maintained in MEM with 10% FBS.
Cytosol Preparation and PDE-Activity Assay. Cells were harvested, washed once in cold PBS, and suspended in lysis buffer containing 50 mM Tris, pH 7.5/100 μM EDTA/1 mM benzamidine/0.1 mM sodium orthovanadate/1 mM DTT and supplemented with a protease inhibitor mixture (Sigma). Cells were ruptured by sonication, and PDE activities were measured in the cell homogenates as described (14, 29). Calmodulin-stimulated PDE1 activity was determined by subtracting cGMP PDE activity in the presence of 1 mM EGTA from activity in the presence of 1 mM CaCl2 and 4 μg/ml calmodulin.
Preparation of PDE1B1 and PDE1B2 Expression Constructs and Transfection in HEK Cells. Sequences encoding the PDE1B1 and PDE1B2 proteins were inserted into the pcDNA3.1/V5-His-TOPO vector from Invitrogen. However, a stop codon sequence was added to prevent expression of the V5-His tag. The sequences for PDE1B1 and PDE1B2 were amplified from a human brain cDNA library (Clontech) and HL-60 cell cDNA, respectively. The plasmids were sequenced and verified to match earlier sequences reported for PDE1B1 and PDE1B2 (GenBank accession nos. NM_000924 and AJ401609, respectively). Recombinant PDE1B proteins were expressed in HEK 293T cells. Cells were transfected with the PDE1B expression plasmids, and after 48 h, cells were harvested, and lysate was prepared as described above. Cytosol was obtained by centrifugation at 16,000 × g for 10 min. Both PDE1B1 and PDE1B2 were found in the cytosol, and cytosolic fractions were used for determination of kinetic constants.
Immunoprecipitation and RT-PCR. PDE1B was immunoprecipitated by using the ACC-1 mAb, as described in ref. 29 and explained in detail in Supporting Materials and Methods, which is published as supporting information on the PNAS web site. Total RNA was isolated from 5 × 106 cells, and RT-PCR was performed as described (14).
PDE1B2 5′-RACE. The 5′-UTR sequence of PDE1B2 mRNA was determined by 5′-RACE using the First-Choice RLM-RACE kit from Ambion (Austin, TX). Total RNA from HL-60 cells differentiated with PMA was isolated, and cDNA with an adaptor ligated to the 5′ ends was generated by using the kit. Two PCRs were performed to amplify a product containing the 5′ end of the PDE1B2 mRNA. The second PCR was run on an agarose gel, and a single band was detected. The PCR product was ligated into the pcDNA4 HisMax topo vector (Invitrogen) for sequencing.
Preparation of Luciferase Reporter Constructs and Measurement of PDE1B Promoter Activity. Genomic DNA was isolated from HL-60 cells by using the DNeasy kit from Qiagen (Valencia, CA). The region 2 kb directly upstream of the translational start sites for the PDE1B1 and PDE1B2 first exons was amplified and inserted into the pCR 2.1 topo TA vector (Invitrogen). The inserts were then cut from the topo TA vector, purified, and ligated into the pGL3 Basic vector (Promega) upstream of the firefly luciferase reporter gene. The luciferase reporter plasmids were transfected into HL-60 cells by electroporation with 10 μg of reporter plasmid by using the ECM 630 Electro Cell Manipulator (BTX, Holliston, MA) at 280 V with a capacitance of 1,050 μF. Luciferase activity was measured by using luficerase reagent (Promega), and activities were normalized by protein concentration.
Results
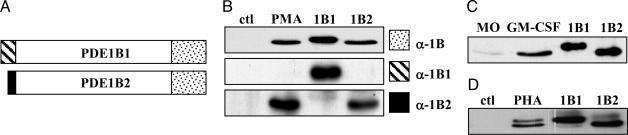
Induction of PDE1B2 mRNA and Protein. In previous work, we found that an undefined form of PDE1B is up-regulated upon monocyte differentiation with GM-CSF (14). To investigate this finding further, we sought to determine which PDE1B variant was expressed in these cells. This identification is important because PDE variants can have significantly different properties. To address this question, we used Abs specific to different regions of the PDE1B1 and PDE1B2 proteins (Fig. 1A). First, we wanted to determine whether PDE1B could also be up-regulated in promyelocytic leukemia HL-60 cells, which can be differentiated to a monocyte/macrophage phenotype by treatment with PMA (30, 31). PMA-induced differentiation of HL-60 cells triggered an increase in PDE1 activity that was maximal by day 3 (see Fig. 5, which is published as supporting information on the PNAS web site). When PDE1 was immunoprecipitated from PMA-differentiated cells, a single band with an electrophoretic mobility similar to recombinant PDE1B2 (59 kDa) and not recombinant PDE1B1 (63 kDa) was detected by using an anti-PDE1B Ab (Fig. 1B). To confirm that only PDE1B2 protein was up-regulated, Abs against the unique N termini of PDE1B1 and PDE1B2 were also used for Western blotting (Fig. 1B). Induction of PDE1B2 in various cell lines by artificial stimuli does not necessarily relate to what happens in primary monocytes exposed to physiological cytokines; therefore, we tested primary monocytes treated with GM-CSF. The data shown in Fig. 1C indicate that PDE1B2 protein is selectively increased when primary human monocytes are differentiated to macrophages with GM-CSF.
Fig. 1.
PDE1B2 protein is up-regulated with monocyte differentiation. (A) The unique N-terminal sequences of PDE1B1 and PDE1B2 are shown as the hatched (PDE1B1) or black (PDE1B2) areas. (B) ACC-1 was used to immunoprecipitate PDE1 from HL-60 cells (ctl), HL-60 cells treated for 3 days with 50 nM PMA (PMA), rPDE1B1 (1B1), or rPDE1B2 (1B2). The immunoprecipitates were separated by SDS/PAGE and Western blotted by using an Ab recognizing a portion of the common C-terminal portion of PDE1B (the epitope is indicated by the dotted region in A) or Abs recognizing either the unique N-terminal sequence of PDE1B1 or PDE1B2 shown in A. (C) The Ab recognizing the C-terminal portion of PDE1B was used to identify the PDE1B variant immunoprecipitated from monocytes (MO) and GM-CSF-treated monocytes (GM-CSF). (D) The same α-PDE1B C-terminal Ab was used also to detect the different PDE1B variants in primary T cells (ctl) and T cells treated with PHA. Western blots are representative of three separate experiments.
PDE1 activity has been reported to be induced in T cells upon activation with PHA (32, 33). Furthermore, PDE1B1 mRNA has been detected in human T cells (33, 34), and a PDE1B protein was found in bovine mononuclear cells after PHA activation (32). However, at the time of the publication of these reports, PDE1B2 had not been identified, and consequently, these researchers (unaware of the existence of PDE1B2) did not test for its presence. We now find that both PDE1B1 as well as PDE1B2 protein (Fig. 1D) are expressed in human T cells activated with PHA. PDE1B1 has been implicated to be important for T cell survival because treatment of lymphoblastoid cell lines with an antisense oligode-oxynucleotide specific to PDE1B1 induced apoptosis (34). Also, it appears that PDE1B2 may also play a role in T cell biology.
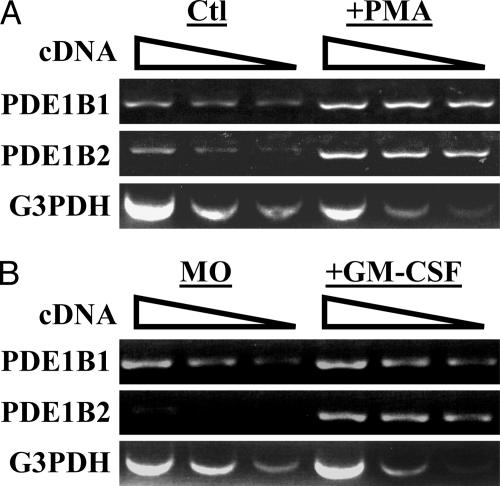
It was of interest to determine whether the changes in PDE1B2 protein that occur with monocyte differentiation were caused by increased levels of PDE1B2 mRNA. Therefore, RT-PCR analysis was applied to total RNA isolated from HL-60 cells or HL-60 cells differentiated for 3 days with PMA. Different concentrations of cDNA template were used to perform a semiquantitative measurement of PDE1B mRNA levels. G3PDH was used as a control to ensure that nearly equal amounts of cDNA template were used from the different treatment conditions. The RT-PCR products were quantitated by using densitometry (see Fig. 6, which is published as supporting information on the PNAS web site). PMA treatment of HL-60 cells was found to cause a 5-fold induction of PDE1B2 mRNA (Fig. 2A). Somewhat surprisingly, PDE1B1 mRNA also was detected in control cells, and it was increased 2-fold by PMA treatment. This finding is strange because PDE1 activity in untreated cells is very low and PDE1B1 protein is not detected in PMA-treated cells. This result suggests that the PDE1B1 message can be regulated posttranscriptionally or that it has very poor translational efficiency. More important, GM-CSF treatment of monocytes was also found to substantially up-regulate PDE1B2 mRNA (Fig. 2B). These results imply that the mechanism for increased PDE1B2 protein expression may be increased transcription.
Fig. 2.
PDE1B2 mRNA is up-regulated by PMA treatment of HL-60 cells or GM-CSF treatment of primary monocytes. (A) RT-PCR was used to detect PDE1B1, PDE1B2, and G3PDH mRNA from HL-60 cells (Ctl) or cells treated for 3 days with PMA (+PMA). (B) RT-PCR was used to detect mRNA in monocytes and monocytes differentiated with GM-CSF. Decreasing amounts of cDNA template were used in the reactions to ensure a linear and semiquantitative comparison of PDE1B2 mRNA levels. cDNA amount decreased, as shown from left to right by the sloped line. G3PDH was used as a control to ensure that similar amounts of cDNA were used. Data are representative of three separate experiments.
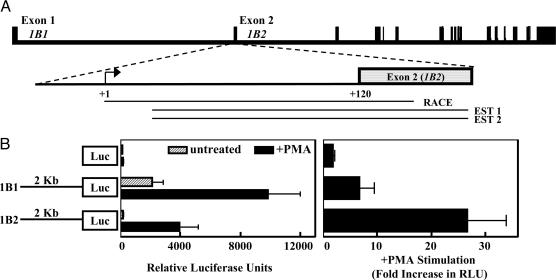
PDE1B1 and PDE1B2 Have Separate Transcriptional Start Sites and Promoters. The origin of the unique N-terminal sequence of PDE1B2 has not been elucidated. To determine the source of the different N-terminal sequences of the two PDE1B variants, we aligned their mRNA sequences with the PDE1B genomic sequence. The resulting alignment reveals that the different N termini of PDE1B1 and PDE1B2 are products of separate first exons. In Fig. 3A, the organization of the PDE1B gene is diagrammed. The unique PDE1B1 and PDE1B2 exons are indicated, and the remaining exons are common between the two variants.
Fig. 3.
PDE1B1 and PDE1B2 have separate transcriptional start sites. (A) The exon and intron organization of the human PDE1B gene is shown. The PDE1B1 and PDE1B2 variants have unique N-terminal sequences that correspond to separate exons and the remaining exons are common. The product obtained from a 5′-RACE reaction (RACE) was found to match the PDE1B2 first exon and genomic sequence directly upstream and is shown aligned to the corresponding genomic region. Two EST sequences (EST 1 and EST 2) that were identified by blast search match the PDE1B2 first exon and are shown aligned as well. (B) We inserted 2 kb of genomic sequence exactly 5′ to the PDE1B1 and PDE1B2 translational start sites in front of a luciferase reporter gene in the pGL3 basic vector, and constructs were electroporated into HL-60 cells. After 24 h, cells from each electroporation were divided into two groups and received no treatment or were treated with 100 nM PMA. At 2 days after treatment, cells were harvested and luciferase activity was measured. Values are means ± SEM and were determined for four separate experiments.
Because the reported PDE1B2 sequence (28) did not contain a 5′-UTR sequence, we sought to identify this sequence to determine whether the transcriptional start site for PDE1B2 is distinct from that of PDE1B1. We used 5′-RACE to identify the PDE1B2 5′-UTR. The sequence obtained from PMA-differentiated HL-60 cells is shown in Fig. 3A (RACE), and the nucleotide sequence alignment is shown in Fig. 7, which is published as supporting information on the PNAS web site. The RACE product obtained for PDE1B2 contained a 5′-UTR sequence that matched exactly the genomic sequence found directly upstream of the PDE1B2 translational start site. Also, two ESTs (GenBank accession nos. CA397265 and BG390932) were found to also contain sequences that matched the genomic sequence directly upstream of the PDE1B2 translational start site. We did not obtain any ESTs or RACE products for PDE1B2 that contained sequence that matched any regions 5′ to the PDE1B1 first exon. Hence, it was probable that the transcriptional start site for PDE1B2 is 3′ of the PDE1B1 first exon and that the two variants have different promoters.
To demonstrate that PDE1B1 and PDE1B2 have separate promoters, we amplified 2 kb of genomic sequence upstream of the PDE1B1 and PDE1B2 first exons and inserted the sequences in front of a firefly luciferase reporter gene. The constructs were transfected into HL-60 cells to determine whether the sequences exhibit promoter activity. We found that the 2-kb-upstream sequence of PDE1B1 was constitutively active (Fig. 3B) in HL-60 cells and was activated by PMA treatment. This result is consistent with our earlier finding of PDE1B1 mRNA expression in HL-60 cells and primary monocytes. Because the PDE1B1 promoter is active and PDE1B1 mRNA is present despite the absence of PDE1B1 protein, it can be inferred that the PDE1B1 mRNA is regulated posttranscriptionally. The regulation of the PDE1B1 mRNA must be cell-type-specific because the induction of PDE1B1 protein can be observed with PHA treatment of T cells. In contrast to PDE1B1, the PDE1B2 promoter sequence was minimally active before PMA treatment and activated >20-fold after treatment with PMA. Again, this result is consistent with our earlier discovery of increased PDE1B2 mRNA and protein levels after differentiation of HL-60 cells and primary monocytes. These results suggest that the sequences directly upstream of the unique PDE1B1 and PDE1B2 first exons both are functional promoters but are regulated differentially in monocytes. Although the 2-kb-upstream sequence tested for PDE1B2 was found to serve as a functional promoter, it may also be regulated in vivo by additional elements upstream in the PDE1B1 promoter. These findings confirm that transcriptional activation is a major mechanism for PDE1B2 up-regulation.
To explore the mechanism of the transcriptional activation of the PDE1B2 promoter by GM-CSF further, we searched a portion of the identified promoter sequence for potential binding sites of transcription factors that are likely to be relevant to GM-CSF-induced differentiation. The 2,000 bp directly upstream of the PDE1B2 first exon were searched by using Yutaka Akiyama's (Kyoto University, Kyoto) tfsearch program with the TRANSFAC (35) database and the signal scan program with the TFD (36) and TRANSFAC databases. By using these tools, multiple sites were identified, including STAT, AP-1, and PU.1 binding sequences that have been shown to be activated by GM-CSF (37–40). Only sites located in the 800 bp directly upstream of the PDE1B2 translational start site are shown in Fig. 8B, which is published as supporting information on the PNAS web site. The PU.1 site may be especially important because PU.1 has been shown to be crucial for GM-CSF regulated macrophage development (38, 41). For comparison, we also analyzed the PDE1B1 first exon promoter region for the same sites. In Fig. 8A, we show that, in the 2,000 bp upstream of the PDE1B1 translational start site, some of the same sites found in the PDE1B2 promoter were present but mostly in lower abundance (for example, C/EBP and c-Ets). These differences could explain the observation that the PDE1B1 promoter is regulated differently than the PDE1B2 promoter.
PDE1B1 and PDE1B2 Have Similar Kinetic Properties. Because PDE1B2 has been discovered only recently and has not been characterized previously, we analyzed its catalytic properties to determine whether its unique N-terminal sequence imparts enzymatic characteristics dissimilar from PDE1B1. PDE1B1 has been characterized previously and was found to be activated by calmodulin at low concentrations and to prefer cGMP as substrate over cAMP at low substrate levels but to have a Vmax that is approximately equal for both cGMP and cAMP (27, 42). We found recombinant PDE1B2 has kinetic constants similar to PDE1B1 (Table 1). The Km for cGMP is nearly equal for PDE1B1 and PDE1B2, whereas PDE1B2 has a slightly lower Km for cAMP (9.96 μM) than PDE1B1 (23.77 μM). The ratio of cGMP Vmax to cAMP Vmax is nearly equal for the two enzymes.
Table 1. PDE1B1 and PDE1B2 have similar kinetic properties.
| Kinetic property | PDE1B1 | PDE1B2 |
|---|---|---|
| Calmodulin EC50 | 3.55 ± 0.36 nM | 1.21 ± 0.29 nM |
| Km cGMP | 5.90 ± 1.27 μM | 4.26 ± 1.32 μM |
| Km cAMP | 23.8 ± 3.87 μM | 9.96 ± 0.34 μM |
| Vmax cGMP/cAMP | 1.61 ± 0.29 | 2.37 ± 0.39 |
PDE1B1 and PDE1B2 were expressed in HEK 293T cells, and the kinetic constants for calmodulin-stimulated hydrolysis of cAMP and cGMP were determined. Values are means ± SEM, and they were determined for three separate transfections of HEK cells.
The EC50 for activation by calmodulin was found to be ≈3-fold lower for PDE1B2 (1.21 nM) than for PDE1B1 (3.55 nM). Fidock et al. (28) found that the N-terminal sequences of PDE1A1 and PDE1B2 align well, whereas the N termini of PDE1A2 and PDE1B1 are homologous. As with the two PDE1B forms described here, sequence variation at the N terminus of PDE1A did not affect Vmax or Km for cAMP and cGMP (43). Because PDE1A1 had been found to have a 10-fold higher affinity for calmodulin than PDE1A2 (18–20), the authors logically predicted that PDE1B2 might have a higher affinity for calmodulin than PDE1B1. Confirming this idea, we find that PDE1B2 has a higher affinity for calmodulin than PDE1B1, although the difference between the PDE1B isoforms is only ≈3-fold.
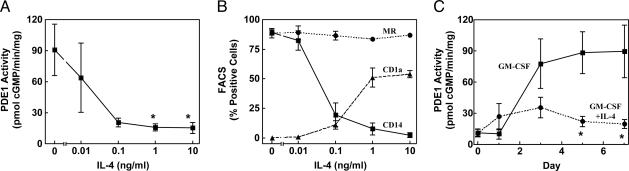
The Effect of IL-4 on PDE1 Induction and Dendritic Cell Formation. When monocyte differentiation is initiated by GM-CSF in the presence of IL-4, the resulting cells have a dendritic cell instead of a macrophage phenotype (9, 10). Fluorescence-activated cell sorting was used to determine expression of cell-surface proteins as markers for differentiation. We found that IL-4 suppresses PDE1B induction (Fig. 4A) under conditions in which it shifts the differentiation phenotype to a dendritic cell, as determined by expression of the dendritic cell marker CD1a and the macrophage marker CD14 (Fig. 4B). This finding is consistent with an earlier study reporting low PDE1 expression levels in dendritic cells (12). A dose-response for IL-4 revealed that very low concentrations of IL-4 are required to inhibit PDE1 induction (Fig. 4A). We found that an IL-4 concentration as low as 0.1 ng/ml is sufficient for a nearly maximal effect.
Fig. 4.
IL-4 suppresses PDE1 induction. Monocytes were differentiated with GM-CSF (100 ng/ml) in the absence or presence of increasing concentrations of IL-4. Cells were harvested after 7 days for measurement of PDE1 activity (A) or for analysis of expression of the cell surface markers mannose receptor (MR), CD14, and CD1a by using fluorescence-activated cell sorting (B). *, P < 0.05, compared with treatment with GM-CSF alone. Values are means ± SEM of four separate donor preparations. (C) Monocytes were differentiated with 100 ng/ml GM-CSF in the absence (squares and solid line) or presence (circles and dotted line) of 25 ng/ml IL-4, and cells were harvested at different time points for measurement of PDE1 activity. *, P < 0.01, compared with treatment with GM-CSF alone.
To gain further insight into the function of PDE1B in monocyte differentiation, we followed the time course for up-regulation of PDE1 activity in response to GM-CSF treatment in the absence or presence of IL-4. The effect of IL-4 on PDE1 takes time to occur. On day 1, the PDE1 activity seems to be increased with IL-4 treatment, and the suppressive effect of IL-4 is not seen until later time points (Fig. 4C). It has been shown that IL-4 can alter the differentiation of osteoclasts as well (44, 45), and prolonged IL-4 treatment was necessary for the effect (44).
Discussion
In this article, we provide evidence that the mRNA, protein and activity of the previously uncharacterized PDE1B2 are up-regulated with monocyte-to-macrophage differentiation. We have explored the mechanism of the induction, and our data strongly suggest that transcription of PDE1B1 and PDE1B2 mRNAs are regulated in vivo at different start sites by separate promoters. Two important issues to consider based on our findings are the rationale for PDE1B1 and PDE1B2 being regulated independently from separate promoters and the functional role of PDE1B2 in macrophage biology.
The use of separate promoters has several implications. First, it confers a different N-terminal coding sequence to the mRNA. In theory, this sequence difference could alter catalytic or regulatory properties. There is some precedent that the N-terminal sequences of PDEs make important regulatory interactions because the UCR1 and UCR2 domains of PDE4D3 provide sites for dimerization (46) and phosphorylation (47, 48). However, no regulatory protein binding or phosphorylation sites have been demonstrated to be exclusive to the unique N-terminal sequences of either of the PDE1B variants.
As expected from the identical sequences in the catalytic domains, the basic substrate kinetic properties of recombinant PDE1B2 are very similar to those of PDE1B1. The lower Km for cGMP compared with cAMP would suggest that this enzyme is induced, at least in part, to regulate the amplitude and duration of cGMP in the cell. PDE1B2 has a 3-fold higher sensitivity for activation by calmodulin than PDE1B1. Although changes in N-terminal sequence of the PDE1A variants caused large disparities in calmodulin sensitivity (18–20), PDE1C variants with different N-terminal sequences had only small differences in calmodulin affinity (49). The PDE1B variants seem to be most similar to the PDE1C family in this regard. It is difficult to know whether a 3-fold difference in calmodulin sensitivity between the PDE1B variants is physiologically significant, but given the importance of tight regulation of calcium in these cells and also the importance of cAMP and cGMP in regulating macrophage function, it is tempting to speculate that the differences are likely to be significant. Perhaps the advantage of a higher calcium sensitivity is enough of a reason for nature to have favored the development of a separate promoter region as a mechanism of providing an altered higher-affinity calmodulin-binding site.
A second possibility for the existence of the unique PDE1B2 promoter, in addition to its different N-terminal coding sequence, is that it may also have evolved to allow differential regulation of transcription in a tissue-specific manner. The two separate promoters would allow the same gene (PDE1B) to be regulated by different signals and transcription factors in different cell types. In this case, we assume that the PDE1B2 promoter allows GM-CSF to induce a calcium sensitive, cGMP-selective PDE in macrophages but not in other tissues. Several binding sites for transcription factors that have been established to be activated by GM-CSF, such as AP-1 and STATs, were identified in the PDE1B2 promoter sequence activated by monocyte differentiation. Also present in the promoter sequence are two canonical binding sites for PU.1, which has been shown to be an important factor for macrophage development. The transcriptional up-regulation of PDE1B2 is likely to occur by means of GM-CSF-stimulated binding of some combination of factors to these sites. The 2,000-bp PDE1B1 promoter region contains some of the same binding sites, but in lower abundance, and it lacks a PU.1 site. This discrepancy could explain the differential regulation of the two promoters in monocytes and macrophages. Also, it was discovered that PDE1B1 is regulated posttranscriptionally as well as at the transcriptional level.
Our data do not exactly define how PDE1B2 contributes to macrophage biology, but it is likely to be important for the regulation of the monocyte-to-macrophage differentiation process, the function(s) of the differentiated macrophage, or both. In this regard, the observations demonstrating that IL-4 shifts the differentiation of monocytes into dendritic cells (9, 10) and the data in this article showing that IL-4 suppresses PDE1 induction (Fig. 4), imply that PDE1 up-regulation may be, at least in part, responsible for defining the characteristics of the specific macrophage phenotype differentiated from monocytes. It is possible that GM-CSF up-regulates PDE1 to remove a cAMP or cGMP block on the conversion of the monocyte to a more differentiated phenotype because it is known that both cAMP and cGMP block this process for dendritic cells (50). During differentiation, IL-4 could prevent the development of the alveolar macrophage phenotype partly by suppressing PDE1B2 induction. In addition to the ability of IL-4 to suppress the induction of PDE1B2, it also presumably promotes the expression of other genes that allow the cell to develop instead into a dendritic cell phenotype. Because PDE1B2 is also up-regulated with T cell activation, its function may be more generalized and involved in the regulation of other immune cell types as well.
PDE1B2 is also likely to be important for functions of the mature macrophage. For example, it is well known that agents such as NO that increase cGMP will alter neutrophil chemotaxis and migration (51–53). This effect is well established in neutrophils and has been suggested to be the case for macrophages (54, 55). cGMP in macrophages has been shown to modulate inflammatory processes, such as inducible NO synthase induction (56, 57) and TNF-α release (58–61). Also, it is known that intracellular calcium transients are important modulators of macrophage (62–64) function. Therefore, the marked up-regulation of PDE1B2 is likely to be critical for the regulation of these processes in differentiated macrophages. The kinetic and regulatory properties of PDE1B2 shown in this article strongly suggest that this enzyme should provide a dominant mechanism by which calcium transients can control the duration and amplitude of the cGMP signal in these cells. This connection could allow crosstalk between the cGMP and calcium pathways in coordinating inflammation. Thus, it would seem prudent to develop selective inhibitors of PDE1B and assess their effects on inflammation. Low concentrations of such inhibitors may selectively target macrophages and, perhaps, other activated immune cells such as T cells. If a PDE1B2 selective inhibitor could be developed, it might have fewer side effects than currently available antiinflammatory agents.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants DK21723 (to J.A.B.) and ARO49042 (to E.H.W.). A.T.B. is a Trainee under University of Washington Cardiovascular Pathology National Institutes of Health Training Grant HL-07312.
Author contributions: A.T.B., E.H.W., and J.A.B. designed research; A.T.B., C.L.O., and E.H.W. performed research; A.T.B., E.H.W., and J.A.B. analyzed data; and A.T.B. and J.A.B. wrote the paper.
Abbreviations: PDE, phosphodiesterase; GM-CSF, granulocyte–macrophage colony-stimulating factor; PMA, phorbol-12-myristate-13-acetate.
References
- 1.Akagawa, K. S. (2002) Int. J. Hematol. 76, 27-34. [DOI] [PubMed] [Google Scholar]
- 2.Rasko, J. E. J. (1997) in Colony-Stimulating Factors: Molecular and Cellular Biology, eds. Garland, J. M., Queensberry, P. J. & Hilton, D. J. (Dekker, New York), pp. 163-202.
- 3.Andreesen, R., Brugger, W., Scheibenbogen, C., Kreutz, M., Leser, H. G., Rehm, A. & Lohr, G. W. (1990) J. Leukocyte Biol. 47, 490-497. [DOI] [PubMed] [Google Scholar]
- 4.Stanley, E., Lieschke, G. J., Grail, D., Metcalf, D., Hodgson, G., Gall, J. A., Maher, D. W., Cebon, J., Sinickas, V. & Dunn, A. R. (1994) Proc. Natl. Acad. Sci. USA 91, 5592-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishinakamura, R., Nakayama, N., Hirabayashi, Y., Inoue, T., Aud, D., McNeil, T., Azuma, S., Yoshida, S., Toyoda, Y., Arai, K., et al. (1995) Immunity 2, 211-222. [DOI] [PubMed] [Google Scholar]
- 6.Robb, L., Drinkwater, C. C., Metcalf, D., Li, R., Kontgen, F., Nicola, N. A. & Begley, C. G. (1995) Proc. Natl. Acad. Sci. USA 92, 9565-9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kitamura, T., Tanaka, N., Watanabe, J., Uchida, Kanegasaki, S., Yamada, Y. & Nakata, K. (1999) J. Exp. Med. 190, 875-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trapnell, B. C., Whitsett, J. A. & Nakata, K. (2003) N. Engl. J. Med. 349, 2527-2539. [DOI] [PubMed] [Google Scholar]
- 9.Sallusto, F. & Lanzavecchia, A. (1994) J. Exp. Med. 179, 1109-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thurner, B., Roder, C., Dieckmann, D., Heuer, M., Kruse, M., Glaser, A., Keikavoussi, P., Kampgen, E., Bender, A. & Schuler, G. (1999) J. Immunol. Methods 223, 1-15. [DOI] [PubMed] [Google Scholar]
- 11.Gantner, F., Kupferschmidt, R., Schudt, C., Wendel, A. & Hatzelmann, A. (1997) Br. J. Pharmacol. 121, 221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gantner, F., Schudt, C., Wendel, A. & Hatzelmann, A. (1999) Pulm. Pharmacol. Ther. 12, 377-386. [DOI] [PubMed] [Google Scholar]
- 13.Tenor, H., Hatzelmann, A., Kupferschmidt, R., Stanciu, L., Djukanovic, R., Schudt, C., Wendel, A., Church, M. K. & Shute, J. K. (1995) Clin. Exp. Allergy 25, 625-633. [DOI] [PubMed] [Google Scholar]
- 14.Bender, A. T., Ostenson, C. L., Giordano, D., Beavo, J. A. (2003) Cell. Signalling 16, 365-374. [DOI] [PubMed] [Google Scholar]
- 15.Francis, S. H., Turko, I. V. & Corbin, J. D. (2001) Prog. Nucleic Acid Res. Mol. Biol. 65, 1-52. [DOI] [PubMed] [Google Scholar]
- 16.Soderling, S. H. & Beavo, J. A. (2000) Curr. Opin. Cell Biol. 12, 174-179. [DOI] [PubMed] [Google Scholar]
- 17.Juilfs, D. M., Soderling, S., Burns, F. & Beavo, J. A. (1999) Rev. Physiol. Biochem. Pharmacol. 135, 67-104. [DOI] [PubMed] [Google Scholar]
- 18.Hansen, R. S. & Beavo, J. A. (1986) J. Biol. Chem. 261, 14636-14645. [PubMed] [Google Scholar]
- 19.Sharma, R. K. (1991) Biochemistry 30, 5963-5968. [DOI] [PubMed] [Google Scholar]
- 20.Sharma, R. K. & Kalra, J. (1994) Biochem. J. 299 (Pt 1), 97-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martins, T. J., Mumby, M. C. & Beavo, J. A. (1982) J. Biol. Chem. 257, 1973-1979. [PubMed] [Google Scholar]
- 22.Yang, Q., Paskind, M., Bolger, G., Thompson, W. J., Repaske, D. R., Cutler, L. S. & Epstein, P. M. (1994) Biochem. Biophys. Res. Commun. 205, 1850-1858. [DOI] [PubMed] [Google Scholar]
- 23.Rosman, G. J., Martins, T. J., Sonnenburg, W. K., Beavo, J. A., Ferguson, K. & Loughney, K. (1997) Gene 191, 89-95. [DOI] [PubMed] [Google Scholar]
- 24.Bentley, J. K., Kadlecek, A., Sherbert, C. H., Seger, D., Sonnenburg, W. K., Charbonneau, H., Novack, J. P. & Beavo, J. A. (1992) J. Biol. Chem. 267, 18676-18682. [PubMed] [Google Scholar]
- 25.Repaske, D. R., Swinnen, J. V., Jin, S. L., Van Wyk, J. J. & Conti, M. (1992) J. Biol. Chem. 267, 18683-18688. [PubMed] [Google Scholar]
- 26.Yu, J., Wolda, S. L., Frazier, A. L., Florio, V. A., Martins, T. J., Snyder, P. B., Harris, E. A., McCaw, K. N., Farrell, C. A., Steiner, B., et al. (1997) Cell. Signalling 9, 519-529. [DOI] [PubMed] [Google Scholar]
- 27.Sharma, R. K. & Wang, J. H. (1986) J. Biol. Chem. 261, 1322-1328. [PubMed] [Google Scholar]
- 28.Fidock, M., Miller, M. & Lanfear, J. (2002) Cell. Signalling 14, 53-60. [DOI] [PubMed] [Google Scholar]
- 29.Sonnenburg, W. K., Rybalkin, S. D., Bornfeldt, K. E., Kwak, K. S., Rybalkina, I. G. & Beavo, J. A. (1998) Methods 14, 3-19. [DOI] [PubMed] [Google Scholar]
- 30.Rovera, G., Santoli, D. & Damsky, C. (1979) Proc. Natl. Acad. Sci. USA 76, 2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harris, P. & Ralph, P. (1985) J. Leukocyte Biol. 37, 407-422. [DOI] [PubMed] [Google Scholar]
- 32.Hurwitz, R. L., Hirsch, K. M., Clark, D. J., Holcombe, V. N. & Hurwitz, M. Y. (1990) J. Biol. Chem. 265, 8901-8907. [PubMed] [Google Scholar]
- 33.Kanda, N. & Watanabe, S. (2001) Biochem. Pharmacol. 62, 495-507. [DOI] [PubMed] [Google Scholar]
- 34.Jiang, X., Li, J., Paskind, M. & Epstein, P. M. (1996) Proc. Natl. Acad. Sci. USA 93, 11236-11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinemeyer, T., Wingender, E., Reuter, I., Hermjakob, H., Kel, A. E., Kel, O. V., Ignatieva, E. V., Ananko, E. A., Podkolodnaya, O. A., Kolpakov, F. A., et al. (1998) Nucleic Acids Res. 26, 362-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh, D. (1991) Trends Biochem. Sci. 16, 445-457. [DOI] [PubMed] [Google Scholar]
- 37.de Groot, R. P., Coffer, P. J. & Koenderman, L. (1998) Cell. Signalling 10, 619-628. [DOI] [PubMed] [Google Scholar]
- 38.Shibata, Y., Berclaz, P. Y., Chroneos, Z. C., Yoshida, M., Whitsett, J. A. & Trapnell, B. C. (2001) Immunity 15, 557-567. [DOI] [PubMed] [Google Scholar]
- 39.Flaherty, D. M., Monick, M. M., Carter, A. B., Peterson, M. W. & Hunninghake, G. W. (2001) Am. J. Respir. Cell Mol. Biol. 25, 254-259. [DOI] [PubMed] [Google Scholar]
- 40.Barahmand-pour, F., Meinke, A., Kieslinger, M., Eilers, A. & Decker, T. (1996) Curr. Top. Microbiol. Immunol. 211, 121-128. [DOI] [PubMed] [Google Scholar]
- 41.Berclaz, P. Y., Zsengeller, Z., Shibata, Y., Otake, K., Strasbaugh, S., Whitsett, J. A. & Trapnell, B. C. (2002) J. Immunol. 169, 6332-6342. [DOI] [PubMed] [Google Scholar]
- 42.Zhao, A. Z., Yan, C., Sonnenburg, W. K. & Beavo, J. A. (1997) in Advances in Second Messanger Phosphoprotein Research, eds. Corbin, J. D. & Francis, S. H. (Lippincott–Raven, Philadelphia), Vol. 31, pp. 237-251. [PubMed] [Google Scholar]
- 43.Snyder, P. B., Florio, V. A., Ferguson, K. & Loughney, K. (1999) Cell. Signalling 11, 535-544. [DOI] [PubMed] [Google Scholar]
- 44.Wei, S., Wang, M. W., Teitelbaum, S. L. & Ross, F. P. (2002) J. Biol. Chem. 277, 6622-6630. [DOI] [PubMed] [Google Scholar]
- 45.Moreno, J. L., Kaczmarek, M., Keegan, A. D. & Tondravi, M. (2003) Blood 102, 1078-1086. [DOI] [PubMed] [Google Scholar]
- 46.Richter, W. & Conti, M. (2002) J. Biol. Chem. 277, 40212-40221. [DOI] [PubMed] [Google Scholar]
- 47.Sette, C. & Conti, M. (1996) J. Biol. Chem. 271, 16526-16534. [DOI] [PubMed] [Google Scholar]
- 48.MacKenzie, S. J., Baillie, G. S., McPhee, I., MacKenzie, C., Seamons, R., McSorley, T., Millen, J., Beard, M. B., van Heeke, G. & Houslay, M. D. (2002) Br. J. Pharmacol. 136, 421-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan, C., Zhao, A. Z., Bentley, J. K. & Beavo, J. A. (1996) J. Biol. Chem. 271, 25699-25706. [DOI] [PubMed] [Google Scholar]
- 50.Giordano, D., Magaletti, D. M., Clark, E. A. & Beavo, J. A. (2003) J. Immunol. 171, 6421-6430. [DOI] [PubMed] [Google Scholar]
- 51.Elferink, J. G. & VanUffelen, B. E. (1996) Gen. Pharmacol. 27, 387-393. [DOI] [PubMed] [Google Scholar]
- 52.Wanikiat, P., Woodward, D. F. & Armstrong, R. A. (1997) Br. J. Pharmacol. 122, 1135-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adachi, R., Matsui, S., Kinoshita, M., Nagaishi, K., Sasaki, H., Kasahara, T. & Suzuki, K. (2000) Int. J. Immunopharmacol. 22, 855-864. [DOI] [PubMed] [Google Scholar]
- 54.Pryzwansky, K. B., Kidao, S., Wyatt, T. A., Reed, W. & Lincoln, T. M. (1995) J. Leukocyte Biol. 57, 670-678. [DOI] [PubMed] [Google Scholar]
- 55.Ke, X., Terashima, M., Nariai, Y., Nakashima, Y., Nabika, T. & Tanigawa, Y. (2001) Biochim. Biophys. Acta 1539, 101-113. [DOI] [PubMed] [Google Scholar]
- 56.Kiemer, A. K. & Vollmar, A. M. (1997) Endocrinology 138, 4282-4290. [DOI] [PubMed] [Google Scholar]
- 57.Kiemer, A. K. & Vollmar, A. M. (1998) J. Biol. Chem. 273, 13444-13451. [DOI] [PubMed] [Google Scholar]
- 58.Tamion, F., Richard, V., Lyoumi, S., Hiron, M., Bonmarchand, G., Leroy, J., Daveau, M., Thuillez, C. & Lebreton, J. P. (1999) Cytokine 11, 326-333. [DOI] [PubMed] [Google Scholar]
- 59.Arias-Diaz, J., Vara, E., Garcia, C., Villa, N. & Balibrea, J. L. (1995) Arch. Surg. (Chicago) 130, 1287-1293. [DOI] [PubMed] [Google Scholar]
- 60.Gong, J. H., Renz, H., Sprenger, H., Nain, M. & Gemsa, D. (1990) Immunobiology 182, 44-55. [DOI] [PubMed] [Google Scholar]
- 61.Kiemer, A. K., Hartung, T. & Vollmar, A. M. (2000) J. Immunol. 165, 175-181. [DOI] [PubMed] [Google Scholar]
- 62.Hoyal, C. R., Giron-Calle, J. & Forman, H. J. (1998) J. Toxicol. Environ. Health 1, 117-134. [DOI] [PubMed] [Google Scholar]
- 63.Lowry, M. A., Goldberg, J. I. & Belosevic, M. (1999) Dev. Comp. Immunol. 23, 253-261. [DOI] [PubMed] [Google Scholar]
- 64.Cuschieri, J., Gourlay, D., Garcia, I., Jelacic, S. & Maier, R. V. (2002) J. Trauma 52, 434-442. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.