Abstract
Rationale: The evidence supporting an association between traffic-related air pollution exposure and incident childhood asthma is inconsistent and may depend on genetic factors.
Objectives: To identify gene–environment interaction effects on childhood asthma using genome-wide single-nucleotide polymorphism (SNP) data and air pollution exposure. Identified loci were further analyzed at epigenetic and transcriptomic levels.
Methods: We used land use regression models to estimate individual air pollution exposure (represented by outdoor NO2 levels) at the birth address and performed a genome-wide interaction study for doctors’ diagnoses of asthma up to 8 years in three European birth cohorts (n = 1,534) with look-up for interaction in two separate North American cohorts, CHS (Children’s Health Study) and CAPPS/SAGE (Canadian Asthma Primary Prevention Study/Study of Asthma, Genetics and Environment) (n = 1,602 and 186 subjects, respectively). We assessed expression quantitative trait locus effects in human lung specimens and blood, as well as associations among air pollution exposure, methylation, and transcriptomic patterns.
Measurements and Main Results: In the European cohorts, 186 SNPs had an interaction P < 1 × 10−4 and a look-up evaluation of these disclosed 8 SNPs in 4 loci, with an interaction P < 0.05 in the large CHS study, but not in CAPPS/SAGE. Three SNPs within adenylate cyclase 2 (ADCY2) showed the same direction of the interaction effect and were found to influence ADCY2 gene expression in peripheral blood (P = 4.50 × 10−4). One other SNP with P < 0.05 for interaction in CHS, rs686237, strongly influenced UDP-Gal:betaGlcNAc β-1,4-galactosyltransferase, polypeptide 5 (B4GALT5) expression in lung tissue (P = 1.18 × 10−17). Air pollution exposure was associated with differential discs, large homolog 2 (DLG2) methylation and expression.
Conclusions: Our results indicated that gene–environment interactions are important for asthma development and provided supportive evidence for interaction with air pollution for ADCY2, B4GALT5, and DLG2.
Keywords: genome-wide interaction study, methylation, gene expression, expression quantitative trait locus, children
At a Glance Commentary
Scientific Knowledge on the Subject
Air pollution exposure early in life has been associated with asthma, but the mechanisms behind this effect are largely unknown. Understanding the biological mechanism that connects air pollutants with asthma and respiratory diseases has the potential to point to new targets for therapeutic intervention and to identify susceptible subgroups in the population.
What This Study Adds to the Field
We performed a genome-wide interaction study followed by functional genomics analyses that indicated involvement of several genes at the genomic, epigenomic, and transcriptomic levels for asthma related to air pollution exposure. Our results support the notion that gene–environment interactions are important for asthma development.
Asthma is the most common chronic disease among children (1). Heredity is a well-known risk factor, exemplified by strong associations between chromosome 17q21 variants and childhood asthma (2), but genetic factors cannot solely explain its increasing prevalence in the past few decades. Exposure to traffic-related air pollution in early childhood (often indicated by the level of nitrogen dioxide [NO2]) has been associated with asthma exacerbations (3) and reduced lung function in children (4–7), but its association with initial asthma development has been less consistent (8–11).
The exact mechanisms by which air pollution may lead to asthma are incompletely understood. Oxidative stress and inflammation represent pathogenic pathways involved in asthma development (3). Interactions between air pollution and allele variants in genes related to antioxidative stress systems, inflammation, and innate immunity have been reported in relation to the incidence of asthma (12, 13). Such gene–environment (G × E) interactions may partially explain the inconsistencies between air pollution and the incidence of asthma. A limitation of these previous studies is that they only included candidate genes, and so far, no genome-wide attempt has been made.
We aimed to identify mechanisms of childhood asthma using genome-wide single-nucleotide polymorphism (SNP) data and individual traffic-related air pollution exposure data, expressed as exposure to NO2 (see Figure E1 in the online supplement). We present genome-wide interaction data from more than 1,500 children with and without asthma from three European birth cohorts in the discovery phase, followed by look-up in two independent North American cohorts with approximately 1,800 children. For each of the SNPs that was nominally significant for interaction in the largest look-up evaluation cohorts (P < 0.05), we evaluated expression quantitative trait locus (eQTL) effects in human lung specimens and traffic-related air pollution–induced gene expression by genotype in peripheral blood cells along with effects of short- and long-term air pollution exposure on peripheral blood DNA methylation patterns. Some of the results of these studies were previously reported in the form of an abstract (14).
Methods
Additional details are available in the online supplement.
Study Subjects
In the discovery phase, meta-analysis was performed based on genome-wide interaction study (GWIS) results from three traffic pollution, asthma, genetics consortium (13) cohorts, including 454 children with asthma and 1,080 children without asthma of European ancestry: BAMSE (Children, Allergy, Milieu, Stockholm, Epidemiological Survey) (6), Stockholm, Sweden (children with asthma: 235; children without asthma: 246), GINIplus (German Infant Study on the Influence of Nutrition Intervention Plus Environmental and Genetic Influences on Allergy Development) (15) and LISAplus (Influence of Life-Style Factors on the Development of the Immune System and Allergies in East and West Germany Plus the Influence of Traffic Emissions and Genetics) (16), Germany (children with asthma: 64; children without asthma: 661), and PIAMA (Prevention and Incidence of Asthma and Mite Allergy) (17), the Netherlands (children with asthma: 155; children without asthma: 173). Detailed cohort descriptions are provided in the online data supplement and elsewhere (13).
The top discovery SNPs were further evaluated in two independent look-up data sets from North America that included a total of 692 children with asthma and 1,096 children without asthma. The birth cohorts of CAPPS (Canadian Asthma Primary Prevention Study; Vancouver and Winnipeg, Canada) (18) and SAGE (Study of Asthma, Genetics and Environment; Manitoba, Canada) (19), both of which included children of white ancestry, had 49 children with asthma and 137 children without asthma; the larger cohort CHS (Children’s Health Study; California) (20), which included children of non-Hispanic white ancestry, had 643 children with asthma and 959 children without asthma. All cohorts obtained ethical approval from their local review boards.
Exposure and Outcome Assessment
For the European birth cohorts, the annual average of NO2 exposure estimates at birth were derived using land use regression modeling (LUR). Site-specific LUR models were developed and validated using the standardized European Study of Cohorts for Air pollution Effects project procedures (www.escapeproject.eu/manuals), as previously described in detail (21). Using a similar methodology, LUR models of birth exposure were developed for CAPPS and SAGE (22, 23). In CHS, NO2 exposure was estimated based on the level in the child’s community at baseline (mean age 8.8 yr) obtained from central site monitors placed in each of the study communities (20, 24). NO2 was used as a proxy for traffic-related air pollution. Exposure data were entered as a continuous nontransformed variable, and the risk estimates were reported per 10 μg/m3 increase in NO2 (Table E1).
Asthma definitions were based on parental reports of an ever doctor’s diagnosis (BAMSE, GINI/LISA, PIAMA and CHS), clinical examinations by a pediatric allergist (CAPPS), or parental reports with confirmation of diagnoses by a pediatric allergist (SAGE) (Table E2).
Genotyping and Quality Control
Genotyping, imputation procedure, and quality control steps for each study are described in the online supplement and Table E2.
SNP × NO2 Interaction and Asthma
Logistic regression analyses for estimation of standard SNP × NO2 interaction effects on asthma (multiplicative interaction model using HapMap-imputed, genome-wide association study [GWAS] data) was performed in each cohort separately as the primary model. A genome-wide significant threshold of P < 7.2 × 10−8 for SNP × NO2 interaction effects was applied (25). Discovery meta-analysis of 2,082,301 overlapping SNPs was conducted using the statistical software METAL with fixed-effect models with default METAL weights (26). In addition to the primary SNP × NO2 GWIS analysis, two other statistical methods to test for genome-wide interaction were used, with the same exposure, outcome, and adjustment factors: a two-step approach, in which in step one, the hypothesis of H0: βSNP = 0 was tested using NO2 as the outcome in a combined set of cases and control subjects; a subset of SNPs that exceeded a given significance threshold (P < 0.05) for the test in step one was further analyzed in step two (in our study, NSNPs = 119,521, equivalent to a genome-wide significance threshold of meta-analysis P < 4 × 10−7 after Bonferroni correction of 119,521 tests); and testing the hypothesis that H0: βSNP*NO2 = 0 (analyzing cases and control subjects; regular G × E interaction test) (27). The second method was the two degree-of-freedom (2 df) test that jointly tested SNP main and SNP × NO2 interaction effects (28).
To avoid false negative findings, an arbitrary cutoff level for look-up of interacting SNPs was set at P < 1 × 10−4 (29) for our primary analysis in the discovery data sets (standard interaction model). Thus, SNPs with a combined interaction P < 1 × 10−4 in the discovery phase were selected for look-up evaluation of standard SNP × NO2 interaction effects on asthma in the CAPPS/SAGE and CHS cohorts. Next, SNPs with P < 0.05 for interaction in the larger CHS cohort (and annotated genes) were included in the functional genomics follow-up described in the following.
Gene Expression Analysis in Lung Tissue and Peripheral Blood Cells
eQTL analyses were performed to evaluate if the SNPs significant in the look-up (P < 0.05) were related to cis-acting lung tissue gene expression. Lung tissue from 1,111 human subjects who underwent lung surgery at three academic sites, Laval University, University of British Columbia, and University of Groningen, were previously analyzed (30, 31). Linear regression models were used separately for each cohort, adjusting for age, sex, and smoking status. Meta-analysis was performed using inverse variance weighting. SNPs were considered an eQTL if they survived 5% Benjamini and Hochberg false discovery rate correction for multiple testing of the number of gene probes tested for each SNP. The Genotype-Tissue Expression (GTEx) portal (http://www.gtexportal.org/home), which provides tissue-specific global gene expression data from genotyped donors, was used next to evaluate whole blood eQTLs (n = 338 samples), analyzing the same SNPs and genes as in the lung eQTL (32). Furthermore, gene expression analyses (Affy HTA 2.0; Affymetrix Inc., Santa Clara, CA) were performed in peripheral blood cells from 263 16-year-olds in the BAMSE cohort as part of the MeDALL (Mechanisms of the Development of Allergy) project (33, 34). A look-up of GTEx-identified eQTLs was performed in 173 BAMSE samples, with GWAS data available using linear regression, adjusting for age, sex, and peripheral blood cell count. In addition, 250 BAMSE samples with exposure data available were used for linear regression association between NO2 at birth and current NO2 exposure at 16 years of age and expression levels of the genes annotated to significant look-up SNPs, with further stratification by genotype.
DNA Methylation in Relation to Long- and Short-Term Air Pollution Exposure
Methylation values for CpG sites within regions ±50 kb upstream and downstream of the identified genes were derived from Illumina 450K (Illumina Inc., San Diego, CA) data sets and investigated for association with air pollution exposure. Methylation data from the BAMSE cohort at 8 years (n = 460 with Illumina 450k data available) were investigated for association with long-term NO2 exposure at birth using robust linear regression, adjusting for age, sex, environmental tobacco smoke exposure during the first year of life, municipality at birth, ever doctor’s diagnosis of asthma up to 8 years of age, cell type, and batch (bisulfite treatment date) (34). The same CpG sites were also investigated for methylation quantitative trait locus (methQTL) effects to evaluate if the significant SNPs in the G × E look-up analyses were associated with methylation changes.
Short-term diesel exhaust exposure (DEP), as a model of particulate air pollution, was next investigated for association with DNA methylation differences in blood samples from 16 19- to 35-year-old nonsmokers with asthma and/or airway hyper-responsiveness using linear mixed effects modeling to compare post-DEP versus pre-DEP, and postfiltered air particles versus prefiltered air particles (35). Adjustment was done using a 5% Benjamini and Hochberg false discovery rate–correction for multiple testing on the set of probes selected for the analysis.
Results
Tables E1 and E2 in the online supplement present the characteristics of the three European and two North American studies, including NO2 exposure assessment, genotyping, and imputation procedures.
SNP × NO2 Interaction and Asthma
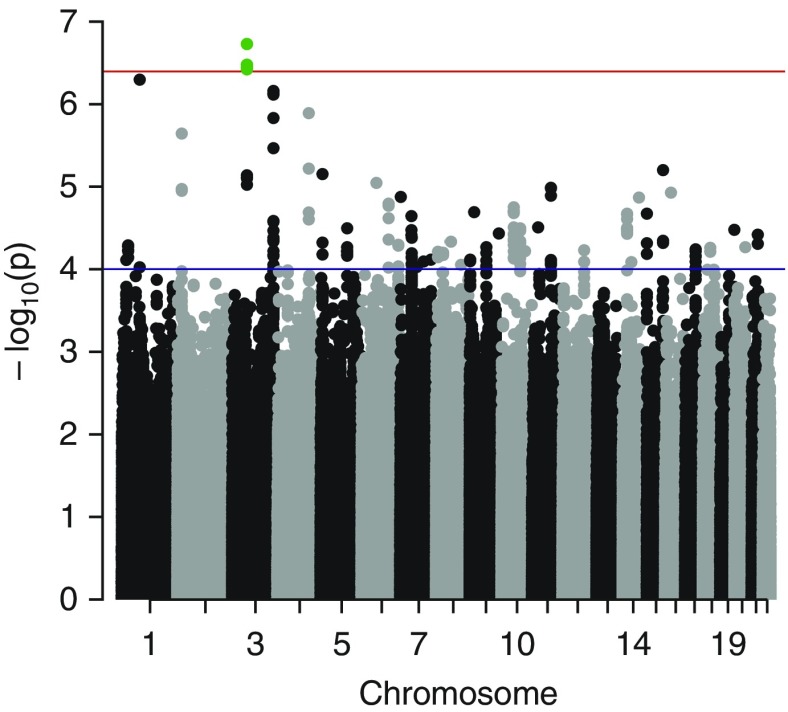
In total, 1,534 children of European ancestry aged 7.4 to 11.3 years were included in the primary GWIS meta-analysis (454 children with asthma and 1,080 children without asthma as control subjects). Figure E2 in the online supplement shows the QQ-plot for the SNP × NO2 interaction analysis on asthma (λ = 1.03). The discovery meta-analysis provided no genome-wide significant hits at the genome-wide significant threshold of P < 7.2 × 10−8. The top SNPs for interaction effects (lowest P = 1.87 × 10−7) are located in chromosome 3p14.1, approximately 244 kb downstream of the membrane-associated guanylate kinase, WW, and PDZ domain–containing 1 (MAGI1) gene (Figure 1 and see Table E3). Next, we used an alternative two-step analytical approach that was suggested to increase the power to detect G × E interactions (27). Four SNPs reached genome-wide significance in this two-step model (P < 4 × 10−7; see Table E4): rs7651862, rs11706125, rs11718057, and rs13066946 close to the MAGI1 gene; these were also identified as top hit SNPs in the primary GWIS meta-analysis. As a third approach, we applied the 2 df test that jointly tested main SNP and SNP × NO2 interaction effects (28). No SNP reached genome-wide significance in this test (lowest P value was 1.08 × 10−6) (see Figures E3 and E4, and Table E5).
Figure 1.
Manhattan plot for the discovery genome-wide interaction meta-analysis of the association between SNP × NO2 and asthma. The horizontal red line indicates the genome-wide significance threshold when using the two-step interaction approach (P < 4 × 10−7). The horizontal blue line indicates the threshold for single-nucleotide polymorphisms selected for look-up (P < 1 × 10−4; n = 186). The locus, near MAGI1 on chromosome 3p14.1, which reached genome-wide significance when using the two-step interaction approach, is marked in green (rs7651862, rs11706125, rs11718057, rs13066946).
Look-up Evaluation
We selected 186 interaction-effect SNPs with P < 1 × 10−4 from our primary model, the discovery GWIS meta-analysis for look-up in two different cohorts (Table E3). Of these 186 SNPs, 172 were available for look-up in the larger CHS-imputed, genome-wide SNP data set (643 children with asthma and 959 control subjects), and 8 SNPs showed nominal significant interaction (P < 0.05) (Table 1 and see Table E9). The SNP with the lowest P value for interaction in CHS (rs686237; P = 0.0016) is located on chromosome 20q13 in a region located 40 and 59 kb upstream of the genes UDP-Gal:betaGlcNAc β-1,4-galactosyltransferase, polypeptide 5 (B4GALT5), and solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 (SLC9A8), respectively. Three SNPs (rs1057251, rs12455842, and rs12457919) are located downstream of, or within the molybdenum cofactor sulfurase (MOCOS), on chromosome 18q12 and were in complete linkage disequilibrium (r2 = 1.0). These three SNPs were also among the top SNPs (P < 1 × 10−4) in the two-step interaction approach meta-analysis (Table E4). Three additional SNPs located within adenylate cyclase 2 (ADCY2) on chromosome 5p15.3 (rs4143882, rs727432, and rs6886921 with high linkage disequilibrium; r2 = 0.93–1.0) and one within discs, large homolog 2 (DLG2) on chromosome 11q14.1 (rs963146) reached nominal significance (P < 0.05) (Table 1). The four SNPs close to the MAGI1 gene and the eight SNPs with P < 0.05 in CHS were also nominally significant in the 2 df test (P value range 5.64 × 10−5 to 0.008) (Table E6). In the smaller Canadian CAPPS- and SAGE-imputed, genome-wide SNP data set (49 children with asthma and 137 control subjects), 122 SNPs were available for look-up evaluation. Two of the SNPs (rs3843891 on chromosome 4q31 and rs17265947 on chromosome 8q12.3) reached nominal significance (P < 0.05) (Table E7), but none of the SNPs were significant in CHS. The top SNPs close to MAGI1 identified in the discovery GWIS meta-analysis and the two-step approach were not significant in CHS or CAPPS/SAGE. No overall significant main effects of the 8 SNPs on asthma were observed (Table E8). NO2 exposure at birth (per 10 μg/m3 increase) was positively associated with asthma up to 8 years of age, but was not statistically significant (meta-analysis adjusted odds ratio, 1.26; 95% confidence interval, 0.61–2.58).
Table 1.
Single-Nucleotide Polymorphisms from the Genome-Wide Interaction Meta-Analysis of the Association between Single-Nucleotide Polymorphism × NO2 Interaction and Asthma That Were Statistically Significant in the Look-up Evaluation
| Discovery GWIS Meta-analysis: BAMSE, GINIplus/LISAplus, PIAMA (n = 1,534) | Look-up |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| CHS (n = 1,602) |
CAPPS/SAGE (n = 186) |
||||||||
| Chr | SNP | MAF | Nearest Gene | Interaction P Value* | Stratification by Genotype†: OR (95% CI) | Interaction P Value‡ | Stratification by Genotype†: OR (95% CI) | Interaction P Value‡ | Stratification by Genotype†: OR (95% CI) |
| 20 | rs686237 | 0.32 | B4GALT5, | 5.43 × 10−5 | CC: 0.77 (0.48–1.24) | 0.0016 | CC: 1.21 (1.04–1.41) | NA | NA |
| SLC9A8 | AC/AA: 1.69 (1.08–2.64) | AC/AA: 0.89 (0.78–1.01) | |||||||
| 18 | rs1057251 | 0.10 | MOCOS | 6.18 × 10−5 | TT: 1.68 (0.85–3.29) | 0.0094 | TT: 0.95 (0.85–1.06) | 0.58 | TT: 2.59 (1.01–6.66) |
| CT/CC: 0.50 (0.22–1.15) | CT/CC: 1.30 (1.03–1.62) | CT/CC: 2.02 (0.06–66.02) | |||||||
| 18 | rs12455842 | 0.10 | MOCOS | 6.10 × 10−5 | TT: 1.70 (0.86–3.39) | 0.010 | TT: 0.95 (0.85–1.06) | 0.55 | TT: 2.59 (1.01–6.66) |
| CT/CC: 0.48 (0.21–1.10) | CT/CC: 1.30 (1.03–1.62) | CT/CC: 2.02 (0.06–66.02) | |||||||
| 5 | rs4143882 | 0.33 | ADCY2 | 4.75 × 10−5 | GG: 0.81 (0.33–1.99) | 0.015 | GG: 0.88 (0.76–1.02) | 0.26 | GG: 4.90 (1.25–19.24) |
| AG/AA: 1.61 (1.04–2.51) | AG/AA: 1.13 (0.98–1.29) | AG/AA: 1.04 (0.26–4.24) | |||||||
| 5 | rs727432 | 0.32 | ADCY2 | 6.67 × 10−5 | GG:0.81 (0.33–1.99) | 0.016 | GG: 0.88 (0.76–1.02) | 0.27 | GG: 4.90 (1.25–19.24) |
| GT/TT: 1.61 (1.04–2.51) | GT/TT: 1.13 (0.98–1.29) | GT/TT: 1.04 (0.26–4.24) | |||||||
| 5 | rs6886921 | 0.34 | ADCY2 | 7.03 × 10−6 | CC:0.76 (0.29–1.99) | 0.016 | CC: 0.88 (0.76–1.02) | NA | NA |
| CT/TT: 1.71 (1.11–2.66) | CT/TT: 1.12 (0.98–1.27) | ||||||||
| 18 | rs12457919 | 0.10 | MOCOS, | 5.52 × 10−5 | AA: 1.68 (0.85–3.29) | 0.012 | AA: 0.95 (0.85–1.06) | NA | NA |
| FHOD3 | AC/CC: 0.39 (0.09–1.75) | AC/CC: 1.30 (1.03–1.62) | |||||||
| 11 | rs963146 | 0.21 | DLG2 | 8.61 × 10−5 | AA: 1.56 (1.04–2.33) | 0.034 | AA: 0.93 (0.83–1.06) | 0.62 | AA: 3.02 (0.84–10.87) |
| AG/GG: 0.67 (0.21–2.18) | AG/GG: 1.12 (0.96–1.32) | AG/GG: 2.75 (0.70-10.79) | |||||||
Definition of abbreviations: ADCY2 = adenylate cyclase 2; B4GALT5 = β-1,4-galactosyltransferase, polypeptide 5; BAMSE = Children, Allergy, Milieu, Stockholm, Epidemiological Survey; CAPPS = Canadian Asthma Primary Prevention Study; Chr = chromosome; CHS = Children’s Health Study; CI = confidence interval; DLG2 = discs, large homolog 2; FHOD3 = formin homology 2 domain–containing 3; GINIplus = German Infant Study on the Influence of Nutrition Intervention Plus Environmental and Genetic Influences on Allergy Development; GWIS = genome-wide interaction study; MAF = minor allele frequency, according to BAMSE; MOCOS = molybdenum cofactor sulfurase; LISAplus = Influence of Life-Style Factors on the Development of the Immune System and Allergies in East and West Germany Plus the Influence of Traffic Emissions and Genetics; NA = not applicable; OR = odds ratio for asthma associated with exposure to traffic-related NO2 for different genotypes; PIAMA = Prevention and Incidence of Asthma and Mite Allergy; SAGE = Study of Asthma, Genetics and Environment; SLC9A8 = solute carrier family 9 member A8; SNP = single-nucleotide polymorphism.
SNPs that were nominally significant in CHS (P < 0.05), ordered by the CHS interaction P value. All P values given are two-sided.
Genome-wide significance threshold, P < 7.2 × 10−8.
Stratification by genotype using dominant model.
Significance threshold for look-up evaluation, P < 0.05.
Direction of Interaction Effect and Asthma Risk
SNPs that showed nominal significance in the larger CHS sample (all 8 SNPs in Table 1) were investigated for their direction and strength of effect in the association between NO2 exposure and childhood asthma. Consistent directions of the interaction effect between the discovery meta-analysis and CHS studies were identified for all three SNPs within ADCY2 (Table 1), with increased risk of asthma associated with NO2 exposure in carriers of the minor alleles. The stratified analyses did not show a consistent pattern of asthma risk for the other SNPs, and the odds ratios for asthma across genotypes varied substantially between the data sets. Because the discovery data sets used exposure at birth and the main look-up study (CHS) used exposure at school age, meta-analyses of the interaction effects were not meaningful, because the odds ratios for asthma represented different measures.
Gene Expression Analysis in Lung Tissue and Peripheral Blood
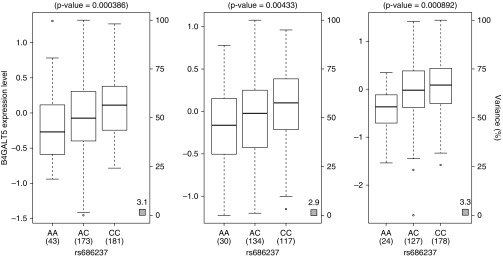
We performed eQTL analyses to evaluate if the eight nominally significant SNPs from the look-up showed cis-acting eQTL associations in lung tissue (n = 1,111). rs686237 was identified as a highly significant cis-eQTL of B4GALT5 (the C allele is associated with increased expression; P = 1.18 × 10−17) (Figure 2 and Table E10). In addition, rs12455842 was a significant cis-eQTL of SLC39A6 (P = 0.003) (Table E10) in lung tissue. No other SNP showed significant cis-eQTL association in lung tissue after 5% FDR correction for multiple testing. GTEx eQTL analyses in whole blood confirmed that rs686237 was a significant cis-eQTL of B4GALT5 (P = 4.00 × 10−4) (Figure E5), but with an opposite effect, in that the C allele was associated with decreased expression. rs6886921, rs727432, and rs4143882 were significant cis-eQTLs for ADCY2 (lowest P = 4.50 × 10−4 for rs6886921, with the T allele associated with decreased expression) (Figure E6 and Table E11). However, these blood eQTLs were not statistically significant in the smaller BAMSE data set (n = 173).
Figure 2.
Gene expression levels of β-1,4-galactosyltransferase, polypeptide 5 (B4GALT5) in lung tissues according to genotyping groups for single-nucleotide polymorphism rs686237 (using an additive model). The left, middle, and right panels are results from Laval University (n = 397; P = 3.86 × 10−4), University of British Columbia (n = 281; P = 0.0043), and Groningen University (n = 329; P = 8.92 × 10−4), respectively, with a meta-analysis P value of 1.18 × 10−17. Expression is presented for probeset 100313047_TGI_at. The left y-axis represents gene expression levels in the lung. The x-axis represents the three genotyping groups for single-nucleotide polymorphism rs686237 (build 37 position 48,370,734) with the number of subjects in parenthesis. The right y-axis presents the percent variance in gene expression levels explained by the genotype. Box boundaries represent the first and third quartiles, with whiskers extending maximum ± 1.5 times the interquartile range, and the median denoted by the center mark in the boxplots. Circles represent outliers.
Next, we explored NO2 exposure association with gene expression in BAMSE (n = 250). NO2 exposure at birth significantly influenced ADCY2, DLG2 and MOCOS expression, with increased expression levels in peripheral blood cells in relation to NO2 (Table 2; similar associations were also seen for exposure at 16 years; Table E12). For the top lung eQTL SNP, rs686237, an interacting SNP × NO2 effect was detected for B4GALT5 expression (interaction P = 0.001, also FDR significant; Table 2), in which the effect of NO2 exposure at birth on gene expression differed depending on genotype status.
Table 2.
Association between NO2 Exposure Levels at Birth and Peripheral Blood Gene Expression Levels at 16 Years of Age in BAMSE (n = 250*)
| Chr | Gene | Probe | Associated SNP | Genotype | Coef | P Value | Interaction P Value |
|---|---|---|---|---|---|---|---|
| 5 | ADCY2 | TC05000054.hg.1 | All (n = 250) | 0.03 | 0.05 | 0.85 | |
| rs6886921 | CC (n = 72) | 0.04 | 0.17 | ||||
| TC (n = 83) | 0.04 | 0.17 | |||||
| TT (n = 18) | −0.07 | 0.19 | |||||
| 5 | ADCY2 | TC05000055.hg.1 | All (n = 250) | 0.04 | 0.09 | 0.17 | |
| rs6886921 | CC (n = 72) | 0.08 | 0.07 | ||||
| TC (n = 83) | −0.05 | 0.53 | |||||
| TT (n = 18) | −0.001 | 0.98 | |||||
| 11 | DLG2 | TC11002159.hg.1 | All (n = 250) | 0.04 | 0.008 | 0.35 | |
| rs963146 | AA (n = 104) | 0.02 | 0.34 | ||||
| AG (n = 64) | 0.05 | 0.08 | |||||
| GG (n = 5) | —— | —— | |||||
| 18 | MOCOS | TC18000149.hg.1 | All (n = 250) | 0.03 | 0.046 | 0.59 | |
| rs1057251 | TT (n = 147) | 0.04 | 0.09 | ||||
| CC (n = 22) | 0.08 | 0.13 | |||||
| CT (n = 4) | —— | —— | |||||
| 20 | B4GALT5 | TC20000928.hg.1 | All (n = 250) | 0.01 | 0.73 | 0.001 | |
| rs686237 | CC (n = 88) | −0.11 | 0.03 | ||||
| AC (n = 66) | 0.17 | 0.02 | |||||
| AA (n = 19) | 0.20 | 0.14 | |||||
| 20 | SLC9A8 | TC20000391.hg.1 | All (n = 250) | −0.01 | 0.53 | 0.18 | |
| rs686237 | CC (n = 88) | −0.06 | 0.09 | ||||
| AC (n = 66) | 0.08 | 0.08 | |||||
| AA (n = 19) | −0.03 | 0.66 |
Definition of abbreviation: ADCY2 = adenylate cyclase 2; B4GALT5 = β-1,4-galactosyltransferase, polypeptide 5; BAMSE = Children, Allergy, Milieu, Stockholm, Epidemiological Survey; Chr = chromosome; Coef = coefficient; DLG2 = discs, large homolog 2; MOCOS = molybdenum cofactor sulfurase; SLC9A8 = solute carrier family 9 member A8; SNP = single-nucleotide polymorphism.
Analyses were adjusted for age, sex, and cell count. Coef is the log-fold change in gene expression per 10 µg/m3 increase in NO2 exposure. P value is the P value for association between NO2 exposure and gene expression. Interaction P value is the P value for association between SNP × NO2 and gene expression using additive effect of SNP. P values in bold have a value ≤0.05.
n = 250 for the NO2 to gene expression association analyses and n = 173 for the SNP × NO2 to gene expression analyses.
DNA Methylation and Air Pollution Exposure
Because air pollution exposure has been associated with differential DNA methylation patterns in peripheral blood cells (36), we explored potential links between NO2 exposure and methylation at the 278 CpG sites identified in a region ±50 kb of the identified genes.
In the BAMSE cohort (n = 460), NO2 exposure at birth was significantly associated with 2.7% decreased methylation in CpG site cg02275784 within DLG2 (per 10 μg/m3 NO2 increase; P = 1.21 × 10−4). Methylation in other CpG sites was not associated with NO2 exposure after 5% FDR correction for multiple testing (data not shown). Minor effects of DNA methylation changes were detected in the methQTL analysis with a level of methylation change up to 1% per allele (nominal P < 0.05). None of the associations remained significant at the 5% FDR level (Table E13).
As a marker for short-term, traffic-related air pollution exposure, 16 adult nonsmokers with asthma were exposed to 2 hours of DEP, at an average concentration of 300 μg/m3, containing high levels of NO2 (0.22 ppm) (35, 37). The difference in DNA methylation level was tested in blood samples pre-exposure versus postexposure. A total of 13 CpG sites were differentially methylated after 5% FDR correction for multiple testing (Table 3). Decreased methylation at eight CpG sites at the DLG2 locus was detected (lowest P = 4.64 × 10−5 for a 2% difference; cg26449294), and increased methylation was detected at two CpG sites close to transcription start sites (lowest P = 1.07 × 10−4 for a 4% difference; cg20275558) (Table 3). Decreased methylation was also identified at one ADCY2 CpG site, and increased methylation was seen at one MOCOS CpG site.
Table 3.
Significant Associations of Short-Term Diesel Exhaust Exposure and CpG Site Methylation Difference (Postexposure − Preexposure) in Adults with Asthma (n = 16)
| Chr | GWIS Locus | Probe | Probe Position (Build 37) | CpG Site Location | ΔFA* (Post − Pre) | ΔDE† (Post − Pre) | DE P Value | DE Adjusted P Value‡ |
|---|---|---|---|---|---|---|---|---|
| 5 | ADCY2 | cg04119977 | 7,826,972 | ADCY2 (Body) | −0.015 | −0.019 | 0.0011 | 0.041 |
| 5 | ADCY2 | cg10995381 | 7,877,198 | MTRR (Body) | −0.013 | −0.032 | 0.0011 | 0.041 |
| 11 | DLG2 | cg26449294 | 83,169,193 | DLG2 (3′UTR) | −0.010 | −0.021 | 4.64 × 10−5 | 0.017 |
| 11 | DLG2 | cg09080874 | 83,284,905 | DLG2 (Body) | −0.015 | −0.027 | 3.01 × 10−4 | 0.029 |
| 11 | DLG2 | cg27373604 | 83,372,714 | DLG2 (5′UTR;Body) | −0.021 | −0.026 | 0.0021 | 0.041 |
| 11 | DLG2 | cg08432013 | 83,393,570 | DLG2 (Body;TSS200) | −0.010 | −0.025 | 5.85 × 10−4 | 0.031 |
| 11 | DLG2 | cg02675969 | 83,526,604 | DLG2 (Body) | −0.010 | −0.017 | 0.0018 | 0.041 |
| 11 | DLG2 | cg05405389 | 84,386,472 | DLG2 (Body) | −0.017 | −0.035 | 0.0016 | 0.041 |
| 11 | DLG2 | cg18023263 | 84,403,466 | DLG2 (Body) | −0.020 | −0.022 | 0.0013 | 0.041 |
| 11 | DLG2 | cg14716968 | 84,635,906 | DLG2 (TSS1500;Body) | −0.003 | −0.037 | 4.28 × 10−4 | 0.029 |
| 11 | DLG2 | cg20275558 | 85,338,473 | TMEM126B; DLG2 (TSS1500;TSS200) | 0.011 | 0.042 | 1.07 × 10−4 | 0.020 |
| 11 | DLG2 | cg06698742 | 85,359,218 | TMEM126A (5′UTR) | 0.0009 | 0.0041 | 0.0014 | 0.041 |
| 18 | MOCOS | cg19453250 | 33,710,783 | SLC39A6; ELP2 (TSS1500;Body) | 0.0076 | 0.024 | 0.0019 | 0.041 |
Definition of abbreviations: ADCY2 = adenylate cyclase 2; Chr = chromosome; DE = diesel exhaust exposure; DLG2 = discs, large homolog 2; ELP2 = elongator acetyltransferase complex subunit 2; FA = filtered air; GWIS = genome-wide interaction study; MOCOS = molybdenum cofactor sulfurase; MTRR = 5-methyltetrahydrofolate-homocysteine methyltransferase reductase; SLC39A6 = solute carrier family 39 member A6; TMEM126A = transmembrane protein 126A; TMEM126B = transmembrane protein 126B; UTR = untranslated region.
P values in bold have a value ≤0.05.
ΔFA: relative methylation change postexposure versus preexposure of filtered air.
ΔDE: relative methylation change postexposure versus preexposure of diesel exhaust.
Adjusted P values using the false discovery rate method for multiple testing at a 5% level.
Discussion
We presented a comprehensive GWIS with functional follow-up integrating genomics and environmental data that identified novel and previously identified loci for childhood asthma in relation to traffic-related air pollution exposure. Identified loci from the genome-wide SNP by NO2 interaction approach, with significant look-up in 1,602 independent samples, were investigated for effects at genomic, epigenomic, and transcriptomic levels. We provided supportive evidence for interaction with air pollution for the novel loci B4GALT5 and the previously lung disease associated loci ADCY2 (38, 39) and DLG2 (40).
The GWIS was used as screening to detect genomic regions with a potential link to traffic-related air pollution exposure and childhood asthma. The SNP with the lowest P value in the look-up evaluation, rs686237 on chromosome 20, was found to be a strong eQTL for expression of B4GALT5 in the lung and was also identified as an eQTL for B4GALT5 in whole blood. These results suggested a potential SNP-mediated effect of the association between NO2 and childhood asthma with a functional consequence as indicated by differential B4GALT5 expression in blood depending on genotype.
The enzyme B4galt5 is involved in the biosynthesis of lactosylceramide, which is a common precursor of glycosphingolipids (41). Previous GWASs have identified a locus on chromosome 17q21, encompassing ORMDL sphingolipid biosynthesis regulator 3 (ORMDL3) and gasdermin B (GSDMB), to be strongly associated with childhood asthma (2). Interestingly, the endoplasmic reticulum transmembrane protein ORMDL3 is involved in the regulation of eosinophil trafficking (42).
ADCY2 encodes a member of the family of adenylate cyclases, which are membrane-associated enzymes involved in G-protein–coupled receptor signaling. Three SNPs in ADCY2 showed statistical significance in the look-up evaluation of SNP × NO2 interaction effects on asthma, and they had all similar direction of effect between the discovery cohorts and the main look-up study, CHS. The three SNPs were also identified as eQTLs for ADCY2 in whole blood. For the ADCY2 eQTL rs6886921, the minor allele T was associated with decreased expression in blood, and CT/TT carriers had the highest risk of asthma associated with NO2 exposure in both the discovery and CHS data sets. ADCY2 was also differentially expressed in relation to air pollution exposure, and decreased methylation levels were found in relation to short-term air pollution exposure. ADCY2 SNPs were previously associated with pulmonary function and chronic obstructive pulmonary disease (COPD) (38, 39, 43).
Using the complementary alternative two-step statistical approach for the genome-wide interaction analysis, genome-wide significance was reached in the discovery data set for four SNPs located near the MAGI1 locus. MAGI1 acts as a scaffolding protein, stabilizing and recruiting various molecules to the cell–cell contacts, and it is widely distributed at tight junctions in epithelial cells (44). Involvement of the airway epithelium is of importance in asthma pathogenesis because disruption of barrier functions could potentially lead to air pollution–related adverse effects. However, the genome-wide significant SNP × NO2 interaction results for MAGI1 did not show statistical significance for interaction in the look-up evaluation, and functional analyses were therefore not pursued. The 2 df test that jointly tested for main genetic and interaction effects is an attractive method in genome-wide interaction studies (28). It was primarily developed to detect main effects while fully taking the environmental exposure into account, and has been successfully used in large-scale lung function studies (45). However, our analyses revealed no statistically significant hits at the genome-wide level, and limited power might have contributed to these results. The choice of method to detect interactions depends on study aims and availability of data, and from our study, it was difficult to draw conclusions about any preferred model. The main focus in our study was to perform functional interaction follow-up analyses on promising hits identified in the GWIS analyses, which we believe, is of crucial importance.
In the diesel exposure study on adults, methylation changes were most notable for CpG sites in the DLG2 gene, with reduced methylation levels at most sites. Analyses of long-term NO2 exposure and DNA methylation profiles also indicated an association between air pollution exposure and DLG2 methylation changes. We did not identify any significant association between the top DLG2 SNP rs963146 and DLG2 methylation, which indicated that the difference in DLG2 methylation levels associated with air pollution was not SNP-mediated. NO2 exposure was associated with higher expression levels of DLG2 in blood cells (Table 2), which provided further evidence that exposure might induce functional changes related to this gene. DLG2 (and MAGI1) belong to the membrane-associated guanylate kinase (MAGUK) family (46). Disruption of Drosophila melanogaster DLG results in acute disorganization of epithelial structure, with disruption of intercellular junction formation (46). DLG2 has recently been associated with COPD (38).
Three MOCOS SNPs were nominally significant in the CHS study, but we did not find convincing data in our functional analyses to support G × E interactions of importance.
FANTOM5 (Functional Annotation of the Mammalian Genome 5) (47) and the Human Protein Atlas (HPA) (48) results showed that the identified genes were expressed at mRNA and protein levels in tissues relevant for asthma, although B4GALT5 could not be evaluated for protein expression in HPA (see the online supplement for additional details).
This study included all available data sets that we are aware of, with the required childhood phenotype, exposure, and genetic data needed for interaction analyses. Nevertheless, it would have been preferable to have larger sample sizes for G × E analyses and functional analyses to decrease the likelihood of both type I and type II errors, and inclusion of non-white populations would have increased the generalizability of our results. We acknowledge that none of the identified SNPs were actually genome-wide significant in the discovery data set, and at the same time, were significant in the look-up data sets. Low statistical power is common in studies using GWIS data, and previous GWIS efforts to detect G × E interaction effects for asthma and lung function indicate that new loci are challenging to discover (45, 49, 50).
For all cohorts, exposures were based on modeled outdoor concentrations of NO2 (a surrogate for traffic-related pollution) at the home and school addresses, but personal exposure to different pollution components, including indoor exposures, were not considered. NO2 level is a good indicator for local air pollution, mainly from motor vehicles, and is highly correlated with other components of motor vehicle emissions, such as exhaust particles (21). However, we observed quite heterogeneous interaction effects in the discovery and look-up data sets, and only ADCY2 SNPs showed similar directions of effect in the discovery and main look-up study. Differences in the levels or constituents of air pollutants, co-exposures, and unmeasured confounding factors between the North American and European cohorts could possibly explain the observed results. We also acknowledge that we used a rather liberal and unspecific definition of asthma [similar to the GABRIEL (A Multidisciplinary Study to Identify the Genetic and Environmental Causes of Asthma in the European Community) GWAS (2)], and the maximum age of asthma definition in CHS was up to 3 years older compared with the other cohorts, which might have contributed to heterogeneous effects (51). Because of these differences, we did not perform meta-analysis of the interaction βs, but we did present interaction βs and P values for each data set.
Previous G × E interaction analyses using candidate gene approaches suggested that genes related to antioxidative stress systems, inflammation, and innate immunity (e.g., GSTP1, TNF and TLR2/4) are important effect modifiers (12, 13). These genes were not among the top hits in our GWIS, but this did not exclude true interaction effects for key SNPs as previously reported.
A key strength of our study was the extensive functional follow-up, and we provided data for asthma that indicated the involvement of identified genes at the genomic, epigenomic, and transcriptomic levels in both lung tissue and peripheral blood cells in relation to air pollution exposure. These results were unlikely to be biased due to ethnic differences in our study populations, because all data were based on an ethnically homogenous (European or non-Hispanic white ancestry) population. In all steps of our study, we corrected for multiple testing to minimize false positive findings.
Our G × E analysis using genome-wide data and multiple functional DNA methylation and gene expression analyses provided promising results for further understanding of the pathogenesis of childhood asthma. Our results supported the notion that G × E interactions are important for asthma development, and that functional genomics analyses in conjunction with detailed environmental exposures provide valuable insight about pathophysiologic mechanisms.
Acknowledgments
Acknowledgment
The authors thank all the families for their participation in the BAMSE, GINIplus, and LISAplus studies. In addition, the authors thank Eva Hallner, André Lauber, and Sara Nilsson at the BAMSE office for invaluable support. The authors further thank all members of the GINIplus and LISAplus Study Groups for their excellent work. The LISAplus Study Group consists of the following: Helmholtz Zentrum Muenchen – German Research Center for Environment and Health, Institute of Epidemiology I, Neuherberg (J. Heinrich, H. E. Wichmann, S. Sausenthaler, and C.-M. Chen); Department of Pediatrics (M. Borte) and Department of Environmental Medicine and Hygiene (O. Herbarth), University of Leipzig; Department of Pediatrics, Marien-Hospital, Wesel (A. von Berg); Bad Honnef (B. Schaaf); UFZ-Centre for Environmental Research Leipzig-Halle, Department of Environmental Immunology (I. Lehmann); IUF – Leibniz Research Institute for Environmental Medicine, Düsseldorf (U. Krämer); Department of Pediatrics, Technical University, Munich (C. P. Bauer and U. Hoffman). The GINIplus Study Group consists of the following: Helmholtz Zentrum Muenchen – German Research Center for Environmental Health, Institute of Epidemiology I, Munich (J. Heinrich, H. E. Wichmann, S. Sausenthaler, C.-M. Chen, E. Thiering, C. Tiesler C, M. Standl, M. Schnappinger, and P. Rzehak); Department of Pediatrics, Marien-Hospital, Wesel (D. Berdel, A. von Berg, C. Beckmann, and I. Groß); Department of Pediatrics, Ludwig Maximilians University, Munich (S. Koletzko, D. Reinhardt, and S. Krauss-Etschmann); Department of Pediatrics, Technical University, Munich (C. P. Bauer, I. Brockow, A., Grübl, and U. Hoffmann); IUF – Leibniz Research Institute for Environmental Medicine, Düsseldorf (U. Krämer, E. Link, and C. Cramer); Centre for Allergy and Environment, Technical University, Munich (H. Behrendt). The PIAMA birth cohort study is a collaboration of the Institute for Risk Assessment Sciences, Utrecht University (B. Brunekreef), Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht (H. A. Smit), Centre for Prevention and Health Services Research, National Institute for Public Health and the Environment, Bilthoven (A. H. Wijga), Department of Pediatrics, Division of Respiratory Medicine, Erasmus MC–Sophia, Rotterdam (J. C. de Jongste), Pulmonology (D. S. Postma) and Pediatric Pulmonology and Pediatric Allergology (G. H. Koppelman) of the University Medical Center Groningen and the Department of Immunopathology, Sanquin Research, Amsterdam (R. C. Aalberse), the Netherlands. The study team gratefully acknowledges the participants in the PIAMA birth cohort study, and all coworkers who helped conduct the medical examinations, field work, and data management. The authors acknowledge Denise Daley and the AllerGen Genetics team for assistance with CAPPS and SAGE data management and transfer.
Footnotes
BAMSE was supported by The Swedish Research Council (G.P. and E. Melén), The Swedish Heart-Lung Foundation, Freemason Child House Foundation in Stockholm, Centre for Allergy Research (E. Melén), Stockholm County Council (ALF), the Strategic Research Program (SFO) in Epidemiology at Karolinska Institutet (E. Melén), MeDALL (Mechanisms of the Development of Allergy) a collaborative project conducted within the European Union (grant agreement No. 261357; J.B. and J.M.A., coordinators), Swedish Foundation for Strategic Research (SSF RBc08-0027) (J.K.), The Swedish Research Council Formas (G.P.), and the Swedish Environment Protection Agency (G.P. and T.B.). GINI and LISA had personal and financial support by the Munich Center of Health Sciences (MCHEALTH) as part of the Ludwig-Maximilians University Munich LMU innovative (J.H.). The PIAMA study was funded by grants from the Dutch Asthma Foundation (grants 3.4.01.26, 3.2.06.022, 3.4.09.081, and 3.2.10.085CO), the ZON-MW Netherlands Organization for Health Research and Development (grant 912-03-031), the Stichting Astmabestrijding and the Ministry of the Environment. Genome-wide genotyping was funded by the European Commission as part of GABRIEL (A Multidisciplinary Study to Identify the Genetic and Environmental Causes of Asthma in the European Community) contract number 018996 under the Integrated Program LSH-2004-1.2.5-1 (Post Genomic Approaches to Understand the Molecular Basis of Asthma Aiming at a Preventive or Therapeutic Control) (D.P., B.B., and G.H.K.). The CHS has been supported in part by grants from the National Institute of Environmental Health Sciences (grants ES #011627, #07048, and #022719) and the NHLBI (grant HL #087680) (W.J.G.). The CAPPS was supported by the Canadian Institutes of Health Research, the British Columbia Lung Association, and the Manitoba Medical Service Foundation. The SAGE was supported by the Canadian Institutes of Health Research. The Traffic Asthma and Genetics collaboration was supported by the AllerGen Networks of Centres of Excellence (M. Brauer, C.C., A. Becker, and A.L.K.). The lung eQTL study at Laval University was supported by the Chaire de pneumologie de la Fondation JD Bégin de l’Université Laval, the Fondation de l’Institut universitaire de cardiologie et de pneumologie de Québec, the Respiratory Health Network of the FRQS, the Canadian Institutes of Health Research (MOP-123369), and the Cancer Research Society and Read for the Cure (Y.B.). The CNRS (N.L., J.P., S.B., and C.A.) and the FP7-MeDALL Consortium were supported by grant agreement FP7 No. 264357. Y.B. is the recipient of a Junior 2 Research Scholar award from the Fonds de recherche Québec – Santé (FRQS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions: E. Melén, G.P., C.C., and M. Brauer designed the study. M. Bottai provided statistical competence in the study design. A. Bergström, G.P., and E. Melén coordinated BAMSE (Children, Allergy, Milieu, Stockholm, Epidemiological Survey). T.B. and G.P. coordinated exposure assessment in BAMSE, and O.G. and M.K. performed exposure assessment. A.K. performed imputation of the BAMSE genome-wide association study (GWAS) data. A.G. performed genome-wide interaction study (GWIS) statistical analysis in BAMSE and CAPPS (Canadian Asthma Primary Prevention Study)/SAGE (Study of Asthma, Genetics and Environment) and meta-analysis. A.S. provided knowledge of statistical analysis of BAMSE GWIS data. J.H. coordinated the German Infant Study on the Influence of Nutrition Intervention Plus Environmental and Genetic Influences on Allergy Development (GINIplus) and the Influence of Life-Style Factors on the Development of the Immune System and Allergies in East and West Germany Plus the Influence of Traffic Emissions and Genetics (LISAplus) study. E.F. performed exposure assessment in GINI/LISA. C.M.T.T. performed imputation of the GINI/LISA GWAS data, and M.S. performed statistical analysis. B.B., G.H.K., and D.P. coordinated PIAMA (Prevention and Incidence of Asthma and Mite Allergy study). J.M.V. contributed with PIAMA data. D.I. performed statistical analysis in PIAMA. W.J.G. coordinated CHS (Children’s Health Study). H.V. performed statistical analysis in CHS. M.C.-Y. coordinated CAPPS. A. Becker and C.C. coordinated CAPPS and SAGE. M. Brauer coordinated exposure assessment in CAPPS/SAGE, and E. MacIntyre and M. Brauer performed exposure data linkage. A.L.K. contributed to CAPPS and SAGE data. J.K. coordinated methylation analysis in BAMSE. C.S. provided BAMSE methylation data, S.K.M., A.K., and O.G. performed quality control, and A.G. and O.G. performed methylation analysis. C.J.-X. and G.H.K. provided methylation quality control and analysis protocols. C.C. coordinated the short-term diesel exhaust exposure study. M.S.K. coordinated the methylation analysis. R.J. and M.J. performed statistical analysis. D.P. coordinated lung expression quantitative trait locus (eQTL) data. M.v.d.B. provided lung eQTL data from Groningen and performed statistical analysis of lung eQTL data. Y.B. provided lung eQTL data from Quebec City, and M.O. provided lung eQTL data from Vancouver. D.N. oversaw genotyping and gene expression measurements for the lung eQTL data set. J.M.A. and J.B. coordinated the MeDALL data. S.B., N.L., J.P., S.J., C.M., and C.A. provided transcriptomics data for BAMSE. S.K.M. performed quality control and statistical analysis. All authors were involved in data interpretation and drafting of the manuscript.
This article has an online data supplement, which is accessible from this issue's table of content online at www.atsjournals.org
Originally Published in Press as DOI: 10.1164/rccm.201605-1026OC on November 30, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Asher MI, Montefort S, Björkstén B, Lai CK, Strachan DP, Weiland SK, Williams H ISAAC Phase Three Study Group. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhinoconjunctivitis, and eczema in childhood: ISAAC Phases One and Three repeat multicountry cross-sectional surveys. Lancet. 2006;368:733–743. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- 2.Moffatt MF, Gut IG, Demenais F, Strachan DP, Bouzigon E, Heath S, von Mutius E, Farrall M, Lathrop M, Cookson WO GABRIEL Consortium. A large-scale, consortium-based genomewide association study of asthma. N Engl J Med. 2010;363:1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlsten C, Melén E. Air pollution, genetics, and allergy: an update. Curr Opin Allergy Clin Immunol. 2012;12:455–460. doi: 10.1097/ACI.0b013e328357cc55. [DOI] [PubMed] [Google Scholar]
- 4.Gauderman WJ, Vora H, McConnell R, Berhane K, Gilliland F, Thomas D, Lurmann F, Avol E, Kunzli N, Jerrett M, et al. Effect of exposure to traffic on lung development from 10 to 18 years of age: a cohort study. Lancet. 2007;369:571–577. doi: 10.1016/S0140-6736(07)60037-3. [DOI] [PubMed] [Google Scholar]
- 5.Gehring U, Gruzieva O, Agius RM, Beelen R, Custovic A, Cyrys J, Eeftens M, Flexeder C, Fuertes E, Heinrich J, et al. Air pollution exposure and lung function in children: the ESCAPE project. Environ Health Perspect. 2013;121:1357–1364. doi: 10.1289/ehp.1306770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultz ES, Hallberg J, Bellander T, Bergström A, Bottai M, Chiesa F, Gustafsson PM, Gruzieva O, Thunqvist P, Pershagen G, et al. Early-life exposure to traffic-related air pollution and lung function in adolescence. Am J Respir Crit Care Med. 2016;193:171–177. doi: 10.1164/rccm.201505-0928OC. [DOI] [PubMed] [Google Scholar]
- 7.Rice MB, Rifas-Shiman SL, Litonjua AA, Oken E, Gillman MW, Kloog I, Luttmann-Gibson H, Zanobetti A, Coull BA, Schwartz J, et al. Lifetime exposure to ambient pollution and lung function in children. Am J Respir Crit Care Med. 2016;193:881–888. doi: 10.1164/rccm.201506-1058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gruzieva O, Bergström A, Hulchiy O, Kull I, Lind T, Melén E, Moskalenko V, Pershagen G, Bellander T. Exposure to air pollution from traffic and childhood asthma until 12 years of age. Epidemiology. 2013;24:54–61. doi: 10.1097/EDE.0b013e318276c1ea. [DOI] [PubMed] [Google Scholar]
- 9.Gehring U, Wijga AH, Hoek G, Bellander T, Berdel D, Brüske I, Fuertes E, Gruzieva O, Heinrich J, Hoffmann B, et al. Exposure to air pollution and development of asthma and rhinoconjunctivitis throughout childhood and adolescence: a population-based birth cohort study. Lancet Respir Med. 2015;3:933–942. doi: 10.1016/S2213-2600(15)00426-9. [DOI] [PubMed] [Google Scholar]
- 10.Mölter A, Simpson A, Berdel D, Brunekreef B, Custovic A, Cyrys J, de Jongste J, de Vocht F, Fuertes E, Gehring U, et al. A multicentre study of air pollution exposure and childhood asthma prevalence: the ESCAPE project. Eur Respir J. 2015;45:610–624. doi: 10.1183/09031936.00083614. [DOI] [PubMed] [Google Scholar]
- 11.Bharadwaj P, Graff Zivin J, Mullins JT, Neidell M. Early life exposure to the great smog of 1952 and the development of asthma. Am J Respir Crit Care Med. doi: 10.1164/rccm.201603-0451OC. [online ahead of print] 8 July 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerkhof M, Postma DS, Brunekreef B, Reijmerink NE, Wijga AH, de Jongste JC, Gehring U, Koppelman GH. Toll-like receptor 2 and 4 genes influence susceptibility to adverse effects of traffic-related air pollution on childhood asthma. Thorax. 2010;65:690–697. doi: 10.1136/thx.2009.119636. [DOI] [PubMed] [Google Scholar]
- 13.MacIntyre EA, Brauer M, Melén E, Bauer CP, Bauer M, Berdel D, Bergström A, Brunekreef B, Chan-Yeung M, Klümper C, et al. TAG Study Group. GSTP1 and TNF gene variants and associations between air pollution and incident childhood asthma: the Traffic, Asthma and Genetics (TAG) study. Environ Health Perspect. 2014;122:418–424. doi: 10.1289/ehp.1307459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gref A, Merid S, Gruzieva O, Ballereau S, Becker A, Bellander T, Bergström A, Bossé Y, Bottai M, Chan-Yeung M, et al. An integrative genomics approach identifies new asthma pathways related to air pollution. Eur Respir J. 2015;46:OA1455. [Google Scholar]
- 15.Berg A, Kramer U, Link E, Bollrath C, Heinrich J, Brockow I, Koletzko S, Grubl A, Filipiak-Pittroff B, Wichmann HE, et al. GINIplus Study Group. Impact of early feeding on childhood eczema: development after nutritional intervention compared with the natural course - the GINIplus study up to the age of 6 years. Clin Exp Allergy. 2010;40:627–636. doi: 10.1111/j.1365-2222.2009.03444.x. [DOI] [PubMed] [Google Scholar]
- 16.Heinrich J, Bolte G, Hölscher B, Douwes J, Lehmann I, Fahlbusch B, Bischof W, Weiss M, Borte M, Wichmann HE LISA Study Group. Allergens and endotoxin on mothers’ mattresses and total immunoglobulin E in cord blood of neonates. Eur Respir J. 2002;20:617–623. doi: 10.1183/09031936.02.02322001. [DOI] [PubMed] [Google Scholar]
- 17.Wijga AH, Kerkhof M, Gehring U, de Jongste JC, Postma DS, Aalberse RC, Wolse AP, Koppelman GH, van Rossem L, Oldenwening M, et al. Cohort profile: the prevention and incidence of asthma and mite allergy (PIAMA) birth cohort. Int J Epidemiol. 2014;43:527–535. doi: 10.1093/ije/dys231. [DOI] [PubMed] [Google Scholar]
- 18.Carlsten C, Dybuncio A, Becker A, Chan-Yeung M, Brauer M. Traffic-related air pollution and incident asthma in a high-risk birth cohort. Occup Environ Med. 2011;68:291–295. doi: 10.1136/oem.2010.055152. [DOI] [PubMed] [Google Scholar]
- 19.Kozyrskyj AL, HayGlass KT, Sandford AJ, Paré PD, Chan-Yeung M, Becker AB. A novel study design to investigate the early-life origins of asthma in children (SAGE study) Allergy. 2009;64:1185–1193. doi: 10.1111/j.1398-9995.2009.02033.x. [DOI] [PubMed] [Google Scholar]
- 20.Peters JM, Avol E, Navidi W, London SJ, Gauderman WJ, Lurmann F, Linn WS, Margolis H, Rappaport E, Gong H, et al. A study of twelve Southern California communities with differing levels and types of air pollution: I. Prevalence of respiratory morbidity. Am J Respir Crit Care Med. 1999;159:760–767. doi: 10.1164/ajrccm.159.3.9804143. [DOI] [PubMed] [Google Scholar]
- 21.Beelen R, Hoek G, Vienneau D, Eeftens M, Dimakopoulou K, Pedeli X, Tsai MY, Kunzli N, Schikowski T, Marcon A, et al. Development of NO2 and NOx land use regression models for estimating air pollution exposure in 36 study areas in Europe: The ESCAPE project. Atmos Environ. 2013;72:10–23. [Google Scholar]
- 22.Henderson SB, Beckerman B, Jerrett M, Brauer M. Application of land use regression to estimate long-term concentrations of traffic-related nitrogen oxides and fine particulate matter. Environ Sci Technol. 2007;41:2422–2428. doi: 10.1021/es0606780. [DOI] [PubMed] [Google Scholar]
- 23.Allen RW, Amram O, Wheeler AJ, Brauer M. The transferability of NO and NO2 land use regression models between cities and pollutants. Atmos Environ. 2011;45:369–378. [Google Scholar]
- 24.McConnell R, Berhane K, Gilliland F, Molitor J, Thomas D, Lurmann F, Avol E, Gauderman WJ, Peters JM. Prospective study of air pollution and bronchitic symptoms in children with asthma. Am J Respir Crit Care Med. 2003;168:790–797. doi: 10.1164/rccm.200304-466OC. [DOI] [PubMed] [Google Scholar]
- 25.Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murcray CE, Lewinger JP, Gauderman WJ. Gene-environment interaction in genome-wide association studies. Am J Epidemiol. 2009;169:219–226. doi: 10.1093/aje/kwn353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kraft P, Yen YC, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered. 2007;63:111–119. doi: 10.1159/000099183. [DOI] [PubMed] [Google Scholar]
- 29.Wu K, Gamazon ER, Im HK, Geeleher P, White SR, Solway J, Clemmer GL, Weiss ST, Tantisira KG, Cox NJ, et al. Genome-wide interrogation of longitudinal FEV1 in children with asthma. Am J Respir Crit Care Med. 2014;190:619–627. doi: 10.1164/rccm.201403-0460OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao K, Bossé Y, Nickle DC, Paré PD, Postma DS, Laviolette M, Sandford A, Hackett TL, Daley D, Hogg JC, et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 2012;8:e1003029. doi: 10.1371/journal.pgen.1003029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamontagne M, Timens W, Hao K, Bossé Y, Laviolette M, Steiling K, Campbell JD, Couture C, Conti M, Sherwood K, et al. Genetic regulation of gene expression in the lung identifies CST3 and CD22 as potential causal genes for airflow obstruction. Thorax. 2014;69:997–1004. doi: 10.1136/thoraxjnl-2014-205630. [DOI] [PubMed] [Google Scholar]
- 32.GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bousquet J, Anto JM, Akdis M, Auffray C, Keil T, Momas I, Postma DS, Valenta R, Wickman M, Cambon-Thomsen A, et al. Paving the way of systems biology and precision medicine in allergic diseases: the MeDALL success story: Mechanisms of the Development of ALLergy; EU FP7-CP-IP; Project No: 261357; 2010-2015. Allergy. 2016;71:1513–1525. doi: 10.1111/all.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruzieva O, Xu CJ, Breton CV, Annesi-Maesano I, Antó JM, Auffray C, Ballereau S, Bellander T, Bousquet J, Bustamante M, et al. Epigenome-wide meta-analysis of methylation in children related to prenatal NO2 air pollution exposure. Environ Health Perspect. 2017;125(1):104–110. doi: 10.1289/EHP36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang R, Jones MJ, Sava F, Kobor MS, Carlsten C. Short-term diesel exhaust inhalation in a controlled human crossover study is associated with changes in DNA methylation of circulating mononuclear cells in asthmatics. Part Fibre Toxicol. 2014;11:71. doi: 10.1186/s12989-014-0071-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gruzieva O, Merid SK, Melén E. An update on epigenetics and childhood respiratory diseases. Paediatr Respir Rev. 2014;15:348–354. doi: 10.1016/j.prrv.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Birger N, Gould T, Stewart J, Miller MR, Larson T, Carlsten C. The Air Pollution Exposure Laboratory (APEL) for controlled human exposure to diesel exhaust and other inhalants: characterization and comparison to existing facilities. Inhal Toxicol. 2011;23:219–225. doi: 10.3109/08958378.2011.562256. [DOI] [PubMed] [Google Scholar]
- 38.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, Franceschini N, van Durme YM, Chen TH, Barr RG, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat Genet. 2010;42:45–52. doi: 10.1038/ng.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castaldi PJ, Cho MH, Litonjua AA, Bakke P, Gulsvik A, Lomas DA, Anderson W, Beaty TH, Hokanson JE, Crapo JD, et al. COPDGene and Eclipse Investigators. The association of genome-wide significant spirometric loci with chronic obstructive pulmonary disease susceptibility. Am J Respir Cell Mol Biol. 2011;45:1147–1153. doi: 10.1165/rcmb.2011-0055OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen W, Brehm JM, Manichaikul A, Cho MH, Boutaoui N, Yan Q, Burkart KM, Enright PL, Rotter JI, Petersen H, et al. A genome-wide association study of chronic obstructive pulmonary disease in Hispanics. Ann Am Thorac Soc. 2015;12:340–348. doi: 10.1513/AnnalsATS.201408-380OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tokuda N, Numata S, Li X, Nomura T, Takizawa M, Kondo Y, Yamashita Y, Hashimoto N, Kiyono T, Urano T, et al. β4GalT6 is involved in the synthesis of lactosylceramide with less intensity than β4GalT5. Glycobiology. 2013;23:1175–1183. doi: 10.1093/glycob/cwt054. [DOI] [PubMed] [Google Scholar]
- 42.Ha SG, Ge XN, Bahaie NS, Kang BN, Rao A, Rao SP, Sriramarao P. ORMDL3 promotes eosinophil trafficking and activation via regulation of integrins and CD48. Nat Commun. 2013;4:2479. doi: 10.1038/ncomms3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panasevich S, Melén E, Hallberg J, Bergström A, Svartengren M, Pershagen G, Nyberg F. Investigation of novel genes for lung function in children and their interaction with tobacco smoke exposure: a preliminary report. Acta Paediatr. 2013;102:498–503. doi: 10.1111/apa.12204. [DOI] [PubMed] [Google Scholar]
- 44.Facciuto F, Cavatorta AL, Valdano MB, Marziali F, Gardiol D. Differential expression of PDZ domain-containing proteins in human diseases: challenging topics and novel issues. FEBS J. 2012;279:3538–3548. doi: 10.1111/j.1742-4658.2012.08699.x. [DOI] [PubMed] [Google Scholar]
- 45.Hancock DB, Soler Artigas M, Gharib SA, Henry A, Manichaikul A, Ramasamy A, Loth DW, Imboden M, Koch B, McArdle WL, et al. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 2012;8:e1003098. doi: 10.1371/journal.pgen.1003098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts S, Delury C, Marsh E. The PDZ protein discs-large (DLG): the ‘Jekyll and Hyde’ of the epithelial polarity proteins. FEBS J. 2012;279:3549–3558. doi: 10.1111/j.1742-4658.2012.08729.x. [DOI] [PubMed] [Google Scholar]
- 47.Severin J, Lizio M, Harshbarger J, Kawaji H, Daub CO, Hayashizaki Y, Bertin N, Forrest AR FANTOM Consortium. Interactive visualization and analysis of large-scale sequencing datasets using ZENBU. Nat Biotechnol. 2014;32:217–219. doi: 10.1038/nbt.2840. [DOI] [PubMed] [Google Scholar]
- 48.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 49.Melén E, Bottai M. On lung function and interactions using genome-wide data. PLoS Genet. 2012;8:e1003174. doi: 10.1371/journal.pgen.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scholtens S, Postma DS, Moffatt MF, Panasevich S, Granell R, Henderson AJ, Melén E, Nyberg F, Pershagen G, Jarvis D, et al. GABRIELA Study Group. Novel childhood asthma genes interact with in utero and early-life tobacco smoke exposure. J Allergy Clin Immunol. 2014;133:885–888. doi: 10.1016/j.jaci.2013.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sbihi H, Koehoorn M, Tamburic L, Brauer M. Asthma trajectories in a population-based birth cohort: impacts of air pollution and greenness. Am J Respir Crit Care Med. 2017;195:607–613. doi: 10.1164/rccm.201601-0164OC. [DOI] [PubMed] [Google Scholar]