Abstract
Purpose
Elevation in D-2-Hydroxyglutarate (D-2HG) has recently emerged as a mandatory byproduct of mutated Isocitrate Dehydrogenase (IDH) genes 1 and 2 in glioma patients. The goal of the present study was to demonstrate the feasibility of detection of elevated levels of D-2HG in the cerebrospinal fluid (CSF) of glioma patients that carry point substitutions in the IDH gene.
Experimental Design
We developed a mass spectrometry (MS)–based platform to detect and quantify the D- and L-forms of 2HG in the CSF of glioma patients. Three independent cohorts of patients were analyzed, comprising a total of 176 samples derived from 84 patients. The levels of D- and L-2HG were used to stratify patients into IDH wild-type or IDH-mutated groups using an empirically obtained threshold of 0.69 μmol/L.
Results
Using this platform, a greater than 17-fold mean increase in D-2HG was observed in the CSF of patients with IDH mutant versus wild-type gliomas. The means for the D-2HG levels in CSF were 0.427 μmol/L in wild-type and 7.439 μmol/L in mutant groups. The C statistic for the receiver operator curve was 0.938, with 84% sensitivity, 90% specificity, and 89% accuracy to detect D-2HG. The levels of D- and L-2HG in CSF from wild-type patients varied by location of CSF draw (cisternal>ventricular>lumbar).
Conclusions
Our findings demonstrate that the CSF of patients harboring IDH mutant gliomas contain increased levels of D-2HG, which can be reliably detected with a MS-based platform.
Introduction
Somatic heterozygous mutations are present in metabolic enzyme genes Isocitrate Dehydrogenase (IDH) 1 and 2 in patients with gliomas and acute myeloid leukemia (AML), as well as a variety of other neoplasms (1–4). The mutations occur in the active site of the enzymes and alter mutant IDH1/2 affinities for their natural substrate, isocitrate, and expected product, α-ketoglutarate (α-KG). The mutant IDH1/2 enzymes acquire a neo-morphic activity, which leads to the reduction of α-KG to the D-enantiomer of 2-Hydroxyglutarate (2HG) (5). The D-2HG metabolic byproduct can accumulate in greater than 100-fold excess in IDH-mutated glioma and AML patients’ tumor cells, with intracellular concentration of D-2HG in the 5 to 35 micromolar range per gram of tumor (5–10). D-2HG behaves as an oncometabolite, promoting oncogenesis by reprogramming the cellular epigenome, collagen synthesis, and cell signaling by inhibiting a class of α-KG–dependent enzymes (11–13). Furthermore, glioma patients bearing IDH1/2 mutations represent a distinct clinical entity, with a unique biology, and display improved overall survival, as compared with their wild-type (WT) counterparts (1, 3, 14). IDH1/2 mutations are found in most of World Health Organization (WHO)-defined grade II/III gliomas, and represent about 10% of grade IV glioblastomas (14).
Although the exact role of excess D-2HG amid altered metabolism and malignant transformation is still being actively investigated, the diagnostic and prognostic value of mutated IDH1/2 and D-2HG is already well recognized. Antibodies directed at specific mutants of IDH1 have been successfully developed (15), and are already utilized in clinical laboratories to distinguish gliomas carrying IDH mutations from other lesions. In addition, the feasibility of exploiting D-2HG diagnostically as a noninvasive imaging marker for tumors harboring IDH mutations using magnetic resonance spectroscopy (MRS) has been shown (8, 16–18).
We have developed a mass spectrometry (MS)–based platform for the detection and accurate quantification of D- and L-forms of 2HG in the cerebrospinal fluid (CSF) of brain tumor patients. This method is less invasive than neurosurgical procedures, more robust and cost-effective than imaging in determining tumor IDH status, and amenable for longitudinal follow-up of patients during their course of therapy. We chose to analyze CSF in contrast to blood or urine, as it is of low complexity, is largely shielded from circulating metabolites from other organs through the blood–brain and blood–CSF barriers, and represents a direct “window” on the brain tumor metabolome due to the absence of barrier between brain tumor tissue and CSF (19). Moreover, brain-secreted metabolites accumulate in a total volume of 120 to 140 mL of CSF rather than being diluted in 5 to 6 liters of blood and subsequently urine. We were able to assess the levels of D-2HG in three independent cohorts of glioma patients and stratify them based on this information into IDH WT or IDH mutated groups.
Translational Relevance.
The diagnosis of neoplasia in the central nervous system is based on clinical symptoms, radiologic imaging, and invasive biopsy. There is an unmet need for the quantitative detection of biomarkers in biofluids of brain tumor patients to complement clinical examination and neuroradiographic images findings, and obviate the need for invasive neurosurgical procedures. Mutations in the Isocitrate Dehydrogenase (IDH) genes 1 and 2 are present in most of human World Health Organization grade II and III gliomas. The mutated enzymes have a neomorphic activity that results in elevated production of metabolite D-2-Hydroxyglutarate (D-2HG) in the tumors. Here, we demonstrate that D-2HG can be detected through mass spectrometry in the cerebrospinal fluid of glioma patients with mutated IDH1/2, supporting its further development as a clinical test for diagnosis and follow-up.
Methods
CSF and tumor tissue samples collection and processing
A total of 176 CSF samples (N = 84 baseline, plus N = 92 longitudinal) with matching tumor tissue samples (N = 82) or IHC slides (N = 2) from cancer patients were selected for this study (Supplementary Table S1). They include gliomas with astrocytic, oligoastrocytic, and oligodendrocytic features (N = 77), meningiomas (N = 2), ependymomas (N = 2), melanoma (N = 1), adenocarcinoma (N = 1), and mammary gland carcinoma (N = 1), as classified by the World Health Organization (2007 edition). Samples were collected retrospectively from the following institutions: Penn State University College of Medicine (Hershey) and University of Massachusetts Medical School (Worcester, MA) for cohort 1 (N = 29 baseline; N = 92 longitudinal; N total =121), University of Mainz (Germany) for cohort 2 (N = 39), and University of California (San Diego) and Huntsman Cancer Center (Salt Lake City) for cohort 3 (N = 16).
The CSF originated from the ventricular system (N = 113), the cavernous or subarachnoid space of the convexities (cisternal; N = 44) or from the spinal canal (lumbar; N = 15), with four samples of unknown origin. Most CSF samples were collected just before or during the surgery (baseline CSF samples). In select patients, an intrathecal access device (Ommaya reservoir) was placed for installation of intrathecal chemotherapy after standard of care treatments were exhausted. This allowed for serial CSF withdrawal and analysis (longitudinal samples). The Institutional Review Board (IRB) committees at the respective institutions approved the CSF collections, and patients gave informed consent. Due to IRB restrictions, we received de-identified samples accompanied with only limited clinical information, e.g., clinical diagnosis, age at the time of CSF draw, sex, and the site of procurement of the CSF (Supplementary Table S1). Furthermore, due to the retrospective nature of our study, potential variables such as duration of storage, total volume drawn, time of day for drawing, or position of lumbar puncture were not standardized or recorded.
The CSF were spun after collection at 3,000 rpm at 4°C to remove cells and debris, and were stored in polypropylene tubes at −80°C. Matching brain tissue was supplied either frozen or formalin-fixed and paraffin-embedded.
Standards: Preparation of D- and L-2HG
D-2HG standard stock was prepared by mixing 25 mg of D-2HG (Santa Cruz Biotech) in 2.604 mL of 0.02 N hydrochloric acid. L-2HG standard stock was prepared by mixing 25 mg of L-2-Hydroxyglutarate (Santa Cruz Biotech) in 3.376 mL of 0.02 N hydrochloric acid.
Derivatization of D- and L-2HG
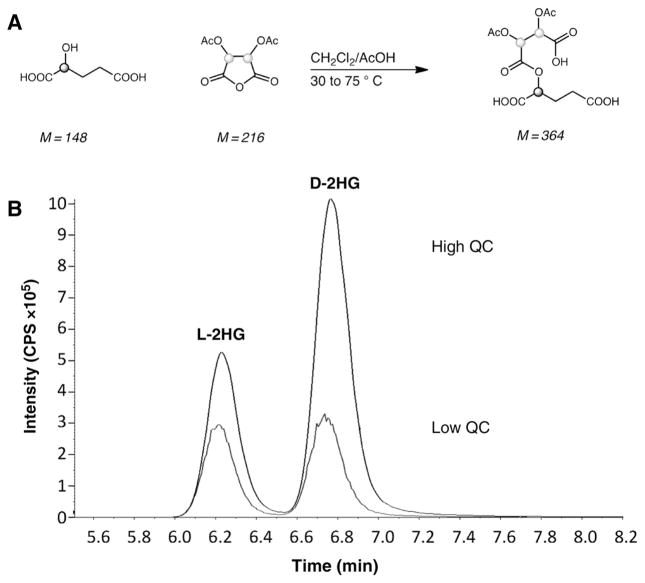
The derivatization step was adapted from a prior study (20). The CSF samples were thawed on ice, filtered using 10 kDa membrane filters, and centrifuged at 10,000 rpm for 10 minutes. 100 μL of neat samples were then transferred to screw cap glass tubes, and 100 μL of 10 μmol/L internal standards were added to each. The solutions were then dried under N2 at 50°C, after which 100 μL of 50 g/L of diacetyl-L-tartaric anhydride (DATAN) was added to each tube (Fig. 1A). The reaction was allowed to proceed for 30 minutes at 75°C, and then dried under N2 and lyophilized overnight. Once completely dry, the mixture was reconstituted in 200 μL of ultrapure water and aliquoted for subsequent MS analysis. The reaction produced D- or L-diastereoisomers capable of separation on an achiral column (Waters XTerra MS C18 5 μm, 3.9 × 150 mm Column).
Figure 1.
Generation and testing of 2HG standards. A, Chemical scheme showing the derivatization of D- or L-2HG with DATAN. B, Overlaid mass fragmentograms for D- and L-2HG from Low and High QCs. The intensity (y-axis) is shown in counts per second (CPS) over the course or time (x-axis) shown in minutes.
Quality control sample preparation and analysis
Quality control (QC) samples were included and analyzed to assess reproducibility of every run as well as the validity of the method. Clinically “normal” pooled CSF was used to prepare the QC samples. QC-High was formulated by spiking pooled CSF with 50 μmol/L of D- or L-2HG, whereas QC-Low by spiking with 0.5 μmol/L of D- or L-2HG. Twenty runs of “Low” and “High” D- and L-2HG levels were collected, and variations of QC measurements between runs recorded. Calibration curves of standards were constructed with 0, 0.1 μmol/L, 0.5 μmol/L, 5 μmol/L, 10 μmol/L, and 50 μmol/L of D- or L-2HG. Acceptance criteria for QC were set within two standard deviations from the historic mean, and the R2 or the Pearson Coefficient of calibration curves were greater than 0.9. Data from a sample run were considered invalid if concurrent QC or calibration curves did not meet the acceptance norms.
LC/MS analysis
LC/MS was used to detect D- or L-2HG in CSF with stable-isotope–labeled internal standards (D, L-[3,3,4,4-2H4]-2-hydroxyglutaric acid). AB-Sciex QTRAP 5500 was used in negative ion mode. The analytes were separated using LC (Shimadzu) with a C18 HPLC column in-line with a C18 HPLC guard column (Waters). IDH1/2 genes’ mutation status was unknown at the time of MS analysis. The MS analysis was done using a Quantitative Gradient Method using 7.0% Acetonitrile in Ammonium Formate buffer (pH = 3.6; flow rate: 0.4 mL/min). Quattro Micro Tune Parameters using negative electro-spray ion mode at 750°C and −4,500 V with −7.5 V collision energy were used. Scan parameters of 200 m/z to 550 m/z were employed.
NMR analyses
Nuclear Magnetic Resonance (NMR) analyses were performed on frozen tumor samples as previously described (16).
Data processing/statistical analysis
Calculations were performed on the Analyst software suite, ABIX. The concentrations of D- and L-2HG in each sample were calculated using an isotope dilution method. Final results were corrected for dilution factors and reported as μmoles/L (μmol/L).
T-test, one-way ANOVA comparisons (Tukey correction), and receiver operator curve (ROC) analyses were performed using GraphPad Prism software (v6.04). The data are expressed as geometric mean with 95% confidence intervals (CI).
Identification of patient tumors with mutated IDH1/2
Sequencing and IHC procedures were previously described (16).
Results
MS-based assay development for D- and L-2HG detection in human CSF
Methodology development for the identification of D- and L-2HG in human CSF using MS
To develop a robust and accurate method for the measurement of D- and L-2HG in human CSF using MS, we adapted a protocol for measurement of urinary D- and L-2HG by LC-MS (20). Using a derivatization step of a racemic 2HG mixture in human CSF with DATAN (Fig. 1A), we were able to achieve robust separation of the D- and L-2HG isomers on the achiral column coupled to the tandem MS/MS instrumentation with retention times of 3.5 minutes for L-2HG and 4.3 minutes for D-2HG (Fig. 1B).
Analytical measurement/sensitivity range determination
Assay sensitivity and analytical range determination were evaluated in triplicate using a 10-point, 2-fold serial dilution of standard D-or L-2HG in MilliQ grade water from 0.0005 to 10,000 μmol/L. The linearity was obtained between 0.05 and 1,000 μmol/L, i.e., low and high limits of detection (LOD/HOD), respectively. The limit of quantification was validated at 0.1 μmol/L with a limit of detection at 0.05 μmol/L. Samples with D-or L-2HG concentration >1 mmol/L were re-measured with dilutions. The coefficient of variation (CV) of 20 measurements of both high and low levels of D- and L-2HG was <20% (Table 1).
Table 1.
Summary of assay properties of the D- and L-2HG in human glioma CSF
| Assay parameter | D-2HG added | Endogenous D-2HG species in CSF | L-2HG added | Endogenous L-2HG species in CSF |
|---|---|---|---|---|
| Analytical reference range (μmol/L) | 0.05–1,000 | 0.05–1,000 | 0.05–1,000 | 0.05–1,000 |
| LLOQ (μmol/L) | 0.1 | 0.1 | 0.1 | 0.1 |
| Inter-assay precision (%CV) | 3.7 | 13.8 | 4.4 | 17.9 |
| Intra-assay precision (%CV) | 0.8 | 4.2 | 0.7 | 2.4 |
| CSF spike recovery (%) | 95.6–107.0 | 93.2–109.0 | ||
| Freeze-thaw stability | 6X thaw cycles | 6X thaw cycles | 6X thaw cycles | 6X thaw cycles |
| Stability at RT for 24 hours | Stable | Stable | Stable | Stable |
| Interference with L-2HG | None (up to 50 μmol/L) |
NOTE: Lowest limit of quantification (LLOQ) is the lowest back-interpolated standard that provides signal 2X over background with percent coefficient of variance <20%. For other measures, mean or range %CV is shown. Spike recovery ranges are reported only for the exogenous D- or L-2HG added. Freeze-thaw stability refers to the number of freeze-thaw cycles that allow <10% change in the signal from the original controls. Stability at room temperature (RT) for 24 hours of a sample was confirmed as “stable” if it met the acceptance criteria of <10% change from the original controls. Samples were run in duplicates.
Stability of samples
CSF sample stability was assessed in two studies using QCs and two representative CSF samples: (1) a freeze-thaw study at −80°C and (2) a stability study at room temperature for 24 hours. To assess the response of 2HG to freeze-thaw stability, CSF samples stored at −80°C were thawed completely at room temperature, then placed back at −80°C until 14X freeze-thaw cycles (over the course of 4 months) were achieved. Our results indicate that D- and L-2HG in CSF can withstand up to 6 freeze-thaw cycles (CV = 9.24%; Table 1). In addition, to define the stability of D- and L-2HG, the samples were assayed on day 1 and then reassayed on day 2 after keeping them at room temperature for 24 hours (CV = 4.81%; Table 1). The results indicate that the samples are stable at room temperature at least over the course of 24 hours.
Recovery of D- and L-2HG in human CSF
Next, we analyzed the recovery of exogenous D- or L-2HG in 4 representative CSF samples ± spiking with 50 μmol/L of D- or L-2HG standards. The percentage of recovery ranged between 95.6%–107.0% and 93.2%–109.0% for D- and L-2HG, respectively (Table 1).
Inter- and intra-assay variability testing
To test intra-assay precision, Low and High QCs and 2 representative samples of 100 μL human CSF were used to measure the concentrations of D- or L-2HG reference compounds. The results of 10 replicates of the intra-day study showed the %CV is between 0.7% and 4.2% (Table 1).
The inter-assay precision was assessed on 10 separate days/runs using duplicate measurements. Low and High QCs and 2 representative CSF samples were utilized. The inter-assay %CV ranged between 3.7 and 17.9 (Table 1).
Both intra- and inter-assay precision thus fell well within the acceptance criteria of CV < 20%.
Effect of L-2HG interference
To test interference of L-2HG in the measurement of D-2HG, we also performed interference experiments in six representative CSF samples spiked with five different levels (0.1 μmol/L, 0.5 μmol/L, 5 μmol/L, 10 μmol/L, 50 μmol/L) of standard L-2HG. Spiked samples were then run together with unspiked (endogenous) CSF samples. Our results demonstrate that the presence of L-2HG in the samples (up to 50 μmol/L L-2HG + endogenous levels) does not interfere with the accurate measurement of D-2HG in human CSF using our assay (Table 1).
Testing of the assay in three cohorts of CSF from glioma patients with or without IDH1 mutations
To test our developed assay with the overall goal of discriminating WT from IDH1/2-mutated gliomas based on 2HG levels in the patient CSF, we obtained three individual human CSF cohorts (total N = 176; baseline N = 84; longitudinal N = 92; Supplementary Tables S1 and S2) in a blind fashion. Cohorts 1, 2, and 3 included N = 29, N = 39, and N = 16 baseline CSF samples, respectively. In addition, 20 of 29 patients from Cohort 1 had repeated CSF draws, adding 92 longitudinal samples. All of the samples were representative of glioma patients at various stages of disease progression (WHO grades II to IV) or control malignancies with unknown IDH1/2 mutation status at the time of 2HG analysis (Table 2). The cohorts were tested independently with Cohort 1 being resampled twice.
Table 2.
Identified mutations from the tissues and quantified D- and L-2HG levels from matched CSF in a combined cohort of 1→3 based on WHO Grade
| Grade # | Mutational status (N) | D-2HG range (μmol/L) | L-2HG range (μmol/L) |
|---|---|---|---|
| II | WT (4) | 0.13–0.76 | 0.15–0.86 |
| mutant (3) | 0.35–1.00 | 0.99–2.94 | |
| III | WT (1) | 0.1 | 0.31 |
| mutant (8) | 0.31–109 | 0.40–4.08 | |
| IV | WT (56) | 0.1–2.51 | 0.14–6.14 |
| mutant (5) | 0.50–55 | 0.45–2.09 | |
| Other | WT (7) | 0.55–1.14 | 0.61–3.01 |
| mutant (0) | N/A | N/A |
Our results indicate that all of the D- and L-2HG values measured in the CSF samples through MS analysis fall within the determined analytical reference range for the developed assay: 0.05–1,000 μmol/L. The absolute levels of D-2HG ranged from 0.11–8.38 μmol/L in Cohort 1; 0.26–22.10 μmol/L in Cohort 2; and 0.10–109 μmol/L in Cohort 3. L-2HG metabolite levels ranged between 0.14–6.14 μmol/L (Cohort 1); 0.4–4.08 (Cohort 2); and 0.246–2.14 μmol/L (Cohort 3).
Confirmation of the mutational status in the matching glioma tissues
To test whether elevated D-2HG levels in the CSF could discriminate between patients with WT versus mutant IHD1/2 gliomas, we next obtained matching tumor tissues from the same patients (N =82 tissues) and identified 16 IDH1-mutated gliomas using DNA sequencing and immunohistochemistry (Supplementary Tables S1 and S2). A single false positive was obtained using IHC, as well as two ambiguous samples, and one sample that tested positive but could not be verified using DNA sequencing.
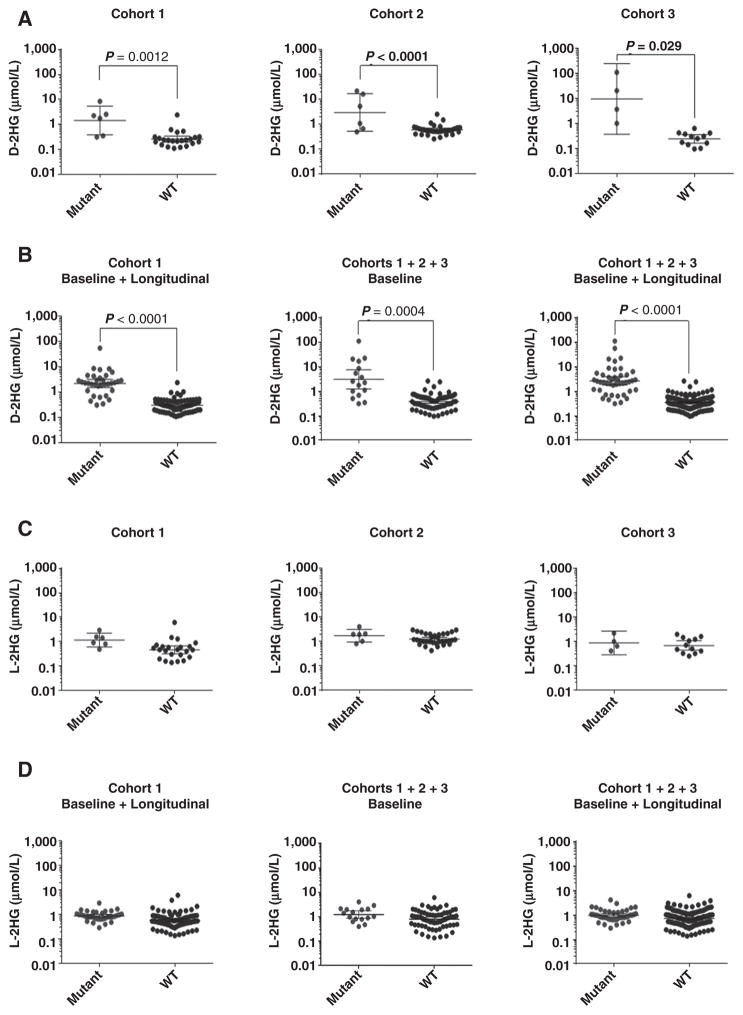
Next, based on the sequencing and immunohistochemical data, we segregated baseline or baseline + longitudinal CSF samples into two groups, based on the WT or mutant IDH status of the corresponding patient’s tumors. We plotted their D- or L-2HG values as a function of identified mutation, either as individual or combined cohorts (Fig. 2). We found significant differences between D-2HG levels in the two groups in all cohorts (sampled individually or in combination; Fig. 2A and B; Table 2; Supplementary Tables S1 and S3). In a combined cohort, 12 of 16 baseline (or 37/44 baseline + longitudinal CSF samples) positive for IDH1/2 mutation demonstrated D-2HG levels of >0.69 μmol/L, whereas 55 of 68 baseline (or 118/132 baseline + longitudinal) WT group CSF demonstrated <0.69 μmol/L levels. These results suggest that the two groups can be separated based on their D-2HG levels using an empirically calculated threshold of 0.69 μmol/L.
Figure 2.
Quantification of D- and L-2HG in CSF samples. A, Levels of D-2HG found in the CSF of Cohorts 1 (baseline; N = 29; left), 2 (N = 39; middle), and 3 (N = 16; right) are plotted as a function of IDH1/2 mutation status. In Cohort 1, patients underwent CSF draws both before (baseline samples) and at different times after surgery (longitudinal samples). B, D-2HG levels of all CSF samples of Cohort 1 (baseline plus longitudinal; N = 121; left), of combined Cohorts 1, 2, and 3 at baseline (N = 84; middle), or at baseline plus longitudinal (N = 176; right) are shown as a function of IDH1/2 mutation status. C, L-2HG levels in same CSF samples as A. D, L-2HG levels in same CSF samples as B. Scatter dot plots with geometric mean and 95% CIs are indicated. “Mutant” and “WT” represent samples with a mutated or WT IDH1/2 gene in the matching tumor, respectively.
The D-2HG levels in the WT group (N = 132) ranged between 0.10 and 2.57 μmol/L, and in the mutant group (N = 44) from 0.31 to 109 μmol/L. The means for the D-2HG levels were 0.427 μmol/L in the WT group and 7.439 μmol/L in the mutant group, comprising a 17-fold increase (mean difference −7.012; 95% CI, −10.11 to −3.916; P < 0.0001; Table 2; Supplementary Table S3). In addition, we consider the 0.6 to 0.7 μmol/L range as a “gray zone,” where the D-2HG level cannot accurately predict whether the matching patient tumors contain IDH1/2 mutations. Note that 7 of 176 samples fell in this region.
With regard to L-2HG levels, the differences between the two groups sampled in a combined fashion were not significant, with ranges for the WT group of 0.14–6.14 μmol/L and mutant group between 0.40 and 4.08 μmol/L. The mean values were 0.87 and 0.99 for WT and mutant groups, respectively (Fig. 2C and D; Table 2; Supplementary Table S3).
Receiver operator characteristic analysis was performed on the D-2HG data from the combined cohorts pool. It demonstrated an area under the curve of 0.877 (0.738–1.02; 95% CI, 0.772–0.983; P < 0.0001) with a sensitivity of 81% and specificity of 85% for baseline CSF (Supplementary Fig. S1A). The overall accuracy of being able to distinguish the baseline WT from IDH1-mutated controls was calculated to be 85%. By combining the three cohorts, including the longitudinal samples (N total = 176), the sensitivity, specificity, and accuracy improved to 84%, 90%, and 89%, respectively, with an area under the curve of 0.938 (0.882–0.993; 95% CI, 0.896–0.980; P < 0.0001; Supplementary Fig. S1B). For comparison, the ROC curves for L-2HG were not significant (Supplementary Fig. S1C and S1D).
D-2HG CSF levels related to tumor grade
A trend toward higher D-2HG levels in tumors of higher WHO grade was observed, even though it was not significant due to the limited number of samples per group (Supplementary Fig. S2; Table 2). Of note, 2 of 3 mutated CSF from grade II patients had D-2HG levels below our empirical threshold of 0.69 μmol/L, which prevented the discrimination between WT and mutant grade II gliomas.
D-2HG CSF levels related to patient gender and age
There was no correlation between patient gender and D-2HG levels (Supplementary Fig. S3). A significant negative correlation between D-2HG levels and age was observed (R = −0.575; P = 0.0033), likely reflecting the younger age of patients with IDH1/2 mutated tumors (40) as no correlation with age was present in mutated-only samples.
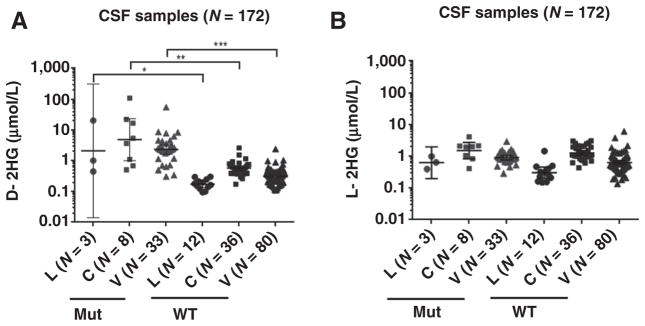
Comparison of D-2HG levels in CSF from different anatomical sources
Comparison of CSF obtained through ventricular (VT), cisternal (CT), or lumbar (LP) draws showed that the D-2HG levels were elevated in all samples derived from patients with IDH mutant gliomas, regardless of anatomical origin (Fig. 3; Table 3). The data were significant for ventricular [P = 0.0002; N (WT:Mut) = 80:33], cisternal [P = 0.0022; N (WT:Mut) = 36:8], and lumbar [P = 0.0283; N (WT:Mut) = 12:3] draws with P values proportional to number of samples. L-2HG values showed no differences between WT and Mut groups regardless of site of CSF draw.
Figure 3.
The levels of D-2HG detected in CSF from lumbar (L), cisternal (C), or ventricular (V) draws were significantly different between WT and mutant IDH patient groups. A, Student t test comparisons show significant differences in the levels of D-2HG found in the CSF of patients with WT or Mut IDH gliomas from lumbar, P = 0.0283, cisternal, P = 0.0022, and ventricular, P = 0.0002, draws. B, There are no significant differences in L-2HG levels between WT and mutant groups. Geometric means with 95% CI are indicated. The number (N) of samples is indicated in the parentheses.
Table 3.
D/L-2HG concentrations segregated by the site of CSF draw (t-test statistics)
| Metabolite | Site of CSF draw | Geometric mean ± SEM (μmol/L)
|
95% CI of geometric mean
|
P value difference between WT and Mut | ||
|---|---|---|---|---|---|---|
| WT | Mutant | WT | Mutant | |||
| D-2HG | Lumbar | 0.173 | 2.084 | 0.135–0.221 | 0.014–308.9 | 0.0283 |
| Cisternal | 0.568 | 4.824 | 0.483–0.668 | 0.100–23.28 | 0.0022 | |
| Ventricular | 0.314 | 2.317 | 0.279–0.353 | 1.613–3.329 | 0.0002 | |
| L-2HG | Lumbar | 0.303 | 0.626 | 0.202–0.455 | 0.199–1.971 | NSa |
| Cisternal | 1.259 | 1.491 | 1.065–1.489 | 0.818–2.716 | NS | |
| Ventricular | 0.622 | 0.888 | 0.540–0.716 | 0.761–1.036 | NS | |
NS, not significant.
Interestingly, we observed that the levels of both D- and L-2HG were site-dependent (Supplementary Fig. S4), suggesting heterogeneous distribution of metabolites among different CSF compartments. The highest levels were observed in cisternal, and the lowest in lumbar-derived CSF (CT>VT>LP). This finding prompted us to re-stratify 2HG thresholds based on the anatomical origin of CSF yielding the following new cut-offs for D-2HG: CT~1.46 μmol/L; VT~0.95 μmol/L; LP~0.33 μmol/L (Supplementary Table S4). These new and better-defined cut-offs enabled us to calculate site-specific values for sensitivity, specificity, and accuracy (Supplementary Table S5). The overall accuracy of distinguishing WT from mutant IDH glioma patients on the basis of D-2HG concentration in lumbar CSF improved to 100% because it “rescued” an originally false-negative sample. These measurements provide a foundation for a prospective study in a larger cohort of site-specific samples.
Comparison of MS-based to HRMAS NMR-based methodologies to detect D-2HG in human brain tumor tissues
Finally, we attempted to see how the MS-based methodology compares to that of NMR (16). To this purpose, we measured D-2HG levels in parallel by both techniques in an independent set of 12 glioma tissues with IDH mutations (Supplementary Fig. S5; Supplementary Table S6). We found a significant correlation between both methods (r = 0.6639; P = 0.0185), with the sensitivity of the MS-based methodology in the order of 200-fold higher than NMR.
Together, our findings demonstrate the feasibility of a MS-based platform for the specific quantification of the D-enantiomer of 2HG in CSF and provide a complementary biomarker methodology in biofluid (by MS) to that in tissues (via NMR).
Discussion
There is a pressing need for the quantitative detection of biomarkers in patients with neoplastic diseases of the central nervous system. Currently, brain lesions are initially identified through neurological examination and imaging procedures (CT and/or MRI scans), followed by pathologic examination of tissues obtained by invasive stereotactic biopsy or tumor resection. There is no laboratory test that predicts tumor recurrence, and imaging studies are often confounded by postsurgical and posttreatment changes, including radiation, corticosteroids, pseudoprogression, antiangiogenic agents, immunotherapies, and targeted therapies (21–25). A biomarker-based and easy to administer method to monitor tumor growth, response to therapy, and recurrence would be of great complementary benefit.
To determine the potential utility of the D-2HG metabolite as such a marker for the stratification and follow-up of patients with gliomas, we developed a robust and quantitative MS-based assay that can differentially detect and quantify D- and L-2HG metabolites in the CSF of patients with gliomas with an analytical range of 0.05 to 1,000 μmol/L. The assay was subsequently extensively characterized, including inter- and intra-assay variability, recovery, linearity, stability, and interference experiments, with the results suggesting that D-2HG and L-2HG calculated using this assay represent robust measurements.
Applying the MS-based D-2HG detection assay to three independent glioma CSF cohorts collected at several collaborating institutions, we were able to discriminate glioma patients with IDH1/2 mutations with an overall empirical D-2HG threshold of 0.69 μmol/L. The CSF of patients with WT IDH1 tumors had D-2HG levels below 0.69 μmol/L, while that of patients with IDH1/2-mutated tumors consistently displayed higher levels. These values are consistent with the published range for “normal” levels for D-2HG in human CSF of 0.07 to 0.3 μmol/L (26). The sensitivity, specificity, and accuracy of this discrimination using this threshold were 84%, 90%, and 89%, respectively. It is important to note that there was a “gray” zone (0.6–0.7 μmol/L), and one IDH1-mutated and six WT group CSF samples had D-2HG levels in this range, suggesting that the accurate cut-off values in discriminating WT from IDH1/2-mutated CSF may need to be further refined with testing larger annotated collections of CSF.
A prior report has measured physiologic levels of L-2HG between 0.3 and 2.3 μmol/L in CSF (26), which agrees with our measurements. Most of our samples contained L-2HG levels of <2.3 μmol/L (N = 169), 6 samples were found to have L-2HG between 2.3 and 3.1 μmol/L, and three outliers contained L-2HG at 3.8, 4.08, and 6.14 μmol/L.
Our study assumes the presence of mutated IDH1/2 enzymes is the major driver of elevated D-2HG/L-2HG ratios in tumors; however, cancer-related alterations in other genes may also lead to moderately increased D- or L-2HG. For example, in tumors with elevated Myc, mitochondrial glutaminase is activated, increasing the flux from glutamine to glutamate to ketoglutarate, and then to 2HG. Likewise, it is conceivable that other mutations occurring in the background of IDH mutant tumors may modify the flux.
Due to limited sample numbers for each grade, we were unable to determine whether there is a statistically significant relationship between D-2HG levels in CSF and WHO grade of IDH-mutant tumors. The data appear to suggest that IDH-mutant Grade II gliomas release less D-2HG in CSF, which may impede discrimination between WT and mutant groups, but this needs further study due to low sample number. Density of tumor cells in tumor tissue, amount of enzyme synthesized per tumor cell, and activity of mutant enzyme in different cellular contexts will all affect D-2HG production and could vary between grades. Other confounders most likely influence the transport of metabolites from the bulk tumor to CSF: for example, edema and bulk flow from solid tumor to adjacent brain, distance from the lesions to the brain surface, and brain anatomy including a sulcus, and the inter-hemispheric and sylvian fissures. This all may affect how fast and how many tumor-produced substances enter in the CSF. Absence of clinical, imaging, and treatment information also precluded us from analyzing possible correlations of D-2HG levels with tumor volume, proximity to the ventricles, and response to therapy. The present study, nonetheless, provides impetus to analyze larger well-characterized longitudinal collections in an effort to answer these important questions.
To determine whether site of CSF sampling affects 2HG concentration, we stratified our overall threshold, and found that L- and D-2HG levels are, in fact, site-dependent, with the highest levels observed in central (cisternal or ventricular), and the lowest in distal (lumbar) CSF (CT>VT>LP). Measurements in a single patient with an IDH1-mutant tumor where both lumbar and ventricular CSF were available confirmed the existence of a gradient. The lumbar CSF sample had 3- to 10-fold lower D-2HG levels than repeated ventricular draws (Sample # 7; Cohort 1). Yet, all measurements were above our site-specific thresholds of 0.33 μmol/L and 0.95 μmol/L, for LP and VT, respectively, highlighting the importance of stratifying the samples by the site of origin. Despite these differences, the amounts of D-2HG found in the CSF were significantly different between IDH WT and mutant groups, regardless of CSF site of origin.
While the CSF circulates from ventricles to the spinal canal and then comes back to the brain cisternae before draining into the circulation at the arachnoid villi, it is important to realize there is a connection between the ventricles and the cisterna magna, so the anatomy influences CSF circulation and the distribution of tumor-derived products in the CSF. While studies comparing lumbar and centrally collected CSF are limited, the available literature suggests that gradients of protein and metabolite concentration exist between these samples (27–34). For instance, IgG contents decrease progressively as the CSF moves from the site of intracranial inflammation to the lumbar sac (29). Similar gradients have been noted for other proteins and metabolites in patients suffering from bacterial meningitis (32) or cysticercosis (30). The presence of such gradients is relevant to molecular diagnostics, given results presented here, and a previous study showing that mutated IDH1 mRNA can be more readily detected in cisternal CSF samples relative to lumbar samples (35). The origin of such gradients may stem from the complexity of CSF physiology (36), and they warrant careful consideration in the design of future CSF sample-based molecular diagnostics. The circadian gradient, patient position, as well as the total volume of CSF collected may also affect measurement of D-2HG in CSF by lumbar puncture. It is also possible that patients with partially impaired CSF flow through the aqueduct of sylvius or with arachnoid adhesions over the convexities of the brain due to tumor effects would have abnormal CSF flow resulting in asymmetry in 2HG concentrations. It will be important to further address these issues, as lumbar collection is conceivable as a moderately invasive diagnostic procedure, whereas ventricular or cisternal collection involves placing intrathecal access devices that are more invasive. The latter may be unpractical for routine screening, but could be helpful to guide the treatment such as distinguishing between pseudo- and true tumor progression.
We also assessed 2HG levels in plasma and urine from glioma patients (data not shown), as did others in serum (37, 38), but no correlation between D-2HG levels and the presence of IDH1/2 mutations was found. A recent study also suggested that CSF is better than plasma for the detection of cell-free circulating tumor DNA (39).
We did not observe any correlation between patient gender and D-2HG levels. Consistent with the well-documented association between younger age and IDH1/2 mutated tumors (40), we did find a significant negative correlation between D-2HG levels and age (R = −0.575; P = 0.0033).
Another important question we attempted to answer is the correlation between repeated CSF draws in the same patients and D-2HG levels. We were not able to establish any correlation in the 20 patients where longitudinal samples were available, possibly related to the short intervals between individual sample draws or other nonstandardized confounding factors.
Importantly for clinical practice, we have demonstrated that the levels of D-2HG in CSF are stable, even with multiple freeze/thaws; as such it makes it possible to send frozen samples to a central clinical laboratory for analysis.
Taken together, our findings demonstrate that D-2HG can be detected in the CSF drawn from various sites, and its potential use in the clinic can be further considered. The quantification of D-2HG in lumbar CSF could be used for diagnostic and prognostic purposes by defining the genetic background of the patient’s brain tumor, without the need for invasive biopsy or surgical resection of the tumor. Elevated D-2HG can stratify the lesion as a glioma of the proneural subtype as they carry IDH1/2 mutations (14, 41). While collection of CSF though lumbar puncture is not entirely noninvasive, and is currently not part of the standard procedures in the clinical care of glioma patients, it could be implemented if justified, as it is safe in the majority of brain tumor patients even in the setting of increased intracranial pressure (42–45). Exceptions are patients with large midline shifts, or large posterior fossa masses, in which needle puncture of the dura can lead to continued escape of CSF into the epidural space, and cause brain herniation. Patients with impaired CSF flow due to ventricular obstruction or with marked epidural spinal cord compression should also be excluded. Coagulopathy or thrombocytopenia may be further contraindications, but these are not specific or even particularly common in patients with primary brain tumors. Recent studies have shown that MRS methods specific to detect and measure D-2HG can now be incorporated in the standard clinical MRI scanner, which is safer and noninvasive compared with lumbar punctures (8, 16–18). However, the sensitivity of D-2HG detection using MRS is 25- to 200-fold lower than by the current MS-based methodology. While MS is more sensitive than NMR, this advantage is offset by the significant drop in concentration of D-2HG from tumor tissue to what is released and diluted in the CSF. Although this will need further study, we believe that both MRS data on tumor and MS data on CSF will be complementary, depending on the clinical question and availability of the samples and methods.
Detection of D-2HG in the CSF using the highly sensitive MS method described here holds perhaps the most promise for the longitudinal monitoring of glioma patients, especially in postoperative patients where the intra-cranial tumor pressure has subsided after tumor debulking. Periodical CSF draws will provide a window into the biology of the tumor, and recent studies have shown this liquid can be informative not only for tumor metabolite production, but also for the presence of metastatic tumor cells, tumor-derived exosomes, nucleic acids, and protein (19, 39, 46–49). Ultimately, diagnosis and treatment will benefit from integrating information from both MRS and lumbar CSF depending on the clinical situation.
In summary, our findings establish a clinical association between D-2HG levels in the CSF and the presence of IDH1/2 mutation in the gliomas of three independent patient cohorts. Our study demonstrates the feasibility of an MS-based platform for the specific quantification of the D-enantiomer of 2HG in CSF, without interference from the L-isoform. A future prospective longitudinal study with well-defined inclusion/exclusion criteria and whose samples and data are collected in a coherent fashion to validate this technology is warranted. With that information in hand, a case can be made for the translation of this technique to standard practice as a complementary diagnostic and surveillance tool in the clinic.
Supplementary Material
Acknowledgments
We thank Narra Sarojini Devi, Milton Merchant, Lawrence Recht and Zhaobin Zhang for help with patient sample banking.
Grant Support
This work was supported in part by grants from the NIH (CA086335, CA116804, NS096236 to E.G. Van Meir, CA169937 to H. Mao, CA138292 to Walter Curran, Winship Cancer Institute), NINDS training grant 2T32NS007480-11 (to J. Kalinina and Allan I. Levey), the Brain Tumor Funders Collaborative (to E.G. Van Meir), and the Georgia Cancer Coalition (to J. Kalinina and E.G. Van Meir).
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
J.J. Olson reports receiving commercial research grants from Genentech and Millennium and other commercial research support from Merck. R.L. Jensen is a consultant/advisory board member for Medtronic, Pharmacokinesis, and Varian. No potential conflicts of interest were disclosed by the other authors.
Authors’ Contributions
Conception and design: J. Kalinina, J. Ahn, H. Mao, E.G. Van Meir
Development of methodology: J. Kalinina, J. Ahn, Y. Li, H. Mao, M. He
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): J. Kalinina, J. Ahn, Y. Li, M. Glantz, T. Smith, A. Giese, R.L. Jensen, C.C. Chen, B.S. Carter, H. Mao
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J. Kalinina, J. Ahn, L. Wang, Y. Li, M. Glantz, T. Smith, A. Giese, R.L. Jensen, C.C. Chen, H. Mao, M. He, E.G. Van Meir
Writing, review, and/or revision of the manuscript: J. Kalinina, J. Ahn, J.J. Olson, M. Glantz, T. Smith, A. Giese, R.L. Jensen, C.C. Chen, B.S. Carter, H. Mao, M. He, E.G. Van Meir
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): J. Kalinina, J. Ahn, N.S. Devi, L. Wang, E.G. Van Meir
Study supervision: J. Kalinina, H. Mao, E.G. Van Meir
References
- 1.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–12. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–66. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–73. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krell D, Mulholland P, Frampton AE, Krell J, Stebbing J, Bardella C. IDH mutations in tumorigenesis and their potential role as novel therapeutic targets. Future Oncol. 2013;9:1923–35. doi: 10.2217/fon.13.143. [DOI] [PubMed] [Google Scholar]
- 5.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465:966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frezza C, Tennant DA, Gottlieb E. IDH1 mutations in gliomas: When an enzyme loses its grip. Cancer Cell. 2010;17:7–9. doi: 10.1016/j.ccr.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 7.Rakheja D, Konoplev S, Su M, Wheeler D, Muzny DM, Ruvolo VR, et al. High incidence of IDH mutations in acute myeloid leukaemia with cuplike nuclei. Br J Haematol. 2011;155:125–8. doi: 10.1111/j.1365-2141.2011.08646.x. [DOI] [PubMed] [Google Scholar]
- 8.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med. 2012;18:624–9. doi: 10.1038/nm.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–34. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, et al. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–44. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–8. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Losman JA, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–5. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki M, Knobbe CB, Itsumi M, Elia AJ, Harris IS, Chio II, et al. D-2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 2012;26:2038–49. doi: 10.1101/gad.198200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Cancer Genome Atlas Research Network. Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–98. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Capper D, Zentgraf H, Balss J, Hartmann C, von Deimling A. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009;118:599–601. doi: 10.1007/s00401-009-0595-z. [DOI] [PubMed] [Google Scholar]
- 16.Kalinina J, Carroll A, Wang L, Yu Q, Mancheno DE, Wu S, et al. Detection of “oncometabolite” 2-hydroxyglutarate by magnetic resonance analysis as a biomarker of IDH1/2 mutations in glioma. J Mol Med (Berl) 2012;90:1161–71. doi: 10.1007/s00109-012-0888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andronesi OC, Kim GS, Gerstner E, Batchelor T, Tzika AA, Fantin VR, et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4:116ra4. doi: 10.1126/scitranslmed.3002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elkhaled A, Jalbert LE, Phillips JJ, Yoshihara HA, Parvataneni R, Srinivasan R, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4:116ra5. doi: 10.1126/scitranslmed.3002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khwaja FW, Van Meir EG. Proteomic discovery of biomarkers in the cerebrospinal fluid of brain tumor patients. In: Van Meir EG, editor. CNS cancer: Models, markers, prognostic factors, targets and therapeutic approaches. 1. New York: Humana Press (Springer); 2009. pp. 577–614. [Google Scholar]
- 20.Struys EA, Jansen EE, Verhoeven NM, Jakobs C. Measurement of urinary D- and L-2-hydroxyglutarate enantiomers by stable-isotope-dilution liquid chromatography-tandem mass spectrometry after derivatization with diacetyl-L-tartaric anhydride. Clin Chem. 2004;50:1391–5. doi: 10.1373/clinchem.2004.033399. [DOI] [PubMed] [Google Scholar]
- 21.de Wit MC, de Bruin HG, Eijkenboom W, Sillevis Smitt PA, van den Bent MJ. Immediate post-radiotherapy changes in malignant glioma can mimic tumor progression. Neurology. 2004;63:535–7. doi: 10.1212/01.wnl.0000133398.11870.9a. [DOI] [PubMed] [Google Scholar]
- 22.Taal W, Brandsma D, de Bruin HG, Bromberg JE, Swaak-Kragten AT, Smitt PA, et al. Incidence of early pseudo-progression in a cohort of malignant glioma patients treated with chemoirradiation with temozolomide. Cancer. 2008;113:405–10. doi: 10.1002/cncr.23562. [DOI] [PubMed] [Google Scholar]
- 23.Watling CJ, Lee DH, Macdonald DR, Cairncross JG. Corticosteroid-induced magnetic resonance imaging changes in patients with recurrent malignant glioma. J Clin Oncol. 1994;12:1886–9. doi: 10.1200/JCO.1994.12.9.1886. [DOI] [PubMed] [Google Scholar]
- 24.Huang RY, Neagu MR, Reardon DA, Wen PY. Pitfalls in the neuroimaging of glioblastoma in the era of antiangiogenic and immuno/targeted therapy - detecting illusive disease, defining response. Front Neurol. 2015;6:33. doi: 10.3389/fneur.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen PY, Macdonald DR, Reardon DA, Cloughesy TF, Sorensen AG, Galanis E, et al. Updated response assessment criteria for high-grade gliomas: Response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–72. doi: 10.1200/JCO.2009.26.3541. [DOI] [PubMed] [Google Scholar]
- 26.Gibson KM, ten Brink HJ, Schor DS, Kok RM, Bootsma AH, Hoffmann GF, et al. Stable-isotope dilution analysis of D- and L-2-hydroxyglutaric acid: Application to the detection and prenatal diagnosis of D- and L-2-hydroxyglutaric acidemias. Pediatr Res. 1993;34:277–80. doi: 10.1203/00006450-199309000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Cutler RW, Murray JE, Cornick LR. Variations in protein permeability in different regions of the cerebrospinal fluid. Exp Neurol. 1970;28:257–65. doi: 10.1016/0014-4886(70)90234-7. [DOI] [PubMed] [Google Scholar]
- 28.Del Bigio MR. Neuropathological changes caused by hydrocephalus. Acta Neuropathol. 1993;85:573–85. doi: 10.1007/BF00334666. [DOI] [PubMed] [Google Scholar]
- 29.Ramos-Kuri M, Montoya RM, Padilla A, Govezensky T, Diaz ML, Sciutto E, et al. Immunodiagnosis of neurocysticercosis. Disappointing performance of serology (enzyme-linked immunosorbent assay) in an unbiased sample of neurological patients. Arch Neurol. 1992;49:633–6. doi: 10.1001/archneur.1992.00530300069012. [DOI] [PubMed] [Google Scholar]
- 30.Rubalcava MA, Sotelo J. Differences between ventricular and lumbar cerebrospinal fluid in hydrocephalus secondary to cysticercosis. Neuro-surgery. 1995;37:668–71. doi: 10.1227/00006123-199510000-00009. discussion 71–2. [DOI] [PubMed] [Google Scholar]
- 31.Weisner B, Bernhardt W. Protein fractions of lumbar, cisternal, and ventricular cerebrospinal fluid. Separate areas of reference. J Neurol Sci. 1978;37:205–14. doi: 10.1016/0022-510x(78)90204-6. [DOI] [PubMed] [Google Scholar]
- 32.Merritt HH, Smith FF. The cerebrospinal fluid. Philadelphia & London: W.B. Saunders Co; 1937. [Google Scholar]
- 33.Glantz MJ, Cole BF, Glantz LK, Cobb J, Mills P, Lekos A, et al. Cerebrospinal fluid cytology in patients with cancer: minimizing false-negative results. Cancer. 1998;82:733–9. doi: 10.1002/(sici)1097-0142(19980215)82:4<733::aid-cncr17>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 34.Fishman RA. Cerebrospinal fluid in diseases of the nervous system. 2. Philadelphia: W.B. Saunders; 1992. [Google Scholar]
- 35.Chen WW, Balaj L, Liau LM, Samuels ML, Kotsopoulos SK, Maguire CA, et al. BEAMing and droplet digital PCR analysis of mutant IDH1 mRNA in glioma patient serum and cerebrospinal fluid extracellular vesicles. Mol Ther Nucleic Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brinker T, Stopa E, Morrison J, Klinge P. A new look at cerebrospinal fluid circulation. Fluids Barriers CNS. 2014;11:10. doi: 10.1186/2045-8118-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capper D, Simon M, Langhans CD, Okun JG, Tonn JC, Weller M, et al. 2-Hydroxyglutarate concentration in serum from patients with gliomas does not correlate with IDH1/2 mutation status or tumor size. Int J Cancer. 2012;131:766–8. doi: 10.1002/ijc.26425. [DOI] [PubMed] [Google Scholar]
- 38.Lombardi G, Corona G, Bellu L, Della Puppa A, Pambuku A, Fiduccia P, et al. Diagnostic value of plasma and urinary 2-hydroxyglutarate to identify patients with isocitrate dehydrogenase-mutated glioma. Oncologist. 2015;20:562–7. doi: 10.1634/theoncologist.2014-0266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Mattos-Arruda L, Mayor R, Ng CK, Weigelt B, Martinez-Ricarte F, Torrejon D, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015;6:8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–74. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 41.Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koren J, Cravioto H, Leicach M. Reevaluation of lumbar puncture. A study of 129 patients with papilledema or intracranial hypertension. Neurology. 1959;9:290–7. doi: 10.1212/wnl.9.4.290. [DOI] [PubMed] [Google Scholar]
- 43.Lubic LG, Marotta JT. Brain tumor and lumbar puncture. Arch Neurol Psychiatr. 1954;72:568–72. doi: 10.1001/archneurpsyc.1954.02330050038006. [DOI] [PubMed] [Google Scholar]
- 44.Sencer W. The lumbar puncture in the presence of papilledema. J Mt Sinai Hosp. 1956;23:808–15. [PubMed] [Google Scholar]
- 45.DeAngelis Lisa M, Posner Jerome B., editors. Neurologic Complications of Cancer. 2. Oxford University Press; New York, USA: 2008. Contemporary Neurology Series. [Google Scholar]
- 46.Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS, et al. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med. 2012;10:5. doi: 10.1186/1479-5876-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sullivan JP, Nahed BV, Madden MW, Oliveira SM, Springer S, Bhere D, et al. Brain tumor cells in circulation are enriched for mesenchymal gene expression. Cancer Discov. 2014;4:1299–309. doi: 10.1158/2159-8290.CD-14-0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Y, Springer S, Zhang M, McMahon KW, Kinde I, Dobbyn L, et al. Detection of tumor-derived DNA in cerebrospinal fluid of patients with primary tumors of the brain and spinal cord. Proc Natl Acad Sci U S A. 2015;112:9704–9. doi: 10.1073/pnas.1511694112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akers JC, Ramakrishnan V, Kim R, Skog J, Nakano I, Pingle S, et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One. 2013;8:e78115. doi: 10.1371/journal.pone.0078115. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.