Abstract
Objectives
Genome-wide association studies (GWAS) have identified >100 loci independently contributing to type 2 diabetes (T2D) risk. However, translational implications for precision medicine and for the development of novel treatments have been disappointing, due to poor knowledge of how these loci impact T2D pathophysiology. Here, we aimed to measure the expression of genes located nearby T2D associated signals and to assess their effect on insulin secretion from pancreatic beta cells.
Methods
The expression of 104 candidate T2D susceptibility genes was measured in a human multi-tissue panel, through PCR-free expression assay. The effects of the knockdown of beta-cell enriched genes were next investigated on insulin secretion from the human EndoC-βH1 beta-cell line. Finally, we performed RNA-sequencing (RNA-seq) so as to assess the pathways affected by the knockdown of the new genes impacting insulin secretion from EndoC-βH1, and we analyzed the expression of the new genes in mouse models with altered pancreatic beta-cell function.
Results
We found that the candidate T2D susceptibility genes' expression is significantly enriched in pancreatic beta cells obtained by laser capture microdissection or sorted by flow cytometry and in EndoC-βH1 cells, but not in insulin sensitive tissues. Furthermore, the knockdown of seven T2D-susceptibility genes (CDKN2A, GCK, HNF4A, KCNK16, SLC30A8, TBC1D4, and TCF19) with already known expression and/or function in beta cells changed insulin secretion, supporting our functional approach. We showed first evidence for a role in insulin secretion of four candidate T2D-susceptibility genes (PRC1, SRR, ZFAND3, and ZFAND6) with no previous knowledge of presence and function in beta cells. RNA-seq in EndoC-βH1 cells with decreased expression of PRC1, SRR, ZFAND6, or ZFAND3 identified specific gene networks related to T2D pathophysiology. Finally, a positive correlation between the expression of Ins2 and the expression of Prc1, Srr, Zfand6, and Zfand3 was found in mouse pancreatic islets with altered beta-cell function.
Conclusions
This study showed the ability of post-GWAS functional studies to identify new genes and pathways involved in human pancreatic beta-cell function and in T2D pathophysiology.
Keywords: EndoC-βH1, Expression analysis, Genome-wide association study, Insulin secretion, RNAi screening, Type 2 diabetes
Highlights
-
•
Expression of genes located nearby T2D associated signals is enriched in β cells.
-
•
Knockdown of 7 T2D genes with known role in β cell changes insulin secretion.
-
•
Knockdown of 4 T2D genes with unknown role in β cell impairs insulin secretion.
-
•
RNA-seq in cells with knockdown of these 4 genes detected T2D-related networks.
1. Introduction
Type 2 diabetes (T2D) is a complex, multifactorial disorder with an estimated heritability ranging between 40% and 70% [1]. Genetic studies have identified 28 genes causing monogenic diabetes due to impaired insulin secretion. These genes are critically important for pancreatic beta-cell lineage, phenotype, and function [1]. Genome-wide association studies (GWAS) have shown that common T2D is highly polygenic, with more than 100 loci contributing to T2D risk. GWAS of related quantitative traits have shown that a significant part of the T2D-associated single nucleotide polymorphisms (SNPs) also modulates fasting plasma glucose levels and the homeostatic model assessment (HOMA) of beta-cell function in non-diabetic individuals, suggesting that their main diabetogenic impact was on insulin secretion and not on insulin resistance [2]. Surprisingly indeed, only a few T2D-associated SNPs were found to have an effect on fasting serum insulin levels, the HOMA of insulin resistance, or the insulin sensitivity index in general populations. About half of the T2D susceptibility SNPs did not have any apparent effect on glucose homeostasis in non-diabetics [2].
Elucidating the function of T2D-associated SNPs (and target genes) and their involvement in T2D pathophysiology may have major translational implications for precision medicine and for the development of novel treatments. Post-GWAS studies are challenging due to the fact that the vast majority of these SNPs are non-coding and often intergenic, which has not facilitated functional investigations of genes located in these chromosome regions. In this context, a recent study has demonstrated that T2D-associated SNPs are significantly enriched in clusters of enhancers that are active in human pancreatic islets, and most of these enhancers map less than 500 kb from transcription start sites (TSS) of nearby genes [3]. Moreover, it has been recently shown that most genes located nearby loci associated with blood pressure (systolic, diastolic, pulse pressure) are highly expressed in vascular tissues [4]. These data suggested that focusing the expression studies and functional investigations on genes nearby GWAS-identified SNPs may be a good first strategy, as they may drive pathways involved in disease pathophysiology.
Here, we aimed to identify novel genes contributing to T2D risk through a dominant effect on insulin secretion in human. To do so, we first performed a comprehensive expression study of candidate T2D susceptibility genes closest to all GWAS-identified T2D SNPs in a large panel of human organs, tissues, and cells, followed by the functional analysis of the knockdown of these genes in human pancreatic beta-cell lines. Using this strategy, we first found that the expression of tested candidate T2D susceptibility genes was significantly and specifically enriched in pancreatic beta cells, and we reported functional evidence for a role in insulin secretion of four T2D susceptibility genes (PRC1, SRR, ZFAND3, and ZFAND6) with previously unknown presence and function in pancreatic beta cells.
2. Material and methods
2.1. Samples included in the panel expression study
Total RNA from human colon, liver, kidney, adipose tissue, lung, skeletal muscle, heart, brain, small intestine, substantia nigra, hippocampus, dorsal root ganglion, and insula, and Poly A + RNA from human hypothalamus, pituitary gland, caudate nucleus, and frontal lobe were purchased from Clontech Laboratories (Palo Alto, CA, USA). Pancreatic islets (n = 8; average purity: 70.0%; average viability: 94.3%) and exocrine pancreas (n = 2) were isolated from adult brain-dead donors without diabetes, in accordance with French and Italian local ethics committee approval [5], [6]. Pancreatic beta cells were obtained by laser capture microdissection (LCM beta cells; n = 2) [7] or were sorted by flow cytometry (FACS sorted beta cells; n = 5), as previously described [8]. Primary pre-adipocytes (Lonza, Basel, Switzerland) came from subcutaneous fat of a non-diabetic Caucasian female patient. These pre-adipocytes were differentiated into mature adipocytes in PBM-2 medium supplemented with insulin, dexamethasone, phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine (IBMX), and indomethacin (all supplied by Lonza) for 10 days according to the manufacturer's recommendations. RNA samples from human EndoC-βH1 cells (n = 3), pre-adipocytes, and mature adipocytes were extracted as described below (see Section 2.9).
2.2. Probe design and selection for the panel expression study
Capture probes (containing a biotin affinity tag) and reporter probes (containing a color-coded molecular barcode) were designed to detect 148 targets, including five housekeeping genes for normalization, genes whose expression is known to be enriched (‘markers’) in pancreatic islet including beta cell (n = 13), gut (n = 3), kidney (n = 4), lung (n = 3), and adipose tissue (n = 3), 28 genes known to be involved in monogenic diabetes, and 104 genes located nearby GWAS-identified SNPs associated with T2D risk (Table A) [1]. The expression of the present housekeeping genes was previously shown to be uniform across a large panel of human tissues using RNA-sequencing (RNA-seq) [9]. Here, we found that the expression of the five housekeeping genes was highly correlated across the present panel of human tissues (R2 > 0.85). The genes for which expression is known to be enriched in specific tissues were chosen according to the literature, the GTEx portal [10] and/or the BioGPS portal [11].
2.3. Panel gene expression study and statistical analysis
The detection of transcripts was carried out in multiplexed hybridization reactions using NanoString Technologies (Seattle, WA, USA), following manufacturer's protocol. Each hybridization reaction contained 25–100 ng total RNA or 5 ng Poly A + RNA at a final concentration of 0.8–3.3 ng/ul and 0.16 ng/ul respectively. Reagents were mixed and incubated at 65 °C in a thermocycler block with a heated lid for 16 h. All post-hybridization steps were handled robotically on a custom liquid-handling robot (Prep Station, NanoString Technologies). Finally, samples were loaded into a microfluidic chamber (nCounter Cartridge, NanoString Technologies) and imaged in a Digital Analyzer (NanoString Technologies). Barcodes were counted in 555 fields of view per sample. Experimental quality control was performed with nSolver (NanoString Technologies) to flag failed samples. Normalized expression values were first obtained by considering the logarithm of the ratio of the expression of a given gene over the average expression of the set of five housekeeping genes (Table A) in corresponding samples. The expression profiles of relevant gene sets were analyzed through heat map representations. A double hierarchical clustering of the tissues and gene expressions in each set was performed, using Ward's method. Log2 expression was centered and scaled. Each cell was colored to quantitatively reflect the relative expression: from green if the gene is over-expressed in the tissue compared to all other tissues, to red otherwise. For enrichment analyses, we established an expression threshold to classify ‘under-expressed’ and ‘over-expressed’ sets of genes (i.e. genes involved in monogenic diabetes, candidate susceptibility genes for T2D, and the different markers) in each panel tissue. For this purpose, the threshold was defined as the average gene expression across all tissues, plus 1.5 standard deviation (SD). We built a contingency table to count the number of genes from each set that were under- and over-expressed in the tested tissue and in the rest of the panel tissues. Fisher's exact test was applied to test whether the gene set was significantly over-expressed in each tested tissue from the panel.
2.4. Culture of EndoC-βH1 cells
EndoC-βH1 cells [12] were cultured in low-glucose (5.6 mM) Dulbecco's modified Eagle's medium (Sigma–Aldrich, St. Louis, MO, USA) with 2% BSA fraction V (Roche Diagnostics, Basel, Switzerland), 50 μM 2-mercaptoethanol, 10 mM nicotinamide (Calbiochem, Merck Millipore, Billerica, MA, USA), 5.5 mg/ml human transferrin (Sigma–Aldrich), 6.7 ng/ml sodium selenite (Sigma–Aldrich), 100 U/ml penicillin, and 100 mg/ml streptomycin (Life Technologies, Carlsbad, CA, USA). Cells were seeded at a density of 2.5 × 106 on Matrigel (1%)/fibronectin (2 mg/ml; Sigma–Aldrich) coated plates and cultured at 37 °C and 5% CO2.
2.5. Transfection of siRNA into EndoC-βH1 cells
SiRNA were transfected into EndoC-βH1 cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Freshly trypsinized EndoC-βH1 (1.5 × 106 cells) were incubated in suspension with Lipofectamine-siRNA complex in Opti-MEM (Invitrogen) for 10 min and then were plated onto Matrigel-fibronectin-coated culture wells. Six hours later, the medium was replaced. We used ON-TARGETplus siRNA SMARTpool for each gene (20–30 nM) and ON-TARGETplus nontargeting pool for controls (siNTP; Dharmacon, Thermo Fisher Scientific, Waltham, MA, USA). Cells were analyzed 72 h post transfection.
2.6. Assessment of insulin secretion from EndoC-βH1 cells
Transfected EndoC-βH1 cells were seeded onto Matrigel-fibronectin-coated 96-well plates at 5 × 104 cells/well. Three days post transfection, cells were incubated overnight in culture medium that contained 2.8 mM glucose and then in HEPES-buffered Krebs–Ringer Buffer (KRB; 116 mmol/l NaCl, 5.06 mmol/l KCl, 1.007 mmol/l CaCl2, 1.01 mmol/l MgCl2, 23.96 mmol/l NaHCO3, 10 mmol/l HEPES, pH 7.4, and 0.2% BSA solution) that contained 0.5 mM glucose ± 0.5 mM IBMX (Sigma Aldrich) for 60 min. This supernatant was subsequently collected (supernatant 1) and replaced with 16.7 mM glucose KRB ± 0.5 mM IBMX for 60 min at 37 °C and then collected (supernatant 2). For insulin content measurement, cells were lysed with TETG buffer (20 mM Tris–HCl pH 8.0, 137 mM NaCl, 1% Triton X-100, 10% Glycerol, 2 mM EGTA with protease inhibitors; Roche). Lysate and supernatants were centrifuged for 5 min at 700 g. Samples were kept frozen at −20 °C before use. Insulin concentration in the supernatants and intracellular content of the EndoC-βH1 cells were measured by ELISA according to manufacturer's instructions using the Human Insulin Kit (Mercodia, Uppsala, Sweden). Insulin content was used for normalization.
2.7. Statistical analysis of insulin secretion from EndoC-βH1 cells
Absorbance data were measured, with technical duplicates of experimental triplicates, leading to six measurements maximum per experimental conditions (that were subsequently repeated). To ensure a reduced technical bias from the absorbance data, the technical duplicates' average absorbance was kept when the relative error was lower than 20% among the technical duplicates (this threshold being based on the observed distribution of the technical relative errors over 100 experiments). Fold changes of insulin secretion (i.e. secretion at stimulatory glucose levels divided by secretion at basal glucose levels) were then computed for each siRNA. Fold change of insulin secretion for each siRNA was analyzed using a linear regression adjusted for experimental conditions (operator and date).
2.8. Measurement of cell viability of EndoC-βH1 cells
Cell viability was measured by quantifying the amount of ATP present, which indicates the presence of metabolically active cells, through the CellTiter-Glo 2.0 Assay (Promega, Madison, WI, USA), following manufacturer's protocol. Luminescence measurement was performed at baseline and 72 h after transfection of siRNA.
2.9. Mice and mouse islet isolation
Control female C57BL/6J mice and obese (ob/ob) female mice of the C57BL/6J background were obtained from Elevage Janvier (Le Genest-Saint-Isle, France). Isolation and islet cultures were performed as previously described [13]. Briefly, mouse islets were maintained in islets medium (RPMI 1640; GIBCO Life Technologies) with serum (FCS; PAA laboratories, GE Healthcare, Velizy-Villacoublay, France), 1 mM sodium pyruvate, 50 μM β-mercaptoethanol, 10 mM Hepes and 100 U/ml penicillin, and 100 mg/ml streptomycin (Life Technologies). For the streptozotocin (STZ) treated islets (from control mice), 2 mM of Streptozocin Sigma S0130 (Sigma–Aldrich) were added in the islet medium for 24 h. All animal procedures were conducted in accordance with the institutional guidelines approved by the institutional Animal Care and Ethical Use Committee of the University of Lille, France.
2.10. RNA isolation, reverse transcription, and quantitative RT-PCR
Total RNA was extracted 72 h post transfection from EndoC-βH1 cell using the NucleoSpin RNA II kit (Macherey–Nagel, Düren, Germany) following the manufacturer's instructions. Total RNA from mouse pancreatic islets was extracted using the RNeasy Mini Kit (Qiagen, Courtabœuf, France).
The assessment of purity and quantity of extracted RNA was performed using NanoDrop 2000 spectrophotometer (Thermo Fisher). cDNA was synthetized from 400 ng of total RNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions.
RT-qPCR was performed using Brilliant III QRT-PCR SYBR Green Low ROX Master Mix (Agilent, Santa Clara, CA, USA) and was analyzed through the ViiA 7 Real-Time PCR System (Applied Biosystems). Knockdown efficiency was assessed by RT-qPCR of the target genes using the following PrimeTime qPCR Primers or designed primers (Integrated DNA Technologies): ZFAND3 (Hs.PT.58.1283673), ZFAND6 (Hs.PT.58.14433903), PRC1 (Hs.PT.58.50434811), SRR (Hs.PT.58.25800883), KCNK16 (Hs.PT.58.20909681), CDKN2A (Hs.PT.58.40743463g), SLC30A8 (Hs.PT.56a.14618432), TBC1D4 (Hs.PT.58.45499), TCF19 (Hs.PT.58.23138597), HNF4A (Forward: 5′-GCC ATC ATC TTC TTT GAC CCA-3′; Reverse: 5′-GAT GTA GTC CTC CAA GCT CAC-3′), GCK (Forward: 5′-TGA AGG TGG GAG AAG GTG AG-3′; Reverse: 5′-GAT GCA CTC AGA GAT GTA GTC G-3′), MPHOSPH9 (Hs.PT.58.3849657), SSR1 (Hs.PT.58.2437881), FAF1 (Hs.PT.58.39097947), KLHDC5 (Hs.PT.58.21065180), in triplicate on 384-well plates. PSMB2 (Forward: 5′-TTG TCC AGA TGA AGG ACG ATC A-3′; Reverse: 5′-AGC CTC TCC AAC ACA CAG GA-3′) endogenous control gene was used to normalize gene expression by ΔΔCt method, where the final normalized quantity was expressed as 2(Ct target – Ct siNTP). For the gene expression study in mouse pancreatic islets, we followed the same methods and used the following primers: Prc1 (Forward: 5′-AGC CTG TGG AGG CAA TTATG-3′; Reverse: 5′-AAC CGT ACA ATC TCG GCA TC-3′), Srr (Forward: 5′-CCT GCA GTG ATA GCT GGA CA-3′; Reverse: 5′-AAG CCA ATG CTG GAT TTG AC-3′), Zfand3 (Forward: 5′-GGG GTC CAG CAA GAC TAT GA-3′; Reverse: 5′-CTA CTA GTC GCG GTC GCT TC-3′), Zfand6 (Forward: 5′-GAG ACA GAA GAC CTG CAA GGA-3′; Reverse: 5′- ACG GTG CAC ACC ACA GTA AA-3′) and Ins2 (Forward: 5′-TCT AGT TGC AGT AGT TCT CCA-3′; Reverse: 5′-TGG CTT CTT CTA CAC ACC CA-3′). Actb (Forward: 5′-CAG CAG ATG TGG ATC AGC AAG-3′; Reverse: 5′-AGC TCA GTA ACA GTC CGC C-3′) endogenous control gene was used to normalize gene expression.
2.11. RNA-seq
RNA-seq was performed in RNA samples that were extracted from EndoC-βH1 cells transfected with either siNTP, siPRC1, siSRR, siZFAND6, or siZFAND3, from at least three independent transfections. RNA libraries were prepared using the TruSeq Stranded mRNA Library Preparation Kit (Illumina, San Diego, CA, USA) following the manufacturer's instructions. The libraries were sequenced using the HiSeq 4000 (Illumina). A mean of 65 million paired-end reads of 75 bp were generated for each sample. More than 90% of the reads for each library were effectively mapped to the hg19 human genome assembly using TopHat2 [14]. Subsequently, both quantification and annotation of the reads were performed using Bioconductor package Rsubread [15]. Finally, the differential gene expression analyses (i.e. EndoC-βH1 cells transfected with siNTP versus EndoC-βH1 cells transfected with either siPRC1, siSRR, siZFAND6, or siZFAND3) were performed using Bioconductor package DESeq2 [16]. The differentially expressed genes were subjected to Ingenuity Pathway Analysis (Qiagen, Hilden, Germany) to decipher the major biological pathways, networks, and diseases emphasized by the significantly deregulated genes (with a p-value < 0.05).
2.12. Immunofluorescence
Isolated human islet clusters or tissue sections from normal human pancreases were collected and fixed in 4% (v/v) paraformaldehyde overnight. Before immunofluorescence, sections were deparaffinized with xylene and rehydrated with a series of alcohol solutions of decreasing concentration (90–70–50%). Sections and cells attached to chambers were then washed with PBS. Antibody dilutions and rinsing steps were performed in PBS. Incubations with primary antibodies were done at 4 °C overnight. For double staining, slides were pre-treated 20 min with 0.5% Triton X-100 and 30 min with 0.1% BSA at room temperature. Slides were then incubated with a guinea pig anti-insulin antibody (1:500; Abcam, Cambridge, UK) and either rabbit anti-PRC1 (1:100; Abcam), rabbit anti-SRR (1:100; Abcam), rabbit anti-ZFAND3 (1:100; Novus Biologicals, Littleton, CO, USA), or rabbit anti-ZFAND6 (1:100; Novus Biologicals) antibodies. After rinsing with PBS, slides were incubated for 1 h with either an Alexa fluor 594-conjugated anti-guinea antibody (1:500; Thermo Fisher Scientific) or Alexa fluor 488-conjugated anti-rabbit antibody (1:500; Thermo Fisher Scientific). For immunofluorescence of EndoC-βH1 cells, after blocking with PBS 5% BSA for 30 min, cells were incubated with the primary antibodies for 1 h. Immunofluorescence staining in tissue sections from normal human pancreases was performed when the immunofluorescence staining in isolated human islet clusters was not clear enough.
3. Results
3.1. Expression of candidate T2D susceptibility genes in a large panel of human organs, tissues, and cells
We analyzed the expression profile of 104 candidate T2D susceptibility genes (Table A) located nearby GWAS-identified SNPs associated with T2D risk [1] in a panel of human organs, tissues, and cells including colon, small intestine, liver, kidney, adipose tissue, pre-adipocytes, mature adipocytes, lung, skeletal muscle, heart, whole brain, substantia nigra, hippocampus, insula, hypothalamus, pituitary gland, caudate nucleus, frontal lobe, dorsal root ganglion, pancreatic islets, LCM beta cells, FACS sorted beta cells, exocrine pancreas, and the human pancreatic beta-cell line EndoC-βH1 [12]. For this purpose, we used the PCR-free NanoString technology enabling the count of messenger RNA molecules without any biases due to amplification step. The heat map representation reporting the expression profile of 104 candidate susceptibility genes for T2D in the panel of human tissues is shown in Figure A.
In order to validate this methodology, we first explored in the same panel of human samples the expression of genes (i.e. ‘markers’) known to be highly (or even specifically) expressed in the gut (Figure B1), adipose tissue (Figure C1), kidney (Figure D1), lung (Figure E1), and pancreatic islet (Figure F1). Furthermore, we investigated the expression of 28 genes known to be involved in monogenic diabetes (Figure G), as the proteins encoded by these genes have a well-known role in the function and/or development of pancreatic beta cells [1]. As expected, we confirmed a significant enrichment of our gut markers expression in small intestine (p = 6.7 × 10−5; Figure B2), of adipose tissue markers expression in mature adipocytes and adipose tissue (p = 3.4 × 10−4; and p = 3.4 × 10−4, respectively; Figure C2), of kidney markers expression in kidney (p = 1.5 × 10−6; Figure D2), and of lung markers expression in lung (p = 1.7 × 10−5; Figure E2). Importantly, we found that the expression of the pancreatic islet markers is significantly enriched in pancreatic islets, FACS sorted beta cells, LCM beta cells, and EndoC-βH1 cells (p = 2.3 × 10−4; p = 1.9 × 10−5; p = 2.3 × 10−11; and p = 5.0 × 10−8, respectively; Figure F2), but is not significantly enriched in exocrine pancreas (p > 0.05; Figure F2).
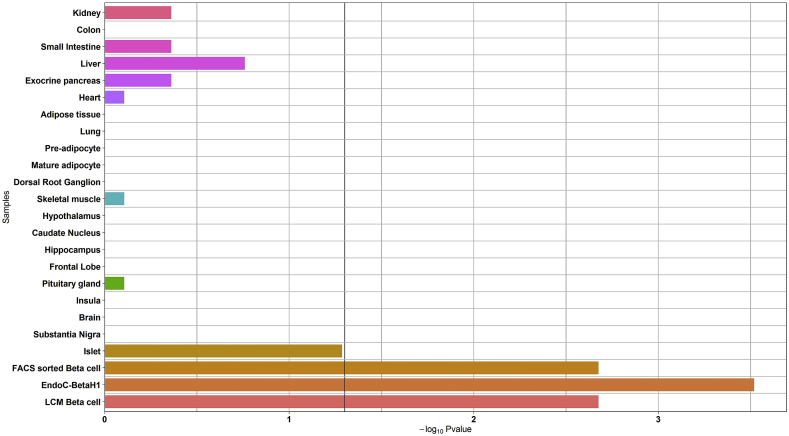
Secondly, we found that the expression of the genes causing monogenic diabetes is significantly enriched in LCM beta cells, FACS sorted beta cells and EndoC-βH1 cells (p = 2.1 × 10−3; p = 2.1 × 10−3; and p = 3.0 × 10−4, respectively; Figure 1). Altogether these results validated the accuracy of our methodology.
Figure 1.
Enrichment analysis of the expression of the genes causing monogenic diabetes in the panel of human organs, tissues, and cells. The black vertical line denotes a p-value of 0.05.
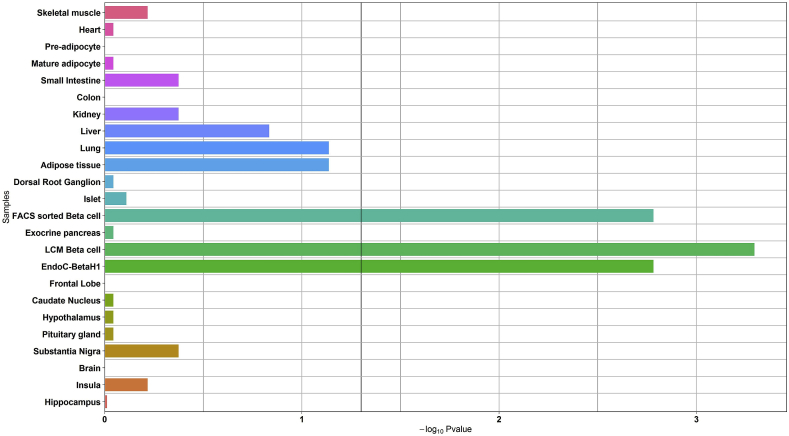
Interestingly, the expression of the 104 candidate T2D susceptibility genes was also significantly enriched in LCM beta cells, FACS sorted beta cells and EndoC-βH1 cells (p = 5.1 × 10−4; p = 1.6 × 10−3; and p = 1.6 × 10−3, respectively; Figure 2), but not in the main insulin-target tissues including liver, adipose tissue, or skeletal muscle (p > 0.05; Figure 2).
Figure 2.
Enrichment analysis of the expression of candidate T2D-susceptibility genes in the panel of human organs, tissues, and cells. The black vertical line denotes a p-value of 0.05.
3.2. Effect of candidate T2D susceptibility genes knockdown on insulin secretion from human EndoC-βH1 cells
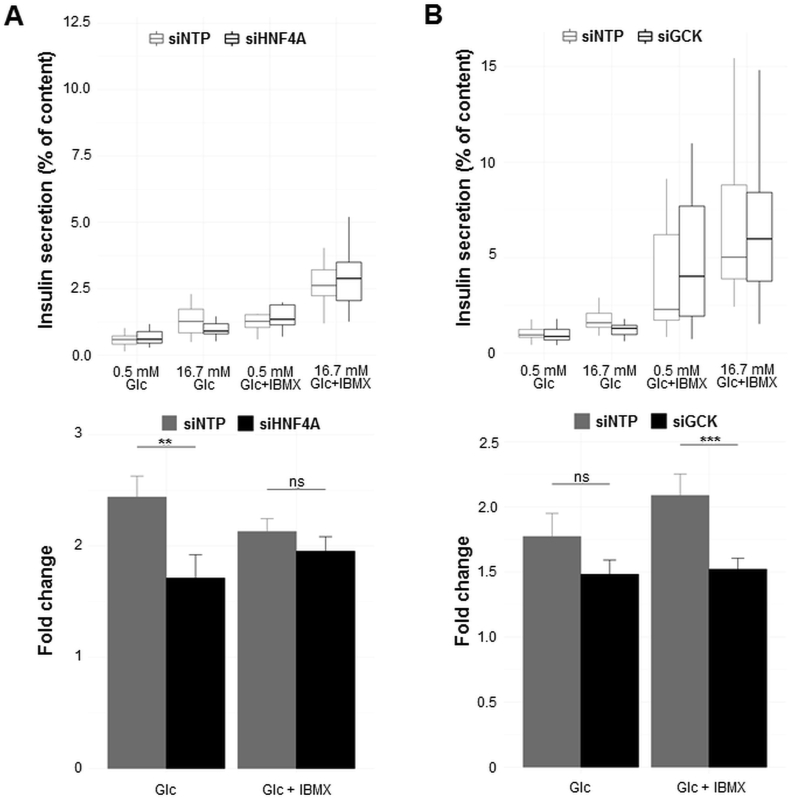
Our present expression results prompted us to investigate the effect of candidate T2D susceptibility gene knockdown by siRNA on insulin secretion from human pancreatic EndoC-βH1 beta-cell line. We selected genes to be silenced based on at least one of these two criteria: i) a high expression in EndoC-βH1 cells, and/or ii) an enriched expression in EndoC-βH1 cells compared to all non-pancreatic tissues. We subsequently obtained 15 genes in which decreased expression by siRNA in EndoC-βH1 cells was above 50% (Figure H). Among these genes, there were GCK encoding glucokinase and HNF4A encoding hepatocyte nuclear factor 4 alpha. These two genes are known to be mutated in patients presenting with monogenic diabetes and to play a key role in insulin secretion from pancreatic beta cells [1]. Furthermore, mice with either a global Gck deletion or beta cell-specific Gck deletion die within a few days of birth from severe hyperglycemia [17]; mice with beta cell-specific Hnf4a deletion exhibit impairment of glucose-stimulated insulin secretion (GSIS) [18]. Therefore, we expected that the decreased expression of GCK or HNFA4 also would have significantly impaired insulin secretion from EndoC-βH1 cells. We found that the decreased expression of HNF4A significantly reduces glucose-stimulated insulin secretion (GSIS; p < 0.01, Figure 3A), and GCK knockdown significantly decreases GSIS evoked by IBMX (p < 0.001; Figure 3B). These results validated our protocol.
Figure 3.
Decreased expression of HNF4A (A) or GCK (B), causing monogenic diabetes, leads to impaired insulin secretion from EndoC-βH1 cells. EndoC-βH1 cells were transfected with control non-targeting pool siRNA (siNTP) or target gene siRNA and were analyzed 72 h post-transfection. Insulin secretion (percentage of secretion of the total insulin content) was analyzed in response to 60 min incubation with 0.5 mM glucose (±0.5 mM IBMX), followed by 60 min incubation with 16.7 mM glucose (±0.5 mM IBMX). Data represent mean values ± SEM of at least three independent experiments. **p < 0.01; ***p < 0.001; ns, not significant. Glc, glucose; IBMX, phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine.
We did not find any significant effect of the decreased beta-cell expression of MPHOSPH9, SSR1, FAF1 or KLHDC5 on GSIS evoked or not by IBMX (p > 0.05; Figure I1–I4, respectively).
In contrast, insulin secretion from EndoC-βH1 cells was significantly changed by the decreased expression of five T2D susceptibility genes already suggested to be expressed and/or to contribute to beta-cell function [19], [20], [21], [22], [23]: the knockdown of TCF19 was found to significantly decrease GSIS (p < 0.01; Figure 4A); the decreased expression of SLC30A8 affected GSIS evoked or not by IBMX (p < 0.05; Figure 4B); the knockdown of TBC1D4 was found to significantly reduce GSIS evoked by IBMX (p < 0.01; Figure 4C); the decreased expression of CDKN2A was found to decrease GSIS evoked by IBMX (p < 0.01; Figure 4D); and the knockdown of KCNK16 was found to stimulate GSIS evoked by IBMX (p < 0.05; Figure 4E).
Figure 4.
Decreased expression of TCF19 (A), SLC30A8 (B), TBC1D4 (C), CDKN2A (D), or KCNK16 (E), already suggested to be expressed in beta cell and/or to contribute to beta-cell function, significantly modifies insulin secretion from EndoC-βH1 cells. EndoC-βH1 cells were transfected with control non-targeting pool siRNA (siNTP) or target gene siRNA and were analyzed 72 h post-transfection. Insulin secretion (percentage of secretion of the total insulin content) was analyzed in response to 60 min incubation with 0.5 mM glucose (±0.5 mM IBMX), followed by 60 min incubation with 16.7 mM glucose (±0.5 mM IBMX). Data represent mean values ± SEM of at least three independent experiments. *p < 0.05; **p < 0.01; ns, not significant. Glc, glucose; IBMX, phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine.
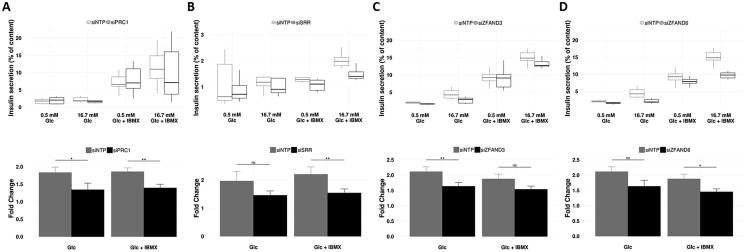
Furthermore, we found that the decreased expression of four other candidate T2D susceptibility genes, with no evidence of expression and role in beta cells, significantly affects insulin secretion from EndoC-βH1 cells: the PRC1 knockdown was found to significantly reduce GSIS evoked or not by IBMX (p < 0.05; Figure 5A); the SRR knockdown was found to significantly decrease GSIS evoked by IBMX (p < 0.01; Figure 5B); the ZFAND3 knockdown was found to significantly affect GSIS (p < 0.01; Figure 5C); and the ZFAND6 knockdown was found to reduce GSIS evoked by IBMX (p < 0.05; Figure 5D).
Figure 5.
Decreased expression of PRC1 (A), SRR (B), ZFAND3 (C), or ZFAND6 (D), with no evidence of expression and role in pancreatic beta cells, leads to impaired insulin secretion from EndoC-βH1 cells. EndoC-βH1 cells were transfected with control non-targeting pool siRNA (siNTP) or target gene siRNA and were analyzed 72 h post-transfection. Insulin secretion (percentage of secretion of the total insulin content) was analyzed in response to 60 min incubation with 0.5 mM glucose (±0.5 mM IBMX), followed by 60 min incubation with 16.7 mM glucose (±0.5 mM IBMX). Data represent mean values ± SEM of at least three independent experiments. *p < 0.05; **p < 0.01; ns, not significant. Glc, glucose; IBMX, phosphodiesterase inhibitor 3-isobutyl-1-methylxanthine.
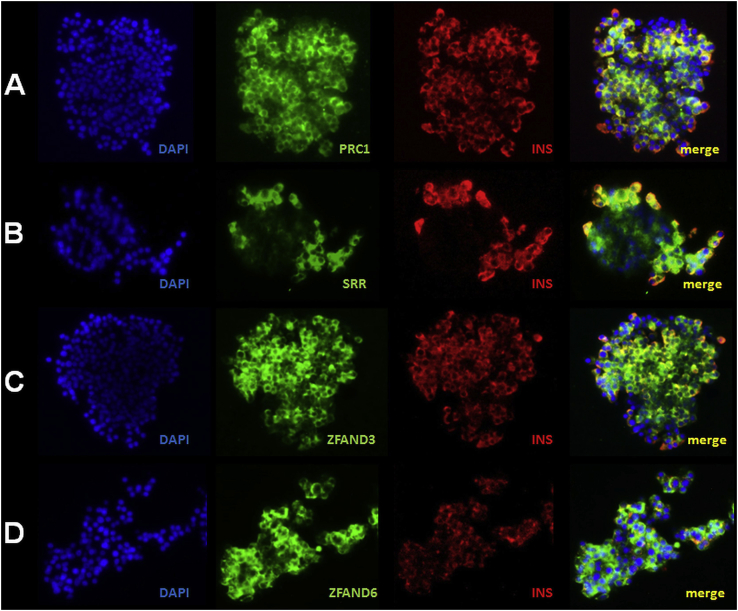
As PRC1 (encoding protein regulator of cytokinesis 1), SRR (encoding serine racemase), ZFAND3 (encoding zinc finger AN1-type containing 3), and ZFAND6 (encoding zinc finger AN1-type containing 6) have never been described in pancreatic islets and beta cells, we confirmed their expression at protein level by immunofluorescence staining in human pancreatic islets (Figure 6A–D, J1 and J2) and in EndoC-βH1 cells (Figure K1–K4).
Figure 6.
PRC1 (A), SRR (B), ZFAND3 (C), and ZFAND6 (D) are expressed in human pancreatic islets and beta cells. Representative images of immunofluorescence staining for PRC1 (green; A), SRR (green; B), ZFAND3 (green; C), ZFAND6 (green, D), and insulin (red) performed on fixed isolated human islet clusters. Yellow (merged images) indicates co-localization of PRC1, SRR, ZFAND3, or ZFAND6 with insulin. Blue, DAPI.
3.3. Deciphering the major biological pathways and networks emphasized by the decreased expression of PRC1, SRR, ZFAND6, and ZFAND3 in EndoC-βH1 cells
We next performed RNA-seq in EndoC-βH1 cells with decreased expression of PRC1, SRR, ZFAND6, or ZFAND3, compared to control cells, to decipher the major biological pathways and networks associated with those genes with yet unknown function in pancreatic beta cells.
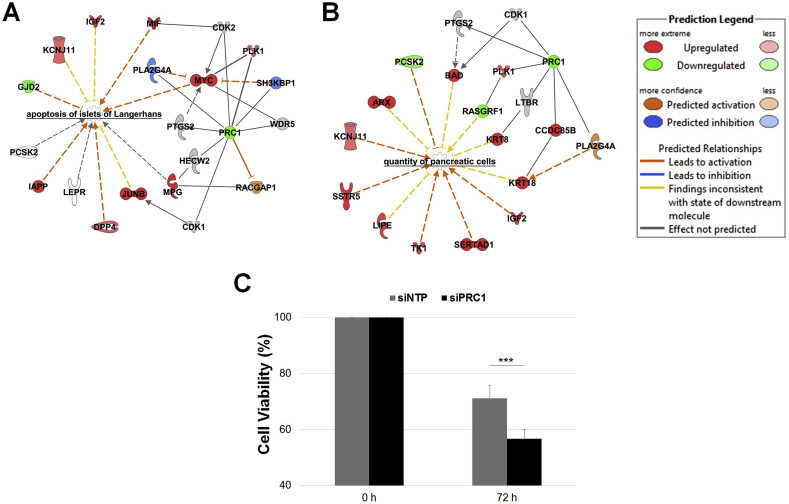
When we analyzed the transcriptome of EndoC-βH1 cells transfected with siPRC1 versus the transcriptome of control cells using Ingenuity Pathway Analysis (IPA), we found that PRC1 was included in a network of 28 molecules entitled “Cell Cycle, Cellular Movement, Cellular Assembly and Organization” (Figure L1). Subsequently, we analyzed the diseases and/or functions emphasized by the PRC1 knockdown in EndoC-βH1 cells. Among the significant outputs, we found networks related to the “quantity of carbohydrates” (p = 2.7 × 10−8; Figure L2), “quantity of insulin in blood” (p = 4.8 × 10−5; Figure L3), or “concentration of d-glucose” (p = 4.6 × 10−8; Figure L4). Furthermore, we found a network related to “apoptosis” (p = 1.1 × 10−4; Table B), in particular “apoptosis of islets of Langerhans” (p = 4.3 × 10−3; Figure 7A), and a network related to “quantity of pancreatic cells” (p = 9.9 × 10−4; Figure 7B). When we analyzed the viability of the EndoC-βH1 cells transfected with siPRC1 compared to control cells, we found that PRC1 knockdown significantly decreased the viability of EndoC-βH1 cells (p = 1.4 × 10−6; Figure 7C).
Figure 7.
Decreased PRC1 expression in EndoC-βH1 cells leads to increased apoptosis and decreased cell viability. List of deregulated genes emphasized by the decreased expression of PRC1 in EndoC-βH1 cells within the network related to apoptosis of islets of Langerhans (A) and the quantity of pancreatic cells (B) through IPA. Cell viability measurement (C) performed at baseline (100%) and 72 h post transfection of siRNA (% compared to baseline). Data at 72 h represent mean values ± SEM of at least five independent experiments. ***p < 0.001.
When we analyzed the diseases and/or functions emphasized by the SRR knockdown in EndoC-βH1 cells, we found a significant network related to “serine phosphorylation” (p = 8.5 × 10−7; Figure M1), in particular “serine phosphorylation of peptide” (p = 6.3 × 10−7; Figure M2), a network related to “endoplasmic reticulum stress response” (p = 8.9 × 10−3; Figure M3) and a network related to “congenital disorders of glycosylation” (p = 2.5 × 10−4; Figure M4).
When we analyzed the transcriptome of EndoC-βH1 cells transfected with siZFAND6 versus the transcriptome of control cells, we found that ZFAND6 was included in a network of 27 molecules entitled “Cell Signaling, Lipid Metabolism, Small Molecule Biochemistry” (Figure N1). Furthermore, among the top diseases and/or functions emphasized by the ZFAND6 knockdown in EndoC-βH1 cells, we found significant networks related to “glycolysis” (p = 8.0 × 10−3; Figure N2) and to “quantity of insulin in blood” (p = 7.3 × 10−3; Figure N3). We found a significant network related to “abnormal morphology of pancreas” (p = 2.2 × 10−3; Figure N4), in particular “abnormal morphology of small islets of Langerhans” (p = 1.8 × 10−2; Figure N5).
We subsequently analyzed the transcriptome of EndoC-βH1 cells transfected with siZFAND3 versus the transcriptome of control cells. We found that ZFAND3 is included in a network of 16 molecules entitled “Cellular Assembly and Organization, Lipid Metabolism, Molecular Transport” (Figure O1). Furthermore, among the top diseases and/or functions emphasized by the ZFAND3 knockdown in EndoC-βH1 cells, we found a significant network related to “transport of alpha-amino acid” (p = 8.9 × 10−4; Figure O2).
3.4. Positive correlation between the expression of Ins2 and the expression of Prc1, Srr, Zfand6, and Zfand3 in mouse pancreatic islets with altered beta-cell function
We finally investigated the expression of the four genes in mouse models with defective pancreatic beta-cell function due to STZ treatment or with compensatory islet cell function in ob/ob mice carrying a non-sense mutation in the leptin gene. In two pools of pancreatic islets treated or not with STZ from eight 10-week old mice, we found a strong positive correlation between the expression of Ins2 and the expression of Prc1, Srr, Zfand3, and Zfand3: the expression of these five genes was significantly reduced in pancreatic islets treated with STZ (Figure P1 and P2). Furthermore, in pancreatic islets from five 5-week old mice or from five 8-week old mice, we also found this positive correlation: the expression of Ins2, Prc1, Srr, Zfand3, and Zfand6 was strongly increased in ob/ob mice when compared to control mice (Figure P3 and P4).
4. Discussion
4.1. Enrichment of GWAS-identified candidate T2D susceptibility genes in human pancreatic beta cells
Our first objective was to select candidate genes for putative insulin secretion defect associated with T2D from the list of genes closed to GWAS-identified SNPs associated with T2D risk. During the course of this analysis, we demonstrated that the expression of these 104 genes was enriched in human pancreatic beta cells. Surprisingly perhaps, we did not find any similar enrichment of their expression in insulin sensitive skeletal muscle, liver, adipose tissue, and in the gut and brain. An obvious limitation of our specific analysis is the fact that we analyzed the expression of the genes closest to GWAS-identified T2D SNPs, while one cannot rule out that a given T2D-associated SNP may also impact expression of gene(s) located several megabases away from the SNP, through a loop between a distal enhancer (where this SNP maps) and the target gene promoter. However, the majority of GWAS-identified T2D genes map in DNA sequences of enhancer specifically active in human pancreatic islets, which are usually located in the vicinity of the cis-regulated gene transcription start sites [3]. In support of our result, when we assessed in the same panel of human tissues the expression of candidate obesity susceptibility genes closed to GWAS-identified obesity SNPs, we did not identify any enrichment of gene expression in beta cells while we found significant enrichment of gene expression in several regions of the brain (data not shown). Therefore, we believe that our conclusion on the enrichment of the expression of our selection of T2D genes in beta cells is not artefactual and is probably representative of other T2D genes, if they exist.
4.2. Decreased expression of seven genes (GCK, HNF4A, TCF19, SLC30A8, TBC1D4, CDKN2A, and KCNK16) already suggested to be expressed in beta cells and/or to contribute to beta-cell function significantly modifies insulin secretion from EndoC-βH1 cells
Subsequently, in the human EndoC-βH1 beta-cell line, we investigated the effect on insulin secretion of the knockdown of the 15 T2D susceptibility genes with the highest and/or the most specific expression in beta cells (with successful RNA interference). We first found that the decreased expression of the seven genes that were already suggested to be expressed and/or to contribute to beta-cell function (including GCK, HNF4A, TCF19, SLC30A8, TBC1D4, CDKN2A and KCNK16) is significantly associated with modified insulin secretion.
Indeed, as discussed above (Section 3.2), both GCK and HNF4A are known to be mutated in patients presenting with monogenic diabetes and to play a key role in insulin secretion from pancreatic beta cells [1].
TCF19 encodes the transcription factor 19 and has been reported to be expressed in human pancreatic islets [21]. Furthermore, TCF19 was found to be a transcription factor necessary for proliferation and survival in the rat pancreatic INS-1 beta-cell line [21]. Our data complement the current knowledge about this gene role, as we found that TCF19 knockdown significantly decreased GSIS in human beta-cell lines.
SLC30A8 encodes the solute carrier family 30 member 8, which is a zinc transporter almost exclusively expressed in pancreatic alpha cells and beta cells and involved in the accumulation of zinc within insulin or glucagon secretory granules [24]. Here, we found that SLC30A8 knockdown significantly decreases GSIS in human beta cells. Importantly, this result was in line with previous data showing that the GWAS-identified nonsynonymous SNP associated with increased T2D risk [25] was deleterious for SLC30A8 zinc transport activity [26], [27]. Our data also supported previous results based on mice models which showed that mice deleted for Slc30a8 present with significant impairments in glucose tolerance and that, in contrast, transgenic mice over-expressing Slc30a8 display improved glucose homeostasis [22]. However, our result did not support the hypothesis generated through the sequencing or genotyping of ∼150,000 individuals that suggested that low-frequency loss-of-function mutations in SLC30A8 protect against T2D [28]. In order to reconcile this apparent contradiction, Rutter and Chimienti have recently suggested that the degrees of overall inhibition of SLC30A8 activity might be different according to the variant frequency, and that the resulting deleterious effect might be dependent on age or hypoxic beta-cell stress [24]. Still, our data showed that decreasing SLC30A8 expression by 70% in human beta cells is apparently deleterious for insulin secretion.
TBC1D4 encodes the Tre-2/BUB2/CDC16 domain family member 4 that is a Rab-GTPase-activating protein. This protein has been actively explored in skeletal muscle and adipose tissue and was found to play a key role in insulin-stimulated glucose uptake in these metabolic tissues [29]. Importantly, the GWAS-identified T2D-associated TBC1D4 SNP in Greenlandic individuals was shown to cause insulin resistance in skeletal muscle [30]. However, this protein was also identified in human and mouse pancreatic beta cells, and TBC1D4 mRNA expression was found to be downregulated in pancreatic islets from individuals with T2D [19]. Furthermore, Tbc1d4 knockdown by siRNA strongly decreased GSIS from mouse pancreatic MIN6 beta-cell line [19], which was in line with our present result in human EndoC-βH1 cells. In conclusion, our data on TBC1D4 have fully complemented previous observational studies in human islets and experimental studies in animal models and cell lines.
CDKN2A encodes the cyclin dependent kinase inhibitor 2A, also known as the tumor suppressor protein P16-INK4A. Here, we found that CDKN2A knockdown significantly decreases GSIS evoked by IBMX from EndoC-βH1 cells. This result was inconsistent with a previous study, which reported that CDKN2A knockdown significantly enhances GSIS evoked by IBMX from the same cell model [31]. These opposite results might be explained by the different experimental conditions. In the present study, we performed a stimulation step (16.7 mM glucose ± IBMX) subsequent to basal incubation (0.5 mM glucose ± IBMX) in the same wells, while Pal and colleagues performed static insulin secretion assays, namely basal incubation and stimulation step were performed in different wells [31]. As EndoC-βH1 cells are not always sensitive to glucose, the static protocol may lead to artefactual positive results. The same team did not confirm their results on CDKN2A knockdown in their recent systematic functional screening of candidate T2D susceptibility genes in the same EndoC-βH1 cells [32]. In support of our conclusions, mice beta-cell specific activation of Cdkn2a enhanced GSIS, and increased Cdkn2a expression was found to be a marker of beta-cell senescence, associated with enhanced GSIS during mice normal aging [20]. Importantly, Helman and colleagues also showed that CDKN2A-induced senescence in human EndoC-βH2 cells enhances GSIS [20]. Thus, we believe from our data and others that decreased CDKN2A expression probably decreases GSIS and increases T2D risk (and not the opposite).
KCNK16 encodes the potassium two-pore domain channel subfamily K member 16 (also known as TALK1). This channel was shown to be expressed in mouse and human pancreatic beta cells and to play a key role in cell electrical excitability and in GSIS [23]. The GWAS-identified nonsynonymous SNP associated with increased T2D risk was found to enhance the channel activity, and Kcnk16 knockout in mouse beta cells enhanced Ca2+ influx and second-phase GSIS [23], which was in line with our present results in EndoC-βH1 cells demonstrating that KCNK16 knockdown increases insulin secretion in humans.
4.3. Decreased expression of four genes (PRC1, SRR, ZFAND6, and ZFAND3) with no evidence of expression and role in pancreatic beta cells significantly modifies insulin secretion from EndoC-βH1 cells
We also found for the first time that the decreased expression of four T2D susceptibility genes (PRC1, SRR, ZFAND6, and ZFAND3, respectively), with no evidence of expression and role in beta cells, significantly affects insulin secretion from human EndoC-βH1 cells. Although PRC1, SRR, ZFAND6, and ZFAND3 were ubiquitously expressed according to our panel of human tissues (Figure Q1–Q4, respectively) and to the Genotype-Tissue Expression (GTEx) portal [10], they were highly expressed and/or enriched in FACS sorted human beta-cells and in EndoC-βH1. Furthermore, the investigation of mouse pancreatic islets with altered beta-cell function demonstrated a positive correlation between the expression of insulin and the expression of Prc1, Srr, Zfand3, and Zfand6, confirming a role of these genes in pancreatic beta-cell function.
4.3.1. PRC1, a new gene involved in beta-cell function
PRC1 is a key regulator of cytokinesis and involved in many cancers. In hepatocellular carcinoma, PRC1 has an oncogenic effect through the enhancement of cancer proliferation, metastasis, and tumorigenesis [33]. Importantly, the GWAS-identified PRC1 SNP associated with T2D risk was also found to contribute to breast cancer risk [34]. Our RNA-seq data from EndoC-βH1 cells with PRC1 knockdown compared to control human cell line identified a network of genes related to cell cycle, cellular movement, cellular assembly and organization, as well as other gene networks related to glucose homeostasis and two networks related to apoptosis, especially one specific to pancreatic islets. We believe these results based on next-generation sequencing are sound as we found that PRC1 knockdown in EndoC-βH1 cells significantly decreased their viability compared to control cells, which validated our RNA-seq analyses.
4.3.2. SRR, a new gene involved in beta-cell function
SRR is an enzyme generating d-serine from l-serine that is an endogenous co-agonist of the N-methyl-d-aspartate (NMDA) receptor. Adult mice deleted for Srr presented with schizophrenia-like behavioral abnormalities, which could be rescued by d-serine administration [35]. Our RNA-seq data from EndoC-βH1 cells with SRR knockdown compared to control EndoC-βH1 cells emphasized a network related to serine phosphorylation (in particular serine phosphorylation of peptides). Importantly, metabolomics studies have suggested that serine predicted T2D incidence [36] and gestational diabetes mellitus incidence [37]. Furthermore, the transcriptome of EndoC-βH1 cells with SRR knockdown identified a network related to endoplasmic reticulum stress response, which has been reported to be involved in decreased insulin secretion and in T2D development [38]. Altogether, our data emphasized the interest of serine metabolism in T2D and human beta cells, opening potential avenues for prevention, in particular in high risk populations such as women with history of gestational diabetes mellitus.
4.3.3. ZFAND3 and ZFAND6, two new genes involved in beta-cell function
Very little is known about ZFAND3 and ZFAND6. Interestingly, the GWAS-identified SNP at ZFAND3 locus associated with T2D risk was precisely altering the related human islet enhancer activity bringing mechanistic explanation of its functional impact [3].
4.4. Conclusion
In conclusion, our staged post-GWAS functional study has several implications for further T2D pathophysiological investigations. First, it demonstrated that the expression of candidate T2D susceptibility genes closest to GWAS-identified T2D SNPs (usually named as T2D genes by consortia) is strongly enriched in human beta cells. We believe our study is useful for T2D pathophysiology, but we acknowledge that our study was far from being exhaustive as we did not, for feasibility reasons, systematically test the several hundreds of genes located in the same topologically associating domain (TAD) as GWAS-identified T2D SNPs, namely all genes that may, in theory, be regulated by these SNPs (as located in the same regulatory loop). Furthermore, we chose not to systematically test the genes located in the so-called linkage disequilibrium (LD) blocks around T2D SNPs, as regulatory regions are different from LD blocks that are only units of similar genetic recombination. Nevertheless, our data provide original information about the consequences of the partial inactivation of analyzed T2D genes on human beta-cell function, which contribute to the elucidation of conflicting functional data obtained for some T2D genes such as SLC30A8 or CDKN2A. We agree that further validation in human mature islets would be of importance, but, unfortunately, it was impossible for us to have enough islet material for a study of this scale. We hope that the progress made in the protocol of beta-cell differentiation of human induced pluripotent stem cells soon will unlock this limitation. In addition, we showed the interest of unbiased multi-tissue gene expression analysis to prioritize challenging human beta-cell investigation. Furthermore, our RNA-seq analyses of the partial invalidation of four T2D susceptibility genes with no previously known function in beta cell identified specific gene networks related to key physiology pathways. When extended to other T2D susceptibility genes, this approach may bring progress towards better patient stratification and future precision medicine based on the genotyping of SNPs in genes sharing same functional signatures. A recent study has also reported the feasibility of systematic screening of T2D susceptibility genes in human beta cell lines without prior step of selection on enriched beta cell genes and suggested some plausible T2D causing genes [32]. As different protocols have been used, the two studies provide complementary results and guarantee that future in-depth investigations of T2D susceptibility genes in pancreatic beta cell should be very fruitful.
Acknowledgments
This work was supported by grants from the French National Research Agency (ANR-10-LABX-46 [European Genomics Institute for Diabetes] and ANR-10-EQPX-07-01 [LIGAN-PM], to PF), from the European Research Council (ERC GEPIDIAB – 294785, to PF), from FEDER (to PF) and from the ‘Région Nord Pas-de-Calais’ (to PF and to FKN). AB was supported by Inserm.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2017.03.011.
Contributor Information
Philippe Froguel, Email: p.froguel@imperial.ac.uk.
Amélie Bonnefond, Email: amelie.bonnefond@inserm.fr.
Conflict of interest
The authors have declared that no conflict of interest exists.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Bonnefond A., Froguel P. Rare and common genetic events in type 2 diabetes: what should biologists know? Cell Metabolism. 2015;21(3):357–368. doi: 10.1016/j.cmet.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 2.Marullo L., El-Sayed Moustafa J.S., Prokopenko I. Insights into the genetic susceptibility to type 2 diabetes from genome-wide association studies of glycaemic traits. Current Diabetes Reports. 2014;14(11):551. doi: 10.1007/s11892-014-0551-8. [DOI] [PubMed] [Google Scholar]
- 3.Pasquali L., Gaulton K.J., Rodríguez-Seguí S.A., Mularoni L., Miguel-Escalada I., Akerman I. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nature Genetics. 2014;46(2):136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warren H.R., Evangelou E., Cabrera C.P., Gao H., Ren M., Mifsud B. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nature Genetics Advance Online Publication. 2017 doi: 10.1038/ng.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnefond A., Lomberk G., Buttar N., Busiah K., Vaillant E., Lobbens S. Disruption of a novel Kruppel-like transcription factor p300-regulated pathway for insulin biosynthesis revealed by studies of the c.-331 INS mutation found in neonatal diabetes mellitus. The Journal of Biological Chemistry. 2011;286(32):28414–28424. doi: 10.1074/jbc.M110.215822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martino L., Masini M., Bugliani M., Marselli L., Suleiman M., Boggi U. Mast cells infiltrate pancreatic islets in human type 1 diabetes. Diabetologia. 2015;58(11):2554–2562. doi: 10.1007/s00125-015-3734-1. [DOI] [PubMed] [Google Scholar]
- 7.Marselli L., Thorne J., Dahiya S., Sgroi D.C., Sharma A., Bonner-Weir S. Gene expression profiles of beta-cell enriched tissue obtained by laser capture microdissection from subjects with type 2 diabetes. PLoS One. 2010;5(7):e11499. doi: 10.1371/journal.pone.0011499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukowiak B., Vandewalle B., Riachy R., Kerr-Conte J., Gmyr V., Belaich S. Identification and purification of functional human-cells by a new specific zinc-fluorescent probe. Journal of Histochemistry & Cytochemistry. 2001;49(4):519–527. doi: 10.1177/002215540104900412. [DOI] [PubMed] [Google Scholar]
- 9.Eisenberg E., Levanon E.Y. Human housekeeping genes, revisited. Trends in Genetics: TIG. 2013;29(10):569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 10.Melé M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M. The human transcriptome across tissues and individuals. Science. 2015;348(6235):660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu C., Orozco C., Boyer J., Leglise M., Goodale J., Batalov S. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biology. 2009;10(11):R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravassard P., Hazhouz Y., Pechberty S., Bricout-Neveu E., Armanet M., Czernichow P. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. Journal of Clinical Investigation. 2011;121(9):3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonner C., Kerr-Conte J., Gmyr V., Queniat G., Moerman E., Thévenet J. Inhibition of the glucose transporter SGLT2 with dapagliflozin in pancreatic alpha cells triggers glucagon secretion. Nature Medicine. 2015;21(5):512–517. doi: 10.1038/nm.3828. [DOI] [PubMed] [Google Scholar]
- 14.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao Y., Smyth G.K., Shi W. The subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Research. 2013;41(10):e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postic C., Shiota M., Niswender K.D., Jetton T.L., Chen Y., Moates J.M. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. The Journal of Biological Chemistry. 1999;274(1):305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 18.Miura A., Yamagata K., Kakei M., Hatakeyama H., Takahashi N., Fukui K. Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells. The Journal of Biological Chemistry. 2006;281(8):5246–5257. doi: 10.1074/jbc.M507496200. [DOI] [PubMed] [Google Scholar]
- 19.Bouzakri K., Ribaux P., Tomas A., Parnaud G., Rickenbach K., Halban P.A. Rab GTPase-activating protein AS160 is a major downstream effector of protein kinase B/Akt signaling in pancreatic-cells. Diabetes. 2008;57(5):1195–1204. doi: 10.2337/db07-1469. [DOI] [PubMed] [Google Scholar]
- 20.Helman A., Klochendler A., Azazmeh N., Gabai Y., Horwitz E., Anzi S. p16(Ink4a)-induced senescence of pancreatic beta cells enhances insulin secretion. Nature Medicine. 2016;22(4):412–420. doi: 10.1038/nm.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krautkramer K.A., Linnemann A.K., Fontaine D.A., Whillock A.L., Harris T.W., Schleis G.J. Tcf19 is a novel islet factor necessary for proliferation and survival in the INS-1-cell line. AJP: Endocrinology and Metabolism. 2013;305(5):E600–E610. doi: 10.1152/ajpendo.00147.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitchell R.K., Hu M., Chabosseau P.L., Cane M.C., Meur G., Bellomo E.A. Molecular genetic regulation of Slc30a8/ZnT8 reveals a positive association with glucose tolerance. Molecular Endocrinology. 2016;30(1):77–91. doi: 10.1210/me.2015-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vierra N.C., Dadi P.K., Jeong I., Dickerson M., Powell D.R., Jacobson D.A. The type-2 diabetes-associated K+ channel TALK-1 modulates beta-cell electrical excitability, 2nd-phase insulin secretion, and glucose homeostasis. Diabetes. 2015 doi: 10.2337/db15-0280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutter G.A., Chimienti F. SLC30A8 mutations in type 2 diabetes. Diabetologia. 2015;58(1):31–36. doi: 10.1007/s00125-014-3405-7. [DOI] [PubMed] [Google Scholar]
- 25.Sladek R., Rocheleau G., Rung J., Dina C., Shen L., Serre D. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 26.Kim I., Kang E.S., Yim Y.S., Ko S.J., Jeong S.-H., Rim J.H. A low-risk ZnT-8 allele (W325) for post-transplantation diabetes mellitus is protective against cyclosporin A-induced impairment of insulin secretion. The Pharmacogenomics Journal. 2011;11(3):191–198. doi: 10.1038/tpj.2010.22. [DOI] [PubMed] [Google Scholar]
- 27.Nicolson T.J., Bellomo E.A., Wijesekara N., Loder M.K., Baldwin J.M., Gyulkhandanyan A.V. Insulin storage and glucose homeostasis in mice null for the granule zinc transporter ZnT8 and studies of the type 2 diabetes-associated variants. Diabetes. 2009;58(9):2070–2083. doi: 10.2337/db09-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flannick J., Thorleifsson G., Beer N.L., Jacobs S.B.R., Grarup N., Burtt N.P. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nature Genetics. 2014;46(4):357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sano H., Kane S., Sano E., Mîinea C.P., Asara J.M., Lane W.S. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. The Journal of Biological Chemistry. 2003;278(17):14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 30.Moltke I., Grarup N., Jørgensen M.E., Bjerregaard P., Treebak J.T., Fumagalli M. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512(7513):190–193. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 31.Pal A., Potjer T.P., Thomsen S.K., Ng H.J., Barrett A., Scharfmann R. Loss-of-function mutations in the cell-cycle control gene CDKN2A impact on glucose homeostasis in humans. Diabetes. 2016;65(2):527–533. doi: 10.2337/db15-0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomsen S.K., Ceroni A., van de Bunt M., Burrows C., Barrett A., Scharfmann R. Systematic functional characterization of candidate causal genes for type 2 diabetes risk variants. Diabetes. 2016;65(12):3805–3811. doi: 10.2337/db16-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen J., Rajasekaran M., Xia H., Zhang X., Kong S.N., Sekar K. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/β-catenin signalling pathway. Gut. 2016;65(9):1522–1534. doi: 10.1136/gutjnl-2015-310625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao Z., Wen W., Michailidou K., Bolla M.K., Wang Q., Zhang B. Association of genetic susceptibility variants for type 2 diabetes with breast cancer risk in women of European ancestry. Cancer Causes & Control. 2016:1–15. doi: 10.1007/s10552-016-0741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hagiwara H., Iyo M., Hashimoto K. Neonatal disruption of serine racemase causes schizophrenia-like behavioral abnormalities in adulthood: clinical rescue by d-serine. PLoS One. 2013;8(4):e62438. doi: 10.1371/journal.pone.0062438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cobb J., Eckhart A., Perichon R., Wulff J., Mitchell M., Adam K.-P. A novel test for IGT utilizing metabolite markers of glucose tolerance. Journal of Diabetes Science and Technology. 2015;9(1):69–76. doi: 10.1177/1932296814553622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bentley-Lewis R., Huynh J., Xiong G., Lee H., Wenger J., Clish C. Metabolomic profiling in the prediction of gestational diabetes mellitus. Diabetologia. 2015;58(6):1329–1332. doi: 10.1007/s00125-015-3553-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter P., Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334(6059):1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.