Abstract
ATP interacts with the two nucleotide-binding domains (NBDs) of CFTR to control gating. However, it is unclear whether gating involves ATP binding alone, or also involves hydrolysis at each NBD. We introduced phenylalanine residues into nonconserved positions of each NBD Walker A motif to sterically prevent ATP binding. These mutations blocked [α-32P]8-N3-ATP labeling of the mutated NBD and reduced channel opening rate without changing burst duration. Introducing cysteine residues at these positions and modifying with N-ethylmaleimide produced the same gating behavior. These results indicate that normal gating requires ATP binding to both NBDs, but ATP interaction with one NBD is sufficient to support some activity. We also studied mutations of the conserved Walker A lysine residues (K464A and K1250A) that prevent hydrolysis. By combining substitutions that block ATP binding with Walker A lysine mutations, we could differentiate the role of ATP binding vs. hydrolysis at each NBD. The K1250A mutation prolonged burst duration; however, blocking ATP binding prevented the long bursts. These data indicate that ATP binding to NBD2 allowed channel opening and that closing was delayed in the absence of hydrolysis. The corresponding NBD1 mutations showed relatively little effect of preventing ATP hydrolysis but a large inhibition of blocking ATP binding. These data suggest that ATP binding to NBD1 is required for normal activity but that hydrolysis has little effect. Our results suggest that both NBDs contribute to channel gating, NBD1 binds ATP but supports little hydrolysis, and ATP binding and hydrolysis at NBD2 are key for normal gating.
Keywords: cystic fibrosis, ion channel, Walker motif
The cystic fibrosis transmembrane conductance regulator (CFTR) is a Cl- channel in the ABC transporter family of membrane proteins (1, 2). It contains two membrane-spanning domains that form the channel pore, an R domain through which phosphorylation regulates channel activity, and two highly conserved nucleotide-binding domains (NBD1 and NBD2) that control channel opening and closing through their interaction with ATP.
Structural studies have suggested how nucleotides interact with ABC transporter NBDs (3–9). Each NBD contains a conserved phosphate-binding loop (P-loop or Walker A motif) that wraps around the ATP phosphates to form part of an ATP-binding pocket. Each NBD also contains a “signature” LSGGQ motif. Some structures suggest that the two NBDs form a dimer clamping two nucleotides at the NBD:NBD interface (7, 10). In the dimeric structures, ATP binds between the P-loop of one NBD and the signature motif of the other NBD. (In this article, we will refer to an NBD1 ATP-binding site as that which involves the P-loop of NBD1, and the NBD2 site as that which involves the P-loop of NBD2.) Our recent findings have provided functional evidence for NBD dimerization (11).
Earlier work showed that ATP binds both CFTR NBDs and that the protein hydrolyzes ATP (12–15). Clues to the role that the two individual NBDs play in gating have come from several studies. Mutation of conserved Walker A motif Lys residues (K464 in NBD1 or K1250 in NBD2) inhibited ATP hydrolysis (16). K464 mutations reduced open-state probability (Po) and opening rate in some but not all studies (17–19). K1250 mutations also altered channel function, reducing the Po by slowing the opening rate while prolonging the burst duration (16–18, 20, 21). These observations suggest that ATP hydrolysis by the NBDs is important for gating activity. However, it has been difficult to attribute the gating effects of the Lys mutations solely to disruption of enzymatic activity because these mutations may also reduce ATP binding. Although equilibrium nucleotide binding has not been measured directly, the Lys mutations were reported to prevent photolabeling with [α-32P]8-N3-ATP (14, 15).
Consequently, during normal gating it is still not well understood whether both NBDs must bind ATP, whether both NBDs hydrolyze ATP, or whether some combination of the two occurs. Moreover, there has been uncertainty in attributing various gating steps to specific nucleotide-bound states of NBD1 and NBD2. Therefore, the goal of this work was to distinguish the gating effects of disrupting ATP binding vs. inhibiting ATP hydrolysis. In an earlier study, we introduced a Cys residue into a nonconserved site of the Walker A motif of each NBD (residues A462 and S1248) (22). Subsequent N-ethylmaleimide (NEM) modification of either Walker A motif partially inhibited channel activity by decreasing the opening rate, suggesting that NEM modification reduced ATP binding. However, direct biochemical evidence was lacking, and we could not be sure that Cys was modified in all channels. Therefore, we asked whether modification of these residues with a bulky side chain would disrupt ATP binding. When we found that it did, we used this as a tool to learn more about how ATP interacts with the two NBDs to regulate CFTR gating.
Methods
Reagents. Catalytic subunit of cAMP-dependent protein kinase (PKA) was from Promega. [α-32P]8-N3-ATP was from Affinity Laboratory Technologies (Lexington, KY). Genenase I was from New England Biolabs. Antibodies 13-1 and 24-1 were from R & D Systems. MM13-4 and M3A7 antibodies were from Upstate Biotechnology (Lake Placid, NY).
Site-Directed Mutagenesis and Transfection. CFTR mutants were prepared as described in ref. 23. Constructs contained a His6-tag inserted between residues 2 and 3; the tag did not alter Po or expression. Cys insertion and the “WT control” (Fig. 5) also contained the C832A substitution, which prevents NEM modification of C832 (24). Constructs were expressed in HeLa cells by using the vaccinia virus/T7 expression system (17).
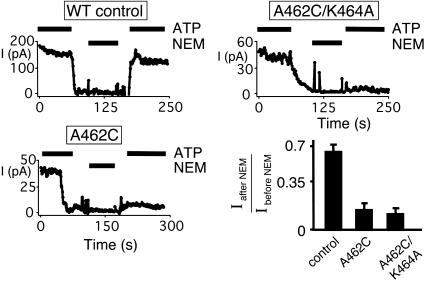
Fig. 5.
Effect of blocking ATP binding to NBD1 on current. Examples of data from excised membrane patches containing many CFTR channels are shown, along with the fraction of current recovered after NEM treatment. ATP (1 mM) and NEM (200 μM) were present during times indicated by bars (n = 3 for each channel type).
Electrophysiology. Methods for excised, inside-out patch-clamp recording and data analysis were as described in ref. 23. Burst duration for patches containing two to three channels were analyzed as described in ref. 17. NEM treatment of CFTR was performed by removing ATP and then adding between 100 μM and 1 mM NEM for 1 min. Transient removal of ATP caused some channel rundown (25); however, when measured this was generally <30%. An effect of this size would not affect our interpretations.
CFTR Biochemistry and Immunoblotting. His6 CFTR was prepared by using methods adapted from refs. 14 and 26. Cells were harvested in Tris buffer (40 mM Tris/5 mM MgCl, pH 7.4) containing 2 μg/ml leupeptin, aprotinin, and benzamidine and 0.1 mM phenylmethylsulfonyl fluoride. Cells were lysed mechanically, and membranes were isolated (200,000 × g) and resuspended in 100 μl of Tris buffer containing 25 μM [α-32P]8-N3-ATP. Membranes were incubated at 37°C for 5 min, followed by 30°C for 5 min, and then exposed to a UV lamp (680 μW/cm2) for 2 min at 30°C. Membranes were then solubilized in Tris buffer with 1% Triton X-100. Detergent-soluble CFTR was subjected either to immunoprecipitation (27) or metal affinity chromatography with Talon beads (Clontech). The product was washed with Tris buffer containing 1% Triton X-100 and three washes without Triton X-100. Beads were resuspended in 50 μl of Tris buffer and incubated for 4 h at room temperature with 1 μg of Genenase I as indicated.
PAGE analyses with 10% gels provided resolution of the relatively small ≈42-kDa NBD2 fragment but did not resolve band C from band B of CFTR (28). The 6% gels revealed that CFTR-A462F and -K464A produced little band C protein, whereas other variants produced predominantly the band C form. Quantitation was performed with digital autoradiography. Western blotting was performed as described in ref. 27. Quantitative radioactive labeling was normalized for amount of CFTR in each experiment by using chemiluminescence of an immunoblot signal. Linearity of chemiluminescence was verified by using serial dilutions of WT CFTR.
Results
Phenylalanine Substitutions in the Walker A Motif Block ATP Binding. Through empirical studies with various proteases, we found that the engineered subtilisin protease Genenase I (29) cut CFTR into separate fragments containing NBD1 and NBD2 (Fig. 1A). Western blot analysis showed an ≈95-kDa fragment that contained epitopes for both the N terminus (antibody MM13-4) and R domain (antibody 13.1); thus, it included all of NBD1. Genenase I also generated an ≈42-kDa fragment containing NBD2 sequence (antibody M3A7) and the C terminus (antibody 24-1). The apparent molecular mass of ≈42 kDa predicted that the fragment extended from the C terminus through transmembrane helices 11 and 12, and thus encompassed the entire NBD2. Although we did not attempt to identify specific Genenase I cleavage sites, CFTR contains a single consensus Genenase I site, Phe-1078–His-1079 that lies in the intracellular loop between transmembrane helices 10 and 11. This site would explain the ≈42-kDa NBD2 fragment. These data also suggest the presence of an additional cryptic Genenase I protease site. Val-855–His-856 is related to the Genenase I consensus sequence, and based on the molecular mass of the NBD1-containing fragment, it is a candidate site.
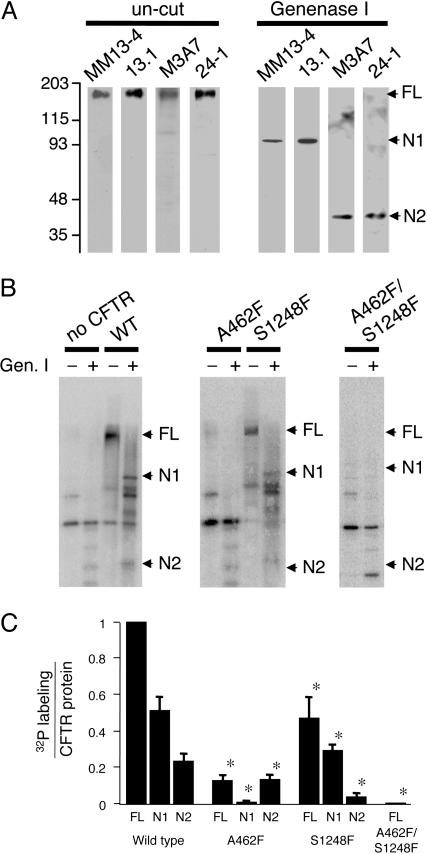
Fig. 1.
Labeling CFTR NBDs with [α-32P]8-N3-ATP. (A) CFTR was purified and blotted with antibody MM13-4 (N-terminal epitope), 13.1 (R domain), M3A7 (NBD2), and 24-1 (C terminus). Protein was uncut or treated with Genenase I. Migration of full-length CFTR (FL), NBD1-containing fragment (N1), and NBD2-containing fragment (N2) are indicated. (B) Autoradiograms of [α-32P]8-N3-ATP labeling of CFTR variants. Membranes were labeled as described in Methods. CFTR was then immunoprecipitated, digested with Genenase I, and run on SDS/PAGE. Background bands represent cellular proteins that label with [α-32P]8-N3-ATP and coimmunoprecipitate with CFTR. Because these background bands were variable between experiments, we confirmed the identity of CFTR bands by Western blotting for each experiment. Digital autoradiography of WT CFTR showed that the sum of the radioactivity in the N1 and N2 bands was 82 ± 9% of the radioactivity in the undigested (FL) CFTR (n = 5 independent labeling reactions). (C) Quantified [α-32P]8-N3-ATP labeling normalized to amount of protein measured by immunoblot. Asterisks indicate P < 0.05 (ANOVA) compared with WT labeling (n = 5).
Based on structural studies of ABC transporter NBDs (3–9), we hypothesized that introducing bulky hydrophobic residues into the Walker A motif at A462 and S1248 would sterically block ATP binding. Supporting this prediction, modifying cysteines at the analogous locations in WT P-glycoprotein disrupted ATPase activity in a nucleotide-dependent manner (30, 31). Moreover, in an earlier study, we found that introducing cysteine at A462 or S1248 did not alter CFTR gating (22). However, NEM modification of either Walker A motif reduced channel activity, and ATP slowed the rate of modification. Therefore, we introduced a phenylalanine at that site and asked whether it altered the interaction with ATP.
[α-32P]8-N3-ATP labeled both NBD1 and NBD2 (Fig. 1B), consistent with previous reports (13–15). The A462F mutation prevented labeling of NBD1; S1248F blocked NBD2 labeling; and the double mutant A462F/S1248F abolished labeling at both NBDs. Quantitative analysis confirmed these observations (Fig. 1C). Blocking nucleotide binding at one NBD tended to reduce [α-32P]8-N3-ATP labeling at the contralateral NBD. These data suggest the possibility of cooperative interactions between the two NBDs. They are also consistent with reports that the two NBDs interact (32) and that disrupting NBD2 by C-terminal truncation impairs NBD1 labeling (33).
The ability to block ATP binding provided an opportunity to answer several questions about the mechanisms of gating.
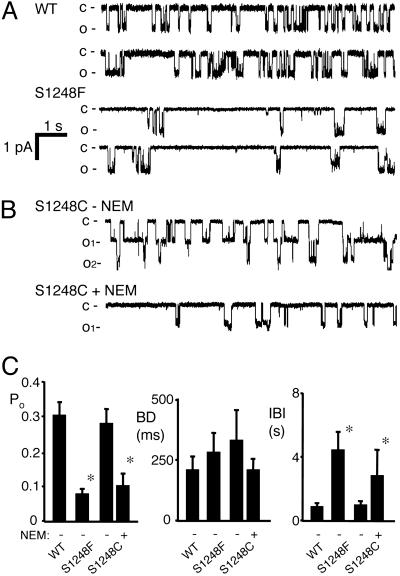
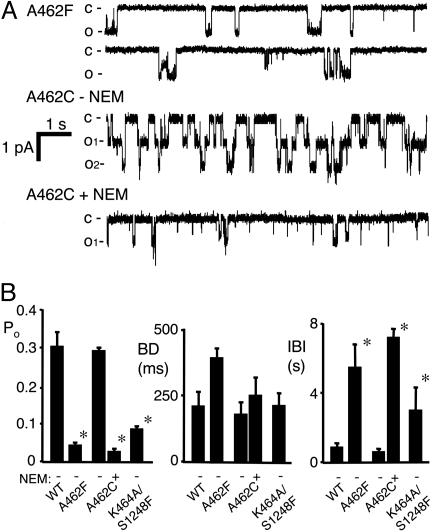
Preventing ATP Binding to NBD2 Reduces Channel Opening. The S1248F mutation reduced Po by prolonging the interburst interval without altering burst duration (Fig. 2 A and C). To directly compare these data with earlier results, we studied S1248C channels (22) and found that NEM altered gating in a similar way (Fig. 2 B and C). These data indicate that NEM modified all or nearly all of the channels that contained the Walker A Cys substitution. In addition, correspondence of the S1248F and NEM-modified S1248C data indicate that the gating effects of the S1248F substitution were not due to misfolding that might have occurred during channel biosynthesis. For WT and NEM-modified S1248C CFTR, increasing the ATP concentration from 1 mM to 10 mM produced only a small increase in current that was not affected by NEM modification (data not shown), indicating that the effects of NEM represented a block in ATP binding rather than a decreased affinity that could be overcome by increasing ATP concentration.
Fig. 2.
Effect of blocking ATP binding to NBD2 on CFTR gating. (A) Examples of single-channel recordings for WT and S1248F CFTR. (B) Recording from a membrane patch containing a small number of S1248C channels before and after treatment with 200 μM NEM. (C) Single-channel gating kinetics. Asterisks indicate P < 0.05 compared with WT CFTR (n = 3–6 for each construct).
These results allow two conclusions. First, inhibiting ATP binding to NBD2 impaired channel opening. Second, an interaction of ATP with NBD1 alone could support some channel activity.
ATP Binding and Hydrolysis at NBD2 Contribute to Gating. Earlier studies showed that mutating the Walker A Lys in CFTR NBD2 (K1250) abolished ATP hydrolysis (16), just as similar mutations abolish hydrolysis in other ABC transporters (34). The K1250 mutation also had two important effects on gating: It reduced the channel opening rate and prolonged the burst duration (16–18, 20, 21). However, the interpretation of these studies has been limited because it has not been clear whether one or both gating effects result from impaired ATP hydrolysis or reduced ATP binding (16, 19). The possibility that the K1250A mutation might reduce ATP binding is based on the observation that Walker A Lys mutations can reduce the interaction with ATP in some ABC transporters (35–37). In CFTR, it has been reported that the K1250A and K464A mutations prevented [α-32P]8-N3-ATP photolabeling of the respective NBD (14, 15), whereas another study found that K464A did not prevent [α-32P]8-N3-ATP NBD1 photolabeling (36).
We found that neither the K1250A nor K464A mutations prevented [α-32P]8-N3-ATP photolabeling of the NBDs (Fig. 3A). These data indicate that the nucleotide can interact with the NBDs in these variants. However, because [α-32P]8-N3-ATP modification is covalent and irreversible after UV irradiation, these data do not tell us about equilibrium binding nor do they assess binding affinity. This is also a limitation of previous studies of CFTR photolabeling (14, 26, 36, 38). Thus, our data, considered together with that in the literature, suggest that K1250A does not abolish nucleotide binding at NBD2, although it might reduce binding affinity.
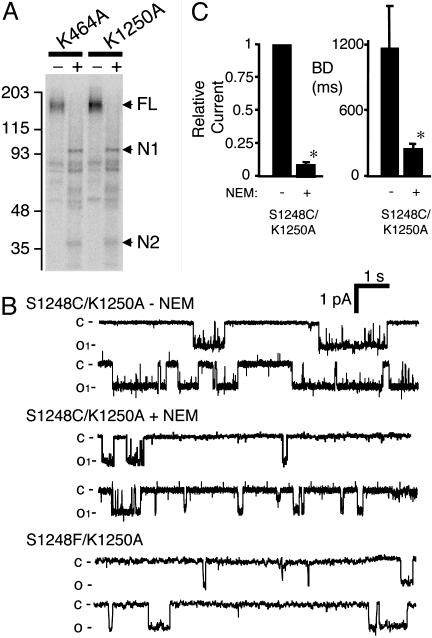
Fig. 3.
Effect of blocking ATP binding to NBD2 on the gating of CFTR-K1250A. (A) Autoradiogram of [α-32P]8-N3-ATP labeling of CFTR-K464A and K1250A; labeling of both NBDs was observed for each mutant. (B) Example of recording of an S1248C/K1250A channel before and after NEM treatment and an S1248F/K1250A channel. (C) Effect of NEM modification of CFTR-S1248C/K1250A on relative current and burst duration. Because we were not able to accurately assess the number of channels in a patch before adding NEM (K1250A has a long interburst interval), Po and the interburst interval were not determined. Asterisks indicate P < 0.05 compared with WT (n = 4).
The finding that channels unable to bind nucleotide at NBD2 (S1248F and NEM-modified S1248C) had a normal burst duration suggested that the prolonged burst duration of K1250A (16–18, 20, 21) arose when ATP bound NBD2 but then did not undergo hydrolysis. To test this hypothesis, we combined the K1250A mutation with S1248C. CFTR-S1248C/K1250A showed the prolonged burst duration typical of CFTR-K1250A (Fig. 3 B and C). However, when we modified NBD2 with NEM to prevent nucleotide binding, burst duration fell into the range of WT or S1248F channels (compare Figs. 2 and 3). Likewise, CFTR-S1248F/K1250A showed a burst duration of 338 ± 50 ms (n = 4) similar to NEM-modified S1248C/K1250A (Fig. 3 B and C).
The photolabeling and functional data suggest three conclusions about the role of NBD2 in gating. First, ATP binds to NBD2. Second, blocking binding markedly reduced the opening rate. This result suggests that ATP binding is associated with channel opening. Third, ATP binds the K1250A variant, and while bound it generated a long burst duration. Evidence that this mutant does not hydrolyze ATP (16) suggests that hydrolysis facilitates some step leading to channel closure. Without hydrolysis, the ATP-bound state (the open state) is prolonged and dissociation (or a very low rate of hydrolysis) may ultimately lead to closure.
Preventing ATP Binding at NBD1 Reduces Channel Opening. To assess the gating effects of ATP binding to NBD1, we studied the A462F variant, which blocks NBD1 nucleotide binding. Po was lower than in WT channels because of a reduced opening rate without much change in burst duration (Fig. 4). A462F reduced Po, as did treating A462C channels with NEM. Po fell because the opening rate decreased; although not statistically significant, burst duration tended to increase which would have the opposite effect on Po. These data are similar to our previous results and indicate that abnormal synthesis and/or folding were not responsible for the gating effects. Increasing the ATP concentration to 10 mM failed to significantly increase current (data not shown). These data suggest that ATP binding to NBD1 is required for normal channel opening.
Fig. 4.
Effect of blocking ATP binding to NBD1 on CFTR gating. (A) Examples of recordings from CFTR-A462F, and of A462C channels before and after NEM treatment. Although the A462F tracing shown has a higher Po than the mean, we chose this example to show the bursts. (B) Average data for channel gating kinetics. Asterisks indicate P < 0.05 compared with WT (n = 3–6 for each construct).
ATP Binding to NBD1 Has a Greater Effect on Gating than Hydrolysis. Previous studies showed that mutating K464 blocked ATP hydrolysis and slowed the opening rate, although to a lesser extent than the corresponding mutation in NBD2 (16, 17). However, because the K464 mutation could have also impaired ATP binding, it has been uncertain whether the gating effects resulted from disrupted ATP hydrolysis or binding. We found that the K464A mutation did not prevent [α-32P]8-N3-ATP photolabeling of NBD1 or NBD2 (Fig. 3A), a finding consistent with some (36) but not all earlier work (14, 15). Our finding indicates that ATP can bind NBD1 bearing the K464A mutation, although for the reasons described above, the ATP binding affinity might have been reduced.
We reasoned that if ATP binds CFTR-K464A to increase opening, then NEM modification of A462C/K464A should block binding and reduce activity. On the other hand, if the K464A mutation prevents ATP binding (contrary to our labeling results), then NEM modification should have no additional effect on current. Fig. 5 shows that NEM treatment markedly reduced both A462C and A462C/K464A current. Therefore, we conclude that ATP binding to NBD1 played an important role in channel activity even when it contained the K464A mutation.
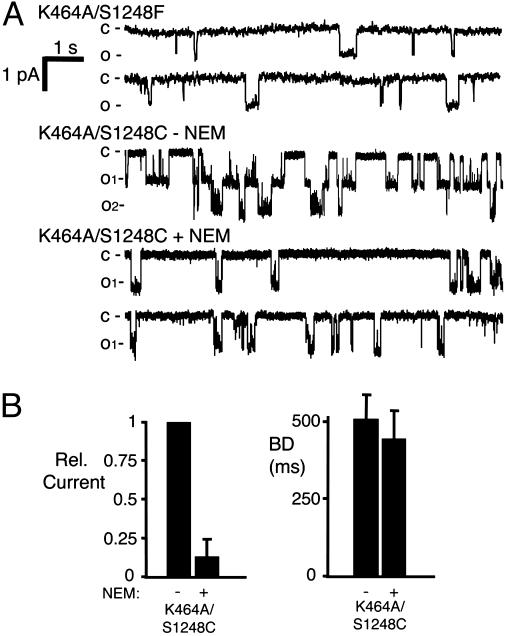
These data suggest ATP binding at NBD1 has a much greater effect on gating than does hydrolysis. To further test this hypothesis, we studied K464A/S1248F. The S1248F mutation will block NBD2 ATP binding, thereby limiting ATP interactions to NBD1, and the K464A mutation will prevent NBD1 ATP hydrolysis. The K464A/S1248F channel was not active without ATP (data not shown). However with ATP, the Po, interburst interval, and burst duration were similar to those obtained with the S1248F mutation alone (Figs. 4B and 6A). We confirmed these results by using CFTR-K464A/S1248C; NEM treatment reduced current with little effect on burst duration (Fig. 6 A and B). Thus, when ATP binding to NBD2 was eliminated, we found that the K464A mutation had little effect on gating. These findings suggest that CFTR can open when ATP binds to NBD1, even though NBD1 cannot hydrolyze ATP.
Fig. 6.
Effect of blocking ATP binding at NBD2 and hydrolysis at NBD1 on gating. (A) Examples of K464A/S1248F and K464A/S1248C channels before and after NEM treatment. (B) Relative current and burst duration from K464A/S1248C channels. Because we were not able to accurately estimate the number of channels in a patch before adding NEM, Po and the interburst interval could not be accurately determined, so we show relative current (n = 3).
Discussion
ATP Binding to a Single NBD Can Open the Channel. Our data show that specific mutations blocked ATP binding to either NBD1 or NBD2. Yet even with only one functional NBD, ATP still opened the channel. This conclusion requires the assumption that the interventions abolished ATP binding. Although our data support this premise, we cannot completely exclude infrequent interactions. Moreover, the finding that ATP binding to NBD1 alone was sufficient to open the channel is consistent with earlier studies of CFTR containing C-terminal truncations (33); the deletions blocked 8-N3-ATP labeling at NBD2, but channels still opened with a reduced Po. Although ATP interaction with one NBD can open the channel, the markedly reduced opening rate indicates that normal opening requires interaction with both NBDs.
At NBD1, ATP Binding Plays a More Important Role than Hydrolysis in Gating. Several observations have suggested that NBD1 binds ATP but has little enzymatic activity. This conclusion was first suggested by its primary sequence. Several residues that are highly conserved in ABC transporter NBDs and that are thought to be involved in hydrolysis are not conserved in CFTR NBD1. For example, residue 573 is a Ser rather than the consensus Glu, residue 605 is a Ser rather than the conserved His, and residue 1348 is a His instead of a Gly (see below for discussion of contribution of NBD2 sequences to the NBD1 ATP-binding site). Photolabeling with ATP analogs has also suggested that NBD1 may have a lower rate of hydrolysis than NBD2 (13, 15, 36). Although we cannot exclude some hydrolysis at NBD1, our data together with earlier studies suggest that if it occurs, hydrolysis at NBD1 has little effect on gating. Instead, ATP binding to NBD1 is the more important event. First, blocking ATP binding sharply decreased current in a channel in which hydrolysis did not occur, i.e., NEM reduced current in the A462C/K464A mutant. Second, the K464A/S1248F mutant behaved similarly to the S1248F single mutation. Thus, under conditions where ATP binding to NBD2 was blocked, preventing NBD1 hydrolysis had little effect on gating. Third, earlier observations showed that the K464A mutation, which blocks hydrolysis, had relatively minor effects on CFTR gating (16–19, 21). Moreover, the gating effects that did occur might have resulted from a quantitative reduction in ATP binding rather than an effect on hydrolysis. For example, our results and those of Basso et al. (36) showed that [α-32P]8-N3-ATP labeled the K464A NBD1, although perhaps with a reduced affinity, whereas Aleksandrov et al. (14, 15) reported that it ablated labeling. Although these labeling studies do not assess equilibrium binding, the differences in results do suggest reduced binding affinity.
At NBD2, both ATP Binding and Hydrolysis Contribute to Gating. Our data indicate that mutating the Walker A Lys in NBD2 did not abolish [α-32P]8-N3-ATP labeling, and earlier work showed that it eliminated ATP hydrolysis (16). However, Aleksandrov et al. (14, 15) reported that it prevented labeling. As we argued above, although these labeling studies do not assess equilibrium binding, taken together they suggest that K1250A probably reduced binding affinity. The functional consequences of the K1250A mutation have also been reported; the mutation reduced the rate of channel opening, and once the channel opened, it delayed its closure (16–18, 20, 21).
What are the consequences of ATP binding to NBD2? Our current data and our earlier work (22) indicate that blocking binding markedly slows channel opening. These findings suggest that ATP binding of NBD2 is intimately coupled to opening. They also suggest that the reduced opening rate of CFTR-K1250A might result from reduced binding affinity.
What causes the long burst duration of K1250A? There are two potential explanations. First, preventing ATP binding to NBD2 could cause the long burst duration. Our results argue against this explanation because blocking NBD2 ATP binding (with either the S1248F mutation or NEM-modification of S1248C) did not prolong burst duration. Second, ATP could bind the K1250A mutant (perhaps with reduced affinity), but impaired hydrolysis could slow channel closing. Our finding that NEM modification of CFTR-S1248C/K1250A shortened the long burst duration supports this interpretation. This conclusion is supported by the observation that substituting Ca2+ for Mg2+ to reduce hydrolysis also lengthened burst duration (39, 40). It is also consistent with the finding that hydrolysis-resistant nucleotides bind infrequently, but that when they do, they generate a long burst duration (17, 19, 41). These results also indicate that the ATP-bound state at NBD2 is open.
Blocking ATP Binding Did Not Alter Channel Open Time. It has been unclear what determines the rate at which CFTR closes. In WT CFTR, the rate of channel closing is relatively independent of ATP concentration (19, 42, 43), although a small effect has been reported (12, 20, 44, 45). These results suggest that ATP binding does not close the channel. That conclusion is consistent with the finding that disrupting ATP hydrolysis at NBD2 prolonged bursts (16–18, 20, 21, 39, 40, 46). Thus, hydrolysis at NBD2 precedes channel closure. However, it seems surprising that blocking ATP binding to NBD2 (with the S1248F mutation or the NEM-modified S1248C mutation) did not change the rate of channel closure compared to WT. Thus, channel openings stimulated by ATP binding to NBD1 without ATP binding or hydrolysis by NBD2 do not have an altered burst duration. Likewise, preventing ATP binding to NBD1 did not alter burst duration. Perhaps relatively similar burst durations for all these conditions is a coincidence, but further investigation seems warranted.
We speculate that ATP hydrolysis per se does not close the channel. Instead, after hydrolysis or ATP dissociation, the burst duration is determined by an ATP-independent mechanism. In such a model, the burst duration would remain unchanged regardless of whether the channel had opened with ATP bound to both or only a single NBD. Interestingly, CFTR can open infrequently without ATP, and the burst duration in the absence of ATP is only slightly shorter than the burst duration in the presence of ATP (45). Also consistent with this hypothesis are thermodynamic studies that reveal only a small energy barrier to channel closure (47, 48), but this energy barrier need not be related to ATP hydrolysis or dissociation.
Implications for NBD Dimerization in Channel Function. Some x-ray crystallographic studies suggest that ABC transporter NBDs form dimers with ATP bound at the interface between the two NBDs (7, 9, 10). Our earlier work suggested that the CFTR NBDs may dimerize to affect gating (11). If the adenine ring of ATP is sandwiched between the two CFTR NBDs, then [α-32P]8-N3-ATP might potentially label the NBD containing the P-loop or the contralateral NBD containing the LSGGQ motif. However, that is not what we observed; the A462F mutation completely blocked NBD1 labeling, and S1248F blocked NBD2 labeling. It is possible that the protein structure places the azido group of 8-N3-ATP in a position that can label only the NBD containing the Walker A motif. Alternatively, if CFTR itself is a dimer (49, 50), then NBD1:NBD1 and NBD2:NBD2 dimers might exist; our studies would not be able to detect such structures. Nevertheless, we did find that the A462F mutation in NBD1 diminished labeling of NBD2 and that S1248F slightly reduced NBD1 labeling. These results suggest cooperative interactions between the NBDs; dimerization would be one potential mechanism for such cooperativity.
Acknowledgments
We thank Pary Weber, Phil Karp, Dan Vermeer, Tamara Nesselhauf, and Theresa Mayhew for excellent assistance. This work was supported by National Heart, Lung, and Blood Institute (NHLBI) Grants HL29851 and HL61234 and Cystic Fibrosis Foundation Grant R458-CR02. We acknowledge the support of the Diabetes Endocrinology Research Center DNA core [supported by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant DK25295] and the In Vitro Models and Cell Culture Core (supported by the NHLBI, Cystic Fibrosis Foundation Grants R458-CR02 and ENGLH9850, and NIDDK Grant DK54759). A.L.B. was supported by NIDDK Grant DK062938. M.I. was an Associate and M.J.W. is an Investigator of the Howard Hughes Medical Institute.
Author contributions: A.L.B., M.I., and M.J.W. designed research; A.L.B. and M.I. performed research; A.L.B., M.I., and M.J.W. analyzed data; and A.L.B. and M.J.W. wrote the paper.
Abbreviations: NBD, nucleotide-binding domain; NEM, N-ethylmaleimide.
References
- 1.Riordan, J. R., Rommens, J. M., Kerem, B. S., Alon, N., Rozmahel, R., Grzelczak, Z., Zielenski, J., Lok, S., Plavsic, N., Chou, J. L., et al. (1989) Science 245, 1066-1073. [DOI] [PubMed] [Google Scholar]
- 2.Welsh, M. J., Ramsey, B. W., Accurso, F. & Cutting, G. R. (2001) in The Metabolic and Molecular Basis of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S., Valle, D., Childs, B. & Vogelstein, B. (McGraw–Hill, New York), pp. 5121-5189.
- 3.Hung, L. W., Wang, I. X., Nikaido, K., Liu, P. Q., Ames, G. F. L. & Kim, S. H. (1998) Nature 396, 703-707. [DOI] [PubMed] [Google Scholar]
- 4.Diederichs, K., Diez, J., Greller, G., Müller, C., Breed, J., Schnell, C., Vonrhein, C., Boos, W. & Welte, W. (2000) EMBO J. 19, 5951-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaudet, R. & Wiley, D. C. (2001) EMBO J. 20, 4964-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yuan, Y. R., Blecker, S., Martsinkevich, O., Millen, L., Thomas, P. J. & Hunt, J. F. (2001) J. Biol. Chem. 276, 32313-32321. [DOI] [PubMed] [Google Scholar]
- 7.Smith, P. C., Karpowich, N., Millen, L., Moody, J. E., Rosen, J., Thomas, P. J. & Hunt, J. F. (2002) Mol. Cell 10, 139-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis, H. A., Buchanan, S. G., Burley, S. K., Conners, K., Dickey, M., Dorwart, M., Fowler, R., Gao, X., Guggino, W. B., Hendrickson, W. A., et al. (2004) EMBO J. 23, 282-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locher, K. P., Lee, A. T. & Rees, D. C. (2002) Science 296, 1091-1098. [DOI] [PubMed] [Google Scholar]
- 10.Hopfner, K.-P., Karcher, A., Shin, D. S., Craig, L., Arthur, L. M., Carney, J. P. & Tainer, J. A. (2000) Cell 101, 789-800. [DOI] [PubMed] [Google Scholar]
- 11.Randak, C. & Welsh, M. J. (2003) Cell 115, 837-850. [DOI] [PubMed] [Google Scholar]
- 12.Li, C., Ramjeesingh, M., Wang, W., Garami, E., Hewryk, M., Lee, D., Rommens, J. M., Galley, K. & Bear, C. E. (1996) J. Biol. Chem. 271, 28463-28468. [DOI] [PubMed] [Google Scholar]
- 13.Szabo, K., Szakacs, G., Hegeds, T. & Sarkadi, B. (1999) J. Biol. Chem. 274, 12209-12212. [DOI] [PubMed] [Google Scholar]
- 14.Aleksandrov, L., Mengos, A., Chang, X., Aleksandrov, A. & Riordan, J. R. (2001) J. Biol. Chem. 276, 12918-12923. [DOI] [PubMed] [Google Scholar]
- 15.Aleksandrov, L., Aleksandrov, A. A., Chang, X. & Riordan, J. R. (2002) J. Biol. Chem. 277, 15419-15425. [DOI] [PubMed] [Google Scholar]
- 16.Ramjeesingh, M., Li, C., Garami, E., Huan, L. J., Galley, K., Wang, Y. & Bear, C. E. (1999) Biochemistry 38, 1463-1468. [DOI] [PubMed] [Google Scholar]
- 17.Carson, M. R., Travis, S. M. & Welsh, M. J. (1995) J. Biol. Chem. 270, 1711-1717. [DOI] [PubMed] [Google Scholar]
- 18.Gunderson, K. L. & Kopito, R. R. (1995) Cell 82, 231-239. [DOI] [PubMed] [Google Scholar]
- 19.Vergani, P., Nairn, A. C. & Gadsby, D. C. (2003) J. Gen. Physiol. 121, 17-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeltwanger, S., Wang, F., Wang, G. T., Gillis, K. D. & Hwang, T. C. (1999) J. Gen. Physiol. 113, 541-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Powe, A. C., Jr., Al-Nakkash, L., Li, M. & Hwang, T. C. (2002) J. Physiol. 539, 333-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cotten, J. F. & Welsh, M. J. (1998) J. Biol. Chem. 273, 31873-31879. [DOI] [PubMed] [Google Scholar]
- 23.Berger, A. L., Ikuma, M., Hunt, J. F., Thomas, P. J. & Welsh, M. J. (2002) J. Biol. Chem. 277, 2125-2131. [DOI] [PubMed] [Google Scholar]
- 24.Cotten, J. F. & Welsh, M. J. (1997) J. Biol. Chem. 272, 25617-25622. [DOI] [PubMed] [Google Scholar]
- 25.Dahan, D., Evagelidis, A., Hanrahan, J. W., Hinkson, D. A., Jia, Y., Luo, J. & Zhu, T. (2001) Pflügers Arch. 443, Suppl. 1, S92-S96. [DOI] [PubMed] [Google Scholar]
- 26.Travis, S. M., Carson, M. R., Ries, D. R. & Welsh, M. J. (1993) J. Biol. Chem. 268, 15336-15339. [PubMed] [Google Scholar]
- 27.Ostedgaard, L. S., Rich, D. P., DeBerg, L. G. & Welsh, M. J. (1997) Biochemistry 36, 1287-1294. [DOI] [PubMed] [Google Scholar]
- 28.Cheng, S. H., Gregory, R. J., Marshall, J., Paul, S., Souza, D. W., White, G. A., O'Riordan, C. R. & Smith, A. E. (1990) Cell 63, 827-834. [DOI] [PubMed] [Google Scholar]
- 29.Carter, P., Nilsson, B., Burnier, J. P., Burdick, D. & Wells, J. A. (1989) Proteins 6, 240-248. [DOI] [PubMed] [Google Scholar]
- 30.Al-Shawi, M. K., Urbatsch, I. L. & Senior, A. E. (1994) J. Biol. Chem. 269, 8986-8992. [PubMed] [Google Scholar]
- 31.Loo, T. W. & Clarke, D. M. (1995) J. Biol. Chem. 270, 22957-22961. [DOI] [PubMed] [Google Scholar]
- 32.Kidd, J. F., Ramjeesingh, M., Stratford, F., Huan, L. J. & Bear, C. E. (2004) J. Biol. Chem. 279, 41664-41669. [DOI] [PubMed] [Google Scholar]
- 33.Gentzsch, M., Aleksandrov, A., Aleksandrov, L. & Riordan, J. R. (2002) Biochem. J. 366, 541-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schneider, E. & Hunke, S. (1998) FEMS Microbiol. Rev. 22, 1-20. [DOI] [PubMed] [Google Scholar]
- 35.Shyamala, V., Baichwal, V., Beall, E. & Ames, G. F. L. (1991) J. Biol. Chem. 266, 18714-18719. [PubMed] [Google Scholar]
- 36.Basso, C., Vergani, P., Nairn, A. C. & Gadsby, D. C. (2003) J. Gen. Physiol. 122, 333-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panagiotidis, C. H., Reyes, M., Sievertsen, A., Boos, W. & Shuman, H. A. (1993) J. Biol. Chem. 268, 23685-23696. [PubMed] [Google Scholar]
- 38.Allard, M., Geoffre, S., Legendre, P., Vincent, J. D. & Simonnet, G. (1989) Brain Res. 500, 169-176. [DOI] [PubMed] [Google Scholar]
- 39.Harrington, M. A., Gunderson, K. L. & Kopito, R. R. (1999) J. Biol. Chem. 274, 27536-27544. [DOI] [PubMed] [Google Scholar]
- 40.Ikuma, M. & Welsh, M. J. (2000) Proc. Natl. Acad. Sci. USA 97, 8675-8680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aleksandrov, A. A., Chang, X., Aleksandrov, L. & Riordan, J. R. (2000) J. Physiol. 528, 259-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venglarik, C. J., Schultz, B. D., Frizzell, R. A. & Bridges, R. J. (1994) J. Gen. Physiol. 104, 123-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Winter, M. C., Sheppard, D. N., Carson, M. R. & Welsh, M. J. (1994) Biophys. J. 66, 1398-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai, Z. & Sheppard, D. N. (2002) J. Biol. Chem. 277, 19546-19553. [DOI] [PubMed] [Google Scholar]
- 45.Hennager, D. J., Ikuma, M., Hoshi, T. & Welsh, M. J. (2001) Proc. Natl. Acad. Sci. USA 98, 3594-3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dousmanis, A. G., Nairn, A. C. & Gadsby, D. C. (2002) J. Gen. Physiol. 119, 545-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathews, C. J., Tabcharani, J. A. & Hanrahan, J. W. (1998) J. Membr. Biol. 163, 55-66. [DOI] [PubMed] [Google Scholar]
- 48.Aleksandrov, A. A. & Riordan, J. R. (1998) FEBS Lett. 431, 97-101. [DOI] [PubMed] [Google Scholar]
- 49.Ramjeesingh, M., Kidd, J. F., Huan, L. J., Wang, Y. & Bear, C. E. (2003) Biochem. J. 374, 793-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Raghuram, V., Mak, D. D. & Foskett, J. K. (2001) Proc. Natl. Acad. Sci. USA 98, 1300-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]