Abstract
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) analysis can detect protein/DNA-binding and histone-modification sites across an entire genome. Recent advances in sequencing technologies and analyses enable us to compare hundreds of samples simultaneously; such large-scale analysis has potential to reveal the high-dimensional interrelationship level for regulatory elements and annotate novel functional genomic regions de novo. Because many experimental considerations are relevant to the choice of a method in a ChIP-seq analysis, the overall design and quality management of the experiment are of critical importance. This review offers guiding principles of computation and sample preparation for ChIP-seq analyses, highlighting the validity and limitations of the state-of-the-art procedures at each step. We also discuss the latest challenges of single-cell analysis that will encourage a new era in this field.
Keywords: chromatin immunoprecipitation, large-scale analysis, experimental design, quality management, differential analysis, single-cell analysis
Introduction
Genome-wide investigations into cooperative interactions among genomic functions, e.g. DNA replication, segregation, translation, repair and rearrangement, are vital for systematically elucidating all biological activities on the genome. To this end, chromatin immunoprecipitation followed by sequencing (ChIP-seq) analysis was developed to understand the cooperation and interactions that occur in a wide variety of organisms using next-generation sequencing (NGS) [1–3]. ChIP-seq analysis is a mainstream method in genomics and epigenomics, and has led to important discoveries related to disease-associated transcriptional regulation [4–7], tissue-specificity of epigenetic regulation [8, 9] and chromatin organization [10–13].
ChlP-seq protocols have many steps involving sample preparation and computational analysis (Figure 1). In brief, cross-linked chromatin is sonicated, and purified with and without immunoprecipitation (ChIP and corresponding input DNA fragments, respectively). DNA fragments are sequenced as reads, which are then mapped onto the reference genome, and the genomic regions that are significantly enriched for ChIP reads, compared with input reads, are detected as peaks. Other genomic regions are regarded as non-specific background. Called peaks, which represent candidates of targeted protein/DNA-binding and histone modification sites, can be used to identify associated functional annotations, including binding motifs [14, 15] and gene ontology [16, 17]. ChIP-seq results can also be integrated with other types of genomic assays, including gene expression, DNA methylation and chromatin conformation, to understand mechanisms of genomic functions from multiple aspects [18–20].
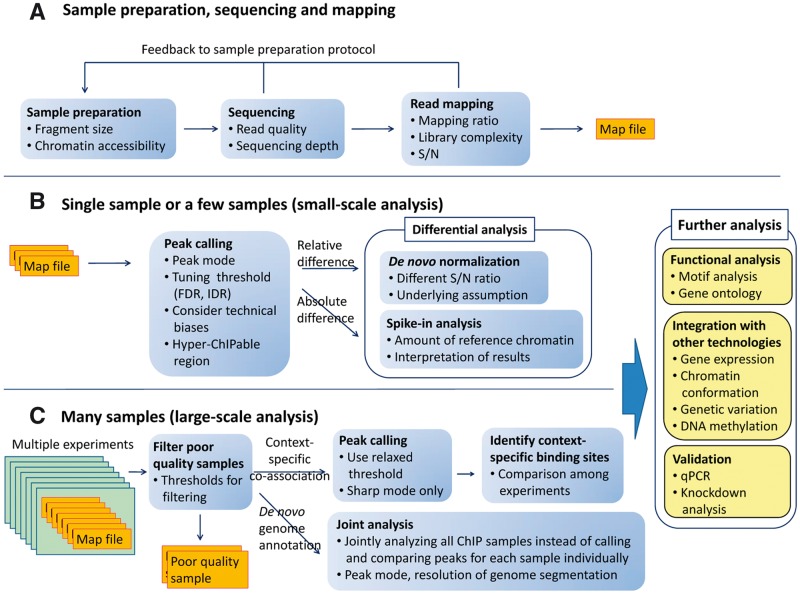
Figure 1.
ChIP-seq analysis workflow. Boxes indicate the steps involved in ChIP-seq analyses for various aims discussed in this review. The considerations for each step are itemized. (A) Sample preparation, sequencing and mapping. This procedure is common to both (B) and (C). (B) Small-scale analysis (single or a few samples). In this case, adjusting peak-calling strategy and parameters to each sample’s property is possible. (C) Large-scale analysis (many samples). Left rectangles indicate the different experiments (e.g. same analysis for different cell types). Because integrative analysis is sensitive to the quality of input samples and one-by-one adjusting is difficult, objective quality metrics for multilateral quantitative assessment is necessary to filter poor-quality data automatically.
The shapes of the peaks vary among proteins, and are classified into three modes [1]: ‘sharp mode’, located at specific positions in the genome; ‘broad mode’, associated with large genomic domains; and ‘mixed mode’, which involves both peak modes. As most point-source transcription factors (TFs) and localized chromatin markers (e.g. H3K4me3) have sharp modes, a large majority of peak-calling algorithms have been designed for this mode, even though other proteins (e.g. heterochromatin protein HP1 [21]) and some histone modifications (e.g. H3K9me3) have broad modes. The mixed mode is observed for RNA polymerase II (Pol II) and transcription elongation factors [22]. The read distribution of Pol II-related NGS analyses, such as Nascent RNA-seq [23] and NET-seq [24], are also classified as mixed mode. Different peak-calling strategies are required for each shape.
Recent advances in sequencing technologies and analyses enable us to handle hundreds of ChIP samples simultaneously; such large-scale analyses revealed the high-dimensional interrelationship for regulatory elements [25–27] and annotate novel functional genomic regions de novo [28, 29]. Because a large-scale analysis is sensitive to the quality of input samples and adjusting the protocols for each sample’s quality is difficult, samples which have insufficient quality should be rejected automatically. As there are various factors (including antibody quality) during sample preparation that affect the quality of the obtained results [30, 31], multilateral quality assessments during the computational procedures are essential. Despite great efforts to streamline this process, no single workflow that is optimal under all circumstances exists, and there are many experimental considerations that are relevant to the method choice for a ChIP-seq analysis. Consequently, to obtain high-quality, unbiased and reasonable data, the overall protocol design and quality management, which are adjusted to the studies’ properties, are of critical importance.
In this review, we describe the computational protocol and sample preparation for a ChIP-seq analysis, and discuss the validity and limitations of emerging programs and quality measures currently available for specific analytical tasks by providing concrete examples. There are ChIP-seq-extended methods that can detect DNA-protein binding sites with base-pair resolution [32, 33]. For simplicity, we have limited the scope of this review to ChIP-seq protocols, for which the computational protocols are similar. The key issues described here also underpin protocols for other NGS-based analyses [34–37]. Finally, we discuss the latest challenges of single-cell analysis that will encourage a new era in this field.
Sample preparation and sequencing
Fragmented DNAs (150–500 bp) from ChIP-seq samples are sequenced as reads (36–100 bp). Single-end reads are often used for typical ChIP-seq analyses, while paired-end ones improve the library complexity and increase mapping efficiency at repetitive regions [38]. When research focuses on repetitive regions, longer and/or paired-end reads are preferred. While paired-end reads can be used to obtain the fragment size distribution, several methods exist that estimate it from single-end mapped data [12, 39, 40].
The chromatin accessibility during fragmentation is not uniform across the genome. In some open-chromatin regions (e.g. actively transcribed promoter regions), DNA is amenable to fragmentation and thereby preferentially represented in the fragmented sample, which causes false-positive read enrichment [41]. Tightly packed regions, e.g. heterochromatin, are sheared to a lesser extent by DNA fragmentation, thereby confounding weak enrichment of true binding sites for heterochromatin markers [38]. These fragmentation biases in a genome-wide read distribution profile should be taken into account when using null model analysis to obtain meaningful conclusions. One way to mitigate this fragmentation bias is to shear longer DNA fragments (350∼800 bp) further using ultrasonication after the immunoprecipitation step [4]. Although including longer fragments widens the obtained peaks, peak-summit resolution is not strongly affected [38].
Read mapping
Sequenced reads are mapped onto the genome using mapping tools [42, 43]. Most ChIP-seq experiments do not require gapped alignments that consider insertions and deletions (indels) because the sequenced reads do not contain them, unlike exon junctions in RNA-seq analyses. The exception is cross-species analysis, which maps reads onto different species’ genomes. If the information about heterozygous variants (e.g. single nucleotide polymorphisms and indels) in the reference genome is available, the allele-specific regulation analysis (personalized genome analysis) can be applied [3, 44, 45].
An important issue concerns the inclusion of multiple mapped reads (reads mapped to multiple loci on the reference genome). Allowing for multiple mapped reads increases the number of usable reads and the sensitivity of peak detection; however, the number of false positives may also increase [46]. In general, uniquely mapped reads are sufficient to analyze typical TFs, except for in-repeat analyses [47]. Considering the percentage of mapped reads (mapping ratio) is important, and desirable rate depends on the species and the read lengths.
Mappability
Recent central ChIP-seq studies [29, 45] used uniquely mapped reads. Instead of including multiple mapped reads, they considered the mappability [48] to correct for the loss of true signals in low-mappable regions. Theoretically, the mappability of a reference genome depends on the read length, read type (color or nucleotide space) and the mapping tool and parameters used [49], but calculating the genome-wide mappability for each is often time-consuming. Moreover, it is difficult to calculate the mappability of paired-end and gapped alignment data, although there has been an effort to calculate the former [50]. Consequently, it is practical to use the mappability data publicly available for similar parameter sets. When low-mappable regions (e.g. a ratio < 0.25) are of interest, it might be better to include multiple mapped reads or use paired-end reads.
Library complexity
Library complexity is measured by the non-redundant fraction (NRF), the fraction Nnonred/Nall, where Nnonred and Nall are the numbers of non-redundant reads and the total number of mapped reads, respectively. The non-redundant reads are defined as reads mapped on the same genomic positons T times or less, where T is the threshold for redundant reads. The redundant reads (Nall − Nnonred) should be filtered from further analysis. For human, T is typically set 1 because the expected number of mapped reads per base pair (sequencing depth) ≪ 1. When it becomes >1, due to the small genome under investigation (e.g. yeast), or in enriched regions for the very high signal-to-noise ratio (S/N) of the antibody (e.g. Pol II), it may be appropriate to relax the threshold T because stringent filtering has small effect on the sensitivity of the peak detection [38]. Moreover, when studying highly repetitive regions (e.g. rDNA regions in Saccharomyces cerevisiae), filtering redundant reads should be omitted.
Because the NRF score depends on the total number of mapped reads, read sampling is necessary when comparing NRF scores among samples. The ENCODE consortium endorses an NRF > 0.8 for 10 million reads (T = 1) [30]. A low library complexity often occurs when samples are prepared from a small amount of starting materials. Even if the number of sequenced reads is sufficient after polymerase chain reaction (PCR) amplification, the substantial read number may be small, resulting in a poor significance power.
Sequencing depth
The number of called peaks increases with the sequencing depth, because weaker sites become statistically significant with a greater number of reads [51]. Although early ChIP-seq analyses produced <10 million reads per sample, it was reported that some highly active regions displayed a modest ChIP enrichment [30] and that weak protein binding by other factors may have important subfunctions [52]. Therefore, deep sequencing is required to include all functional sites. Broad markers that cover large genomic areas require more depth than those of sharp-mode peaks.
A sufficient sequencing depth depends on the S/N of the antibody. To determine sufficient sequencing depths, a ‘saturation analysis’ can be used, which subsamples the original read set in a stepwise fashion and calculates the proportion of identified peaks that overlap the original ones for each depth [1]. The point where the proportion is saturated is defined as sufficient depth. Although this approach is useful, there is no clear saturation point for most histone modifications [53]. Therefore, an agreeable depth has been determined empirically. For human samples, the ENCODE consortium suggested that at least two biological replicates with 10 million uniquely mapped reads for each replicate (providing at least 20 million reads per factor) is a minimum for typical TFs (sharp mode) [30]. Chen et al. [38] suggested that up to 60 million reads may be required for broad histone markers, and Jung et al. [53] suggested 40–50 million reads as a practical minimum for most broad histone markers. If the saturation point has not been detected at the available depth, it is still possible to apply tools for a sufficient depth estimation using a power analysis [54] or for predicting the benefits of additional sequencing [55].
Signal-to-noise ratio (S/N)
The S/N is evaluated by the number and strength of peaks obtained for each ChIP sample. This measure can also be used to assess the degree of noises in the input sample. The ENCODE consortium proposed two metrics, fraction of reads in peaks (FRiP) and cross-correlation profiles (CCPs) to measure the S/Ns [30]. The FRiP value is calculated as FRiP = Npeak/Nnonred, where Npeak is the number of reads falling within peak regions. This value correlates positively with the number and intensity of the identified peaks. ChIP samples that have too few peaks can be filtered using a cutoff for this score. However, because the FRiP score obviously depends on the sequencing depth and the parameters set for peak calling, it is not a perfectly objective metric. Conversely, CCPs assess the read-clustering levels without calling peaks beforehand [30]. This analysis plots the Pearson cross-correlations (CCs) between mapped read densities of positive and negative strands (y-axis) with shifting one strand (x-axis). Samples with large and small S/Ns typically have high CCs at the shift points corresponding to the fragment length (Cfrag) and the read length (Cread), respectively. Based on this observation, two quantitative measures are scored, the normalized strand coefficient NSC = Cfrag/Cmin and relative strand correlation RSC = (Cfrag − Cmin)/(Cread − Cmin), where Cmin is the minimum CC observed. The ENCODE consortium recommends an NSC ≥ 1.05 and an RSC ≥ 0.8 for typical TFs (sharp mode). Conversely, input and negative control samples should have low scores. Using this criteria, Marinov et al. [56] reported that a substantial minority (20%) of vertebrate ChIP-seq data sets for TFs in the Gene Expression Omnibus (GEO) were of insufficient quality, suggesting the necessity of quality check even for published data. Hansen et al. [12] has proposed the Hamming distance plot analogous to CCPs. CCPs are helpful; however, they have mainly been tested using only a few species and a more extensive investigation is necessary to understand the applicability of this approach to many other species. Moreover, a large S/N does not guarantee that the identified peaks are genuine binding sites—a large score merely means that there are many read-enriched regions in the genome. Samples that have many false-positive sites (e.g. non-specific binding sites) also have large S/Ns.
Example of the visualization and quality check
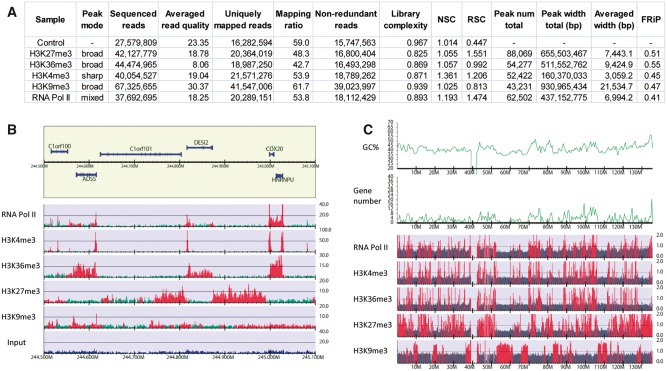
In Figure 2 we show an example of human ChIP-seq data obtained from the ENCODE project [45]. This example includes sharp mode (H3K4me3), broad mode (H3K36me3, H3K27me3 and H3K9me3) and mixed mode (RNA Pol II) samples with the control. Figure 2A summarizes the statistics of the quantitative quality metrics. The differences in average peak widths reflect the binding modes of each sample. The larger mapping ratio of H3K9me3 compared with H3K36me3 is mainly a consequence of the greater read quality. Conversely, the low affinity of the H3K9me3 antibody is indicated by the small RSC score and a comparable FRiP score against a much larger total peak width. These metric scores are used to automatically assess sample quality, leading to the rejection of poor-quality samples from further analyses.
Figure 2.
Statistics and visualization of ChIP-seq analysis for human K562 cells. A representative data set of ENCODE consortium [45]. The sequenced read files (fastq) and the reference peak lists (detected by Scripture [57] under the assumption of uniform background signal) were downloaded from GEO under accession number GSE29611. The fastq files were mapped onto the human genome (UCSC hg19) using Bowtie version 1.1.0 [42], allowing uniquely mapped reads only. (A) Summary statistics for each sample. The averaged read quality was obtained using fastqc version 0.11.4 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). The number of non-redundant reads, library complexity for 10 million mapped reads and FRiP scores were calculated using DROMPA3 version 3.0.0 [58]. Normalized strand coefficient (NSC) and relative strand correlation (RSC) scores were obtained using phantompeakqualtools version 1.1 (https://code.google.com/p/phantompeakqualtools). (B) The non-redundant read distribution for each sample with a RefSeq gene annotation (chromosome 1, 244.5–245.1 Mb). For the gene line (yellow box), genes in the upper and lower halves are on forward and reverse strands, respectively. The green and blue histograms represent the read distribution of ChIP and the input samples for 100-bp bins, respectively. The reference peak regions are highlighted in red. Note that the y axis indicates the read number normalized for the number of non-redundant reads, whereas the reference peak lists were identified based on raw read numbers. The gene reference was obtained from the UCSC genome browser [59]. (C) Visualization of the ChIP/Control enrichment distribution for 100-kb bins (chromosome 10). Bins with ChIP/control >1 are highlighted in red, and those with ChIP/control ≤1 are in gray. The GC contents and gene numbers for 500 kb windows are also plotted. The figures (B) and (C) were generated by DROMPA3.
Figure 2B shows the read distribution of each sample normalized for total non-redundant reads. The visualization of the read distribution is useful at the first step for judging the validity of the experimental results. In this figure, the active regions occupied by H3K36me3 and the silent regions occupied by H3K27me3 are distinguishable. On the other hand, H3K9me3 also has peaks, whereas their reads are not highly enriched. Considering H3K9me3 is a heterochromatin marker and generally not enriched in active gene regions [29], these peaks might be false positives. This is possibly because the mapped read number of H3K9me3 is more than twice that of the other samples (Figure 2A), resulting in a peak-calling threshold that is not stringent enough. In fact, because the S/N of the H3K9me3 antibody is smaller than those of H3K36me3 and H3K27me3, a high sequencing depth may be required to identify enriched regions. However, the chromosome-wide visualization of ChIP/input enrichment with a 100-kb bin clearly shows the exclusivity between H3K9me3 and H3K27me3 (Figure 2C). This large bin size enables us to directly use the ChIP/control distribution, even for large genomes, and provides an important insight.
Peak-calling
Establishing a definitive algorithm for peak detection has been the central topic in ChIP-seq analysis, resulting in the development of a plethora of programs. As the space is limited, we will just introduce here 20 representative programs.
Progress in the development of peak-calling algorithms
Peaks detection of a ChIP sample generally uses a corresponding input sample to estimate the background distribution at any genomic locus. Naked genomic DNA is less appropriate as a control because the input sample reflects the GC bias and chromatin structure rather than naked DNA [38, 41]. It was also reported that histone H3 ChIP-seq data can be used as a control [60]. ChIP samples for non-DNA-binding proteins, e.g. IgG, are often used to detect nonspecific binding sites. See [30] for a detailed discussion regarding control samples.
Early programs adopted the Poisson model, which assumes that the background reads are uniformly distributed along the genome (e.g. SICER [61] and CCAT [62]). However, a greater variation in the read distribution than allowed by the Poisson model is typically experimentally observed, and the negative binomial model that is an extension of the Poisson model was adopted to approximate such an overdispersion (e.g. CisGenome [63] and BayesPeak [64]). This model was extended to a zero-inflated negative binomial model to account for the zero-inflated read distribution caused by a lack of sequencing depth and low-mappable regions (e.g. MOSAiCS [65] and ZINBA [66]). For other strategies, MACS [39] uses the local Poisson model that estimates the parameter λ for each local genomic position. GPS [67] and PICS [68] predict protein-binding events using an EM algorithm. SISSRs [69], Peakzilla [70] and Q [12] focus on the equivalence between the read numbers of positive and negative strands to improve peak resolution. PePr [71] and JAMM [72] integrate information from multiple replicates to identify consistent or differential binding sites. The multiple hypothesis correction is performed to calculate false-discovery rates (FDRs) using the Benjamini–Hochberg procedure or the empirical method that calculates the peak number of the input sample compared with that of the ChIP sample.
Broad mode and mixed mode peaks depict weak and widespread enriched regions compared with the sharp mode, and there are no clear peak summits and sequence specificity. Although several peak-calling programs for the broad mode have been developed [61, 66, 73–75], and some peak-calling programs also have parameter settings for the broad mode, detection of such enrichment is still challenging. For proteins that are expected to be distributed within genic regions (e.g. Pol II and H3K36me3), a gene-annotation-based method is also useful, e.g. an aggregation plot around active genes and methods for differential gene expression analyses [76, 77]. For the characterization of Pol II occupancy, a travelling ratio (or pausing index) has been proposed [78]. The traveling ratio for gene i is defined as TRi = dpp/dgene, where dpp and dgene are the Pol II density in the promoter-proximal region and the gene body of gene i, respectively. This score indicates whether the promoter-proximal Pol II stalled at the gene. MUSIC discriminates between the binding modes, and between stalled and elongating forms of Pol II [74]. When investigating broad markers distributed in intergenic regions (e.g. H3K9me3), the gene-annotation-based method cannot be used. In such cases, it is still possible to use genome-wide visualization (Figure 2C) and compare the results with other public annotations, e.g. genome-wide maps of histone modifications [29].
While large genomes (e.g. human) require a statistical framework for peak calling owing to the low density and high variance of the reads, it is effective for small genomes (e.g. yeast [13, 79–81]) to inspect a genome-wide ChIP/input enrichment distribution itself (Figure 3).
Figure 3.
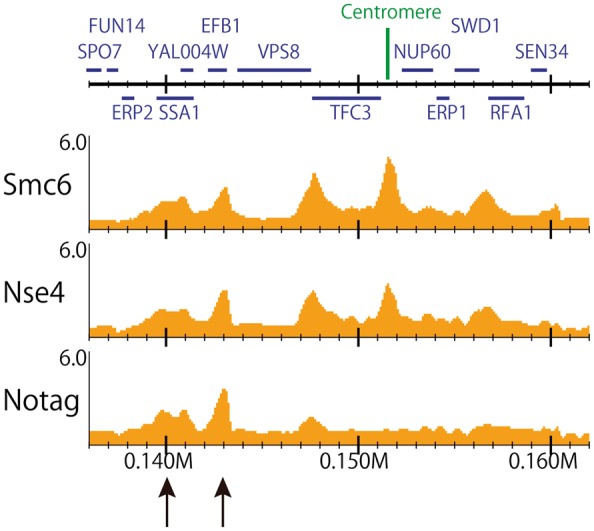
ChIP/input enrichment distribution of S. cerevisiae (chromosome I, 136–162 kb). Data from [11]. Smc6, Nse4 and ‘No tag (negative control)’ ChIP-seq data for a 100-bp bin with gene annotation obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org). The reads were mapped onto the genome, allowing multiple mapped reads. For the yeast genome, inspecting the genome-wide ChIP/input enrichment distribution is effective because a read depth is large enough (>10-fold) and the division with the input sample can minimize the technical and biological biases of the conditions. The enriched regions of Smc6 and Nse4 that overlap those of the ‘No tag’ sample (black arrows) suggest false positives (e.g. hyper-ChIPable regions).
Which program is best for our analysis?
A performance comparison with a large set of programs in various aspects is of interest. However, several issues make such large-scale comparative study difficult. First, installing dozens of programs with multiple prerequired data (e.g. mappability) and assembling their results are often problematic owing to a large variety of existing file formats, and the availability and version-control of required tools in different computing environments. Second, because the programs have different underlying assumptions (e.g. using mappability and/or multiple mapped reads, subdividing peaks and necessity of replicates), the simultaneous comparison of them is difficult to be fair. Finally, the substantial evaluation of obtained peaks is difficult due to the lack of annotation for ‘true’ binding sites (see the next subsection). Although there are early studies of performance evaluations for peak-calling programs, based on the number, width and the distribution of peaks [82, 83], the reproducibility across replicates [38] and the accuracy against the known binding motif sites [84] or the manually curated benchmark data sets [85], the number of programs compared and quality metrics used in each study were limited. Moreover, the comparison with latest programs is lacked.
The appropriate method depends on the species, sample conditions and target proteins. Even though there is no clear consensus on which is best, the latest and widely used programs may be satisfactory for our needs.
How reliable are the obtained results?
Owing to the lack of annotation for true binding sites, the development of computational methods to evaluate the identified peaks has been limited. Motif-based evaluations are not applicable for proteins that do not have sequence specificity (e.g. histone modifications). Even for proteins with canonical motifs, there can be many tissue-specific binding sites recruited by other factors that do not involve the motif sequence [52, 86]. Another way of assessing the validity of the identified peaks is to focus on reproducibility. Cross-correlation coefficients of peak regions or whole-genomes measure the global similarity between two biological replicates [87]. The irreproducible discovery rate (IDR) assesses the rank consistency of common peaks between two replicates [88]. Based on a copula mixture model, IDR estimates the reproducibility of each peak pair, and reports the expected rate of irreproducible discoveries in the obtained peaks in a similar way to the FDR. In contrast to the CC coefficient, the IDR can assess each peak separately and therefore, can be used as a threshold robust for the technical variance owing to the analysis protocols and the choice of a peak-calling program. However, the poorest quality samples can become the bottleneck. When many true peaks do not appear in a replicate with a low S/N, they will be rejected as non-reproducible. Furthermore, several genomic regions tend to show artificially high enrichment levels, resulting in ‘reproducible’ false-positive peaks that cannot be filtered by the IDR. The ENCODE consortium summarizes empirically identified ‘blacklist regions’ for several species, which include repetitive and low-mappable regions [45]. Moreover, there are ‘hyper-ChIPable’ regions in the genome, in which peaks of unrelated proteins, including negative controls, overlap [79] (see Figure 3). These regions are positively correlated with promoters of well-expressed genes and are unchanged in cells containing the mutant protein of interest [89]. This result indicates that the peaks obtained by current methods may contain some (or a large) amount of unrecognizable false positives. Therefore, when investigating a protein with an unknown DNA-binding pattern, the obtained peaks should be validated carefully with a negative control (e.g. IgG), especially around transcription start sites (TSSs) of expressed genes.
How to treat low-quality samples?
There are various factors that can affect the data quality of the sample preparation step, including the quality of the antibody—e.g. its affinity and specificity, over-crosslinking, DNA fragmentation and overamplification by PCR and ChIP conditions. Different antibodies, even for the same protein (and even biological replicates of the same antibody), often produce completely different peak distributions. Despite an investigation of sequencing biases present in NGS data [90], it is still difficult to ascertain the exact sources of each bias in a sample preparation protocol [31]. In our experience, when a sample has a poor score for a quality measure, it often has other problems (e.g. low complexity causes a strong GC bias). As it is difficult to rescue poor-quality samples and include them in the analysis pipeline even with read normalization, fine-tuning sample preparation protocols to produce high-quality samples may be necessary for each project.
An efficient way to allow the use of ChIP-seq data of modest quality, while suppressing the noise, is by limiting the genomic regions to be investigated to a few candidate regions that satisfy the working hypothesis, and then validating them using other biological experiments, as suggested in [30]. This is a practical approach, rather than seeking the most accurate method by minutely tuning the parameters for each sample. Adding samples from related proteins and biological replicates increases reliability.
Normalization for a differential analysis
Relative-level difference (de novo normalization)
A typical procedure after obtaining peak sets is to summarize peak similarities and differences among samples in a binary (common or unique) or quantitative (variety of peak intensity) manner, which necessitates read normalization. The simplest normalization approach is to scale reads using the total read number (Nnonred) within the whole genome or background region, which assumes that the differences in the mapped reads among samples are small enough compared with the total read number. Instead of scaling reads using a constant factor, Taslim et al. [91] proposed the nonlinear method using a locally weighted regression (LOESS) to remove the effects of bias and systematic errors. However, the underlying assumption that the genome-wide distribution of read counts has an equal mean and variance across samples may not be valid in most cases (e.g. the different S/Ns between samples). Maehara et al. [92] proposed the co-localization score that measures the global similarity between two samples based on the common peaks, while it does not aim to identify individual differential peaks. Methods for differential gene expression analysis [76, 77] can be used to directly compare more than two groups, and these methods do not consider the different S/Ns among samples either. In contrast, MAnorm [93] and ChIPcomp [94] are designed to consider different S/Ns. MAnorm scales the reads of peaks common to two samples using a robust linear regression based on the MA plot. ChIPcomp performs quantitative comparison of multiple ChIP samples, which measures genomic background using control data and considers multiple-factor experimental designs. These tools assume that most of the common peaks should have similar binding intensities among the samples (e.g. same antibody for different conditions). See reference [94] for a detailed discussion regarding these de novo normalization programs.
Absolute-level difference (spike-in analysis)
In cases where the genome-wide peak distribution changes drastically (e.g. knock down analysis or stimulated versus non-stimulated) the aforementioned assumptions do not hold. Moreover, those normalizations are essentially limited to investigating relative differences in protein binding among samples [31]. For example, when the reads are relatively enriched in euchromatic regions and depressed in heterochromatic regions in a sample, it is difficult to discern whether protein binding was increased in the euchromatin or decreased in the heterochromatin in vivo. Recently, to investigate the absolute differences, several studies adopted spike-in analysis for human [95–97] and yeast [98, 99]. This analysis adds same quantities of chromatin DNA to all samples compared before or after immunoprecipitation. Because the number of reads derived from this reference chromatin should be same across samples, this number can be used as an internal control for read normalization. Thus, the spike-in analysis can detect global differences that cannot be identified by aforementioned de novo normalization methods. To discern reference reads from sample ones, the reference chromatin should be derived from a different genome.
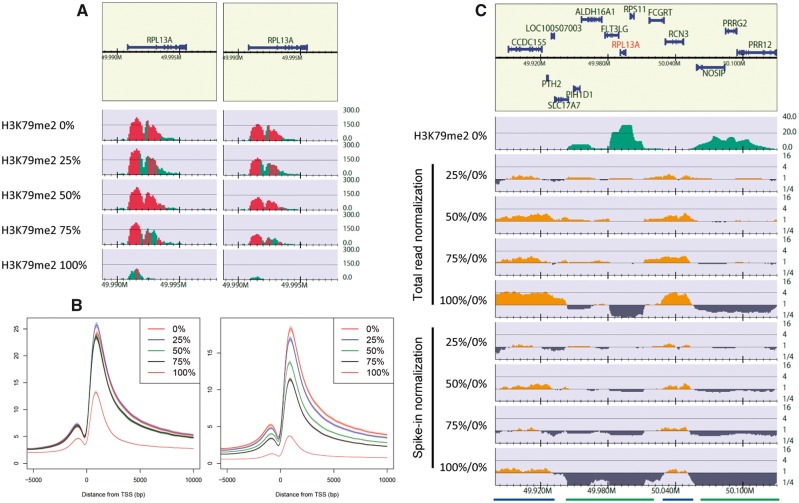
Herein, we show an example of the spike-in data of H3K79me2 for EPZ5676-treated Jurkat cells from Orlando et al. (Figure 4). As shown in the original paper, spike-in normalization revealed a decrease in the H3K79me2 enrichment (Figure 4A and B). Next, the relative enrichment of H3K79me2 against that of 0% cells was visualized (Figure 4C). In the total read normalization for 100%/0% (fifth line), the read depth in the background (blue bars) was about four times greater owing to the genome-wide substantial decrease in H3K79me2-enriched regions (green bars). Using spike-in normalization (bottom line), the relative enrichment of the background declined to one, which indicates there is little difference within the background at an absolute level. A similar tendency can be observed for three other sample pairs, but they were less significant.
Figure 4.
Spike-in analysis of H3K79me2 ChIP-seq data for 0%, 25%, 50%, 75% and 100% EPZ5676-treated Jurkat cells. Data from [96] (GEO under accession number GSE60104). Spike-in normalization was implemented using the number of reads uniquely mapped onto the fly genome (UCSC dm3). (A) Read distribution near the RPL13A gene locus for 100-bp bins. Left: total read normalization, right: spike-in normalization. (B) Aggregation plots of total read normalization (left) and spike-in normalization (right) from 5-kb upstream to 10-kb downstream of the TSSs of the RefSeq genes. Shaded regions indicate a 95% confidence interval. (A) and (B) are identical visualizations of Figure 3C and E in reference [96], respectively. (C) Log-scale relative enrichment of H3K79me2 for 25%, 50%, 75% and 100% treated cells against 0% treated cells near the RPL13A gene locus (chromosome 19,49.88–50.13 Mb), with a 100-kb bin and 20-kb smoothing window. The top green line displays a H3K79me2 read distribution for 0% treated cells to roughly identify H3K79me2-enriched (green bars) and background regions (blue bars). Regions in which the enrichment (y-axis) is > 1 and <1 indicate a relative increase and decrease, respectively.
Although a spike-in analysis should be powerful and useful, its applicability and limitations, e.g. balancing the amount of spike-in relative to the chromatin of interest, are still not clear [31]. Moreover, we have occasionally observed a decrease in the read density owing to protein knockdowns in both the peak regions and in the background. It is challenging to determine whether a decrease in the read density of the background indicates that the protein of interest is also distributed in the background, and if we can conclude that the peak decreases owing to the knockdown when the read depth in the background decreases as much as in the peak regions. To answer these questions, extensive testing may be necessary, e.g. using a mock knockdown analysis. In addition, spiking genomes of multiple species may make the analysis more robust. The optimal normalization method should be chosen based on prior knowledge of the system and the statistics of the sequenced samples. The obtained results of a differential analysis should be evaluated using another method, e.g. quantitative PCR.
Integrative analysis for a de novo genome annotation
Although it is now possible to produce many ChIP-seq data sets at reasonable costs, their comparison and integration are not trivial. For example, when examining the differential ChIP-seq analysis of four proteins obtained under two conditions, and the knockdown effects on these data sets are of interest, it is necessary to investigate the differences among four proteins under the two conditions and/or between wild-type and knockdown cells simultaneously. As the results strongly depend on the peak-calling result of each sample [87], of which the protocol should be selected individually, the extensive work needed for tuning the protocol and integrating all results will be challenging. Consequently, there is a great demand for tools that jointly analyze all samples simultaneously.
Context-specific co-association identification
In the ENCODE consortium, Gerstein et al. [25] applied a machine-learning framework and examined the genome-wide co-association of 119 TFs that contained over 450 ChIP-seq samples. In this framework, peak calling is used with relaxed thresholds merely to obtain candidate regions for investigation. The identified sample peak sets are integrated into co-binding maps, and then ‘context-specific’ co-associations are identified, which are subsets of peaks binding to different TF sets in other genomic regions. These combinatorial patterns provide biological information on the high-dimensional interrelationship level for regulatory elements, which is difficult to ascertain using typical genome-wide pairwise comparisons. This concept has also been used for cross-species analyses of regulatory information [100–102]. Because this approach targeted the binding sites of point-source TFs, the results did not contain regions enriched in broad markers and background regions. The targeted regions may differ among experiments, and therefore it is difficult to directly compare the results across multiple experiments.
Joint analysis for a de novo genome annotation
Recently, integrative methods were developed to segment, classify and annotate a whole-genome sequence de novo, based on unsupervised machine-learning methods. These methods directly receive all ChIP sample data and analyze them simultaneously, instead of calling peaks and comparing them individually.
ChromHMM [103] and Segway [104] were developed to systematically identify the specific combination patterns of histone modifications as a chromatin state, which can detect large-scale variations of histone marks across the genome. ChromHMM is the most widely used tool, which models binary vectors (1 or 0) for each 200-bp bin converted from raw read counts using a sample-specific threshold as an independent Bernoulli random variable. Segway, by contrast, transforms the counts into real values and uses a dynamic Bayesian network at a 1-bp resolution. It can incorporate more complex relationships among samples in each region, although it requires a magnitude larger computational cost. Using these tools, high-quality chromatin-state maps for many cell lines are available [28, 29]. Several methods also exist that expand the methodologies of these tools to improve accuracy, computational cost or the interactive navigation [105–108]. Although this chromatin-state segmentation is not suitable for quantitative analysis among multiple experiments, several tools were developed to integrate and compare chromatin state sets from different experiments [109, 110]. hiHMM jointly infers chromatin state maps across multiple genomes and cell types [111].
There are also various joint analysis tools designed for TFs, based on several probabilistic models, such as the generalized EM algorithm and the Markov random field model [112–115]. These tools jointly model the dependencies among ChIP samples to identify global and local combinatorial enrichment patterns in whole-genome or specific functional regions. The number of classified enrichment patterns is not necessarily optimal for distinct functions of each protein, but these powerful approaches do efficiently reveal context-specific co-associations, and it may be possible to find out the false positives derived from hyper-ChIPable regions from total peak sets. Although the machine-learning approach for a large number of TFs is computationally daunting, these data-driven approaches may positively impact large-scale analyses in the near future.
Future potential: single-cell analysis
A limitation of ChIP-seq analysis is the requirement for large amounts of starting material (∼105 cells); accordingly, ChIP-seq analysis focuses on ensemble (averaged) features across large number of cells. To elucidate the internal heterogeneity within complex tissues and cell populations, the development of single-cell methodology is desired. In spite of rapid development of single-cell technologies in other genomic fields (reviewed in [116, 117]), there has been so far no report for single-cell ChIP-seq analysis. Recently, the first method for collection of chromatin data at single-cell resolution was published [118]. This ‘Drop-ChIP’ method adopts a droplet-based microfluidics system [119] for labeling chromatin from single cells before immunoprecipitation, which is also employed for single-cell RNA-seq analysis [120–122]. Labeled chromatins from all cells are sequenced, mapped and partitioned into single-cell reads by their barcode sequences. The single-cell profiles are classified by an unsupervised hierarchical clustering, and aggregated into profiles for subpopulations. The experiment using H3K4me2 antibody demonstrated that, although just a few hundred peaks were identified per cell owing to low sequencing depth (∼10 000 reads), this method could distinguish individual three cell types (mouse embryonic stem (ES) cells, embryonic fibroblasts and hematopoietic progenitors cells) with nearly 100% accuracy, and identify subpopulations in ES cells using the differences in chromatin signatures of pluripotency and differentiation priming.
This method involves several important considerations. First, because of low sensitivity and high technical read variance owing to low depth, this method can accept only antibodies for sharp mode peaks with high S/N so far. It is difficult to directly apply exiting techniques and quality metrics for traditional ChIP-seq to single-cell ChIP-seq data. Therefore, it is better to combine the result with typical ChIP-seq analysis, if possible. Second, this single-cell analysis aims to unveil internal heterogeneity by classifying patterns of subpopulations, rather than examine an individual cell (e.g. single-cell-level comparison with gene expression data). In this respect, this method is not interchangeable with low-input analyses [123–127], which aims to reduce the cell number required by a typical ChIP-seq workflow for precious biological samples (e.g. primary cells and clinical samples). Finally, identification of small subpopulation requires large number of sample cells [118]. Significance power of this method depends on the number of input cells and the performance of systems. Anyway, this first innovative challenge will encourage a new era in this field.
Concluding remarks
In this review, we discuss the computational aspects of ChIP-seq analysis and highlight key points associated with each step. We emphasize that the design of a ChIP-seq experiment is of critical importance and that a quality check of the data at each step is important, even when using published ChIP-seq data. By addressing a wide range of relevant subtopics within ChIP-seq analysis, which include integrative large-scale analysis and single-cell analysis, this review expands the topic and enhances the value of previous reviews. We believe that this review benefits researchers in related fields.
There are high-quality genome-wide maps of TFs and histone modifications provided by many consortia, and the open web servers (e.g. UCSC genome browser [59] and WashU Epigenome Browser [128]) enable researchers to effectively refer and track these resources for their own projects. Growth of publicly available resources will be a superb driving force for research progresses in the biological and bioinformatics field.
Remaining challenges for the future is to classify direct and indirect binding, capture temporary and non-site-specific (drifting) TF binding and investigate highly repetitive regions, e.g. centromeres. Finally, integration with other technologies, e.g. human genetic variation analyses [129–131], genome editing [132] and de novo assembly [133, 134], will make ChIP-seq analysis more fruitful and provide us with knowledge regarding the underlying mechanisms of genome function and evolution. Such comprehensive analyses facilitate the systematic elucidation of diverse biological activities intricately cooperating in the genome.
Acknowledgments
We are grateful to Drs. H. Kimura and M. Suyama for their valuable comments. We would also like to thank our laboratory’s members and collaborators.
Funding
This work was supported by a Grant-in-Aid for Scientific Research from MEXT (15K18465 to R.N., 15H02369, 15H05970 to K.S.); Core Research for Evolutional Science and Technology; The Japan Agency for Medical Research and Development; and Platform for Drug Discovery, Informatics and Structural Life Science.
Key Points
Recent advances in sequencing technologies and analysis tools enable us to compare hundreds of ChIP-seq samples simultaneously; such large-scale analysis can reveal the high-dimensional interrelationship level for regulatory elements and annotate novel functional genomic regions de novo.
Despite great efforts to streamline the ChIP-seq procedure, no single workflow that is optimal under all circumstances exists, and there are many experimental considerations that are relevant to the method choice for a ChIP-seq analysis.
This review highlights important points using concrete examples to provide guiding principles for the design and management of various computational ChIP-seq analyses.
The emerging machine-learning approaches that jointly analyze all samples simultaneously have potential to positively impact large-scale analyses.
The first innovative challenge of single-cell ChIP-seq analysis will encourage a new era in this field.
Ryuichiro Nakato is a Research Associate at the University of Tokyo. His research interests involve the computational approaches using high-throughput sequencers, including data analysis and program development.
Katsuhiko Shirahige is a Professor at the University of Tokyo. His research interests involve the cooperative interactions among biological functions on the genomes.
References
- 1. Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat Rev Genet 2009;10:669–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pepke S, Wold B, Mortazavi A.. Computation for ChIP-seq and RNA-seq studies. Nat Methods 2009;6:S22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Furey TS. ChIP-seq and beyond: new and improved methodologies to detect and characterize protein-DNA interactions. Nat Rev Genet 2012;13:840–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deardorff MA, Bando M, Nakato R, et al. HDAC8 mutations in Cornelia de Lange syndrome affect the cohesin acetylation cycle. Nature 2012;489:313–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schaub MA, Boyle AP, Kundaje A, et al. Linking disease associations with regulatory information in the human genome. Genome Res 2012;22:1748–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zuin J, Franke V, van Ijcken WF, et al. A cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLoS Genet 2014;10:e1004153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Izumi K, Nakato R, Zhang Z, et al. Germline gain-of-function mutations in AFF4 cause a developmental syndrome functionally linking the super elongation complex and cohesin. Nat Genet 2015;47:338–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mikkelsen TS, Xu Z, Zhang X, et al. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010;143:156–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 2011;473:43–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shang WH, Hori T, Martins NM, et al. Chromosome engineering allows the efficient isolation of vertebrate neocentromeres. Dev Cell 2013;24:635–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jeppsson K, Carlborg KK, Nakato R, et al. The chromosomal association of the smc5/6 complex depends on cohesion and predicts the level of sister chromatid entanglement. PLoS Genet 2014;10:e1004680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hansen P, Hecht J, Ibrahim DM, et al. Saturation analysis of ChIP-seq data for reproducible identification of binding peaks. Genome Res 2015;25:1391–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sutani T, Sakata T, Nakato R, et al. Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nat Commun 2015;6:7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Machanick P, Bailey TL.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics 2011;27:1696–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thomas-Chollier M, Herrmann C, Defrance M, et al. RSAT peak-motifs: motif analysis in full-size ChIP-seq datasets. Nucleic Acids Res 2012;40:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McLean CY, Bristor D, Hiller M, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010;28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Welch RP, Lee C, Imbriano PM, et al. ChIP-Enrich: gene set enrichment testing for ChIP-seq data. Nucleic Acids Res 2014;42:e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012;485:376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li G, Ruan X, Auerbach RK, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 2012;148:84–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Agirre X, Castellano G, Pascual M, et al. Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers. Genome Res 2015;25:478–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Alekseyenko AA, Gorchakov AA, Zee BM, et al. Heterochromatin-associated interactions of Drosophila HP1a with dADD1, HIPP1, and repetitive RNAs. Genes Dev 2014;28:1445–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin C, Garrett AS, De Kumar B, et al. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev 2011;25:1486–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Churchman LS, Weissman JS.. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 2011;469:368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nojima T, Gomes T, Grosso AR, et al. Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 2015;161:526–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gerstein MB, Kundaje A, Hariharan M, et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012;489:91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yan J, Enge M, Whitington T, et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 2013;154:801–13. [DOI] [PubMed] [Google Scholar]
- 27. Griffon A, Barbier Q, Dalino J, et al. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic Acids Res 2015;43:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hoffman MM, Ernst J, Wilder SP, et al. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res 2013;41:827–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kundaje A, Meuleman W, Ernst J, et al. ; Roadmap Epigenomics Consortium. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Landt SG, Marinov GK, Kundaje A, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 2012;22:1813–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meyer CA, Liu XS.. Identifying and mitigating bias in next-generation sequencing methods for chromatin biology. Nat Rev Genet 2014;15:709–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rhee HS, Pugh BF.. Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 2011;147:1408–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He Q, Johnston J, Zeitlinger J.. ChIP-nexus enables improved detection of in vivo transcription factor binding footprints. Nat Biotechnol 2015;33:395–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kelly TK, Liu Y, Lay FD, et al. Genome-wide mapping of nucleosome positioning and DNA methylation within individual DNA molecules. Genome Res 2012;22:2497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buenrostro JD, Giresi PG, Zaba LC, et al. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 2013;10:1213–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Muller CA, Hawkins M, Retkute R, et al. The dynamics of genome replication using deep sequencing. Nucleic Acids Res 2014;42:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. He HH, Meyer CA, Hu SS, et al. Refined DNase-seq protocol and data analysis reveals intrinsic bias in transcription factor footprint identification. Nat Methods 2014;11:73–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Y, Negre N, Li Q, et al. Systematic evaluation of factors influencing ChIP-seq fidelity. Nat Methods 2012;9:609–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kharchenko PV, Tolstorukov MY, Park PJ.. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol 2008;26:1351–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Auerbach RK, Euskirchen G, Rozowsky J, et al. Mapping accessible chromatin regions using Sono-Seq. Proc Natl Acad Sci USA 2009;106:14926–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009;10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H, Durbin R.. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rozowsky J, Abyzov A, Wang J, et al. AlleleSeq: analysis of allele-specific expression and binding in a network framework. Mol Syst Biol 2011;7:522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Encode Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chung D, Kuan PF, Li B, et al. Discovering transcription factor binding sites in highly repetitive regions of genomes with multi-read analysis of ChIP-Seq data. PLoS Comput Biol 2011;7:e1002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Day DS, Luquette LJ, Park PJ, et al. Estimating enrichment of repetitive elements from high-throughput sequence data. Genome Biol 2010;11:R69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rozowsky J, Euskirchen G, Auerbach RK, et al. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat Biotechnol 2009;27:66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Koehler R, Issac H, Cloonan N, et al. The uniqueome: a mappability resource for short-tag sequencing. Bioinformatics 2011;27:272–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Derrien T, Estelle J, Marco Sola S, et al. Fast computation and applications of genome mappability. PLoS One 2012;7:e30377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sims D, Sudbery I, Ilott NE, et al. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet 2014;15:121–32. [DOI] [PubMed] [Google Scholar]
- 52. Schmidt D, Schwalie PC, Ross-Innes CS, et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 2010;20:578–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jung YL, Luquette LJ, Ho JW, et al. Impact of sequencing depth in ChIP-seq experiments. Nucleic Acids Res 2014;42:e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zuo C, Keles S.. A statistical framework for power calculations in ChIP-seq experiments. Bioinformatics 2014;30:753–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Daley T, Smith AD.. Predicting the molecular complexity of sequencing libraries. Nat Methods 2013;10:325–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marinov GK, Kundaje A, Park PJ, et al. Large-scale quality analysis of published ChIP-seq data. G3 (Bethesda) 2014;4:209–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guttman M, Garber M, Levin JZ, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol 2010;28:503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nakato R, Itoh T, Shirahige K.. DROMPA: easy-to-handle peak calling and visualization software for the computational analysis and validation of ChIP-seq data. Genes Cells 2013;18:589–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rosenbloom KR, Armstrong J, Barber GP, et al. The UCSC Genome Browser database: 2015 update. Nucleic Acids Res 2015;43:D670–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Flensburg C, Kinkel SA, Keniry A, et al. A comparison of control samples for ChIP-seq of histone modifications. Front Genet 2014;5:329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zang C, Schones DE, Zeng C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics 2009;25:1952–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xu H, Handoko L, Wei X, et al. A signal-noise model for significance analysis of ChIP-seq with negative control. Bioinformatics 2010;26:1199–204. [DOI] [PubMed] [Google Scholar]
- 63. Ji H, Jiang H, Ma W, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol 2008;26:1293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Spyrou C, Stark R, Lynch AG, et al. BayesPeak: Bayesian analysis of ChIP-seq data. BMC Bioinformatics 2009;10:299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kuan PF, Chung DJ, Pan GJ, et al. A statistical framework for the analysis of ChIP-Seq data. J Am Stat Assoc 2011;106:891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rashid NU, Giresi PG, Ibrahim JG, et al. ZINBA integrates local covariates with DNA-seq data to identify broad and narrow regions of enrichment, even within amplified genomic regions. Genome Biol 2011;12:R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Guo Y, Papachristoudis G, Altshuler RC, et al. Discovering homotypic binding events at high spatial resolution. Bioinformatics 2010;26:3028–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang X, Robertson G, Krzywinski M, et al. PICS: probabilistic inference for ChIP-seq. Biometrics 2011;67:151–63. [DOI] [PubMed] [Google Scholar]
- 69. Jothi R, Cuddapah S, Barski A, et al. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res 2008;36:5221–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bardet AF, Steinmann J, Bafna S, et al. Identification of transcription factor binding sites from ChIP-seq data at high resolution. Bioinformatics 2013;29:2705–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhang Y, Lin YH, Johnson TD, et al. PePr: a peak-calling prioritization pipeline to identify consistent or differential peaks from replicated ChIP-Seq data. Bioinformatics 2014;30:2568–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ibrahim MM, Lacadie SA, Ohler U.. JAMM: a peak finder for joint analysis of NGS replicates. Bioinformatics 2015;31:48–55. [DOI] [PubMed] [Google Scholar]
- 73. Wang J, Lunyak VV, Jordan IK.. BroadPeak: a novel algorithm for identifying broad peaks in diffuse ChIP-seq datasets. Bioinformatics 2013;29:492–3. [DOI] [PubMed] [Google Scholar]
- 74. Harmanci A, Rozowsky J, Gerstein M.. MUSIC: identification of enriched regions in ChIP-Seq experiments using a mappability-corrected multiscale signal processing framework. Genome Biol 2014;15:474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xing H, Mo Y, Liao W, et al. Genome-wide localization of protein-DNA binding and histone modification by a Bayesian change-point method with ChIP-seq data. PLoS Comput Biol 2012;8:e1002613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhou X, Lindsay H, Robinson MD.. Robustly detecting differential expression in RNA sequencing data using observation weights. Nucleic Acids Res 2014;42:e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rahl PB, Lin CY, Seila AC, et al. c-Myc regulates transcriptional pause release. Cell 2010;141:432–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Teytelman L, Thurtle DM, Rine J, et al. Highly expressed loci are vulnerable to misleading ChIP localization of multiple unrelated proteins. Proc Natl Acad Sci USA 2013;110:18602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Foltman M, Evrin C, De Piccoli G, et al. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep 2013;3:892–904. [DOI] [PubMed] [Google Scholar]
- 81. Kubota T, Katou Y, Nakato R, et al. Replication-Coupled PCNA Unloading by the Elg1 Complex Occurs Genome-wide and Requires Okazaki Fragment Ligation. Cell Rep 2015;12:774–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Laajala TD, Raghav S, Tuomela S, et al. A practical comparison of methods for detecting transcription factor binding sites in ChIP-seq experiments. BMC Genomics 2009;10:618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Malone BM, Tan F, Bridges SM, et al. Comparison of four ChIP-Seq analytical algorithms using rice endosperm H3K27 trimethylation profiling data. PLoS One 2011;6:e25260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wilbanks EG, Facciotti MT.. Evaluation of algorithm performance in ChIP-seq peak detection. PLoS One 2010;5:e11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Rye MB, Saetrom P, Drablos F.. A manually curated ChIP-seq benchmark demonstrates room for improvement in current peak-finder programs. Nucleic Acids Res 2011;39:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Faure AJ, Schmidt D, Watt S, et al. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome Res 2012;22:2163–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bardet AF, He Q, Zeitlinger J, et al. A computational pipeline for comparative ChIP-seq analyses. Nat Protoc 2012;7:45–61. [DOI] [PubMed] [Google Scholar]
- 88. Li Q, Brown JB, Huang H, et al. Measuring reproducibility of high-throughput experiments. Ann Appl Stat 2011;5:1752–79. [Google Scholar]
- 89. Jain D, Baldi S, Zabel A, et al. Active promoters give rise to false positive 'Phantom Peaks' in ChIP-seq experiments. Nucleic Acids Res 2015;43:6959–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Diaz A, Park K, Lim DA, et al. Normalization, bias correction, and peak calling for ChIP-seq. Stat Appl Genet Mol Biol 2012;11:article 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Taslim C, Wu J, Yan P, et al. Comparative study on ChIP-seq data: normalization and binding pattern characterization. Bioinformatics 2009;25:2334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Maehara K, Odawara J, Harada A, et al. A co-localization model of paired ChIP-seq data using a large ENCODE data set enables comparison of multiple samples. Nucleic Acids Res 2013;41:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Shao Z, Zhang Y, Yuan GC, et al. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol 2012;13:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Chen L, Wang C, Qin ZS, et al. A novel statistical method for quantitative comparison of multiple ChIP-seq datasets. Bioinformatics 2015;31:1889–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Bonhoure N, Bounova G, Bernasconi D, et al. Quantifying ChIP-seq data: a spiking method providing an internal reference for sample-to-sample normalization. Genome Res 2014;24:1157–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Orlando DA, Chen MW, Brown VE, et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 2014;9:1163–70. [DOI] [PubMed] [Google Scholar]
- 97. Grzybowski AT, Chen Z, Ruthenburg AJ.. Calibrating ChIP-Seq with nucleosomal internal standards to measure histone modification density genome wide. Mol Cell 2015;58:886–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Hu Z, Chen K, Xia Z, et al. Nucleosome loss leads to global transcriptional up-regulation and genomic instability during yeast aging. Genes Dev 2014;28:396–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hu B, Petela N, Kurze A, et al. Biological chromodynamics: a general method for measuring protein occupancy across the genome by calibrating ChIP-seq. Nucleic Acids Res 2015;43:e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cheng Y, Ma Z, Kim BH, et al. Principles of regulatory information conservation between mouse and human. Nature 2014;515:371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Boyle AP, Araya CL, Brdlik C, et al. Comparative analysis of regulatory information and circuits across distant species. Nature 2014;512:453–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yue F, Cheng Y, Breschi A, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature 2014;515:355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ernst J, Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods 2012;9:215–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hoffman MM, Buske OJ, Wang J, et al. Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat Methods 2012;9:473–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Nielsen CB, Younesy H, O'Geen H, et al. Spark: a navigational paradigm for genomic data exploration. Genome Res 2012;22:2262–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Biesinger J, Wang Y, Xie X.. Discovering and mapping chromatin states using a tree hidden Markov model. BMC Bioinformatics 2013;14(Suppl 5):S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Song J, Chen KC.. Spectacle: fast chromatin state annotation using spectral learning. Genome Biol 2015;16:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mammana A, Chung HR.. Chromatin segmentation based on a probabilistic model for read counts explains a large portion of the epigenome. Genome Biol 2015;16:151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yen A, Kellis M.. Systematic chromatin state comparison of epigenomes associated with diverse properties including sex and tissue type. Nat Commun 2015;6:7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Ernst J, Kellis M.. Large-scale imputation of epigenomic datasets for systematic annotation of diverse human tissues. Nat Biotechnol 2015;33:364–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Sohn KA, Ho JW, Djordjevic D, et al. hiHMM: Bayesian non-parametric joint inference of chromatin state maps. Bioinformatics 2015;31:2066–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zeng X, Sanalkumar R, Bresnick EH, et al. jMOSAiCS: joint analysis of multiple ChIP-seq datasets. Genome Biol 2013;14:R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bao Y, Vinciotti V, Wit E, et al. Joint modeling of ChIP-seq data via a Markov random field model. Biostatistics 2014;15:296–310. [DOI] [PubMed] [Google Scholar]
- 114. Mahony S, Edwards MD, Mazzoni EO, et al. An integrated model of multiple-condition ChIP-Seq data reveals predeterminants of Cdx2 binding. PLoS Comput Biol 2014;�10:e1003501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wong KC, Li Y, Peng CB, et al. SignalSpider: probabilistic pattern discovery on multiple normalized ChIP-Seq signal profiles. Bioinformatics 2015;31:17–24. [DOI] [PubMed] [Google Scholar]
- 116. Bheda P, Schneider R.. Epigenetics reloaded: the single-cell revolution. Trends Cell Biol 2014;24:712–23. [DOI] [PubMed] [Google Scholar]
- 117. Schwartzman O, Tanay A.. Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet 2015;16:716–26. [DOI] [PubMed] [Google Scholar]
- 118. Rotem A, Ram O, Shoresh N, et al. Single-cell ChIP-seq reveals cell subpopulations defined by chromatin state. Nat Biotechnol 2015;33:1165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Mazutis L, Gilbert J, Ung WL, et al. Single-cell analysis and sorting using droplet-based microfluidics. Nat Protoc 2013;8:870–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Klein AM, Mazutis L, Akartuna I, et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 2015;161:1187–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Rotem A, Ram O, Shoresh N, et al. High-Throughput Single-Cell Labeling (Hi-SCL) for RNA-Seq using drop-based microfluidics. PLoS One 2015;10:e0116328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 2015;161:1202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Adli M, Bernstein BE.. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat Protoc 2011;6:1656–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Shankaranarayanan P, Mendoza-Parra MA, Walia M, et al. Single-tube linear DNA amplification (LinDA) for robust ChIP-seq. Nat Methods 2011;8:565–7. [DOI] [PubMed] [Google Scholar]
- 125. Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science 2014;345:943–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Greenleaf WJ. Assaying the epigenome in limited numbers of cells. Methods 2015;72:51–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Schmidl C, Rendeiro AF, Sheffield NC, et al. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods 2015;12:963–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Zhou X, Li D, Zhang B, et al. Epigenomic annotation of genetic variants using the Roadmap Epigenome Browser. Nat Biotechnol 2015;33:345–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Gulko B, Hubisz MJ, Gronau I, et al. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat Genet 2015;47:276–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Brookes AJ, Robinson PN.. Human genotype-phenotype databases: aims, challenges and opportunities. Nat Rev Genet 2015;16:702–15. [DOI] [PubMed] [Google Scholar]
- 131. Church DM, Schneider VA, Steinberg KM, et al. Extending reference assembly models. Genome Biol 2015;16:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Sander JD, Joung JK.. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 2014;32:347–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. He X, Cicek AE, Wang Y, et al. De novo ChIP-seq analysis. Genome Biol 2015;16:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Simpson JT, Pop M.. The theory and practice of genome sequence assembly. Annu Rev Genomics Hum Genet 2015;16:153–72. [DOI] [PubMed] [Google Scholar]