Abstract
Mate choice can play a pivotal role in the nature and extent of reproductive isolation between species. Mating preferences are often dependent on an individual's social experience with adult phenotypes throughout development. We show that olfactory preference in a swordtail fish (Xiphophorus malinche) is affected by previous experience with adult olfactory signals. We compare transcriptome-wide gene expression levels of pooled sensory and brain tissues between three treatment groups that differ by social experience: females with no adult exposure, females exposed to conspecifics and females exposed to heterospecifics. We identify potential functionally relevant genes and biological pathways differentially expressed not only between control and exposure groups, but also between groups exposed to conspecifics and heterospecifics. Based on our results, we speculate that vomeronasal receptor type 2 paralogs may detect species-specific pheromone components and thus play an important role in reproductive isolation between species.
Keywords: olfaction, sexual imprinting, learning, Xiphophorus, reproductive isolation, synaptic plasticity
1. Introduction
Mate preferences can be influenced by social learning throughout life [1]. Experience with a particular phenotype can elicit preferences either for or against that trait, which can have important ecological and evolutionary consequences by promoting assortative mating [2,3] or outcrossing by what Darwin called ‘antipathy’ [4], respectively. Individuals can be genetically predisposed to favour early learning of familiar stimuli [5] or may favour traits that are rare or novel [6,7]. In certain scenarios, learned preferences can strengthen reproductive isolation between species [8–11] and can be an important mechanism in preventing hybridisation. For example, in sticklebacks learned preferences for familiar odour and colour act to minimize hybridisation between benthic and limnetic ecotypes [8]. Alternately, in certain contexts learned preferences can promote hybridisation (reviewed in [12]). For example, in Darwin's finches, mis-imprinting on heterospecific songs promotes hybridisation [13]. In zebra finches, mating preferences for hybrid individuals can further be specified through exposure to a mixed social environment early in life [14]. These studies show that learned mate preferences can have a major impact on reproductive isolation between species. Determining the proximate mechanisms underlying learned preferences will help us understand their evolutionary impact.
In vertebrates, notably fishes, olfactory cues are important to conspecific mating preferences. In insects, odorant receptors at the periphery play a determining role in mate choice. The vertebrate olfactory periphery has the potential to play a similar role: it includes a relatively large repertoire of olfactory receptors (OR), allowing for the evolution of specialized receptor families, such as olfactory receptors and vomeronasal receptors (vomeronasal receptor type 1, V1R and vomeronasal receptor type 2, V2R). This receptor diversity at the olfactory periphery allows for the evolution of specific behavioural responses to unique cues.
As with other cues, response to olfactory cues depends on previous experience. A compelling mechanism for experience-dependent plasticity in olfactory responses is the ‘tuning’ of olfactory receptor neurons (ORNs) in the sensory periphery. Electrophysiological studies in salmon [15] and zebrafish [16] found that exposure to artificially supplied chemicals during development altered sensitivity to these odorants later in life. In zebrafish, learned sensitivity to new odours was accompanied by increased expression of genes involved in odorant receptor cell neurogenesis [16]. In mice, fear conditioning with acetophenone results in an increased number of ORNs expressing an acetophenone receptor. This increase can even be passed on to offspring through epigenetic mechanisms [17]. More generally, olfactory sensitivity at the periphery can be mediated by differential proliferation of ORNs sensitive to specific odorants, which is associated with stable differential expression (DE) of the OR genes [18]. In zebrafish, OR mRNA levels are highly correlated with their respective sensory neuron counts [19]. We therefore sought to test whether learned differences in olfactory mating preferences are associated with DE of odorant receptor genes with RNAseq.
ORNs project to the olfactory bulb, which further project throughout the forebrain and other brain regions. Individuals have been shown to have a neuronally plastic response in the forebrain to varying levels of social exposure [20–22]. For example, visual familiarization to a potential mating partner activates terminal nerve gonadotropin releasing hormone 3 neurons, leading to preference for familiar individuals in medaka fish [23]. Hence, environmental cues detected by the sensory periphery often have downstream effects at the structural and transcriptomic levels. Therefore, we included a portion of the forebrain in our genetic analysis to examine the effect of social exposure on the gene expression profile of first-order processing brain regions such as the olfactory bulb.
Swordtail fishes (genus Xiphophorus) rely primarily on olfactory signalling for conspecific mate preference [24–27]. In the naturally hybridising sister species [28] Xiphophorus malinche and Xiphophorus birchmanni, wild-caught females prefer the chemical cues of conspecific males [29]. For X. birchmanni, the preference for conspecifics is dependent on both early [16] and recent [30] experience with conspecifics; females prefer familiar olfactory cues regardless of whether they are conspecific or heterospecific. Short-term exposure of X. malinche has the opposite effect, with familiarity breeding contempt: females exposed to X. birchmanni preferred X. malinche scent [31]. This stark difference in how these sister species respond to social experience may suggest distinct neural processes beyond the sensory periphery and has motivated us to test whether X. malinche females showed similar disdain for cues learned before sexual maturity and to explore the neurogenomic processes responsible for modulating the effects of social experience on mating preference.
In the present study, we perform long-term exposure of female X. malinche to different visual and olfactory adult cues starting early in life through sexual maturity. We examine the effects of this exposure on behavioural mating preferences and use RNA sequencing to quantify the effects of learning on gene expression in the forebrain and sensory periphery. We show that social experience induces major gene expression changes in X. malinche. A smaller subset of genes was differentially expressed between conspecific and heterospecific treatments, indicating differential neurogenomic responses to species-typical adult cues. These genes were enriched for gene ontology (GO) categories tied to behaviour, neurogenesis and synaptic transmission. Also, we found enrichment of three biological pathways with previously established roles in addiction, emotional learning and preference for familiar individuals. Intriguingly, we found upregulation of a rapidly evolving pheromone receptor gene in response to conspecific exposure.
2. Material and methods
(a). Fish collection and exposure treatments
Offspring of 16 wild-caught X. malinche females were reared under one of three treatment groups: (i) M-EXP (starting N = 33 both sexes, females surviving to adulthood N = 17) visually and chemically exposed to two males and two females of adult X. malinche from the Chicayotla locality [29]; (ii) B-EXP (starting N = 34 both sexes, surviving females N = 18) exposed to two males and two females of adult X. birchmanni from the Garces locality [29] and (iii) controls (starting N = 55 both sexes, surviving females N = 26), which did not receive adult stimulus exposure. Once all females had matured, we tested female preference for visual and olfactory cues of X. birchmanni and X. malinche. After all behavioural trials were concluded, females were rinsed in tank water and returned to their respective treatment for an additional two months before sample collection to minimize possible short-term effects from behavioural trials. To compare the preferences of lab-reared and naturally raised individuals, we also tested mate preference of wild-caught X. malinche females. For detailed methods on rearing conditions, see electronic supplementary material, I.
(b). Preference trials
To assess visual preference, we used previously described computer animation playback techniques [27,30,32] and a well-established dichotomous choice design [27,30,32] to test focal females' preferences for X. birchmanni versus X. malinche visual cues, using association time as a proxy for preference. We tested 26 (19 responsive) control, 18 (18 responsive) B-EXP and 17 (13 responsive) M-EXP females.
Two days after the visual trials, we tested female preference for conspecific versus heterospecific male odours following a protocol described in previous studies [24,26,27]. Briefly, olfactory cues obtained from model conspecific and heterospecific males were dripped on either side of the trial tank according to species. To account for potential side biases, each female was tested twice back-to-back with the location of each cue switched. We averaged the association time in the two trials for data analysis. If the female was unresponsive (did not visit both cue zones within 5 min), we only included association time from the responsive trial in analysis. We tested 26 (18 responsive) control, 18 (17 responsive) B-EXP and 17 (17 responsive) M-EXP females. Wild-caught females described above were also tested for olfactory preference (N = 10 responsive).
We tested the normality of association time datasets using Shapiro–Wilk tests. For datasets fitting the normality assumption, we used paired Student's t-tests to detect differences in mean association time between two stimuli for each group. Unpaired t-tests assuming equal variance were used to test for group-wise differences. A one-way ANOVA on preference index (time with X. malinche—time with X. birchmanni) was used to test whether exposure experience significantly affected mate preference. We used non-parametric tests (Wilcoxon signed-rank tests and Kruskal–Wallis rank-sum tests) for datasets violating the normality assumption. For detailed methods on preference trials and analysis, see electronic supplementary material, II.
(c). Tissue sample collection
We randomly selected four females from each of the treatment groups (M-EXP, B-EXP and control, 12 samples). Females were euthanized with an overdose of MS-222; whole heads were preserved in Trizol solution and stored at −80°C until use. Females were dissected to ensure that females carried unfertilized eggs (and thus were sexually mature and showed no evidence of physical contact with males) and showed no sign of parasitic infection. Chemoreceptive tissue was dissected from the head by making a single 45° cut with dissection scissors in front of the anterior edge of the orbits. This tissue sample includes the lips, tongue, olfactory epithelium, olfactory bulb and the anterior portion of the telencephalon (electronic supplementary material, figure S1). Specifically, this sample includes the medial and lateral zones of the dorsal telencephalon (Dm and Dl), regions previously known to have important roles in the mesolimbic reward system [33] and mate choice decision-making [34] in teleosts. Pooling different tissues in the same library was required to obtain sufficient RNA for library preparation. Our results on differential gene expression should be considered conservative because pooling decreases power due to the dilution of transcript copies [35].
(d). RNA extraction and library preparation
Total RNA extraction followed a Trizol protocol. Sequencing library was constructed with Illumina's TruSeq mRNA Sample Prep Kit with minor modifications. Trimmomatic [36] was used to clean the raw reads (electronic supplementary material, III).
(e). Read mapping
Reads were mapped to a pseudogenome assembly (heterozygous sites hard-masked) of X. malinche at approximately 35X coverage [28,37] with TopHat 2.0.10 (electronic supplementary material, III). We repeated mapping to the Xiphophorus maculatus reference genome (a de novo assembly [38]) to confirm robustness of results (electronic supplementary material, IV). We also confirmed results by choosing a different mapping algorithm (electronic supplementary material, V).
(f). Odorant receptor identification and molecular evolution
We followed a previously described workflow [39,40] to annotate odorant receptor genes in the X. maculatus genome (electronic supplementary material, VI). Briefly, teleost V2R [39], OR [39], trace amine-associated receptor (TAAR) [39] and V1R (from GenBank) sequences (electronic supplementary material, files 1–4) were aligned to the X. maculatus genome with megablast-dc [41], followed by Genewise v. 2.2.0 [42] to predict gene models. In most cases, observed RNAseq reads mapped to these predicted gene models more accurately than to Ensembl (version 74) [43] gene predictions (electronic supplementary material figure S2). The number of odorant receptors found through this method is well within the range of odorant receptor families found throughout teleost fish [44]. Newly predicted odorant receptors were added back to the alignment for the next iteration. Two iterations were performed before no more new V2R, OR, TAAR or V1R sequences were identified (GTF file in electronic supplementary material, file 5, matched to X. maculatus genome, Ensembl version 74). Phylogenies of the identified sequences were reconstructed with RAxML v. 7.2.6 [45] with GTR + Gamma model with 100 rapid bootstraps for each family (electronic supplementary material, files 6–8) except for V1R because we found only two paralogs.
We extracted orthologous odorant receptor sequences from polymorphism-masked pseudogenomes of five Xiphophorus species [28,37,38] using the exon structure identified by Genewise. In case multiple paralogs are mapped to the same reference locus, all differing sites are masked and thus conservative for non-synonymous divergence/synonymous divergence ratio (dN/dS) analysis. Sequences containing premature stop codons were discarded from further analysis (11 ORs, 8 TAARs, 0 V1R and 8 V2Rs excluded). We test for diversifying selection using a likelihood ratio test approach in Codeml, comparing M8 versus M8a, and M8 versus M7 models [46,47], with gene trees separately built for each paralog using RAxML [48] (details in electronic supplementary material, VI).
(g). Differential expression analyses
Gene models (v. 74) for X. maculatus were downloaded from Ensembl. Because this file does not contain information for manually annotated genes (described above), we added entries into the gene annotation file (GTF file) for the V2R, OR, TAAR and V1R receptors identified. With this updated GTF file, we counted the number of reads uniquely mapping to each gene using the python package htseq-count (strand-specific: no, mode: union, counted feature: exon), requiring a mapping quality of 20. These raw counts were imported into the edgeR 3.0.8 [49] and DESeq 1.20.0 [50] packages (for DESeq analysis, see electronic supplementary material, VII) for DE analysis. We visualized gene expression profiles of individuals by conducting a constrained correspondence analysis on edgeR normalized gene counts using the vegan package in R [51].
Using edgeR, we performed two sets of analyses for DE by olfactory treatment (i) restricting our analysis to OR genes and (ii) a transcriptome-wide analysis. For the first analysis, we extracted expression levels of transcripts that we had annotated as members of the V2R, OR, TAAR or V1R gene families from edgeR results. We calculated q values using the R package ‘Qvalue’ [52] for all the raw p-values for these genes given by edgeR. Three comparisons were made: B-EXP to M-EXP, B-EXP to Control and M-EXP to Control.
For the second analysis, we followed a previously described analysis protocol [53]. The only modification of this workflow was that we required that at least four individuals had to surpass a 0.5 counts per million threshold in order for a given transcript to be analysed.
Lastly, the inclusion of non-neuronal tissue into our sample could have driven the differentially expressed patterns. To rule out such possibilities, we repeated the edgeR and GO analyses after excluding zebrafish orthologues showing higher expression in the skin and muscle tissue compared to the olfactory epithelium and the brain (see electronic supplementary material, VIII).
(h). qPCR validation of candidate genes
We validated the following genes using qPCR with increased sample size (n = 8 per group): V2R6 and V2R40. Primers were designed with the NCBI primer design tool on predicted coding sequences. We determined the primer efficiency and the total cDNA input to each reaction (electronic supplementary material, IX). We normalize CT values by total input cDNA, and we compare the means between groups using non-parametric tests (Wilcoxon for pairwise and Krusalis–Wallis for three groups; electronic supplementary material, X).
where E is the primer efficiency, Ct is the cycle number to reach the target threshold and D is the amount of cDNA (ng) used in the qPCR reaction.
(i). Gene ontology enrichment and PANTHER pathway analysis
To determine whether particular functional categories and pathways were enriched in genes differentially expressed between the treated and control groups and the birchmanni-exposed and malinche-exposed females, we performed GO enrichment analysis. We used the annotated X. maculatus genome to assign Human Genome Organization (HUGO) gene symbols to each gene. All genes that passed coverage filtering in edgeR, excluding all manually predicted genes (see above), were included as part of the gene universe and we tested for significant enrichment (FDR < 0.05) of different biological processes and pathways by comparing the gene universe to lists of significant genes using the PANTHER Classification System [54] (release 20160321). Briefly, genes are organized into families and subfamilies according to sequence homology and functional similarity. They are then assigned GO terms (‘GO biological processes complete’) and placed within one of 177 ‘PANTHER Pathways’ (for detailed methods, see [55]). Gene lists are compared to the gene universe to find GO terms or pathways that are statistically over-represented or under-represented using a binomial test. We used REVIGO [56] (http://revigo.irb.hr/) to visualize GO categories, clustered by semantic similarities (SimRel).
3. Results
(a). Female preference behaviour
Regardless of learning experience (one-way ANOVA, F1,2 = 0.36, p = 0.70), all three groups of X. malinche females showed preferences for heterospecific X. birchmanni visual cues (control: t18 = 2.85, p = 0.010; X. birchmanni exposed: t17 = 2.34, p = 0.030; X. malinche exposed: t12 = 2.22, p = 0.046, figure 1). Only females exposed to adult X. birchmanni showed a significant olfactory preference for conspecific (X. malinche) male water (Wilcoxon signed-rank, T = 5, p < 0.001, N = 17); other females did not show significant olfactory preferences (control: T = 58.5, p = 0.25, N = 18; M-EXP: T = 73, p = 0.89, N = 17). Early social experience had a significant effect on olfactory preference (Kruskal–Wallis rank-sum test, 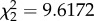 , p = 0.0082), driven by the fact that the B-EXP group preferred X. malinche (conspecific) cues more than the control group (Wilcoxon rank sum, W = 92, p = 0.046) and the M-EXP group (Wilcoxon rank sum, W = 53.5, p = 0.0018, figure 1).
, p = 0.0082), driven by the fact that the B-EXP group preferred X. malinche (conspecific) cues more than the control group (Wilcoxon rank sum, W = 92, p = 0.046) and the M-EXP group (Wilcoxon rank sum, W = 53.5, p = 0.0018, figure 1).
Figure 1.
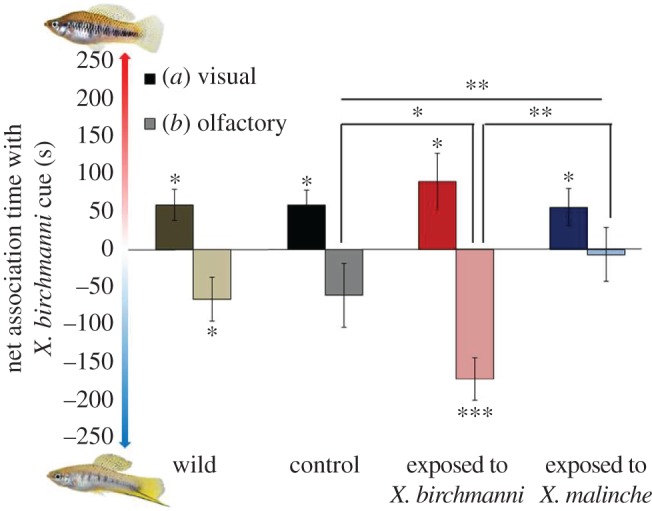
Preferences of female X. malinche varying in early social experience for different stimulus pairs. (a) Visual cues: computer animations representing mean male X. birchmanni and X. malinche phenotypes; (b) olfactory cues: water containing male pheromones of each species. Bar height represents net association time with the heterospecific X. birchmanni ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
(b). Transcriptome-wide differential expression
Social environment had a dramatic effect on gene regulation (figure 2). Following quality trimming, an average of 24 million reads were retained per individual, and 81.3% of these were mapped by TopHat [57] (electronic supplementary material table S1). After coverage filtering with edgeR, 17765 genes were retained. The control group showed gene expression patterns that were clearly distinct from both social exposure groups (figure 2). After false discovery correction at FDR = 0.05, 2248 and 1939 genes were significantly differentially expressed in B-EXP and M-EXP groups compared to the control group, respectively (electronic supplementary material, table S2). Most genes differentially expressed between the treated and control groups were consistent; 58% of genes differentially expressed in the birchmanni-exposed group were shared with the malinche-exposed group and 67% of genes differentially expressed in the malinche-exposed group were shared with the birchmanni-exposed group (electronic supplementary material, figure S3). Of the 1295 genes that were significantly differentially expressed in both olfactory treatment groups, 99% had the same direction of expression relative to the control (e.g. up- or downregulated). Many fewer genes showed differential regulation between conspecific and heterospecific exposures (N = 330, electronic supplementary material, figure S3; table S2). These results remain robust after excluding ‘skin/muscle-specific’ genes identified in zebrafish orthologues (electronic supplementary material, VIII).
Figure 2.
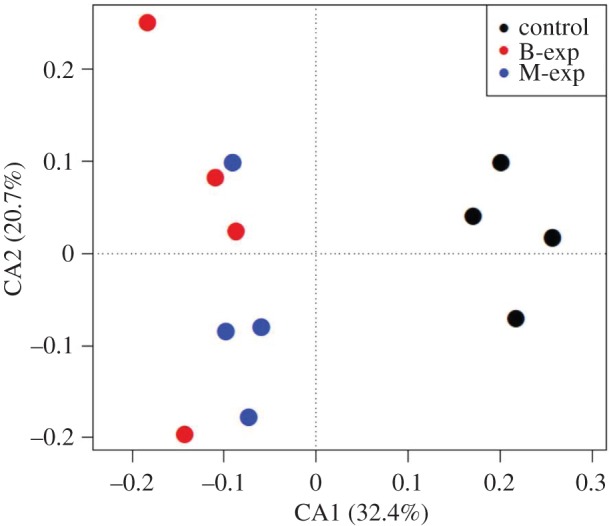
Constrained correspondence analysis plot of gene expression dimensions 1 and 2 including all genes passing 0.5 cpm filter in at least four individuals. Each point represents an individual used for the RNAseq analysis. Standard lengths by group (n = 4): control 35.28 ± 4.43 mm, B-exp 37.25 ± 1.5 mm, M-exp 34.88 ± 2.66 mm. Analysis was generated using the vegan package in R.
(c). Differential expression of olfactory receptors
We separately investigated expression levels of OR candidate genes. Six V2R receptors were significantly differentially expressed between the control and malinche-exposed individuals, five ORs were differentially expressed and one of the ORs survived FDR correction for 83 genes. In the control versus birchmanni-exposed comparison, two ORs, one TAAR and two V2Rs were differentially expressed; one V2R survived FDR correction. Twelve V2R receptors were significantly differentially expressed between malinche and birchmanni-exposed females, though none of these were significant following multiple testing correction (electronic supplementary material, table S3). We used qPCR on an increased number of individuals (n = 8, 4 used in RNAseq) to verify whether the top candidate V2R genes differentially expressed between malinche-exposed and birchmanni-exposed individuals but not surviving FDR correction (V2R_6, V2R_40) are indeed differentially expressed. We found that these two genes generally showed the same trend of expression as observed with RNAseq data, but only V2R_6 remained statistically significant between B-EXP and M-EXP groups (Wilcoxon rank sum, W = 13, p = 0.049, N = 8, figure 3).
Figure 3.
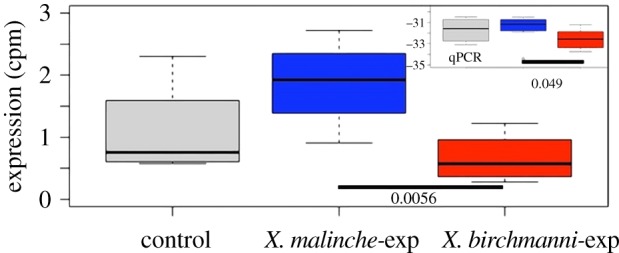
Boxplot showing expression levels of V2R6 (in counts per million reads, N = 4 per group) by treatment; V2R6 is differentially expressed between females exposed to conspecifics and heterospecifics. Insets show qPCR validation with eight individuals per treatment group. Raw p values are indicated. (Online version in colour.)
(d). Gene ontology and PANTHER pathway analysis
To investigate whether particular functional categories were enriched in the differentially expressed genes, we performed GO enrichment analysis. In both comparisons versus control, ‘response to organic substance’ (GO:0010033) was the most significantly enriched process (both FDR < 0.001, electronic supplementary material, tables S5 and S6). In genes differentially regulated between B-EXP and M-EXP females, ‘single-organism behaviour’ (GO:0044708) and ‘behaviour’ (GO:0007610) were the most significantly enriched biological process categories (FDR < 0.001; electronic supplementary material, figure S5). In total, 61 GO categories were significantly enriched or underrepresented at p < 0.05 (after FDR correction) between the two olfactory treatments (electronic supplementary material, table S4). Between treated and control individuals, 133 and 102 GO categories were significantly enriched or underrepresented at p < 0.05 (after FDR correction) for malinche- and birchmanni-exposed groups, respectively (electronic supplementary material, tables S5 and table S6). The major results are robust to the exclusion of muscle-specific and skin-specific genes from these analyses (electronic supplementary material, figure S5b).
PANTHER pathway analyses revealed four significantly enriched biological pathways in the lists of differentially expressed genes across the three exposure groups. When compared to the control group, angiogenesis (P00005) was the only enriched pathway after FDR correction in both the malinche (p = 0.002) and birchmanni (p = 0.035) comparisons. In genes differentially expressed between M-EXP and B-EXP females, cholecystokinin receptor (CCKR) signalling map (P06959: p = 0.002), gonadotropin-releasing hormone (GnRH) receptor pathway (P06664: p = 0.005) and endogenous cannabinoid signalling (P05730: p = 0.046) were significantly enriched after FDR correction.
(e). Amino acid divergence in differentially expressed odorant receptors
One hypothesis for the difference in olfactory responses between X. birchmanni and X. malinche females is that the odorant receptor proteins in the olfactory periphery are functionally divergent between species [58]. Using orthologous sequences from five Xiphophorus species, representing all three major clades [28], we found better support for the M8 model assuming some non-zero proportion of the sites with a dN/dS ratio > 1 (conserved + neutral 0 < ω1 ≤ 1 β distributed, and faster-than-neutral: ω2 > 1) compared to the null model assuming only 0 < ω1 ≤ 1 (model M7) or the null model having ω2 fixed at 1 (M8a) on four V2R receptors (electronic supplementary material, table S8; M8–M7 p < 0.05, M8–M8a p < 0.02, electronic supplementary material, figure S9), one OR receptor (M8–M7 p < 0.05, M8–M8a p < 0.02, electronic supplementary material, figure S10) and five TAAR receptors (M8–M7 p < 0.05, M8–M8a p < 0.02, electronic supplementary material, figure S11). This may suggest diversifying selection on these genes. Both V1R receptors are conserved in the Xiphophorus phylogeny (M8–M7 p > 0.4, M8–M8a p > 0.1).
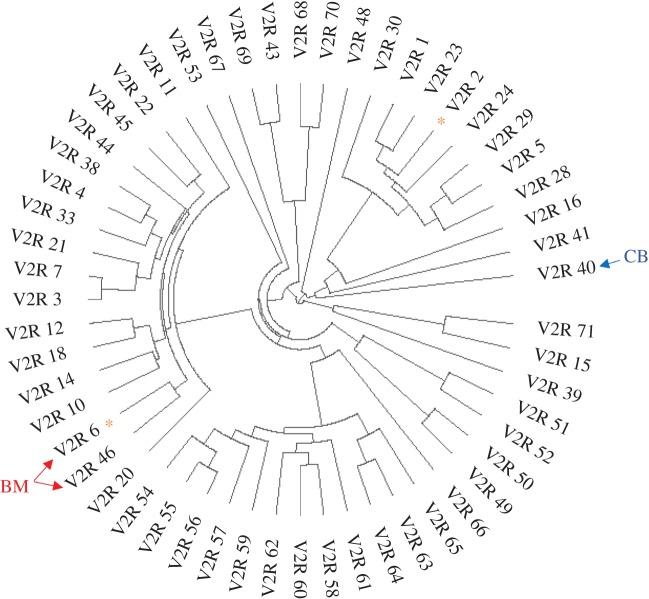
Within the two differentially expressed (raw p < 0.01) V2Rs between M-EXP and B-EXP groups, V2R_46 is conserved while V2R_6 shows a signature of positive selection (electronic supplementary material, figure S8). The differentially expressed V2R between control and B-EXP groups is conserved (electronic supplementary material, figure S8). The differentially expressed (raw p < 0.01) TAAR and OR are both conserved in sequence (electronic supplementary material, figures S9–S11).
4. Discussion
(a). Xiphophorus malinche females avoid familiar olfactory phenotypes
Female Xiphophorus malinche showed experience-independent preferences for visual cues and negative experience-dependent preferences for olfactory cues, in contrast to females of the sister species X. birchmanni that previous work has shown to have positive experience-dependent preferences in both modalities [27]. These patterns mirror the effects of short-term social experience on olfactory preferences in adult X. malinche and X. birchmanni females, where X. malinche females avoided heterospecific males after a one-week exposure to heterospecifics, while X. birchmanni females did not [31].
(b). Social exposure alters gene expression related to behaviour and synaptic plasticity
Our results show a strong gene expression response to social exposure (electronic supplementary material, figure S3), and strikingly, that patterns of gene expression change compared to control are highly similar in response to both conspecific and heterospecific cues. Most differentially expressed genes in both olfactory treatment groups also show the same direction of expression change compared to control. This pattern underscores that not only are similar genes involved in learning based on conspecific and heterospecific cues in X. malinche, but the direction of their response is nearly identical. The similarity in patterns of DE suggests that similar proximate mechanisms are being evoked by social exposure, regardless of species identity of the cue (given that the model species are closely related), and the neurogenetic response to species-specific adult exposure may be relatively small. Though the chemical identity of male pheromone components is unknown in Xiphophorus, in other fish species pheromones contain metabolites or hormonal derivatives [59–63] that may be shared across species and associated with genetic compatibility or body condition. Thus, a strong gene expression response to social exposure likely reflects a proximate response to these components of chemical and visual stimuli shared between closely related species.
The functions of differentially expressed genes are related to neural plasticity. GO analysis reveals an enrichment of genes involved in response to chemical stimulus in all three comparisons (electronic supplementary material, figures S5–S8). Interestingly, we found enrichment of GO terms related to behaviour, cognition, neurogenesis and synaptic transmission between the conspecific and heterospecific olfactory treatments (electronic supplementary material, figure S5). The transcript detected to be the most significantly differentially expressed among the three exposure groups is ΔFosB (electronic supplementary material, figures S6A and S7), an isoform of the fosb gene that is known to be associated with long-term memory [64–67].
Interestingly, all three biological pathways significantly enriched in the differentially expressed genes between X. malinche- and X. birchmanni-exposed females (electronic supplementary material, table S7) are important in shaping experience-dependent female preference or disdain. Cholecystokinin is implicated in avoidance behaviour in rats [68,69], endogenous cannabinoid signalling is involved in emotional associative learning [70] and a gonadotropin-releasing hormone has previously been suggested to play an essential role in shaping visual preferences for familiar males in female medaka [23].
These results suggest that olfactory learning in Xiphophorus triggers different responses in neural plasticity-related genes depending on whether the cue is conspecific or heterospecific. Though behavioural research demonstrates that females can distinguish between con- and heterospecific cues [24,25], it was unknown what the relative contributions are from the sensory periphery and higher-order neuronal processes that contribute to this behaviour. While our results highlight the large gene expression response that social exposure has on the sensory periphery and forebrain, they also show that cues produced by the two species of Xiphophorus elicit different, albeit relatively subtle, responses in X. malinche females, particularly in relation to genes involved in neural plasticity. This study focused on the transcriptomic implications of learning in only a small portion of the forebrain, and future studies are required to determine the impact of differential social exposure on the whole brain. Nevertheless, these results confirm that the region we sampled exhibits a socially sensitive, neural response that functionally pertains to behaviour and cognition. Furthermore, future experiments will focus on localizing these enriched genes in the brain to verify whether candidate brain regions involved in olfactory learning show distinct responses to conspecific versus heterospecific olfactory cues.
(c). Vomeronasal receptors type 2 as candidates for species-specific cue detectors
Vomeronasal receptor families—V1R and V2R—are associated with pheromone detection in vertebrates, especially mammals [71]. Recent studies have suggested that the main olfactory pathways (those expressing ORs) also detect certain components of sex pheromones. Despite V1R being implicated in pheromone detection in fish [72], we detect neither DE (possibly due to low expression levels, as found in other teleosts [18]) nor a signature of positive selection of this gene family. On the other hand, we did detect expression of a large number of V2R receptors. V2R receptors are of particular interest in olfactory preference because they have been implicated in pheromone detection in both mammals and fish. In mice, V2Rs are co-expressed with major histocompatibility proteins (MHC), which have been implicated in individual recognition and compatibility-based mate choice in a wide variety of taxa [73]. V2R neurons detect MHC-1 ligands [74] and respond to proteinaceous pheromone components in mice [75] and fish [76], respectively. In mice, social exposure has been shown to increase the number of classical OR neurons [17], but the effect on V2R neurons is unknown.
Interestingly, the most significant differentially expressed odorant receptors between the conspecific versus heterospecific social treatments are V2Rs (cut-off at raw p < 0.02, q < 0.1; electronic supplementary material, table S3; figures S9–S11). Within 65 odorant receptors that passed coverage cut-off, on average the raw p-values of V2Rs are lower than non-V2Rs (nine ORs and eight TAARs) in the conspecific versus heterospecific comparison compared to the other two comparisons (linear model, p = 0.00082; full model in electronic supplementary material, table S9). qPCR validation showed that the most differentially expressed V2R, V2R_6, is most highly expressed in the conspecific exposure treatment, lowest in heterospecific treatment and intermediate in controls (electronic supplementary material, figure S6B). Intriguingly, this gene also has the highest rate of molecular evolution of V2Rs among 5 Xiphophorus species (figure 4; electronic supplementary material, figure S8). There are 13 amino acid substitutions between X. birchmanni and X. malinche sequences at this gene, 9 of which are located on the N-terminus (electronic supplementary material, figure S8), where the putative ligand-binding domain is located [77]. These results suggest that the V2R paralog may have functionally diverged between X. birchmanni and X. malinche, and its higher expression level in conspecific exposure makes it an interesting candidate for detecting a species-specific component in male olfactory cues. We can also confidently conclude that these observed differences in gene expression profiles are occurring at the sensory periphery, as there is little to no expression of odorant receptor genes within the brain of other teleosts (electronic supplementary material, figure S12).
Figure 4.
DE and signature of positive selection plotted on a V2R phylogeny of five Xiphophorus spp. Because all orthologues are monophyletic for the five species, the tips are pruned to leave a single representative gene. *CodeML dN/dS M8-M8a model p < = 0.01; BM, differentially expressed between birchmanni–malinche exposures; CB, differentially expressed between control and birchmanni-exposed. The phylogeny is rooted at a clade containing V2R_71, corresponding to OlfCc1 (v2r1l) in zebrafish, a previously known basal V2R. (Online version in colour.)
Other V2Rs that are differentially expressed in response to conspecific and heterospecific cues do not show this signature of positive selection (electronic supplementary material, figure S9), which could suggest detection of conserved chemical components [59]. Consistent with the interpretation of a conserved response to social stimuli, differentially expressed odorant receptors in control versus exposure comparisons are all structurally conserved in the five species of Xiphophorus analysed. It is consistent with a recent finding that the hormone derivative prostaglandin F2α signals female reproductive state through detection by the OR family [78]. Together, these observations are consistent with the hypothesis that certain components in the sex pheromone blend are chemically conserved, and information is conveyed through ratio or concentration variations.
(d). Periphery and higher-order neuronal processes in relation to mate choice evolution
Our results suggest that the detection of sexual signals at the periphery has the potential to evolve independently of, or interact with, how these are evaluated at higher levels. In particular, conspecific exposure elicits higher expression of a rapidly diverging V2R receptor, suggesting that this receptor evolves to maximize detection of conspecific signals. However, a putatively greater sensitivity at the periphery does not result in higher preference for conspecifics in this species. A possible explanation is that this species has evolved aversion to learned olfactory cues independent of the sensory periphery. The three biological pathways found to be differentially regulated between exposure treatments have been established as playing functional roles in emotional learning [70], aversion [68,69] and even establishing mating preferences for familiar individuals [23]. Therefore, these pathways could play a role in explaining how learned aversion evolved in this system. Future studies should examine the neurogenomic response to social exposure in the sister species preferring familiar cues, X. birchmanni, which should be expected to have sharply different or opposite neurogenomic responses to social stimuli.
5. Conclusions
There is a large literature on the role that learned mate preferences may play in maintaining reproductive isolation between species by preventing hybridisation [7–10]. However, some learned preferences, particularly preferences for novel traits, may actually promote hybridisation, as is the case with X. malinche. In the X. malinche–X. birchmanni system, natural populations vary in their degree of exposure to heterospecifics and hybrids [79]. Our results suggest that exposure to heterospecifics during development could then play an important role in determining a female's learned preference through changes at both the sensory periphery and central nervous system, ultimately impacting evolutionary processes. Specifically, the results of this experiment predict that hybridisation might be common when X. malinche females have only had contact with conspecific cues during development. However, we note that in natural environments both the learning scenario and dynamics of hybridisation are complex. Some contact zones between X. birchmanni and X. malinche are hybrid swarms, as might be predicted by our olfactory learning and visual preference results. In other hybrid populations, by contrast, X. malinche individuals discriminate against heterospecifics [79]. Ultimately, understanding the neurogenomic mechanisms of sexual imprinting can help biologists make predictions about mate choice and evolutionary outcomes in the wild. By revealing potential candidate genes and biological pathways that connect developmental social environment with mate choice outcomes, this study provides a necessary first step in understanding the neurogenetic mechanisms that regulate reproductive isolation.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Clay Small for advice on RNA sample collection and J. Brad Johnson for advice on fish husbandry and Machteld Verzijden and Adam Jones for comments on an early draft.
Ethics
Ethical approval for experiments involving animals was obtained from the International Animal Care and Use Committee, and all experiments conducted in this study complied with current state, federal and local laws in the United States and Mexico.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material. Aligned reads are deposited in NCBI's SRA SRP103059.
Authors' contributions
R.C. designed the study, carried out fish husbandry, behaviour trials and statistical analysis, RNA extraction, qPCR primer design, read mapping, odorant receptor identification and molecular evolution and DE analysis and drafted the manuscript. P.J.D. carried out DE analysis, GO and PANTHER Pathway enrichment analysis and edited the manuscript. M.S. carried out qPCR and cDNA library preparation for RNAseq and participated in data analysis and editing the manuscript. G.G.R. participated in study design and editing the manuscript. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
This project is supported by an NSF DDIG no. 1210324 to R.C. and by NSF IOS-0923825 to G.G.R. Analysis was performed on the Texas A&M Brazos cluster.
References
- 1.Grant PR, Grant BR. 1997. Hybridization, sexual imprinting, and mate choice. Am. Nat. 149, 1–28. ( 10.1086/285976) [DOI] [Google Scholar]
- 2.Servedio MR, Saether SA, Saetre GP. 2009. Reinforcement and learning. Evol. Ecol. 23, 109–123. ( 10.1007/s10682-007-9188-2) [DOI] [Google Scholar]
- 3.Verzijden MN, Lachlan RF, Servedio MR. 2005. Female mate-choice behavior and sympatric speciation. Evolution 59, 2097–2108. ( 10.1111/j.0014-3820.2005.tb00920.x) [DOI] [PubMed] [Google Scholar]
- 4.Hughes KA, Houde AE, Price AC, Rodd FH. 2013. Mating advantage for rare males in wild guppy populations. Nature 503, 108–110. ( 10.1038/nature12717) [DOI] [PubMed] [Google Scholar]
- 5.Marler P. 1991. The instinct to learn. In The epigenesis of mind: essays on biology and cognition (eds Carey S, Gelman R), pp. 37–66. Mahwah, NJ: Lawrence Erlbaum Associates, Inc. [Google Scholar]
- 6.Witte K, Sawka N. 2003. Sexual imprinting on a novel trait in the dimorphic zebra finch: sexes differ. Anim. Behav. 65, 195–203. ( 10.1006/anbe.2002.2009) [DOI] [Google Scholar]
- 7.Westerman EL, Hodgins-Davis A, Dinwiddie A, Monteiro A. 2012. Biased learning affects mate choice in a butterfly. Proc. Natl. Acad. Sci. USA 109, 10 948–10 953. ( 10.1073/pnas.1118378109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozak GM, Head ML, Boughman JW. 2011. Sexual imprinting on ecologically divergent traits leads to sexual isolation in sticklebacks. Proc. R. Soc. B 278, 2604–2610. ( 10.1098/rspb.2010.2466) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kozak GM, Boughman JW. 2009. Learned conspecific mate preference in a species pair of sticklebacks. Behav. Ecol. 20, 1282–1288. ( 10.1093/beheco/arp134) [DOI] [Google Scholar]
- 10.Verzijden MN, Korthof REM, ten Cate C. 2008. Females learn from mothers and males learn from others. The effect of mother and siblings on the development of female mate preferences and male aggression biases in Lake Victoria cichlids, genus Mbipia. Behav. Ecol. Sociobiol. 62, 1359–1368. ( 10.1007/s00265-008-0564-x) [DOI] [Google Scholar]
- 11.Verzijden MN, ten Cate C. 2007. Early learning influences species assortative mating preferences in Lake Victoria cichlid fish. Biol. Lett. 3, 134–136. ( 10.1098/rsbl.2006.0601) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irwin DE, Price T. 1999. Sexual imprinting, learning and speciation. Heredity 82, 347–354. ( 10.1038/sj.hdy.6885270) [DOI] [PubMed] [Google Scholar]
- 13.Grant BR, Grant PR. 2008. Fission and fusion of Darwin's finches populations. Phil. Trans. R. Soc. B 363, 2821–2829. ( 10.1098/rstb.2008.0051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ten Cate C. 1987. Sexual preferences in zebra finch males raised by two species II. The internal representation resulting from double imprinting. Anim. Behav. 35, 321–330. ( 10.1016/S0003-3472(87)80255-5) [DOI] [Google Scholar]
- 15.Nevitt GA, Dittman AH, Quinn TP, Moody WJ. 1994. Evidence for a peripheral olfactory memory in imprinted salmon. Proc. Natl. Acad. Sci. USA 91, 4288–4292. ( 10.1073/pnas.91.10.4288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whitlock KE, Harden MV, Newton LA, Lloyd RC. 2006. Olfactory imprinting is correlated with changes in gene expression in the olfactory epithelia of the zebrafish. J. Neurobiol. 66, 1452–1466. ( 10.1002/neu.20328) [DOI] [PubMed] [Google Scholar]
- 17.Dias BG, Ressler KJ. 2014. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat. Neurosci. 17, 89–96. ( 10.1038/nn.3594) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bazáes A, Olivares J, Schmachtenberg O. 2013. Properties, projections, and tuning of teleost olfactory receptor neurons. J. Chem. Ecol. 39, 451–464. ( 10.1007/s10886-013-0268-1) [DOI] [PubMed] [Google Scholar]
- 19.Saraiva LR, Ahuja G, Ivandic I, Syed AS, Marioni JC, Korsching SI, Logan DW. 2015. Molecular and neuronal homology between the olfactory systems of zebrafish and mouse. Sci. Rep. 5, 1660 ( 10.1038/srep11487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corotto F, Henegar J, Maruniak J. 1994. Odor deprivation leads to reduced neurogenesis and reduced neuronal survival in the olfactory bulb of the adult mouse. Neuroscience 61, 739–744. ( 10.1016/0306-4522(94)90397-2) [DOI] [PubMed] [Google Scholar]
- 21.Yamaguchi M, Mori K. 2005. Critical period for sensory experience-dependent survival of newly generated granule cells in the adult mouse olfactory bulb. Proc. Natl. Acad. Sci. USA 102, 9697–9702. ( 10.1073/pnas.0406082102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makinodan M, Rosen KM, Ito S, Corfas G. 2012. A critical period for social experience-dependent oligodendrocyte maturation and myelination. Science 337, 1357–1360. ( 10.1126/science.1220845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okuyama T, et al. 2014. A neural mechanism underlying mating preferences for familiar individuals in medaka fish. Science 343, 91–94. ( 10.1126/science.1244724) [DOI] [PubMed] [Google Scholar]
- 24.McLennan DA, Ryan MJ. 1999. Interspecific recognition and discrimination based upon olfactory cues in northern swordtails. Evolution 53, 880–888. ( 10.2307/2640728) [DOI] [PubMed] [Google Scholar]
- 25.Crapon de Caprona M-D, Ryan MJ. 1990. Conspecific mate recognition in swordtails, Xiphophorus nigrensis and X. pygmaeus (Poeciliidae): olfactory and visual cues. Anim. Behav. 39, 290–296. ( 10.1016/S0003-3472(05)80873-5) [DOI] [Google Scholar]
- 26.Fisher HS, Wong BBM, Rosenthal GG. 2006. Alteration of the chemical environment disrupts communication in a freshwater fish. Proc. R. Soc. B 273, 1187–1193. ( 10.1098/rspb.2005.3406) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verzijden MN, Rosenthal GG. 2011. Effects of sensory modality on learned mate preferences in female swordtails. Anim. Behav. 82, 557–562. ( 10.1016/j.anbehav.2011.06.010) [DOI] [Google Scholar]
- 28.Cui R, Schumer M, Kruesi K, Walter R, Andolfatto P, Rosenthal GG. 2013. Phylogenomics reveals extensive reticulate evolution in Xiphophorus fishes. Evolution 67, 2166–2179. ( 10.1111/evo.12099) [DOI] [PubMed] [Google Scholar]
- 29.Culumber Z, Fisher H, Tobler M, Mateos M, Barber P, Sorenson M, Rosenthal G. 2011. Replicated hybrid zones of Xiphophorus swordtails along an elevational gradient. Mol. Ecol. 20, 342–356. ( 10.1111/j.1365-294X.2010.04949.x) [DOI] [PubMed] [Google Scholar]
- 30.Fisher HS, Mascuch SJ, Rosenthal GG. 2009. Multivariate male traits misalign with multivariate female preferences in the swordtail fish, Xiphophorus birchmanni. Anim. Behav. 78, 265–269. ( 10.1016/j.anbehav.2009.02.029) [DOI] [Google Scholar]
- 31.Verzijden MN, Culumber ZW, Rosenthal GG. 2012. Opposite effects of learning cause asymmetric mate preferences in hybridizing species. Behav. Ecol. 23, 1133–1139. ( 10.1093/beheco/ars086) [DOI] [Google Scholar]
- 32.Wong BBM, Rosenthal GG. 2006. Female disdain for swords in a swordtail fish. Am. Nat. 167, 136–140. ( 10.1086/498278) [DOI] [PubMed] [Google Scholar]
- 33.Northcutt RG. 2008. Forebrain evolution in bony fishes. Brain Res. Bull. 75, 191–205. ( 10.1016/j.brainresbull.2007.10.058) [DOI] [PubMed] [Google Scholar]
- 34.Wong RY, Cummings ME. 2014. Expression patterns of neuroligin-3 and tyrosine hydroxylase across the brain in mate choice contexts in female swordtails. Brain Behav. Evol. 83, 231–243. ( 10.1159/000360071) [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Zhou J, White KP. 2014. RNA-seq differential expression studies: more sequence or more replication? Bioinformatics 30, 301–304. ( 10.1093/bioinformatics/btt688) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: A flexible trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schumer M, Cui R, Boussau B, Walter R, Rosenthal G, Andolfatto P. 2013. An evaluation of the hybrid speciation hypothesis for Xiphophorus clemenciae based on whole genome sequences. Evolution 67, 1155–1168. ( 10.1111/evo.12009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schartl M, et al. 2013. The genome of the platyfish, Xiphophorus maculatus, provides insights into evolutionary adaptation and several complex traits. Nat. Genet. 45, 567–572. ( 10.1038/ng.2604) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hashiguchi Y, Furuta Y, Nishida M.. 2008. Evolutionary patterns and selective pressures of odorant/pheromone receptor gene families in teleost fishes. PLoS ONE 3, e4083 ( 10.1371/journal.pone.0004083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oka Y, Saraiva LR, Kwan YY, Korsching SI. 2009. The fifth class of Gα proteins. Proc. Natl. Acad. Sci. USA 106, 1484–1489. ( 10.1073/pnas.0809420106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2008. BLAST+: architecture and applications. BMC Bioinformatics 10, 421 ( 10.1186/1471-2105-10-421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Birney E, Clamp M, Durbin R. 2004. GeneWise and genomewise. Genome Res. 14, 988–995. ( 10.1101/gr.1865504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yates A, et al. 2015. Ensembl 2016. Nucleic Acids Res. 44, D710–D716. ( 10.1093/nar/gkv1157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin Q, et al. 2016. The seahorse genome and the evolution of its specialized morphology. Nature 540, 395–399. ( 10.1038/nature20595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. ( 10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 46.Wong WS, Yang Z, Goldman N, Nielsen R. 2004. Accuracy and power of statistical methods for detecting adaptive evolution in protein coding sequences and for identifying positively selected sites. Genetics 168, 1041–1051. ( 10.1534/genetics.104.031153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z, Bielawski JP. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 15, 496–503. ( 10.1016/S0169-5347(00)01994-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. ( 10.1093/bioinformatics/btl446) [DOI] [PubMed] [Google Scholar]
- 49.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. ( 10.1093/bioinformatics/btp616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S, Huber W. 2012. Differential expression of RNA-Seq data at the gene level–the DESeq package. Heidelberg, Germany: European Molecular Biology Laboratory (EMBL). [Google Scholar]
- 51.Oksanen J, et al. 2015. Vegan: Community ecology Package. 2015. R package Version 2.2-1.
- 52.Bass J, Dabney A, Robinson D.. 2015. Qvalue: Q-value estimation for false discovery rate control. R package version 2.2.0.
- 53.Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8, 1765–1786. ( 10.1038/nprot.2013.099) [DOI] [PubMed] [Google Scholar]
- 54.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. 2003. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141. ( 10.1101/gr.772403) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mi H, Muruganujan A, Casagrande JT, Thomas PD. 2013. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566. ( 10.1038/nprot.2013.092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Supek F, Bošnjak M, Škunca N, Šmuc T. 2011. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 6, e21800 ( 10.1371/journal.pone.0021800) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trapnell C, Pachter L and Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. ( 10.1093/bioinformatics/btp120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leary GP, Allen JE, Bunger PL, Luginbill JB, Linn CE, Macallister IE, Kavanaugh MP, Wanner KW. 2012. Single mutation to a sex pheromone receptor provides adaptive specificity between closely related moth species. Proc. Natl. Acad. Sci. USA 109, 14 081–14 086. ( 10.1073/pnas.1204661109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levesque HM, Scaffidi D, Polkinghorne CN, Sorensen PW. 2011. A multi-component species identifying pheromone in the goldfish. J. Chem. Ecol. 37, 219–227. ( 10.1007/s10886-011-9907-6) [DOI] [PubMed] [Google Scholar]
- 60.Sorensen PW, Fine JM, Dvornikovs V, Jeffrey CS, Shao F, Wang JZ, Vrieze LA, Anderson KR, Hoye TR. 2005. Mixture of new sulfated steroids functions as a migratory pheromone in the sea lamprey. Nat. Chem. Biol. 1, 324–328. ( 10.1038/nchembio739) [DOI] [PubMed] [Google Scholar]
- 61.Sorensen PW, Hara TJ, Stacey NE, Dulka JG. 1990. Extreme olfactory specificity of male goldfish to the preovulatory steroidal pheromone 17α,20β-dihydroxy-4-pregnen-3-one. J. Comp. Physiol. A 166, 373–383. ( 10.1007/bf00204810) [DOI] [Google Scholar]
- 62.Sorensen P, Hara T, Stacey N, Goetz FW. 1988. F prostaglandins function as potent olfactory stimulants that comprise the postovulatory female sex pheromone in goldfish. Biol. Reprod. 39, 1039–1050. ( 10.1095/biolreprod39.5.1039) [DOI] [PubMed] [Google Scholar]
- 63.Dulka J, Stacey N, Sorensen P, Van Der Kraak G. 1987. A steroid sex pheromone synchronizes male–female spawning readiness in goldfish. Nature 325, 251–253. ( 10.1038/325251a0) [DOI] [Google Scholar]
- 64.Hedges V, Chakravarty S, Nestler E, Meisel R. 2009. ΔFosB overexpression in the nucleus accumbens enhances sexual reward in female Syrian hamsters. Genes Brain Behav. 8, 442–449. ( 10.1111/j.1601-183X.2009.00491.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kelz MB, et al. 1999. Expression of the transcription factor ΔFosB in the brain controls sensitivity to cocaine. Nature 401, 272–276. ( 10.1038/45790) [DOI] [PubMed] [Google Scholar]
- 66.Nestler EJ, Barrot M, Self DW. 2001. ΔFosB: a sustained molecular switch for addiction. Proc. Natl. Acad. Sci. USA 98, 11 042–11 046. ( 10.1073/pnas.191352698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pitchers KK, Frohmader KS, Vialou V, Mouzon E, Nestler EJ, Lehman MN, Coolen LM. 2010. ΔFosB in the nucleus accumbens is critical for reinforcing effects of sexual reward. Genes Brain Behav. 9, 831–840. ( 10.1111/j.1601-183X.2010.00621.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fekete M, Lengyel Á, Hegedüs B, Penke B, Zarándy M, Tóth GK, Telegdy G. 1984. Further analysis of the effects of cholecystokinin octapeptides on avoidance behaviour in rats. Eur. J. Pharmacol. 98, 79–91. ( 10.1016/0014-2999(84)90111-0) [DOI] [PubMed] [Google Scholar]
- 69.Itoh S, Katsuura G. 1987. Cholecystokinin octapeptide prevents extinction of active avoidance behavior in the rat. Drug Dev. Res. 10, 171–175. ( 10.1002/ddr.430100305) [DOI] [Google Scholar]
- 70.Laviolette SR, Grace AA. 2006. Cannabinoids potentiate emotional learning plasticity in neurons of the medial prefrontal cortex through basolateral amygdala inputs. J. Neurosci. 26, 6458–6468. ( 10.1523/JNEUROSCI.0707-06.2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rodriguez I, Greer CA, Mok MY, Mombaerts P. 2000. A putative pheromone receptor gene expressed in human olfactory mucosa. Nat. Genet. 26, 18–19. ( 10.1038/79124) [DOI] [PubMed] [Google Scholar]
- 72.Pfister P, Rodriguez I. 2005. Olfactory expression of a single and highly variable V1r pheromone receptor-like gene in fish species. Proc. Natl. Acad. Sci. USA 102, 5489–5494. ( 10.1073/pnas.0402581102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dulac C, Olson R, Bjorkman PJ. 2006. MHC homologs in the nervous system—they haven't lost their groove. Curr. Opin. Neurobiol. 16, 351–357. ( 10.1016/j.conb.2006.05.007) [DOI] [PubMed] [Google Scholar]
- 74.Leinders-Zufall T, et al. 2004. MHC class I peptides as chemosensory signals in the vomeronasal organ. Science 306, 1033–1037. ( 10.1126/science.1102818) [DOI] [PubMed] [Google Scholar]
- 75.Chamero P, Marton TF, Logan DW, Flanagan K, Cruz JR, Saghatelian A, Cravatt BF, Stowers L. 2007. Identification of protein pheromones that promote aggressive behaviour. Nature 450, 899–902. ( 10.1038/Nature05997) [DOI] [PubMed] [Google Scholar]
- 76.Speca DJ, Lin DM, Sorensen PW, Isacoff EY, Ngai J, Dittman AH. 1999. Functional identification of a goldfish odorant receptor. Neuron 23, 487–498. ( 10.1016/S0896-6273(00)80802-8) [DOI] [PubMed] [Google Scholar]
- 77.Yang H, Shi P, Zhang Y-, Zhang J. 2005. Composition and evolution of the V2r vomeronasal receptor gene repertoire in mice and rats. Genomics 86, 306–315. ( 10.1016/j.ygeno.2005.05.012) [DOI] [PubMed] [Google Scholar]
- 78.Yabuki Y, et al. 2016. Olfactory receptor for prostaglandin F2α mediates male fish courtship behavior. Nat. Neurosci. 19, 897–904. ( 10.1038/nn.4314) [DOI] [PubMed] [Google Scholar]
- 79.Culumber ZW, Ochoa OM, Rosenthal GG. 2014. Assortative mating and the maintenance of population structure in a natural hybrid zone. Am. Nat. 184, 225–232. ( 10.1086/677033) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material. Aligned reads are deposited in NCBI's SRA SRP103059.