Abstract
Ubiquitin-specific protease 25 (Usp25) is a deubiquitinase that is involved in multiple biological processes. The N-terminal ubiquitin-binding region (UBR) of Usp25 contains one ubiquitin-associated domain, one small ubiquitin-like modifier (SUMO)-interacting motif and two ubiquitin-interacting motifs. Previous studies suggest that the covalent sumoylation in the UBR of Usp25 impairs its enzymatic activity. Here, we raise the hypothesis that non-covalent binding of SUMO, a prerequisite for efficient sumoylation, will impair Usp25’s catalytic activity as well. To test our hypothesis and elucidate the underlying molecular mechanism, we investigated the structure and function of the Usp25 N-terminal UBR. The solution structure of Usp251–146 is obtained, and the key residues responsible for recognition of ubiquitin and SUMO2 are identified. Our data suggest inhibition of Usp25’s catalytic activity upon the non-covalent binding of SUMO2 to the Usp25 SUMO-interacting motif. We also find that SUMO2 can competitively block the interaction between the Usp25 UBR and its ubiquitin substrates. Based on our findings, we have proposed a working model to depict the regulatory role of the Usp25 UBR in the functional display of the enzyme.
Introduction
Ubiquitin contains seven lysine residues (K6, K11, K27, K29, K33, K48, and K63), which allows for the formation of seven homotypic linkage types and various heterotypic polyubiquitin chains (1). The structural diversity generated by protein ubiquitination is intimately linked to the regulation of distinct biological processes (2). For example, K48-linked polyubiquitin modification usually serves as a signal for protein degradation, whereas K63-linked polyubiquitination plays a role in DNA repair and signal transduction pathways. The attachment of ubiquitin to a protein requires the consecutive action of three enzymes: ubiquitin-activating enzyme, E1, ubiquitin-conjugating enzyme, E2, and ubiquitin ligase, E3, whereas the removal of ubiquitin is promoted by deubiquitinases (DUBs) (3, 4). DUBs are important enzymes that regulate the functional outcomes of ubiquitination. Dysfuntions of DUBs are associated with severe human diseases like cancers and neurodegenerative disorders (5, 6, 7). The human genome encodes ∼100 DUBs, which can be grouped into six subfamilies according to their different structural and functional features: ubiquitin-specific proteases (USPs), ubiquitin C-terminal hydrolases (UCHs), ovarian tumor-like proteases, Machado-Joseph disease proteases, JAMM/MPN-domain-associated metallopeptidases, and the monocyte chemotactic protein-induced protein family (7). More than 50% of DUBs belong to the USP subfamily, and quite a few of these are associated with the lifespan control of oncogenes or the regulation of ubiquitin-mediated signaling (8, 9, 10).
Usp25, a member of the USP subfamily, is involved in multiple biological processes, including the immune response (11), endoplasmic-reticulum-associated protein degradation (12), and cell migration and invasion (13). There are three isoforms of Usp25 in humans: Usp25a, Usp25b, and Usp25m. Usp25a and Usp25b are widely expressed in different human tissues, whereas Usp25m mainly distributes in muscle (14). Apart from the catalytic USP domain, all of the three isoforms of Usp25 contain a ubiquitin-binding region (UBR) composed of one ubiquitin-associated (UBA) domain and two ubiquitin-interacting motifs (UIMs) at their N-terminus. Additionally, a small-ubiquitin-like-modifier (SUMO)-interacting motif (SIM) locates adjacent to the first UIM of Usp25 (Fig. 1 A). The non-catalytic UBR of Usp25 is suggested to play an important role in the substrate recognition of the enzyme. Deletions of one or two UIM motifs impair the deubiquitination activity of Usp25 (15). The enzymatic activity of Usp25 is also inhibited by sumoylation in the Usp25 UBR (15). Noticeably, the non-covalent binding of SUMO is a prerequisite for efficient sumoylation of Usp25 (15, 16). Taking the spatial location of structural motifs in the Usp25 UBR into account, here we raise the hypothesis that not only the covalent conjugation but also the non-covalent binding of SUMO to Usp25 will impair the catalytic activity of the enzyme. To test our hypothesis and elucidate the underlying molecular mechanism, we investigated the structure and function of the N-terminal UBR of human Usp25.
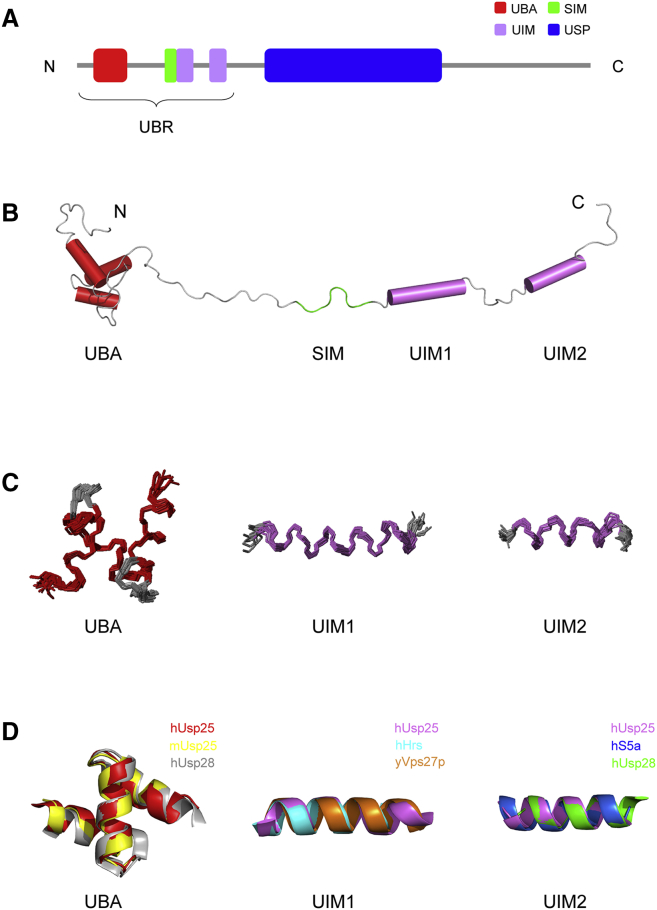
Figure 1.
Solution structure presentations of Usp25 UBR. (A) Schematic view of the domain architecture of Usp25FL. (B) Representation of the solution structure of the Usp25 UBR. (C) Ensembles of the folded parts of 15 calculated Usp25 UBRs. (D) Structural alignments of UBAs and UIM1/2s among human Usp25 and other proteins. Mouse Usp25 (mUsp25 (PDB: 1VDL)), human Usp28 (hUsp28 (PDB: 2LVA)), human Hrs (hHrs (PDB: 2D3G)), yeast Vps27p (yVps27p (PDB: 1O06)), and human S5a (hS5a (PDB: 1P9C)) are used for structural comparisons. To see this figure in color, go online.
Materials and Methods
Plasmid construction and protein expression and purification
Full-length cDNA of Usp25m (Usp25FL) was kindly provided by Dr. Frauke Melchior (Department of Biochemistry, Faculty of Medicine, Georg-August-University of Göttingen). Usp251–146 cDNA was subcloned into the pET28a vector via NdeI and BamHI restriction sites. The mutants for Usp251–146 and Usp25FL were created by polymerase chain reaction. SUMO2 cDNA was subcloned into pET28a (NdeI/SalI) or pGBTNH (BamHI/XholI) vectors. All constructs were validated by DNA sequencing.
Usp251–146 and Usp25FL were expressed in Escherichia coli BL21(DE3) cells and purified as described previously (15, 17). 15N- and 13C-labeled Usp251–146 samples were produced in M9 minimal medium with 15N-labeled ammonium chloride and 13C-labeled glucose as the only nitrogen and carbon sources, respectively. Expression and purification for ubiquitin, Ub-K48R, E1, E2-25K, SUMO2, and UCH-L3 was almost the same as that for Usp251–146, with minor modifications. Protein purity was checked by sodium dodecyl sulfate polyacrylamide gel electrophoresis (15% gel) and protein concentration was determined using ultraviolet and Bradford methods.
Size-exclusion chromatography
Purified proteins were prepared at a final concentration of 1 mg/mL in gel filtration buffer (20 mM Na2HPO4/NaH2PO4 (pH 6.5), 100 mM NaCl, and 5 mM dithiothreitol (DTT)). At 4°C, 500 μL of protein samples were loaded onto a Superdex 200 Increase 10/300 GL column (GE Healthcare, Chicago, IL). The flow rate was set to 0.5 mL/min and the elution process was monitored by A280.
Dynamic light scattering
Dynamic light scattering (DLS) studies were carried out at 25°C on a DynaPro NanoStar instrument (Wyatt Technology, Goleta, CA). Protein samples were prepared at a concentration of 1 mg/mL in DLS buffer (20 mM Na2HPO4/NaH2PO4 (pH 6.5), 100 mM NaCl, and 5 mM DTT) and filtered by 0.22 μm filters before use. Proteins with a volume of 20 μL were added into a 45 μL quartz cuvette. After a waiting time of 1 min, the signals were recorded. Each measurement consisted of 10 acquisitions. To determine whether the overall molecular shape of Usp251–146 deviates from spherical, SrtA60–206, which is a well-known globule-like protein, was selected as the referencing protein, and a globular model was then assumed when processing the DLS data of SrtA60–206. For comparison purposes, the same parameter was applied when processing the DLS data for Usp251–146.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) measurements were performed at 25°C using a MicroCal iTC200 isothermal titration calorimeter (GE Healthcare, Little Chalfont, United Kingdom). ITC buffer (20 mM Na2HPO4/NaH2PO4, 100 mM NaCl, and 15 mM β-mercaptoethanol (pH 6.5)) was degassed by sonication in advance of measurements. SUMO2- or K48-linked diubiquitin samples were prepared at a concentration of 50 μM in ITC buffer and added into the 200 μL sample cell. Usp251–146 at a concentration of 1 mM in ITC buffer was transferred into the 40 μL syringe. A total number of 19 injections (2 μL for each) of Usp251–146 into the sample cell were carried out, and the corresponding heat was recorded. Data were subsequently analyzed using the Origin software package (Origin 7.0).
Solution structure determination of Usp251–146
Uniformly 15N-, 13C-, and 15N/13C-labeled Usp251–146 samples at a final concentration of 0.6 mM in NMR buffer (20 mM Na2HPO4/NaH2PO4 (pH 6.5), 100 mM NaCl, 2 mM DTT, and 10% or 100% D2O) were used for NMR data acquisition at 25°C on a Bruker 600 or 800 MHz spectrometer equipped with cryoprobes. The programs NMRPipe (18) and CARA (19) were used for the processing and analysis of NMR data, respectively. Assignments of 1H, 15N, and 13C resonances of Usp251–146 were obtained using the following experiments: HNCA/HN(CO)CA pair, HNCO/HN(CA)CO pair, HNCACB, HBHA(CO)NH, HCCH total correlated spectroscopy, HCCH correlation spectroscopy, 15N-dispersed nuclear Overhauser effect spectroscopy (NOESY), and 13C-dispersed NOESY. Full assignments for Usp251–146 had been deposited into BioMagResBank : 19111 (17). Distance constraints for structure calculations were obtained using a 3D 15N-dispersed NOESY spectrum on a 15N-labeled sample, a 13C-dispersed NOESY spectrum on a 13C-labeled sample, and a two-dimensional NOESY spectrum on an unlabeled sample. Backbone Φ and Ψ torsion angle restraints were generated from chemical shifts using the program TALOS+ (20). The NOE-derived distance constraints, hydrogen bonds, and dihedral angle constraints (Table 1) were applied in the program Xplor-NIH (21, 22) to determine the solution structure of Usp251–146. Fifteen structures of Usp251–146 were obtained that had no NOE >0.3 Å or dihedral-angle violation >5°, and these structures were further evaluated by PROCHECK-NMR (23). The final refined ensemble of 15 structures was deposited into the Protein Data Bank with an accession code of PDB: 2MUX.
Table 1.
Structural Statistics for NMR Structures of Usp251–146
| NMR Distance and Dihedral Constraints | |
|---|---|
| Distance Restraints | |
| Total NOE | 1251 |
| Intra-residual | 363 |
| Sequential (|i − j| = 1) | 465 |
| Medium range (2 ≤ |i − j| ≤ 4) | 334 |
| Long range (|i − j | > 4) | 89 |
| Hydrogen bonds | 82 |
| Dihedral-angle Restraints | |
| Φ | 63 |
| Ψ | 63 |
| Violations | |
| NOE violations (>0.3 Å) | 0 |
| Torsion angle violations (>5°) | 0 |
| RMSD from Mean Structure (Å) | |
| Backbone for UBA domain (residues 16–56) | 0.59 ± 0.13 |
| Heavy atoms for UBA domain | 1.39 ± 0.19 |
| Backbone for UIM1 (residues 100–116) | 0.62 ± 0.22 |
| Heavy atoms for UIM1 | 1.68 ± 0.28 |
| Backbone for UIM2 (residues 125–137) | 0.71 ± 0.23 |
| Heavy atoms for UIM2 | 1.85 ± 0.31 |
| Ramachandran Statistics | |
| Most favored region (%) | 58.8 |
| Allowed (%) | 29.4 |
| Generous (%) | 11 |
| Disallowed (%) | 0.7 |
15N relaxation measurements
Backbone 15N relaxation parameters, including longitudinal relaxation rate R1, transverse relaxation rate R2, and the 1H-15N heteronuclear steady-state NOE, were measured at 25°C on a Bruker 600 MHz NMR spectrometer equipped with a cryoprobe using uniformly 15N-labeled Usp251–146 samples at a concentration of 0.6 mM. A recycle delay of 3 s was used for both R1 and R2 measurements. Relaxation delays were set to 10, 20, 50, 100, 200, 500, 1000, 1500, and 2000 ms for the R1 experiments and to 17, 34, 51, 68, 85, 102, 136, 170, and 204 ms for the R2 experiments. The relaxation rates and the errors were extracted by a single-exponential curve fitting of the crosspeak intensities using the program Sparky (Goddard and Kneller, Sparky 3, University of California, San Francisco). The 1H-15N steady-state NOE enhancements were recorded in an interleaved manner with or without 1H saturation. A delay of 7.5 s between scans was set to guarantee the longitudinal magnetization recovery. The NOE values were calculated as the ratio of the peak intensities with and without proton saturation. The NOE errors were estimated by repeated experiments.
NMR titrations and chemical-shift perturbation analysis
15N-labeled proteins at a concentration of 0.1–0.2 mM were mixed with unlabeled binding partners with increasing molar ratios. The interactions of Usp251–146 or its variants with monoubiquitin, K48-linked diubiquitin, or SUMO2 were monitored using 1H-15N heteronuclear single-quantum correlation (HSQC) spectra acquired at 25°C on a Bruker 600 MHz NMR spectrometer equipped with a cryoprobe. Backbone resonance assignments of Usp251–146 (BMRB ID: 19111), SUMO2 (BMRB ID: 6801), and ubiquitin (generously provided by Dr. Kylie Walters at the National Cancer Institute) were used in the chemical-shift perturbation (CSP) analysis, and CSP values (Δδ) for 15N and 1H nuclei were calculated using Eq. 1:
| (1) |
where ΔδN and ΔδH represent the CSP values of the amide nitrogen and proton, respectively. Dissociation constants for the binding of monoubiquitin to each domain/motif of Usp251–146 or its variants were evaluated by global fit according to Eq. 2:
| (2) |
in which Δδ is the CSP value, δTOT is the total chemical-shift change between the complexed protein and the free protein, n is the binding stoichiometry, LT is the total ligand concentration, PT is the total protein concentration, and Kd is the dissociation constant.
For NMR competition experiments, 15N-Usp251–146 was first mixed with ubiquitin or K48-linked diubiquitin and then titrated with SUMO2. NMR spectra were analyzed with the program Sparky (Goddard and Kneller, Sparky 3, University of California, San Francisco, San Francisco, CA).
Ub-AMC assay
Ubiquitin with 7-amino-4-methylcoumarin (Ub-AMC) assays were implemented by incubating 50 nM Usp25FL or its mutants with 0.625 μM Ub-AMC in reaction buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 5 mM DTT) at 37°C in the absence or presence of SUMO2 with gradient concentrations. Enzymatic activity analysis was performed by measuring the increase of fluorescence intensity at 460 nm (excitation at 380 nm) in a continuous time mode lasting for 5 min with a total number of 300 time points. Final reported data were derived from three repeated experiments.
K48-linked diubiquitin synthesis and hydrolysis
The synthesis of K48-linked diubiquitin was carried out using ubiquitin, Ub-K48R, E1, E2-25K, and UCH-L3 according to the protocol reported previously (24, 25). Synthesized proteins were further purified by Ni-NTA column and size-exclusion chromatography. Final products were dialyzed into water and lyophilized for further use. The hydrolysis experiments were executed by mixing 2 μM full-length Usp25 (Usp25FL) or its mutants with 20 μM K48-linked diubiquitin substrates in reaction buffer (50 mM Tris-HCl (pH 8.0), 150 mM NaCl, and 5 mM DTT) at 37°C in the absence or presence of 10 μM SUMO2. The hydrolysis efficiencies at different time points were evaluated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (15% gel) and Coomassie blue staining.
Results and Discussion
Solution structure of Usp251–146
To unravel the structural basis for substrate recognition, we characterized the solution structure of the Usp25 N-terminal UBR (the fragment spanning residues 1–146). Compared to the C-terminal domain of Staphylococcus aureus sortase A (SrtA60–206), a well-known globule-like protein of comparable size, Usp251–146 exhibits larger apparent molecular weight (Fig. S1) and hydrated radius (Fig. S2), indicating that its overall molecular shape deviates from sphericity. The NMR-determined structure of Usp251–146 presents as three ordered domains/motifs connected by unfolded linkers (Fig. 1 B). The UBA domain comprises a compact bundle of three α-helices (α1, H16–E26; α2, T33–D42; and α3, L47–A56). Both of the UIMs form a single α-helical structure (α4, D100–A116, and α5, E125–I137). The ensembles of 15 UBA domains or UIM1/2s and the calculated root mean-square deviations (RMSDs) suggest that the folded parts of Usp251–146 are highly ordered (Fig. 1 C; Table 1). We compared UBA/UIM1/UIM2 structures from different proteins and different species (Fig. 1 D). The structures for either UBAs or UIM1/2s overlap very well among proteins, indicating conserved fold patterns in structural evolution. Unlike UIMs, the SIM (the fragment spanning residues N90–D98) of Usp251–146 is totally unstructured (Fig. 1 B). However, Sekiyama et al. (26) have reported that the SIM of MBD1-containing chromatin-associated factor 1 forms a parallel β-sheet pairing with strand β2 of SUMO3. Song et al. (27) have also revealed that the SIM in the protein inhibitor of activated STAT2 (PIAS2) complexed with SUMO1 adopts a short β-sheet structure. Thus, we cannot exclude the possibility that the Usp251–146 SIM may have multiple conformations in molecular recognition. It is worth noting that ∼50% of residues in Usp251–146 are unfolded (Fig. 1 B), which is in agreement with the chemical-shift index data reported previously (17). Due to the lack of NOEs among UBA and UIM domains/motifs, when anchored with the UBA domain, the left regions of Usp251–146 occupy a large space (Fig. S3). Moreover, backbone dynamics data show that most secondary-structure elements of Usp251–146 have typically higher R2/R1 and NOE values compared with unfolded linkers (Fig. S4). Unexpectedly, the R2/R1 or NOE values for the UIM2 motif are comparable to those for the unstructured regions (Fig. S4). It could be inferred that the UIM2 helix is not stable. As well, the unstructured linkers, such as residues 60–80, may form transient secondary structures. Overall, the beads-on-a-string structure of Usp251–146, with high flexibility, is expected to be a key structural feature, in support of recruitment by Usp25 of polyubiquitin chains with various linkage types.
Both UBA domains and UIMs of the Usp25 UBR are involved in ubiquitin recognition
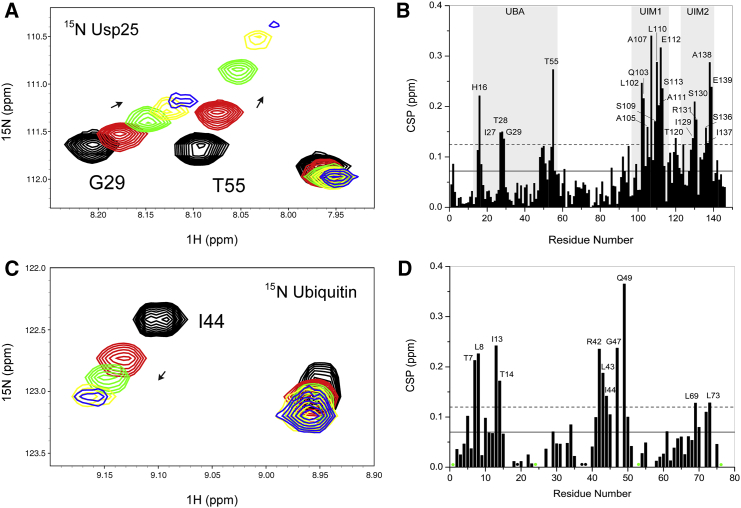
To reveal the binding sites between Usp251–146 and ubiquitin substrates, we performed NMR titration experiments. With the addition of monoubiquitin, a few localized residues of Usp251–146 show dose-dependent CSPs in the 1H-15N HSQC spectra (Fig. 2 A), indicative of a specific interaction between Usp251–146 and monoubiquitin in the fast-exchange regime on the NMR timescale. The calculated CSP values unambiguously disclose that the most disturbed residues are mainly restricted in the UBA domain and UIM1/2 motifs (Fig. 2 B), which indicates that both UBA and UIMs of Usp251–146 are involved in monoubiquitin recognition. In the reverse titration, it can be seen by observing the NMR resonances of monoubiquitin that a few localized residues of monoubiquitin exhibit significant CSPs with the accession of Usp251–146 (Fig. 2 C). The CSP analysis suggests that the canonical hydrophobic patch of monoubiquitin with residue I44 at its center plays a dominant role in its binding to Usp251–146 (Fig. 2 D).
Figure 2.
Characterization of the interaction between the Usp25 UBR and monoubiquitin. (A) Zoomed view of the superimposed 1H-15N HSQC spectra for the 15N-labeled Usp25 UBR titrated with monoubiquitin at Usp25 UBR/monoubiquitin molar ratios of 1:0 (black), 1:5 (red), 1:10 (green), 1:20 (yellow), and 1:30 (blue). (B) CSP analysis for the Usp25 UBR upon its binding to monoubiquitin. (C) Zoomed view of the superimposed 1H-15N HSQC spectra for 15N-labeled monoubiquitin titrated with Usp25 UBR at monoubiquitin/Usp25 UBR molar ratios of 1:0 (black), 1:1 (red), 1:2 (green), 1:5 (yellow) and 1:10 (blue). (D) CSP analysis for monoubiquitin upon its binding to Usp25 UBR. Green dots indicate residues that were undetectable and black dots represent proline residues. Solid and dashed lines in (B) and (D) are indicators for the mean and mean + SD values, respectively. Residues with CSP values greater than the mean + SD value are labeled. To see this figure in color, go online.
Based on the titration data, we evaluated the binding affinities between monoubiquitin and each domain/motif of Usp251–146. The binding of monoubiquitin to UBA, UIM1, or UIM2 of Usp251–146 satisfies a 1:1 binding model. Thus, we fixed n = 1 in Eq. 2 and selected a few representative residues for a global fit (Fig S5, A–C). We determined the dissociation constant (Kd) values for binding of ubiquitin to the UBA domain, UIM1, and UIM2 as 4.01 ± 0.45, 0.26 ± 0.03, and >10 mM, respectively (Table 2). To exclude the effects from other domains/motifs, and to make it clear whether these three domains/motifs recruit monoubiquitin individually or cooperatively, we constructed several variants of Usp251–146. The residues mutated with the purpose of abolishing the binding of the UBA domain and the UIMs to ubiquitin were selected by following the structural hints and the reported references (28, 29, 30, 31). These variants are denoted as UBAm (I27A/I30A/F53A/L54A), UIM1m (I106A/L108A/L110A), UIM2m (I129A/L133A), UBA-UIM1m (I27A/I30A/F53A/L54A/I106A/L108A/L110A), UBA-UIM2m (I27A/I30A/F53A/L54A/I129A/L133A), and UIM1-UIM2m (I106A/L108A/L110A/I129A/L133A). We titrated all the Usp251–146 variants with monoubiquitin and measured the corresponding Kd values (Fig S5, D–L; Table 2). It is clear that a certain domain/motif from the wild-type or the variants of Usp251–146 has comparable affinities for monoubiquitin binding (Table 2), which indicates that the UBA domain, UIM1, and UIM2 act independently in monoubiquitin recognition. Thereby, the rank for affinities between monoubiquitin and each domain/motif of Usp251–146 is UIM1 > UBA > UIM2 (Table 2).
Table 2.
Dissociation Constants for Binding of Each Domain/Motif of Usp251–146 or Its Variants to Monoubiquitin
| WT | UBAm | UIM1m | UIM2m | UBA-UIM1m | UBA-UIM2m | UIM1-UIM2m | |
|---|---|---|---|---|---|---|---|
| UBA | 4.01 ± 0.45 | ND | 3.36 ± 0.14 | 4.56 ± 0.31 | ND | ND | 3.04 ± 0.12 |
| UIM1 | 0.26 ± 0.03 | 0.50 ± 0.02 | ND | 0.48 ± 0.03 | ND | 0.47 ± 0.01 | ND |
| UIM2 | >10 | >10 | >10 | ND | >10 | ND | ND |
All values are represented as the mean ± SD (mM). Kd, dissociation constant; WT, wild-type; ND, not detectable.
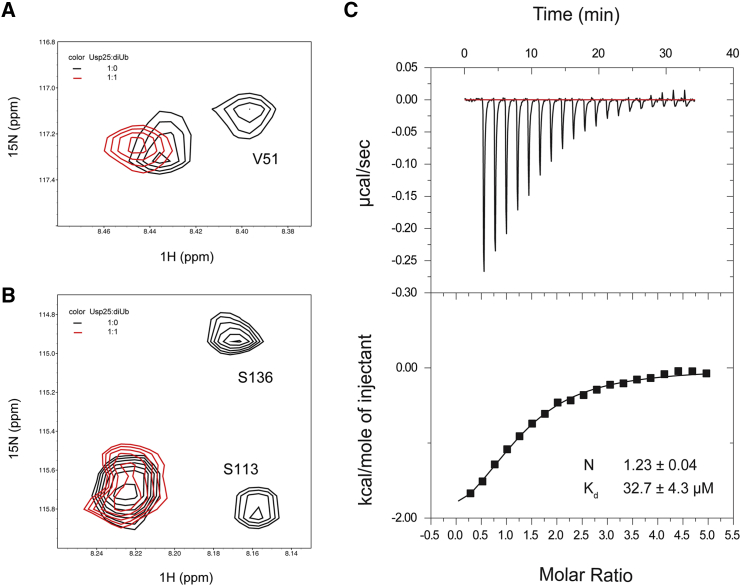
In addition to the monoubiquitinated substrate Ub-AMC, it is reported that Usp25 can catalyze the hydrolysis of polyubiquitin chains efficiently (15). Diubiquitin is the shortest polyubiquitin chain, and Usp25 has robust activities toward all lysine-linked diubiquitin topoisomers (32). Thereby, we further characterized the interaction between Usp251–146 and K48-linked diubiquitin. In the NMR titration using 15N-labeled Usp251–146 and unlabeled K48-diubiquitin, peak intensity attenuations can be clearly observed for representative residues V51 in the UBA domain, S113 in UIM1, and S136 in UIM2 of Usp251–146 (Fig. 3, A and B), which demonstrates that both the UBA domain and the UIMs of Usp251–146 are involved in diubiquitin recognition. In agreement with the NMR observation, the ITC measurement determines the dissociation constant (Kd) as 32.7 ± 4.3 μM (Fig. 3 C) for the Usp25 UBR/K48-linked diubiquitin system.
Figure 3.
Characterization of the interaction between the Usp25 UBR and K48-linked diubiquitin. (A and B) Zoomed view of the superimposed 1H-15N HSQC spectra of the 15N-labeled Usp25 UBR without K48-linked diubiquitin (black) and with equal molar amounts of K48-linked diubiquitin (red). (C) ITC characterization of the interaction between the Usp25 UBR and K48-linked diubiquitin. To see this figure in color, go online.
Usp28 is the closest homolog to Usp25, with a sequence identity of up to 51% (33). The N-terminal UBR of Usp28 contains a UBA domain and a UIM. Our recent study disclosed that the Usp28 UBA domain and UIMs recruit monoubiquitin independently to each other, and the binding stoichiometry of the Usp28 UBR to monoubiquitin is 1:2 (34). In comparison with Usp28, Usp25 UBR has one more UIM that is endowed with the ability to recognize monoubiquitin (Fig. 2 B). Since the UBA domain, UIM1, and UIM2 bind to monoubiquitin independently of each other (Fig S5; Table 2), it could be inferred that the Usp25 UBR interacts with monoubiquitin in a 1:3 binding stoichiometry. On the other hand, different from the monoubiquitin recognition, Usp28 UBR binds to K48-linked diubiquitin at a ratio of 1:1, suggesting a cooperative interaction (34). Our ITC data indicate that the binding stoichiometry for the Usp25 UBR/K48-linked diubiquitin system is 1:1 (Fig. 3 C), which also supports a multivalent binding mode. It is generally believed that the multivalency is an efficient way to achieve physiologically relevant affinities in vivo. In addition, since the UIM1 motif of Usp25 UBR binds to monoubiquitin with the highest affinity (Fig S5; Table 2), we expect that one ubiquitin unit in K48-linked diubiquitin would preferentially interact with the UIM1 motif. The other ubiquitin unit in K48-linked diubiquitin might bind to either the UBA domain or UIM2 motif, and it is likely that a larger population is involved in UBA-domain binding.
SUMO2 non-covalently binds to the Usp25 SIM and impairs the enzymatic activity of the protein
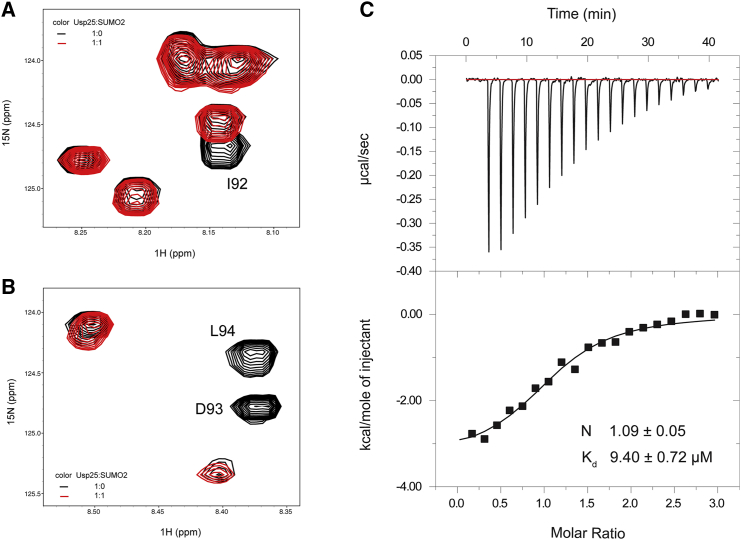
SUMO proteins play an important role in various biological processes, including DNA-damage repair, signal transduction, and cell cycle control (35, 36). Meulmeester et al. (15) have reported that the enzymatic activity of Usp25 is down-regulated by sumoylation. In fact, the non-covalent binding of SUMO is a prerequisite for efficient sumoylation of Usp25 (15, 16). To understand how Usp25 recognizes SUMO molecules, we investigated the non-covalent interaction between Usp251–146 and SUMO2. The NMR titration data show that the resonances for residues V91, I92, D93, L94, and T95 of 15N-labeled Usp251–146 disappear completely in the 1H-15N HSQC spectra with the addition of an equivalent amount of SUMO2 (Fig. 4, A and B). These residues constitute the hydrophobic core of the SIM region (90NVIDLTGDD98), implying that the SIM is responsible for SUMO recognition of the Usp25 UBR. In the reverse titration, using 15N-labeled SUMO2 and unlabeled Usp251–146, plenty of resonances exhibit intensity attenuations (Fig S6), suggesting that the interaction is in the intermediate exchange regime of the NMR timescale. The ITC experiment determines the binding stoichiometry as 1:1 for the Usp25 UBR/SUMO2 system (Fig. 4 C). There is only one SIM in the Usp25 UBR, and hence, the fitted stoichiometry is reasonable and acceptable. The measured Kd value for the interaction between the Usp25 UBR and SUMO2 is 9.40 ± 0.72 μM (Fig. 4 C), which is consistent with our NMR spectral observations.
Figure 4.
Characterization of the interaction between the Usp25 UBR and SUMO2. (A and B) Zoomed view of the superimposed 1H-15N HSQC spectra for 15N-labeled Usp25 UBR without SUMO2 (black) and with equal molar of SUMO2 (red). (C) ITC characterization of the interaction between Usp25 UBR and SUMO2. To see this figure in color, go online.
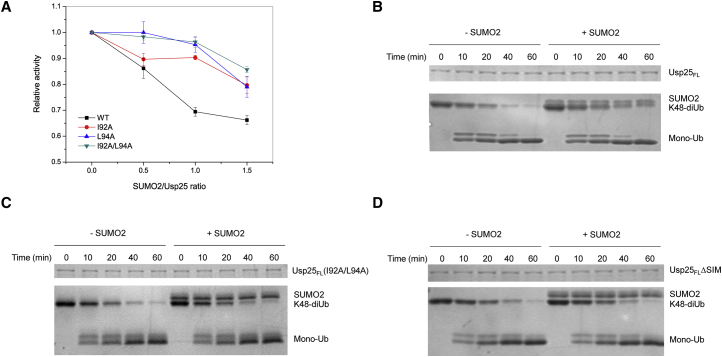
To investigate the effect of SUMO2 binding on the catalytic activity of Usp25, we performed the classic Ub-AMC assay. With the addition of SUMO2, Usp25FL shows significantly reduced enzymatic activity in catalyzing the hydrolysis of Ub-AMC (Fig. 5 A). Based on our structural hints, we then substituted residue I92, residue L94, or both of them with alanine in the SIM of Usp25FL to block SUMO2 binding. In the presence of SUMO2, all of the three variants display much higher catalytic activity in hydrolyzing Ub-AMC compared to the wild-type Usp25FL (Fig. 5 A). We next carried out K48-linked diubiquitin hydrolysis experiments. Consistently, Usp25FL exhibits a lower catalytic efficiency in breaking down the K48-diubiquitin chain when SUMO2 is added (Fig. 5 B). However, for the I92A/L94A variant of Usp25FL, the presence or absence of SUMO2 makes no significant difference in the hydrolysis of K48-linked diubiquitin (Fig. 5 C). A similar result is observed using the other variant of Usp25FL, in which residues 90–98 are deleted (hereby denoted as Usp25FLΔSIM) (Fig. 5 D). All these data suggest that SUMO2 negatively regulates the proteolytic activity of Usp25FL. Taken together, our results demonstrate that the non-covalent binding of SUMO2 to the Usp25 SIM impairs the catalytic activity of the enzyme.
Figure 5.
SUMO2 binding impairs the catalytic activity of Usp25. (A) The catalytic activities of Usp25FL and its variants with and without SUMO2 were characterized by Ub-AMC assay. (B–D) The catalysis efficiencies of (B) Usp25FL, (C) the I92A/L94A variant of Usp25FL, and (D) the Usp25FLΔSIM variant with or without SUMO2 were characterized by K48-diubiquitin hydrolysis assay. To see this figure in color, go online.
SUMO2 prevents the interaction between the Usp25 UBR and ubiquitin substrates in a competitive way
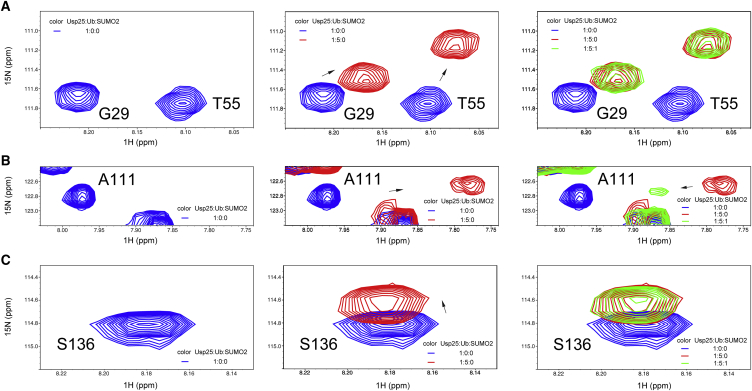
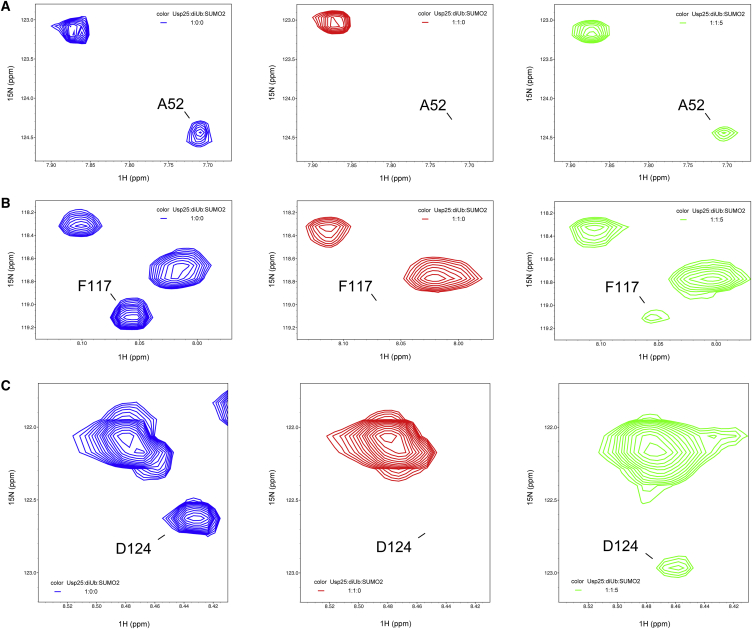
SUMO has a global fold similar to that of ubiquitin. Both ubiquitin substrates and SUMO2 can specifically bind to the N-terminal UBR of Usp25 (Figs. 2, 3, and 4). As the SIM is adjacent to the UIM1, which presents a much higher binding affinity to ubiquitin than do the UBA domain and the UIM2 of Usp25 (Table 2), ubiquitin substrates and SUMO2 may not be able to interact with Usp251–146 simultaneously. In addition, the comparable binding affinities for Usp251–146/K48-linked diubiquitin and Usp251–146/SUMO2 systems (Figs. 3 C and 4 C) suggest a possible competition between K48-linked diubiquitin and SUMO2 when binding to Usp251–146. To clarify whether SUMO2 can competitively block interaction of the Usp25 UBR with ubiquitin substrates, we carried out NMR competition experiments. We first investigated binding of monoubiquitin and SUMO2 to the Usp25 UBR. The representative residues G29 and T55 in the UBA domain, A111 in UIM1, and S136 in UIM2 of 15N-labeled Usp251–146 all exhibit CSPs with the addition of monoubiquitin (Fig. 6). Different from the residues in the UBA domain and UIM2, residue A111 in UIM1 shows a tendency to move back to its initial position in the presence of SUMO2 (Fig. 6). This observation indicates that SUMO2 can block the binding of monoubiquitin to the UIM1 proximal to the SIM, but not the binding to the distal UBA domain and UIM2. We next investigated the binding of K48-linked diubiquitin and SUMO2 to the Usp25 UBR. With the addition of K48-linked diubiquitin, the resonances of representative residues A52 in UBA, F117 in UIM1, and D124 in UIM2 for 15N-labeled Usp251–146 attenuate significantly in the 1H-15N HSQC spectra (Fig. 7). When SUMO2 is sequentially added, these previously attenuated signals display obvious intensity recovery (Fig. 7). Our data strongly suggest that the non-covalent binding of SUMO2 to the Usp25 UBR prevents its interaction with ubiquitin substrates. Such observations well explain why the deubiquitinating activity of Usp25 is inhibited by SUMO2 binding.
Figure 6.
Competitive binding of monoubiquitin and SUMO2 to Usp251–146 detected by 1H-15N HSQC titration experiments. CSPs are presented for the representative residues (A) G29 and T55 in UBA, (B) A111 in UIM1, and (C) S136 in UIM2 of the 15N-labeled Usp25 UBR with the sequential addition of monoubiquitin and SUMO2. To see this figure in color, go online.
Figure 7.
SUMO2 competitively prevents interaction between the Usp25 UBR and K48-linked diubiquitin. Representative residues including (A) A52 in UBA, (B) F117 in UIM1, and (C) D124 in UIM2 of the 15N-labeled Usp25 UBR show intensity attenuation and recovery in 1H-15N HSQC spectra when K48-linked diubiquitin and SUMO2 are added sequentially. To see this figure in color, go online.
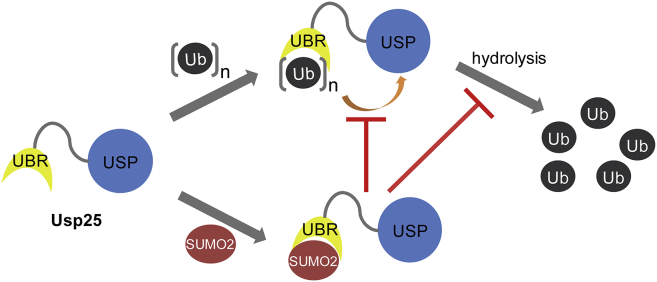
The functions of DUBs are tightly controlled in vivo, which can be achieved in several ways, including substrate-induced conformational changes, binding to adaptor proteins, proteolytic cleavage, and post-translational modifications (32, 37). DUB members in the USP subfamily generally share a conserved papain-like protease domain but contain variable external modulators (38), which indicate that their functions are regulated in different ways. Based on our findings, we proposed a working model to depict the regulatory role of the Usp25 UBR in the functional display of the enzyme (Fig. 8). On one hand, the N-terminal UBR helps Usp25 recruit ubiquitin substrates to its catalytic USP domain, promoting the deubiquitinating reaction of the enzyme. On the other hand, SUMO2 non-covalently binds to the Usp25 UBR and impairs the enzymatic activity of Usp25 by competitively blocking its interaction with ubiquitin substrates. Usp25 adopts such a competitive mechanism to balance its deubiquitinating activity.
Figure 8.
A working model to depict the regulatory role of the Usp25 UBR in the functional display of the enzyme. To see this figure in color, go online.
Interestingly, similar to Usp25, receptor-associated protein 80 (Rap80), which has a critical role in the DNA-damage response, possesses a tandem SIM-UIM-UIM in its N-terminus (39). It is reported that the Rap80 SIM-UIM-UIM region can bind to both K63-linked polyubiquitin and SUMO2 conjugates simultaneously, and that a coordinated ubiquitin and SUMO modification is required for the recruitment of Rap80 to DNA double-strand breaks (39). Noticeably, the SIM and UIM1 motifs in Rap80 are composed of residues 40–47 and 79–96, respectively. The 32-amino-acid-long linker between the two motifs ensures enough space to accommodate ubiquitin substrates and SUMO at the same time. In the case of Usp25, however, there is almost no extra residue between SIM and UIM1. The compact arrangement of motifs in Usp25 determines the incompatibility in the binding of ubiquitin substrates and SUMO. Thus, the SIM acts as a negative modulator rather than a positive stimulator to control the DUB function of Usp25. In cells, the level of SUMO undergoes significant changes under certain circumstances (36, 40, 41). The accompanying activity variation of Usp25 is thought to contribute to subsequent biological responses or pathological changes.
Conclusion
In summary, we have investigated the structure and function of the N-terminal UBR of Usp25 in this study. Our results shed light on the underlying mechanism in the regulation of the DUB function of Usp25 and will be helpful for inhibitor discoveries in the future.
Author Contributions
N.Z., Y.W., and H.-Y.H. designed the research. Y.Y., L.S., and Y.D. prepared the protein samples. Y.Y., L.S., and Y.W., performed the spectroscopic experiments. Y.Y., L.S., Y.S., Y.W., and N.Z. analyzed the data. Y.W. and N.Z. wrote the manuscript.
Acknowledgments
The plasmid of full-length Usp25m was generously given by Dr. Frauke Melchior (Department of Biochemistry, Faculty of Medicine, Georg-August-University of Göttingen, Göttingen, Germany). The resonance assignments for amide groups of ubiquitin were provided by Dr. Kylie J. Walters (National Cancer Institute of the NIH, Bethesda, MA). This work was funded by grants from the Institutes for Drug Discovery and Development, Chinese Academy of Sciences (nos. CASIMM0120164002 and CASIMM0120163013 to Naixia Zhang), the National Natural Science Foundation of China (no. 21272246 to Naixia Zhang and no. 31300608 to Yi Wen), and the National Key Basic Research Program of China (no. 2013CB910900 to Naixia Zhang).
Editor: David Eliezer.
Footnotes
Yuanyuan Yang and Li Shi contributed equally to this work.
Six figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30438-1.
Contributor Information
Yi Wen, Email: wenyi1984112@simm.ac.cn.
Naixia Zhang, Email: nxzhang@simm.ac.cn.
Supporting Material
References
- 1.Ikeda F., Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rape M. Ubiquitin, infinitely seductive: symposium on the many faces of ubiquitin. EMBO Rep. 2009;10:558–562. doi: 10.1038/embor.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Song L., Rape M. Reverse the curse—the role of deubiquitination in cell cycle control. Curr. Opin. Cell Biol. 2008;20:156–163. doi: 10.1016/j.ceb.2008.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reyes-Turcu F.E., Wilkinson K.D. Polyubiquitin binding and disassembly by deubiquitinating enzymes. Chem. Rev. 2009;109:1495–1508. doi: 10.1021/cr800470j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussain S., Zhang Y., Galardy P.J. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle. 2009;8:1688–1697. doi: 10.4161/cc.8.11.8739. [DOI] [PubMed] [Google Scholar]
- 6.Hegde A.N., Upadhya S.C. Role of ubiquitin-proteasome-mediated proteolysis in nervous system disease. Biochim. Biophys. Acta. 2011;1809:128–140. doi: 10.1016/j.bbagrm.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraile J.M., Quesada V., López-Otín C. Deubiquitinases in cancer: new functions and therapeutic options. Oncogene. 2012;31:2373–2388. doi: 10.1038/onc.2011.443. [DOI] [PubMed] [Google Scholar]
- 8.Kim J.H., Park K.C., Chung C.H. Deubiquitinating enzymes as cellular regulators. J. Biochem. 2003;134:9–18. doi: 10.1093/jb/mvg107. [DOI] [PubMed] [Google Scholar]
- 9.Sacco J.J., Coulson J.M., Urbé S. Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB Life. 2010;62:140–157. doi: 10.1002/iub.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi D., Grossman S.R. Ubiquitin becomes ubiquitous in cancer: emerging roles of ubiquitin ligases and deubiquitinases in tumorigenesis and as therapeutic targets. Cancer Biol. Ther. 2010;10:737–747. doi: 10.4161/cbt.10.8.13417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong B., Liu X., Dong C. Negative regulation of IL-17-mediated signaling and inflammation by the ubiquitin-specific protease USP25. Nat. Immunol. 2012;13:1110–1117. doi: 10.1038/ni.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blount J.R., Burr A.A., Todi S.V. Ubiquitin-specific protease 25 functions in endoplasmic reticulum-associated degradation. PLoS One. 2012;7:e36542. doi: 10.1371/journal.pone.0036542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J., Tan Q., Yao M. miRNA-200c inhibits invasion and metastasis of human non-small cell lung cancer by directly targeting ubiquitin specific peptidase 25. Mol. Cancer. 2014;13:166. doi: 10.1186/1476-4598-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bosch-Comas A., Lindsten K., Marfany G. The ubiquitin-specific protease USP25 interacts with three sarcomeric proteins. Cell. Mol. Life Sci. 2006;63:723–734. doi: 10.1007/s00018-005-5533-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meulmeester E., Kunze M., Melchior F. Mechanism and consequences for paralog-specific sumoylation of ubiquitin-specific protease 25. Mol. Cell. 2008;30:610–619. doi: 10.1016/j.molcel.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Denuc A., Bosch-Comas A., Marfany G. The UBA-UIM domains of the USP25 regulate the enzyme ubiquitination state and modulate substrate recognition. PLoS One. 2009;4:e5571. doi: 10.1371/journal.pone.0005571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi L., Wen Y., Zhang N. 1H, 13C and 15N backbone and side-chain resonance assignments of the N-terminal ubiquitin-binding domains of USP25. Biomol. NMR Assign. 2014;8:255–258. doi: 10.1007/s12104-013-9495-1. [DOI] [PubMed] [Google Scholar]
- 18.Delaglio F., Grzesiek S., Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 19.Keller R. Cantina Verlag; Goldau, Switzerland: 2004. The Computer Aided Resonance Assignment Tutorial. [Google Scholar]
- 20.Shen Y., Delaglio F., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwieters C.D., Kuszewski J.J., Clore G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 22.Schwieters C.D., Kuszewski J.J., Clore G.M. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 2006;48:47–62. [Google Scholar]
- 23.Laskowski R.A., Rullmannn J.A., Thornton J.M. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 24.Raasi S., Pickart C.M. Ubiquitin chain synthesis. Methods Mol. Biol. 2005;301:47–55. doi: 10.1385/1-59259-895-1:047. [DOI] [PubMed] [Google Scholar]
- 25.Zhou Z.R., Zhang Y.H., Hu H.Y. Length of the active-site crossover loop defines the substrate specificity of ubiquitin C-terminal hydrolases for ubiquitin chains. Biochem. J. 2012;441:143–149. doi: 10.1042/BJ20110699. [DOI] [PubMed] [Google Scholar]
- 26.Sekiyama N., Ikegami T., Shirakawa M. Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3. J. Biol. Chem. 2008;283:35966–35975. doi: 10.1074/jbc.M802528200. [DOI] [PubMed] [Google Scholar]
- 27.Song J., Zhang Z., Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 28.Bertolaet B.L., Clarke D.J., Reed S.I. UBA domains of DNA damage-inducible proteins interact with ubiquitin. Nat. Struct. Biol. 2001;8:417–422. doi: 10.1038/87575. [DOI] [PubMed] [Google Scholar]
- 29.Chen L., Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol. Cell. Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q., Goh A.M., Walters K.J. Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry. 2003;42:13529–13535. doi: 10.1021/bi035391j. [DOI] [PubMed] [Google Scholar]
- 31.Bai J.J., Safadi S.S., Shaw G.S. Ataxin-3 is a multivalent ligand for the parkin Ubl domain. Biochemistry. 2013;52:7369–7376. doi: 10.1021/bi400780v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faesen A.C., Luna-Vargas M.P., Sixma T.K. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem. Biol. 2011;18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Valero R., Bayes M., Marfany G. Characterization of alternatively spliced products and tissue-specific isoforms of USP28 and USP25. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-10-research0043. RESEARCH0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wen Y., Shi L., Zhang N. The N-terminal ubiquitin-binding region of ubiquitin-specific protease 28 modulates its deubiquitination function: NMR structural and mechanistic insights. Biochem. J. 2015;471:155–165. doi: 10.1042/BJ20150088. [DOI] [PubMed] [Google Scholar]
- 35.Johnson E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 36.Hay R.T. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 37.Kessler B.M., Edelmann M.J. PTMs in conversation: activity and function of deubiquitinating enzymes regulated via post-translational modifications. Cell Biochem. Biophys. 2011;60:21–38. doi: 10.1007/s12013-011-9176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nijman S.M., Luna-Vargas M.P., Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 39.Hu X., Paul A., Wang B. Rap80 protein recruitment to DNA double-strand breaks requires binding to both small ubiquitin-like modifier (SUMO) and ubiquitin conjugates. J. Biol. Chem. 2012;287:25510–25519. doi: 10.1074/jbc.M112.374116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilkinson K.A., Henley J.M. Mechanisms, regulation and consequences of protein SUMOylation. Biochem. J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saitoh H., Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.