Abstract
The α-fetoprotein (AFP) and H19 genes are transcribed at high levels in the mammalian fetal liver but are rapidly repressed postnatally. This repression in the liver is controlled, at least in part, by the Afr1 gene. Afr1 was defined >25 years ago when BALB/cJ mice were found to have 5- to 20-fold higher adult serum AFP levels compared with all other mouse strains; subsequent studies showed that this elevation was due to higher Afp expression in the liver. H19, which has become a model for genomic imprinting, was identified initially in a screen for Afr1-regulated genes. The BALB/cJ allele (Afr1b) is recessive to the wild-type allele (Afr1a), consistent with the idea that Afr1 functions as a repressor. By high-resolution mapping, we identified a gene that maps to the Afr1 interval on chromosome 15 and encodes a putative zinc fingers and homeoboxes (ZHX) protein. In BALB/cJ mice, this gene contains a murine endogenous retrovirus within its first intron and produces predominantly an aberrant transcript that no longer encodes a functional protein. Liver-specific overexpression of a Zhx2 transgene restores wild-type H19 repression on a BALB/cJ background, confirming that this gene is responsible for hereditary persistence of Afp and H19 in the livers of BALB/cJ mice. Thus we have identified a genetically defined transcription factor that is involved in developmental gene silencing in mammals. We present a model to explain the liver-specific phenotype in BALB/cJ mice, even though Afr1 is a ubiquitously expressed gene.
Keywords: development, genetics, positional cloning
The α-fetoprotein (AFP) gene is transcribed abundantly in the fetal liver and is dramatically repressed at birth (reviewed in refs. 1 and 2). In the mid-1970s, AFP became the focus of considerable interest because serum AFP concentration showed such a dramatic decline after birth and because AFP was often reactivated in hepatocellular carcinomas (3). To explore this regulation, Ruoslahti and coworkers (4) tested whether adult serum AFP levels varied among different mouse strains. Of the 27 strains that were analyzed, 26 had low serum AFP levels. The one exception was BALB/cJ, which had ≈5- to 20-fold higher AFP levels than did the other mouse strains. This trait was specific to BALB/cJ; other BALB/c substrains had low adult serum AFP levels. This trait was transmitted by a single autosomal locus, which was originally called raf, for regulator of alpha-fetoprotein, but was later renamed Afr1 (for alpha-fetoprotein regulator 1). The Afr1 allele in BALB/cJ mice (Afr1b) is recessive to the Afr1a allele found in other mouse strains (4).
When the Afp cDNA was cloned, Northern analysis revealed that adult liver steady-state Afp mRNA levels were higher in BALB/cJ than in other mouse strains (5). This difference was not observed in the adult gut, where Afp is also expressed at low levels, or in the fetal liver, suggesting that Afr1 action is limited to the adult liver. This study also showed that Afr1 was unlinked to the Afp gene (5). Studies in adult chimeric mice generated by the aggregation of BALB/cJ and C57BL/6 embryos found that Afp mRNA levels were directly proportional to the percentage of BALB/cJ cells in the liver, indicating that Afr1 acted in a cell-autonomous manner (6).
A molecular genetic screen for additional Afr1 targets identified a single gene, named H19, which was unlinked to Afp and Afr1 (7). H19 was also expressed at low levels in the adult gut and muscle. H19 levels in these two adult tissues were not responsive to Afr1, supporting the idea that Afr1 action is limited to the adult liver. The H19 locus has become an important model in the study of genomic imprinting, but aspects of tissue-specific and developmental control of H19 have remained largely unexplored (8, 9).
Data described above identified some features of Afr1 but provide little insight into the mechanisms by which Afr1 regulates Afp and H19. Nuclear run-on analysis found no difference in the rate of transcription of Afp and H19 between Afr1a and Afr1b mice in adult liver, despite the difference in their steady-state mRNA levels as judged by Northern analysis (10). This observation suggested that Afr1 acts at the posttranscriptional level to affect mRNA processing and/or stability (10). In contrast to these results, we showed that the 250-bp Afp promoter could confer Afr1 regulation to a linked reporter gene (11, 12). These transgenic data argue that Afr1 acts at the level of transcription and does not require Afp coding sequences. These seemingly disparate results have led us to propose that Afr1 regulates gene expression in a manner that couples transcriptional to posttranscriptional events (11).
The murine Afr1 gene was originally mapped to chromosome 15, 2–3 centimorgans from c-myc (13). We confirmed this map position and generated a higher-resolution genetic map of the Afr1 locus (12). Here, we refined further the interval where Afr1 resides and searched mouse and human genomic databases for candidate Afr1 genes in this region. We identified a zinc fingers and homeoboxes (Zhx) gene, which is the mouse orthologue to human ZHX2, as the product of the Afr1 gene. Analysis of this gene in Afr1a and Afr1b mice indicates that reduced Zhx2 expression in BALB/cJ mice is due to a mouse endogenous retroviral (MERV) element inserted into intron 1. Transgene complementation by liver-specific Zhx2 overexpression in BALB/cJ mice results in complete H19 repression, confirming that this gene is responsible for the Afr1 phenotype.
Materials and Methods
Mice/Transgenic Mice. BALB/c, C3H/HeJ, and DBA/2 mice were purchased from Harlan (Indianapolis). BALB/cJ mice were purchased from The Jackson Laboratory. Animals were housed in the University of Kentucky Animal Facility. (DBA/2 × BALB/cJ)F1 males were backcrossed to BALB/cJ females, and the resulting F2 progeny were killed at 3–4 weeks of age. To generate transgenic mice, the entire ORF containing the mouse Zhx2 gene was PCR amplified and cloned into pGEM-T EASY (Promega), sequenced, and inserted into a transthyretin expression vector (14) to generate TTR-Afr1. The transgene was excised from plasmid sequences, purified, and introduced into F2 embryos derived from the matings of (C57BL/6 × C3H) parents. Injections were performed by staff of the University of Kentucky Transgenic Mouse Facility. Three independent founders were crossed to BALB/cJ; resulting transgenic offspring were backcrossed to BALB/cJ. DNA from tail biopsies from the resulting F2 offspring were analyzed by PCR for the presence of the TTR-Afr1 transgene and the endogenous Afr1 genotype (a/b or b/b). Four weeks after birth, animals were killed and liver RNA was prepared. The breeding protocol and all experimental procedures were approved by the University of Kentucky Institutional Animal Care and Use Committee, following guidelines established by the National Institutes of Health.
Genotyping. Genomic DNA was prepared from tail biopsies and was amplified by PCR using primers for polymorphic D15Mit markers as described (12). The products amplified by using D15Mit132 primers are 97 and 95 bp long from BALB/cJ and C3H, respectively; products amplified from the D15Mit121 locus are 158 and 143 bp long from BALB/cJ and C3H, respectively. PCR products were resolved by electrophoresis on polyacrylamide gels and visualized by ethidium bromide staining. Linkage analysis was determined by using map manager (Mouse Genome Informatics, The Jackson Laboratory).
PCR/RACE. Primers for amplification of genomic fragments were designed by using sequences available from the public databases. All PCR amplifications used Therm-os-star Mastermix (ABgene, Surrey, U.K.), Taq DNA polymerase (Qiagen, Valencia, CA), or Pfx Platinum polymerase (Invitrogen) according to the suppliers' recommendations; Pfx polymerase amplicons were A-tailed with Taq DNA polymerase in the presence of 0.2 mM dATP at 70°C for 30 min. All amplified products were cloned into pGEM-T or pGEM-T EASY (Promega). For RT-PCR, 1 μM oligo(dT) primers or 7.5 μM random hexamers in a 20-μl reaction volume were used to synthesize first-strand cDNA from 2 μg of total liver RNA by using the Omniscript RT kit (Qiagen). One-tenth of the reverse transcriptase reaction product was used as a template for PCR. For 5′-RACE, 10 μg of total liver RNA from 3-week-old C3H mice was used with the RLM-RACE kit (Ambion, Austin, TX) according to the supplier's instructions except the reaction used Omniscript reverse transcriptase (Qiagen) that was incubated at 37°C and primed with a gene-specific primer. The 3′-RACE-walk was performed by using BALB/cJ adult liver mRNA and following the protocol of Park et al. (15). The RACE products were cloned into pGEM-T or pGEM-T EASY and sequenced. The cDNA and genomic subclones were sequenced at the University of Kentucky Advanced Genome Technology Center. All primer sequences and cycling conditions are available on request.
Northern/Southern Analysis. RNA was prepared from various tissues by using the lithium chloride procedure (12), and poly(A)+ RNA was isolated on oligo(dT) columns. For Northern analysis, total RNA or poly(A)+ RNA was electrophoresed in 1% agarose/formaldehyde gels, blotted to nitrocellulose membranes (Optitran, Schleicher and Schuell), and processed by standard procedures. For Southern analysis, 10 μg of genomic DNA was digested overnight and separated by electrophoresis on 0.75% agarose gels. Gels were processed, blotted, and UV-crosslinked. The blots were hybridized with random prime-labeled probes by using Quickhyb (Stratagene). Probes were PCR amplified from genomic DNA and subcloned or used directly for labeling using the Decaprime kit (Ambion). Blots were exposed to PhosphorImager screens, and image analysis was performed by using imagequant software (Molecular Dynamics).
S1 Nuclease Protection Assay. S1 nuclease protection assays were done as described in ref. 16. Probes were generated by amplifying corresponding genomic sequences by PCR using one primer that had been end-labeled with [γ-32P]dATP.
Bioinformatics. A contiguous 20-bp stretch from the human ENST00000314393 sequence was used to blast search the mouse genome. A 60,000-bp sequence from mouse chromosome 15 centered on the blast hit region was used to predict ORFs by using grailexp (17). The longest predicted transcript was then used to blast search the Fantom2 database to search for full-length cDNA clones.
Databases and Tools. The following databases and tools were used in our work: http://genome.ucsc.edu, http://blast.wustl.edu, www.ensembl.org, http://ftp.genome.washington.edu/RM/RepeatMasker.html, http://grail.lsd.ornl.gov/grailexp/, http://fantom2.gsc.riken.go.jp, www.ncbi.nlm.nih.gov/BLAST/, and www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html.
Accession Numbers. GenBank accession numbers were as follows: RIKEN clone 6320424E06, AK031782; Zhx1, Z54200; Afr1 5′ EST clones, AI218918, BE989471, BF513650, CF137377, and W48088; and ZHX2, AB083653.
Results and Discussion
High-Resolution Map of Afr1 Region and Identification of Afr1 Candidate Gene. We previously reported a high-resolution map of the Afr1 locus on mouse chromosome 15 based on F2 backcross analysis of (C3H × BALB/cJ)F1 × BALB/cJ progeny (12). This interval was refined further by using a (DBA/2 × BALB/cJ)F1 × BALB/cJ backcross. In F2 animals, the Afr1 genotype was deduced from adult liver H19 mRNA levels; those with high H19 mRNA (BALB/cJ F2 mice 91, 93, and 219) were scored as Afr1b/b, whereas those with low H19 (F2 mice 89, 92, 95, 218, and 220) were scored as Afr1a/b (Fig. 1A). PCR was used to determine the genotype of microsatellite markers in this region, including D15Mit132 and D15Mit121 (Fig. 1B). Mice 89 and 219 show recombination between D15Mit132 and Afr1, and Afr1 and D15Mit121, respectively. These data, along with analysis of >700 F2 animals, indicated that Afr1 was located between D15Mit132 and D15Mit121. At that time, nine characterized or predicted genes resided within this 2.4-megabase region. Two of these were not analyzed further because their encoded proteins were unlikely to cause the Afr1 phenotype. Zhx1 was of interest because it encodes a protein that functioned as a transcriptional repressor in transient transfections (18). However, analysis of adult BALB/cJ and C3H liver RNA by RT-PCR and/or Northern blotting detected no difference in the level or size of transcripts of Zhx1 or the six other candidate genes (data not shown).
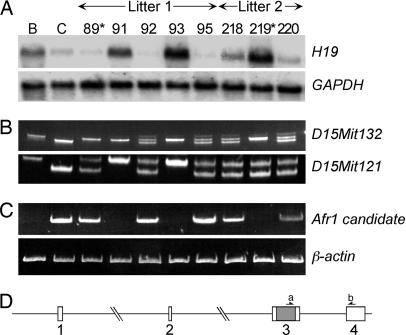
Fig. 1.
Persistent H19 expression in adult liver correlates with reduced mRNA of Afr1 candidate gene Zhx2. (A) Determination of Afr1 genotype by Northern analysis. Ten micrograms of total adult liver RNA was used in each lane from the following mice: BALB/cJ (B), C3H (C), and (BALB/cJ × C3H) × BALB/cJ F2 backcross animals (89, 91, 92, 93, and 95 from litter 1; 218, 219, and 220 from litter 2). Mice 89 and 219 (asterisks) show recombination between D15Mit132 and Afr1, and Afr1 and D15Mit121, respectively. Blots were hybridized with an H19 probe and then reprobed with GAPDH to control for RNA loading. (B) PCR genotyping using tail biopsied DNA from mice described above by using primers for D15Mit132 and D15Mit121 microsatellite markers (designated on right). (C) RT-PCR analysis of total liver RNA from the indicated mice by using primers for RIKEN cDNA clone ID 6320424E06 (labeled a and b in D). Reverse transcription samples were also amplified with primers for the β-actin gene as a control. (D) Structure of the gene encoding RIKEN clone 6320424E06. The gene contains four exons, including an unusually large internal third exon that codes for the entire predicted ZHX2 protein. The first, second, and third introns are 68.4, 58.1, and 14.9 kb in length, respectively.
The region on human chromosome 8 that is syntenic with the mouse Afr1 interval contains two human transcripts, ZHX1 and ENST00000314393 [which is identical to the recently characterized human ZHX2 gene (19)], which were both identified as mouse Zhx1 orthologues (20, 21). By blast analysis, human ZHX1 showed better alignment with mouse Zhx1. Although a mouse orthologue to human ZHX2 was not predicted, our comparison indicated that mouse Zhx2 did exist. Furthermore, a RIKEN cDNA (ID 6320424E06 or E06) corresponded to the candidate mouse Zhx2 gene (22) and, subsequent to our analysis, the mouse Zhx2 cDNA was identified (23). Aligning the E06 cDNA against the mouse genome indicated that it contains four exons, with the entire coding region present within the large third exon (Fig. 1D), a structure that is identical to the human ZHX2 gene (19). RT-PCR of adult liver RNA, using 5′ and 3′ primers from exons 3 and 4, respectively (designated as a and b in Fig. 1D), revealed a band of the expected size in C3H, DBA/2, and BALB/c strains, but not in BALB/cJ mice (Fig. 1C and data not shown). Sequencing confirmed the identity of this PCR product. Analysis of 11 F2 mice with recombination in the Afr1 region showed that the E06 transcript was present only in mice with low H19 mRNA levels and was absent in mice still expressing high H19 levels (Fig. 1 and data not shown). This perfect correlation between H19 repression and E06 expression indicates that Zhx2 likely corresponds to Afr1.
Analysis of Afr1 mRNA Expression in Adult Tissues. The Afr1 phenotype of incomplete Afp and H19 postnatal repression is observed in the adult liver, but not in fetal liver or other adult tissues where Afp and H19 are expressed (7, 11). Analysis of Afr1 in various tissues identified a single transcript of 4.4 kb, the size of the predicted Zhx2 transcript, although levels were substantially lower in liver and testes (Fig. 2, lanes 2–11). Afr1 mRNA is more abundant in the adult liver than in the fetal liver (Fig. 2, compare lanes 1 and 6), which is consistent with its activity as a postnatal repressor of Afp and H19 in this organ. However, because the strain-specific difference in Afp and H19 is liver-specific, we were somewhat surprised to find Afr1 expression to be lower in the liver than in other adult organs.
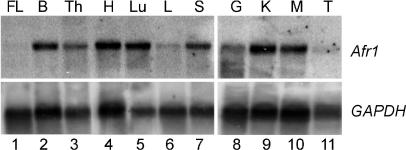
Fig. 2.
Afr1 is ubiquitously expressed in adult tissues and is more abundant in adult liver than in fetal liver. Northern analysis was performed by using 5 μg of poly(A)+ RNA from various BALB/c(Afr1a) tissues with a probe derived from Afr1 exon 3. FL, fetal day 18.5 liver; B, brain; Th, thymus; H, heart; Lu, lung; L, liver; S, spleen; G, gut; K, kidney; M, muscle; T, testes. The blots were stripped and rehybridized with a GAPDH probe to control for RNA loading.
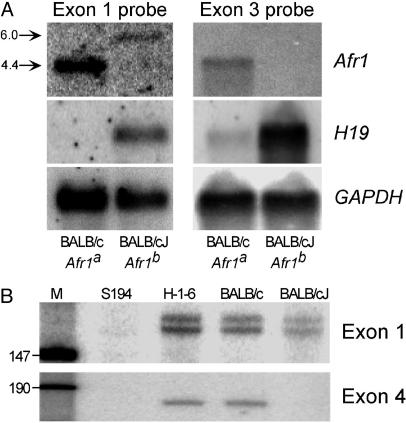
Comparison of Afr1 mRNA Between BALB/c and BALB/cJ Mice. To determine the basis for the dramatic reduction of Afr1 mRNA levels in BALB/cJ liver, we analyzed the Afr1 locus in BALB/c and BALB/cJ mice. The use of these related substrains, which separated ≈65 years ago (24) yet differ in their adult liver Afp and H19 mRNA levels, should minimize the extent of genetic polymorphisms in this comparison. By sequence analysis, the four exons and surrounding splice junctions from BALB/cJ (Afr1b) were found to be identical to corresponding BALB/c sequences (Afr1a) (data not shown). In addition, no sequence differences were found in ≈1,000 bp of DNA upstream of the putative transcription start site (data not shown). Thus, mutations in these regions could not account for the dramatic reduction of Afr1 transcripts in BALB/cJ. To extend our studies beyond RT-PCR analysis shown in Fig. 1C, we performed Northern and S1 nuclease protection analyses. When an exon 3-specific probe was used in a Northern blot, a transcript of 4.4 kb was detected in BALB/c but not in BALB/cJ adult liver (Fig. 3A Right). An identical 4.4-kb band was observed in the BALB/c mRNA when an exon 1 probe was used (Fig. 3A Left). In contrast, this probe hybridized to an ≈6-kb band in BALB/cJ mRNA. Therefore, this aberrant, polyadenylated, transcript contains exon 1 but not exon 3. In S1 nuclease protection analysis using a probe that spans the 3′ splice junction of exon 4, a band of the predicted size was protected with BALB/c but not BALB/cJ liver RNA (Fig. 3B). However, when a probe that spans the putative transcription start site was used, two bands of similar size and intensity were protected with RNA from both BALB/c and BALB/cJ liver. When additional RT-PCRs were performed by using primer combinations from each Afr1 exon, only transcripts from BALB/c liver RNA were readily detectable; normal Afr1 transcripts could be detected in BALB/cJ RNA only by extending the PCR cycles (data not shown). Together, these data suggest that transcription initiates correctly in BALB/cJ mice but that a majority of the RNA synthesized does not include exons 2–4, suggesting that the defect in the Afr1b allele is within the first intron.
Fig. 3.
RNA analysis reveals the presence of a correctly initiated but aberrantly sized Afr1 transcript in BALB/cJ mice. (A) Northern analysis was performed with 5 μg of poly(A)+ RNA from adult BALB/c (Afr1a) and BALB/cJ (Afr1b) liver. In Left, the blot was hybridized with a probe corresponding to the 3′ end of Afr1 exon 1. The arrows denote the 4.4-kb and ≈6.0-kb bands present in BALB/c and BALB/cJ samples, respectively; the blot was overexposed to show the ≈6.0-kb transcript. In Right, the blot was hybridized with a probe corresponding to the 3′ end of Afr1 exon 3. In both Left and Right, the blots were stripped and reprobed with an H19 cDNA probe to confirm the Afr1 phenotype, and stripped again and reprobed for GAPDH to control for RNA loading. (B) S1 nuclease protection assay using RNA derived from the S194 mouse plasmacytoma cell line (S194), Hepa1–6 mouse hepatoma cell line (H-1-6), adult BALB/c liver, and adult BALB/cJ liver. In Upper,5 μg of poly(A)+ was hybridized with a probe that spans the 5′ end of Afr1 exon 1 and putative promoter region. In Lower, 100 μg of total RNA was hybridized with a probe that spans the Afr1 exon 4 splice acceptor junction. M, DNA marker lane.
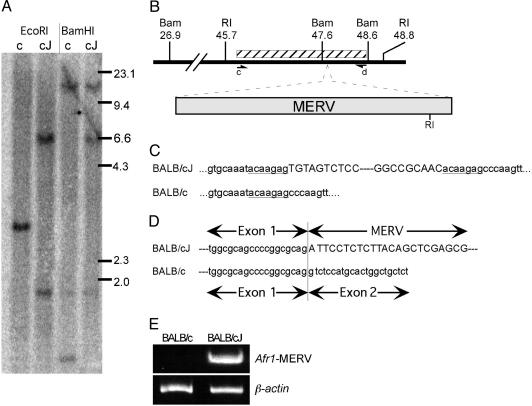
Identification of a MERV Element in Afr1 Intron 1 in BALB/cJ Mice. The first Afr1 intron is ≈68 kb in length. A series of PCRs were performed to amplify overlapping 2.5-kb DNA fragments across this large intron as a means to rapidly identify differences between BALB/c and BALB/cJ Afr1 alleles. The amplified products were identical in size and intensity between these substrains with a single exception; primers that spanned the region 47 kb downstream from exon 1 (primers c and d in Fig. 4B) failed to amplify a product from BALB/cJ DNA, suggesting that this region had been disrupted (data not shown). Southern analysis of BALB/cJ and BALB/c DNA by using a probe from this intronic region was used to further investigate this difference (Fig. 4A). The single 3.1-kb EcoRI fragment and two BamHI fragments (20.7 and 1.0 kb) in the BALB/c samples are identical to what is predicted from the published C57BL/6 mouse sequence. In contrast, two EcoRI fragments (1.9 and 6.7 kb) and two BamHI fragments (20.7 and 6.6 kb) were seen in BALB/cJ samples. These data indicate the presence of an ≈5.6-kb insert that contains a single EcoRI site in the Afr1b allele (depicted in Fig. 4B). This insertion and flanking region was PCR-amplified from BALB/cJ genomic DNA and sequenced. We found that a class II retrotransposon belonging to the MERV-K family had inserted into Afr1b intron 1 in the same transcriptional orientation as the Afr1 gene (Fig. 4C). Consistent with this finding, 7-bp target site duplications are present on either side of the insert; the identical sequence of these repeats and recent divergence of BALB/c substrains indicate that the integration giving rise to the Afr1b allele was a recent event. Sequence analysis also revealed that this MERV element contains two LTRs that are 317 bp in length, as well as an EcoRI site that is 655 bp from its 3′ end, as predicted by Southern analysis.
Fig. 4.
Insertion of a mouse endogenous retroviral element into intron 1 of the BALB/cJ Afr1b allele. (A) Southern analysis of DNA from BALB/c (c) and BALB/cJ (cJ) mice. Ten micrograms of DNA was digested with BamHI or EcoRI, resolved on 0.75% agarose gels, and transferred to nitrocellulose. Blots were probed with a 2.6-kb fragment of intron 1 (shown as a hatched box in B) centered 47 kb downstream of the first exon. (B) Schematic representation of the location of the MERV insertion in the Afr1b allele. The locations of the BamHI (Bam) and EcoRI (RI) sites are shown (in kb) downstream of the transcription start site. The arrowheads designated c and d represent oligonucleotides used for PCR amplification of DNA from this region. (C) Sequence of the site of MERV integration in the Afr1b allele and the corresponding Afr1a allele. BALB/cJ and BALB/c DNA were amplified by PCR using primers c and d in B, cloned, and sequenced. The endogenous sequence is shown in lowercase, with the target site duplications underlined. MERV sequences are in uppercase, with the dashed line representing the intervening MERV region. (D) Sequence of 3′-RACE-walk clone of the chimeric transcript arising from the Afr1b allele. The sequences from the Afr1 exon 1 are lowercase and MERV sequence is uppercase. Sequence of the exon 1–exon 2 junction of the Afr1a cDNA from BALB/c is shown below for comparison. (E) RT-PCR confirms the presence of a chimeric transcript in the Afr1b but not Afr1a allele. Random hexamer-primed reverse transcription was performed by using 0.5 μg of adult liver poly(A)+ RNA from BALB/c and BALB/cJ mice, followed by PCR using a forward primer derived from Afr1 exon 1 and a reverse primer derived from the MERV insertion element. As a control, PCR amplification was performed with the same RT-PCR templates by using β-actin gene primers.
Because S1 nuclease analysis had shown that the Afr1 allele in BALB/cJ was initiated properly, we used 3′-RACE-walk (15) to identify the sequences downstream of exon 1 in the Afr1b mRNA. Multiple clones indicated that transcripts from Afr1b exon 1 were spliced into the retrotransposon (Fig. 4D). The MERV sequence of these clones begins 246 bp into the 5′ LTR and the longest 3′-RACE-walk clone contained 346 contiguous nucleotides from the MERV insert; these transcripts presumably are the ≈6-kb mRNA seen by Northern blot analysis (Fig. 3A). To confirm the presence of these chimeric transcripts from the Afr1b allele, RT-PCR was performed by using forward and reverse primers from Afr1 exon 1 and the retrotransposon, respectively. This analysis confirmed the presence of the chimeric transcript in BALB/cJ but not BALB/c adult liver (Fig. 4E). While as many as 15% of mutations in mice may be due to retroviral insertions, not all affect endogenous gene expression in the straightforward way of creating a chimeric transcript as described here (20).
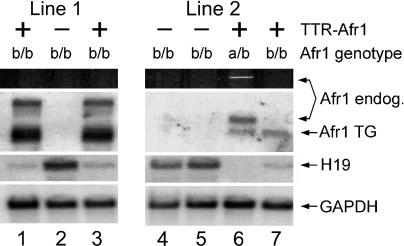
Liver-Specific Zhx2 Transgene Expression Restores H19 Repression in Afr1b/b Mice. To confirm that the mutation in Zhx2 is responsible for the hereditary persistence of H19 and Afp expression, we tested whether transgene complementation would restore normal postnatal repression of Afr1-target genes. A transthyretin expression vector (14) was fused to the entire Zhx2 coding region to generate TTR-Afr1, which should be expressed primarily in hepatocytes. Three founder animals were obtained, one with low and two with high levels of transgene expression. These three founders were then backcrossed to BALB/cJ. In all three lines, Zhx2 expression was able to restore H19 repression in the livers of adult animals containing two mutated copies of the endogenous Zhx2 locus (Fig. 5 and data not shown). This observation confirms that Zhx2 is the gene responsible for the Afr1 phenotype in BALB/cJ mice.
Fig. 5.
Liver-specific expression of a Zhx2 transgene restores H19 repression in an Afr1b/b background. Three founder mice containing the TTR-Afr1 transgene were backcrossed to BALB/cJ; liver RNA from F2 offspring was harvested 4 weeks after birth. Data are shown from F2 littermates from line 1 (high-copy transgene, lanes 1–3) and line 2 (low-copy transgene, lanes 4–7); similar results were seen with the third line. The presence or absence of the TTR-Afr1 transgene (+ or -, respectively) and the endogenous Afr1 genotype (a/b or b/b) were determined by PCR analysis of tail-biopsied DNA from each mouse. RT-PCR (using the same oligonucleotides as those shown in Fig. 1d) confirmed the expression of the endogenous Zhx2 gene in mouse 6. Northern analysis (using an exon 3 probe) confirmed the presence of Zhx2 mRNA from the endogenous locus (4.4-kb band) in mouse 6 and the TTR-Afr1 transgene (Afr1 TG) (3.2-kb band) in mice 1, 3, 6, and 7. H19 levels were determined by Northern analysis and shown to be reduced in the presence of the TTR-Afr1 transgene relative to nontransgenic littermates. GAPDH was used as a control for RNA loading.
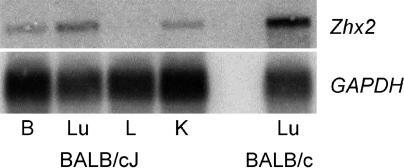
Model to Account for the Liver-Restricted Afr1 Phenotype. Because the Afr1 phenotype is restricted to the liver, even though H19 and Afp are expressed in other tissues, we had expected to find Afr1 expression to be highest in the liver. In contrast, we found that Afr1 is expressed at lower levels in the liver than in most other adult organs. It may be that Afr1-mediated repression of Afp and H19 requires a cofactor that is liver-restricted, or that other ZHX proteins may compensate for Afr1 in organs other than the liver. However, because the retrotransposon insertion in the Afr1b allele reduces, but does not eliminate, wild-type mRNA, we have considered whether the liver-restricted phenotype may be due to a dosage effect. In tissues where Afr1 is more highly expressed, there may be sufficient levels of wild-type protein remaining in BALB/cJ for proper repression. However, because normal Afr1 expression is much lower in the liver, the further reduction in BALB/cJ mice results in protein levels that are below the threshold required to repress target genes. To test this possibility, we used an exon 3-specific probe to analyze Afr1 mRNA levels in several tissues from adult BALB/cJ mice. Although Zhx2 transcripts were not detected in BALB/cJ liver by Northern analysis, transcripts were seen in the brain, lung, and kidney, albeit at levels that are lower than those found in BALB/c (Fig. 6). These data support the idea that it is the lower Afr1 expression in the adult liver that makes target genes in this organ particularly susceptible to the knockdown seen in BALB/cJ mice. One prediction of this model is that a complete Afr1 knockout would result in dysregulation of target genes in multiple organs.
Fig. 6.
Zhx2 transcripts are low but detectable in several adult BALB/cJ tissues but are not detectable in the liver. Northern analysis was performed by using 5 μg of poly(A)+ RNA from various BALB/cJ (Afr1b) tissues and BALB/c lung (Afr1a) with an Afr1 exon 3. The blots were stripped and rehybridized with a GAPDH probe to control for RNA loading. B, brain; Lu, lung; L, liver; K, kidney.
We know of no other genetically defined mammalian transcription factor that has been identified as being involved in developmental transcriptional repression in mammals. Data presented here identify Afr1 as the mouse Zhx2 gene (23), with the predicted mouse and human ZHX2 proteins being 87% identical. All ZHX proteins (ZHX1, ZHX2, and ZHX3) contain two zinc-finger motifs and five homeodomains; the mouse Zhx1 and Zhx2 genes are ≈300 kb apart on chromosome 15, whereas Zhx3 is on chromosome 2. All three proteins function as transcriptional repressors in transient cotransfection assays (18, 19, 23, 25). This activity, and the perinatal increase in Afr1 mRNA levels in the liver that correlates with Afp and H19 silencing, are consistent with Afr1 acting as a postnatal repressor of Afp and H19. Mouse Zhx1 was originally cloned by expression screening (26), and the human counterpart was subsequently identified in a screen for NF-Y-interacting proteins (27). More recently, human ZHX2 and ZHX3 were identified as ZHX1 interacting proteins (19, 25); ZHX2 was also shown to interact with nuclear factor Y (NF-Y) proteins. This raises the possibility that NF-Y may be involved in Zhx2-mediated repression of Afp and H19. However, to date there is no evidence that NF-Y regulates either Afp or H19, although NF-Y has been shown to regulate albumin, which is evolutionarily related to AFP but is not a target of Afr1.
Afr1 was originally identified in 1977 by persistent AFP serum levels in adult BALB/cJ mice (4) because of continued Afp expression in the liver (5). H19 was later identified by using a molecular screen to identify additional targets of Afr1 (7). To date, Afp and H19 are the only genes known to be regulated by Afr1. Other genes that are repressed in the adult liver or show changes in expression during the perinatal period would be potential Afr1 targets. On the basis of the ubiquitous expression of Afr1, it is likely that genes in tissues other than the liver may also be controlled by Afr1. Previous work has suggested that Afr1 acts at both the transcriptional and posttranscriptional levels (10–12), suggesting that ZHX2 and possibly other ZHX proteins may connect these different steps of gene expression.
AFP is a clinically important diagnostic marker in humans for several reasons. It is used as a marker for spina bifida because elevated maternal serum AFP levels are associated with neural tube defects in the developing fetus. Furthermore, although AFP and H19 are normally silent in the adult liver, these genes can be reactivated in hepatocellular carcinomas (HCCs) and certain other cancers (5, 7, 28, 29). As such, serum AFP screening is used as a test for HCC. Screening for AFP has identified several cases of hereditary persistence of AFP (HPAFP), an apparently benign condition in which adult AFP serum levels remain elevated. In several of these families, mutations have been found within the AFP promoter resulting in increased binding of hepatocyte nuclear factor 1 (30, 31). Data presented here raise the possibility that polymorphisms within the human ZHX2 gene may also lead to HPAFP. Furthermore, it will be of great interest to determine whether elevated AFP and H19 in HCC are caused by decreased levels or activity of ZHX2.
Acknowledgments
We thank Miriam Meisler for helpful comments on the manuscript, Gina Bingham for Southern analysis, Michelle Glenn for DNA sequencing, Michael Green for transgenic mouse production, and Debbie McKelvey and Abbie Lemaster for F2 mouse breeding and PCR genotyping. This work was supported by National Institutes of Health Grants DK51600 and DK59866.
Author contributions: S.P., M.L.P., and B.T.S. designed research; S.P. and R.W.C.D. performed research; S.P., R.W.C.D., M.L.P., and B.T.S. contributed new reagents/analytic tools; S.P., R.W.C.D., M.L.P., and B.T.S. analyzed data; and B.T.S. wrote the paper.
Abbreviations: AFP, α-fetoprotein; MERV, mouse endogenous retrovirus; Afr1, AFP regulator 1; ZHX, zinc fingers and homeoboxes.
References
- 1.Tilghman, S. M. (1985) Oxford Surv. Eukaryotic Genes 2, 160-206. [PubMed] [Google Scholar]
- 2.Spear, B. T. (1999) Semin. Cancer Biol. 9, 109-116. [DOI] [PubMed] [Google Scholar]
- 3.Abelev, G. I. (1971) Adv. Cancer Res. 14, 295-358. [DOI] [PubMed] [Google Scholar]
- 4.Olsson, M., Lindahl, G. & Ruoslahti, E. (1977) J. Exp. Med. 145, 819-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belayew, A. & Tilghman, S. M. (1982) Mol. Cell. Biol. 2, 1427-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogt, T. F., Solter, D. & Tilghman, S. M. (1987) Science 236, 301-303. [DOI] [PubMed] [Google Scholar]
- 7.Pachnis, V., Belayew, A. & Tilghman, S. M. (1984) Proc. Natl. Acad. Sci. USA 81, 5523-5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long, L. & Spear, B. T. (2004) Mol. Cell. Biol. 24, 9601-9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartolomei, M. S. & Tilghman, S. M. (1997) Annu. Rev. Genet. 31, 493-525. [DOI] [PubMed] [Google Scholar]
- 10.Vacher, J., Camper, S. A., Krumlauf, R., Compton, R. S. & Tilghman, S. M. (1992) Mol. Cell. Biol. 12, 856-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spear, B. T. (1994) Mol. Cell. Biol. 14, 6497-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peyton, D. K., Huang, M.-C., Giglia, M. A., Hughes, N. K. & Spear, B. T. (2000) Genomics 63, 173-180. [DOI] [PubMed] [Google Scholar]
- 13.Blankenhorn, E. P., Duncan, R., Huppi, C. & Potter, M. (1988) Genetics 119, 687-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu, H., Wade, M., Krall, L., Grisham, J., Xiong, Y. & Van Dyke, T. (1996) Genes Dev. 10, 245-260. [DOI] [PubMed] [Google Scholar]
- 15.Park, D. J., Pask, A. J., Renfree, M. B. & Graves, J. A. M. (2003) BioTechniques 34, 750-756. [DOI] [PubMed] [Google Scholar]
- 16.Seipelt, R. L., Spear, B. T., Snow, E. C. & Peterson, M. L. (1998) Mol. Cell. Biol. 18, 1042-1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu, Y. & Uberbacher, E. C. (1997) J. Comput. Biol. 4, 325-338. [DOI] [PubMed] [Google Scholar]
- 18.Yamada, K., Kawata, H., Matsuura, K., Shou, Z., Hirano, S., Mizutani, T., Yazawa, T., Yoshino, M., Sekiguchi, T., Kajitani, T. & Miyamoto, K. (2002) Biochem. Biophys. Res. Commun. 297, 368-374. [DOI] [PubMed] [Google Scholar]
- 19.Kawata, H., Yamada, K., Shou, Z., Mizutani, T., Yazawa, T., Yoshino, M., Sekiguchi, T., Kajitani, T. & Miyamoto, K. (2003) Biochem. J. 373, 747-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waterston, R. H., Lindblad-Toh, K., Birney, W., Rogers, J., Abril, J. F., Agarwal, P., Agarawala, R., Ainscough, R., Alexandersson, M., An, P., et al. (2002) Nature 420, 520-562. [DOI] [PubMed] [Google Scholar]
- 21.Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 22.Okazaki, Y., Furuno, M., Kasukawa, T., Adachi, J., Bono, H., Kondo, S. Nikaido, I., Osato, N., Saito, R. Suzuki, H., et al. (2002) Nature 420, 563-573.12466851 [Google Scholar]
- 23.Kawata, H., Yamada, D., Shou, Z., Mizutani, T. & Miyamoto, K. (2003) Gene 323, 133-140. [DOI] [PubMed] [Google Scholar]
- 24.Potter, M. (1985) Curr. Top. Microbiol. Immunol. 122, 1-5. [DOI] [PubMed] [Google Scholar]
- 25.Yamada, K., Kawata, H., Shou, Z., Hirano, S., Mizutani, T., Yazawa, T., Sekiguchi, T., Yoshino, M., Kajitani, T. & Miyamoto, K. (2003) Biochem. J. 373, 167-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barthelemy, I., Carramolino, L., Gutierrez, J., Barbero, J. L., Marquez, G. & Zaballos, A. (1996) Biochem. Biophys. Res. Commun. 224, 870-876. [DOI] [PubMed] [Google Scholar]
- 27.Yamada, K., Printz, R. L., Osawa, H. & Granner, D. K. (1999) Biochem. Biophys. Res. Commun. 261, 614-621. [DOI] [PubMed] [Google Scholar]
- 28.Abelev, G. I. & Eraiser, T. L. (1999) Semin. Cancer Biol. 9, 95-107. [DOI] [PubMed] [Google Scholar]
- 29.Ariel, I., Miao, H. Q., Ji, X. R., Schneider, T., Roll, D., de Groot, N., Hochberg, A. & Ayesh, S. (1998) Mol. Pathol. 51, 21-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blesa, J. R., Giner-Duran, R., Vidal, J., Lacalle, M. L., Catalan, I., Bixquert, M., Igual, L. & Hernandez-Yago, J. (2003) J. Hepatol. 38, 541-544. [DOI] [PubMed] [Google Scholar]
- 31.McVey, J. H., Michaelides, K., Hansen, L. P., Ferguson-Smith, M., Tilghman, S., Krumlauf, R. & Tuddenham, E. G. D. (1993) Hum. Mol. Genet. 2, 379-384. [DOI] [PubMed] [Google Scholar]