Highlight
Over 100 fruit volatile QTLs were identified in a RIL population derived from the red-fruited wild species Solanum pimpinellifolium and a fresh market tomato variety ‘Moneymaker’ and subsequently confirmed in introgression lines.
Keywords: Fruit flavor, introgression lines (ILs), quantitative trait loci (QTLs), recombinant inbred lines (RILs), Solanum habrochaites, Solanum pimpinellifolium, SolCap tomato SNP array, tomato, volatiles.
Abstract
Volatile organic compounds (VOCs) are major determinants of fruit flavor, a primary objective in tomato breeding. A recombinant inbred line (RIL) population consisting of 169 lines derived from a cross between Solanum lycopersicum and a red-fruited wild tomato species Solanum pimpinellifolium accession (SP) was characterized for VOCs in three different seasons. Correlation and hierarchical cluster analyses were performed on the 52 VOCs identified, providing a tool for the putative assignation of individual compounds to metabolic pathways. Quantitative trait locus (QTL) analysis, based on a genetic linkage map comprising 297 single nucleotide polymorphisms (SNPs), revealed 102 QTLs (75% not described previously) corresponding to 39 different VOCs. The SP alleles exerted a positive effect on most of the underlying apocarotenoid volatile QTLs—regarded as desirable for liking tomato—indicating that alleles inherited from SP are a valuable resource for flavor breeding. An introgression line (IL) population developed from the same parental genotypes provided 12 ILs carrying a single SP introgression and covering 85 VOC QTLs, which were characterized at three locations. The results showed that almost half of the QTLs previously identified in the RILs maintained their effect in an IL form, reinforcing the value of these QTLs for flavor/aroma breeding in cultivated tomato.
Introduction
Tomato is cultivated worldwide, constituting one of the most important vegetable fruit crops in the world and serving as a model in the study of fruit ripening processes. In the 20th century, substantial effort was invested in tomato breeding, principally to generate high-yielding and disease-resistant varieties. However, fruit quality and consumer preferences have not generally been taken into account in breeding programs, leading to a reduction in the taste appreciated by consumers. Tomato flavor is a complex trait, composed of primary characteristics such as sugar content and acidity and, also, a large number of volatile organic compounds (VOCs) that have an important impact on flavor (Klee and Tieman, 2013).
Breeding for improved flavor by controlling the production of VOCs in the fruit has seldom been attempted for a number of reasons. First, our perception of flavor is influenced by the interaction of a relatively large number of both volatile [~16–20 or more (Buttery and Ling, 1993; Tieman et al., 2012)] and non-volatile compounds (Abegaz et al., 2004; Baldwin et al., 2008; Tieman et al., 2012), making it difficult to identify those that it would be desirable to enhance or reduce. Secondly, volatile composition is under polygenic genetic control, and the genetic basis of the mechanisms controlling their levels is largely unknown. Thirdly, volatile composition also depends on agronomic management, environment, and genetic background. Therefore, we need to identify those regions of the genome that modify volatile levels in the ripe fruit to understand this genetic control and breed for tomato flavor.
Information on the genetic control of tomato fruit volatiles is limited. Quantitative trait loci (QTLs ) for these traits have been identified in experimental populations obtained from crosses between tomato cultivars and different germplasm sources used as donor parents, such as cherry tomato (Saliba-Colombani et al., 2001; Zanor et al., 2009) or the distantly related, green-fruited, wild tomato species Solanum pennellii (Tadmor et al., 2002; Tieman et al., 2006b ) and Solanum habrochaites (Mathieu et al., 2009). In the case of these two species, wild chromosomal regions were introgressed into processing cultivars, whereas in the case of the cherry tomato, two fresh-market varieties were used as the recipient parent.
In the last decade, a number of genes involved in different metabolic pathways in the tomato fruit have been identified, shedding some light on the biosynthesis of volatile compounds. Research has led to the characterization of several genes involved in the biosynthesis of fatty acid-derived volatiles (Speirs et al., 1998; Chen et al., 2004; Matsui et al., 2000, 2007; Shen et al., 2014), apocarotenoids (Simkin et al., 2004), esters (Goulet et al., 2012, 2015), phenylpropanoids (Tieman et al., 2010; Mageroy et al., 2012), and other phenylalanine-derived volatile compounds (Tieman et al., 2006a , 2007), and the role of conjugation in the accumulation and emission of volatiles (Louveau et al., 2011; Tikunov et al., 2013). However, we have only obtained partial knowledge of the metabolic pathways and the genes involved in volatile biosynthesis and regulation.
In the present work, QTLs for a large set of volatile compounds were identified in a recombinant inbred line (RIL) population derived from a cross between the fresh market variety ‘Moneymaker’ and the closely related red-fruited wild species S. pimpinellifolium. Most of the genetic regions containing these loci were validated in introgression lines (ILs) derived from the same parental lines, confirming their value for use in breeding programs for the fresh tomato market. Furthermore, genes and associated functions located within the QTL regions are provided; these will facilitate the identification of candidates for those genes involved in volatile biosynthesis/regulation.
Materials and methods
Plant material
The RIL population was originated from an interspecific cross between S. lycopersicum cv. ‘Moneymaker’ and S. pimpinellifolium accession TO-937, producing F1 seeds. Selfing of a single F1 plant generated an F2 segregating population from which a RIL population of 169 F7:8 lines was generated by the single seed descent (SSD) method (Alba et al., 2009). Five plants of each RIL were grown during the winter–spring cycle in a plastic greenhouse under standard commercial growing conditions. Three biological replicates were analyzed for each line, each consisting of a mixture of 3–5 red ripe fruits obtained from trusses 2–4. This experimental design was repeated over 3 years.
An IL collection was previously generated from the same parents as the RIL population (Barrantes et al., 2014). Twelve ILs covering different regions harboring selected volatile QTLs and the recurrent ‘Moneymaker’ parent were cultivated under a plastic greenhouse during the summer cycle at three locations in Spain: Alginet (Coagri co-operative), Orihuela (Miguel Hernández University), and Málaga (Institute for Mediterranean and Subtropical Horticulture ‘La Mayora,’ IHSM-UMA-CSIC). The experimental design followed a randomized block with eight blocks, with one single plant replicate per IL and six replicates of ‘Moneymaker’ in each block. Eight independent biological replicates were analyzed per genotype, each consisting of between three and five fruits from a single plant.
In addition, four S. habrochaites (accession LA1777) (SH) ILs, in the genetic background of the processing tomato cultivar E6203, were also analyzed. These ILs belonged to a new set of Conserved Ortholog Set II (COSII)-anchored SH ILs, which were developed to improve the previously published SH IL/BIL population (Monforte and Tanksley, 2000) (SG, unpublished data). The four SH ILs were selected to cover two genomic regions where several volatile QTLs had been mapped in the SP RILs. More specifically, two partially overlapping SH ILs—SH IL-9C and SH IL-9F—were selected for chromosome 9 to cover a region of ~50 cM (delimited by markers SNP_340a and SNP_605a); whereas, for chromosome 11, two non-overlapping SH ILs—SH IL11-B and SH IL-11C—were selected, partially covering a region of ~35 cM (defined by flanking markers: SNP_678a and SNO_700a) (see Supplementary Fig. S1 at JXB online). The four SH ILs had been anchored to 131 PCR-based markers: 122 COSII markers, eight cleaved amplified polymorphic sequences (CAPS), and one simple sequence repeat (SSR) marker (SG, unpublished data). The genotypic data based on these markers showed that the ILs carried a single wild introgression.
Analysis of volatile compounds
For volatile compound analysis, red ripe tomato fruits were collected, carefully washed with water, and dried with a paper cloth to avoid external contamination from trichome secretions. A slice of pericarp (avoiding the septa) was excised from each fruit, taking care to remove all locular tissue, and immediately frozen in liquid nitrogen, ground in a cryogenic mill, and stored at –80 ºC until analysis.
Volatile compounds were captured by means of headspace solid phase microextraction (HS-SPME) and separated and detected by means of GC/MS. Samples were processed similarly to as described in Rambla et al. (2015). About 500 mg of frozen tomato powder were introduced into a 7 ml glass vial and incubated at 37 ºC for 10 min in a water bath. A 500 ml aliquot of an EDTA 100 mM, pH 7.5 solution and 1.1 g of CaCl2·2H2O were added, mixed gently, and sonicated for 5 min. A 1 ml aliquot of the resulting paste was transferred to a 10 ml screw cap headspace vial with a silicon/PTFE septum, and analyzed within 12 h. Volatile compound extraction and chromatography conditions were as described in Orzaez et al. (2009) except for the analyses of the RIL population in the first season, which were performed as described in Rambla et al. (2015).
For quantitation, one specific ion was selected for each compound and the corresponding peak from the extracted ion chromatogram was integrated. An admixture reference sample was prepared for each season by thoroughly mixing equal amounts of each sample. A 500 mg aliquot of the admixture was analyzed every 6–7 samples and processed as any other sample as part of the injection series. This admixture was used as a reference to normalize for temporal variation and fiber aging. Finally, the normalized results for a sample were expressed as the ratio of the abundance of each compound in that particular sample to those present in the reference admixture.
Fifty-two compounds were unequivocally identified and quantified in all the samples except in those from the RIL population in the first season, in which only 32 compounds were identified.
Genotyping and map construction
RILs and parental lines were genotyped with the 8K SolCap Illumina Infinium SNP (single nucleotide polymorphism) tomato array (Sim et al., 2012) at the INCLIVA Genotyping and Genetic Diagnosis Unit (Valencia, Spain). Monomorphic markers, as well as markers with a high number of missing values (>30%) or unknown parental genotypes were removed before further analysis. Linkage analysis and map construction were performed with JoinMap 4 mapping software (Kyazma BV, The Netherlands) (Van Ooijen, 2006) using the maximum likelihood mapping algorithm. Markers were assigned to linkage groups at a minimum LOD value of 5.0.
QTL analysis in RILs
To facilitate computational analysis, the genetic map was condensed to 297 markers. QTL analysis was performed independently for each season with log 2-transformed data using Composite Interval Mapping with WinQTLCart software (http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). The confidence interval for QTL position was established at 2–LOD units from the maximum LOD. QTLs detected in at least two seasons were retained for further analysis; QTLs only detected in a single season were discarded. Genetic and QTL maps were drawn with Mapchart 2.2 (Voorrips, 2002).
Other statistical analysis
All data obtained from volatile analyses of the samples were log 2 transformed before the statistical analyses to achieve a normal distribution.
Correlation matrices between metabolites were constructed separately from the individual sample data from seasons 2 and 3, and from the ~1000 samples representing the sum of the two seasons. Data from season 1 were not used as they contained a lower number of compounds. Pearson correlations were calculated using SPSS 16.0 software.
Two-way ANOVA was used to determine the effects of genotype, environment, and genotype×environment interaction on the levels of each of the volatiles being analyzed. For the RIL population, we used data from seasons 2 and 3. For the IL population, data from the three different locations were used. ANOVA was conducted using Statgraphics Centurion XVI.II 64 bit software.
To evaluate the statistical significance of the differences between the average of the volatiles in each of the ILs and those in the recipient parent ‘Moneymaker’, Dunnett’s test (P<0.05) was used. We considered that a QTL previously identified in the RILs was confirmed in the respective IL when the level of that particular compound was significantly different from the control parent from at least one of the three locations evaluated. Our criterion for considering a new volatile QTL was that it was observed in fruits of an IL from at least two of the three locations. Dunnett’s test was conducted with JMP 12 software.
Hierarchical cluster analysis and cluster similarity
Hierarchical cluster analysis (HCA) based on correlations (Eisen et al., 1998) was performed for the combined data set obtained from the RILs of seasons 2 and 3, and also over each data set individually. To assess the similarity between clusterings with statistical significance, the metric proposed by Fowlkes and Mallows (1983) was used, and random data sets were generated by permutation (bootstrapping). Data were analyzed with Matlab (MathWorks).
Results and discussion
In the present study, we used an S. lycopersicum×S. pimpinellifolium RIL population which had previously been employed for the localization of QTLs involved in pest resistance (Salinas et al., 2013) and other fruit quality components (Capel et al., 2015). Line TO-937 was derived from an S. pimpinellifolium accession originally collected in Lambayeque, Peru (Alba et al., 2009), which produces the species’ typical small, red fruits containing higher levels of many flavor compounds and a number of healthy metabolites such as sugars, organic acids, vitamin C, or carotenoids (Capel et al., 2015). ‘Moneymaker’ is a medium-size fruit cultivar for the fresh market with an indeterminate growth habit that is commonly used in genetic, physiological, phytopathological, and developmental studies. The two species are close relatives and, in contrast to wild green-fruited tomato species that have been used for genetic analyses of fruit quality traits, they share common characteristics for fruit ripening, metabolite profiling, etc. Additionally, they present no major cross-incompatibility that might cause substantial segregation distortions or the elimination of introgressed genomic regions in the mapping populations.
The volatile network
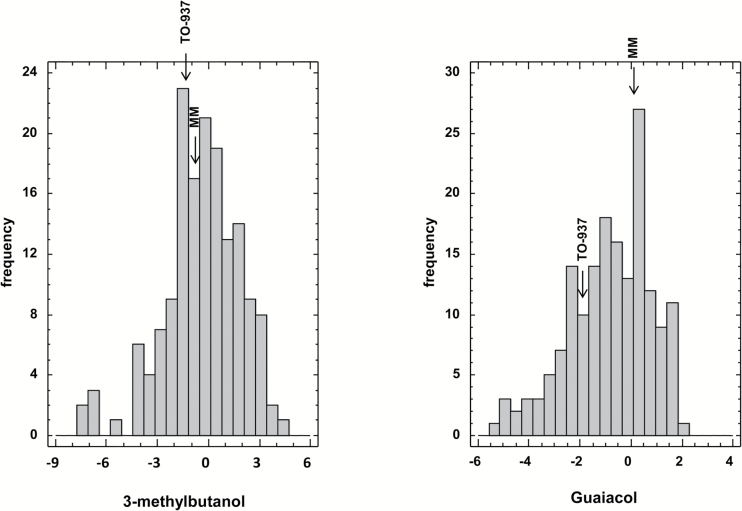
Fifty-two different volatile compounds were unequivocally identified in the ripe fruit samples obtained from the RIL population. A continuous variation was observed, with transgressive segregation in both directions for all the compounds (Supplementary Fig. S2), showing a large degree of variation for most of the volatiles, with up to 15 of them displaying a >100-fold difference in levels between the RILs with extreme values. Compounds displaying the highest degree of variation included phenylpropanoids such as guaiacol, eugenol, or methyl salicylate, other phenylalanine derivatives such as 1-nitro-2-phenylethane, benzylnitrile, or 2-phenylethanol, and branched-chain amino acid-related compounds such as 2-isobutylthiazole, 3-methylbutanol, 2-methylbutanol, 3-methylbutanal, 3-methylbutanoic acid, or 3-methylbutanenitrile. It must be noted that broad variability was detected not only for volatiles that were differentially accumulated between the parents of the population (i.e. guaiacol) but also for those that showed very similar levels in both of them (i.e. 3-methylbutanol) as illustrated in Fig. 1. On the other hand, a minority of volatiles showed a relatively low degree of variation, such as fatty acid derivatives octanal, nonanal, and decanal, with a <5-fold range between the RILs with extreme values (Supplementary Table S1).
Fig. 1.
Histogram of the distribution of 3-methylbutanol and guaiacol in the RIL population. The positions of the parentals S. lycopersicum cv. ‘Moneymaker’ (MM) and S. pimpinellifolium accession TO-937 are indicated with an arrow.
This indicates that by combining the genomes of the two parents in the RIL population, we generated new unpredictable variability in fruit volatile content.
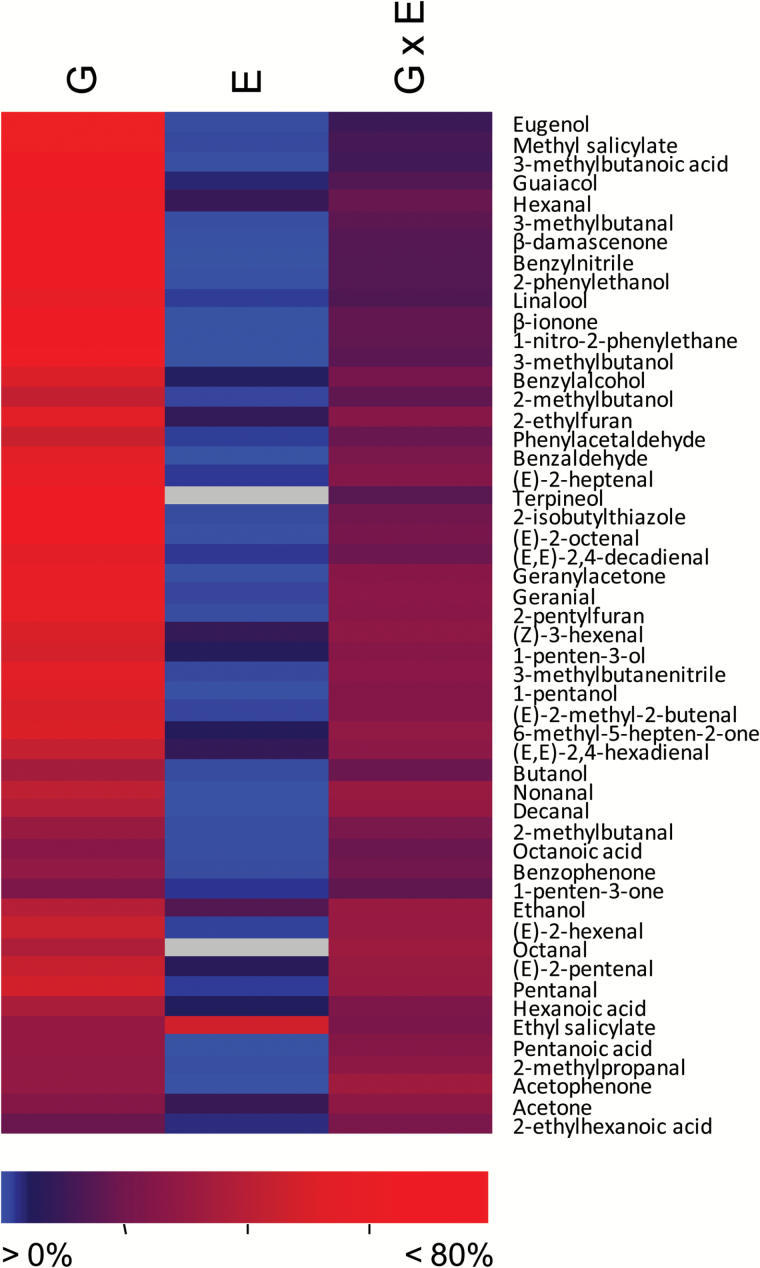
Statistical analysis of the volatile levels revealed that the genotype had a highly significant effect on all the volatiles profiled and was the principal factor explaining the variability found (Supplementary Table S2). The genotypic effect was particularly high in the case of some apocarotenoid volatiles such as β-ionone and β-damascenone, several branched-chain amino acid-related volatiles, phenylalanine derivatives, and particularly phenylpropanoids such as eugenol and methyl salicylate, for which the genotype explained >80% of the total variance. Interestingly, most of the traits with higher heritability were also among those with a larger degree of variation between the RILs with extreme values. For compounds with low heritability, genotypically explained variability scored ~20%, and included volatiles from different metabolic pathways such as fatty acid derivatives 2-ethylhexanoic acid, and 1-penten-3-one. It should be noted that interaction between genotype and environment was also highly significant for almost all the volatiles, and accounted for ~15–30% of total variability for most of the compounds. The effect of the environment was significant but remarkably low for most of the volatiles, with just a few exceptions, such as in the case of ethyl salicylate (Fig. 2; Supplementary Table S2). Therefore, these results suggest that the environment—at least under our experimental conditions—did not have a general effect on variability in volatile biosynthesis, which was modulated mostly by the particular combination of genes in each RIL.
Fig. 2.
Heat map showing the effect of genotype (G), environment (E), and the genotype×environment (G×E) interaction for each volatile in the RIL population. Data correspond to the percentage of variability explained, according to the scale below. High values (up to 80%) are indicated in red and low values in blue. The exact values are detailed in Supplementary Table S2.
Correlation between metabolites
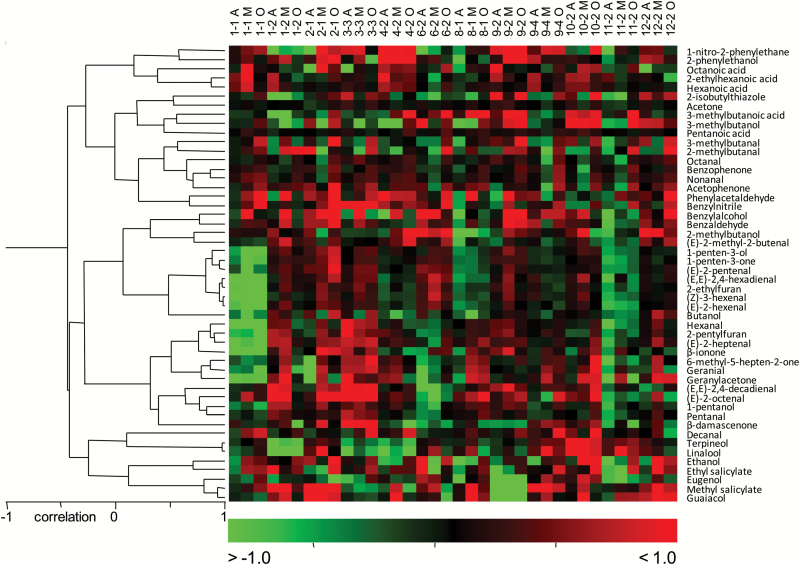
A correlation matrix was constructed from the results of the analysis of volatiles for each of seasons 2 and 3 separately, and for the two seasons together, to determine the relationship between the 52 volatile compounds identified in the ripe fruit (Supplementary Table S3). Statistical analyses were performed on data from >1000 biological samples corresponding to 169 independent RILs derived from the cross between the cultivated tomato and the red-fruited wild relative S. pimpinellifolium, each line harboring about a half of the randomly inherited genome of each parental line. Therefore, the correlation matrix obtained from this set of data can be considered as a reliable, largely unbiased description of the relationship between volatile compounds in tomato. Assuming that S. pimpinellifolium is the closest wild relative of tomato, we could consider that each RIL represents different alterations of the tomato volatile network, but these would not be as drastic as those occurring when combining more distant relatives. On the whole, the correlation results for both seasons matched, as revealed by the independent HCAs of correlations from the data set of one season or the other. Similarity between the hierarchical trees of seasons 2 and 3 was assessed following the metric proposed by Fowlkes and Mallows (1983), which consists of cutting the two hierarchical trees and then counting the number of matching entries in a number of clusters in each tree. We obtained a clustering comparison metric of 0.45 for 10 clusters (note that this metric ranges from 0 to 1; the higher the metric, the greater the degree of similarity). To assess the corresponding statistical significance, we kept the data set for season 3 and generated 1000 random data sets by permuting the values in the data set for season 2 (bootstrapping). By comparing each of the random data sets against the data set for season 3, we obtained a null distribution of the clustering comparison metric, with mean 0.13. This gave a P-value of 0 for the value of 0.45, indicating that trees derived from both seasons are statistically similar. Because this clustering comparison metric depends on the number of clusters selected upon cutting, we decided to reassess the statistical significance for five clusters. The clustering comparison metric between seasons was now 0.53, and the mean of the recalculated null distribution 0.27. This also resulted in a P-value of 0, reinforcing the similarity of the two trees (Supplementary Fig. S3). HCA was also performed using the results of the volatile levels in each of the samples for both seasons together, producing clusters for volatile compounds, thus facilitating their classification according to their respective correlations (Fig. 3). HCA with the average results of the RILs was also performed in order to facilitate their classification according to their volatile profile (Supplementary Fig. S4).
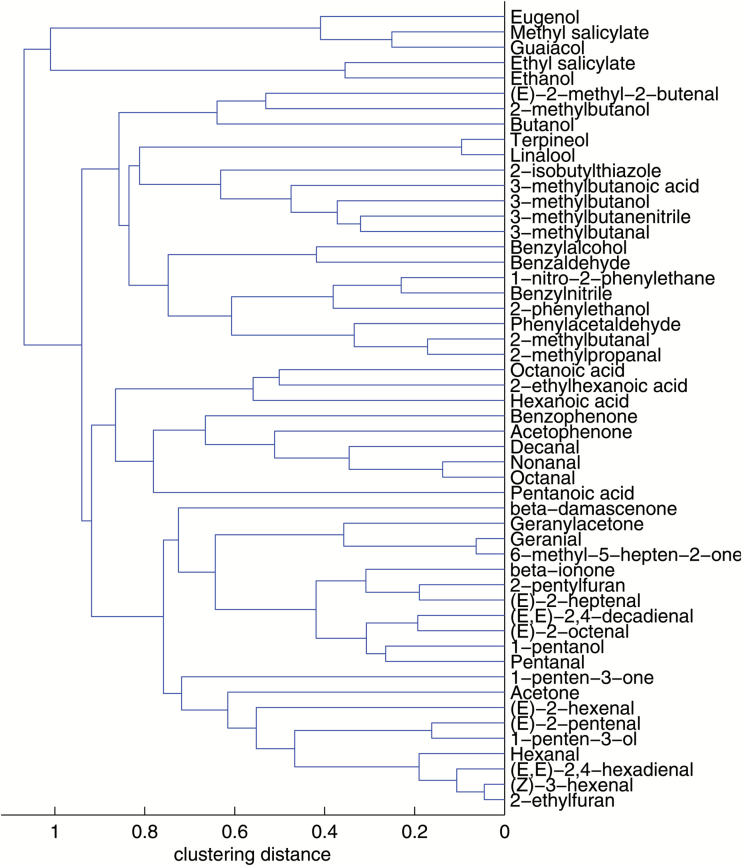
Fig. 3.
Hierarchical cluster of volatiles in the RIL population in seasons 2 and 3. (This figure is available in colour at JXB online.)
A number of factors may have caused the parallel levels of different compounds in the RIL population, such as those shown in both the correlation matrices and the HCA. Two or more genes may possibly control the metabolism of compounds that are physically very closely linked one to another, so that any recombination between them would be rare and the lines with a particular allele for one of the genes may have also inherited the same allele for the other gene(s). Nevertheless, the most appropriate explanation in most cases is that significant correlations between volatiles are due to some common process in their biosynthesis, such as sharing the same precursor, or an enzyme in the pathway, or the existence of an independent process favoring both pathways (i.e. cell wall degradation which would facilitate the availability of different substrates). Consequently, cluster analysis had previously been used to propose an assignment of volatile compounds to metabolic pathways (Tikunov et al., 2005; Mathieu et al., 2009), constituting a valuable tool to explain the biosynthesis of those compounds for which a metabolic pathway has not been clearly established. Thus, the most relevant results obtained from both hierarchical cluster and correlation analyses will be discussed hereafter in these terms.
On the basis of its chemical structure, geranial is a monoterpenic aldehyde and could be expected to cluster with the monoterpenoids linalool and terpineol. However, geranial is located in the apocarotenoid cluster, as previously reported (Tikunov et al., 2005), and has a very strong positive correlation with 6-methyl-5-hepten-2-one (r=0.94), thus suggesting a common substrate in the biosynthesis of both compounds, probably the linear carotenoid lycopene and, perhaps, also a common carotenoid cleavage dioxygenase enzyme with both 5,6 and 7,8 cleavage activities.
In the case of β-damascenone, it has been suggested that β-carotene could be its precursor on the grounds that this compound clusters with β-ionone in fruit volatile data sets obtained from ILs derived from other wild tomato species (Mathieu et al., 2009). However, in our RIL population, levels of β-damascenone did not correlate strongly with β-ionone. On the contrary, it was located in another cluster, with linear apocarotenoids 6-methyl-5-hepten-2-one, geranylacetone. and geranial, with which it even showed a slightly higher correlation (r=0.30–0.33) than with β-ionone (r=0.26). Therefore, our results reinforce the hypothesis that we might consider precursors other than β-carotene, such as neoxanthin, proposed elsewhere (Skouroumounis et al., 1993). Alternatively, the low correlation observed between β-damascenone and all the other apocarotenoids (r<0.33) suggests a different mechanism of synthesis, as reviewed in Sefton et al. (2011) proposing a chemical (non-enzymatic) acid-catalyzed hydrolysis of different plant-derived apocarotenoids as the origin of at least part of the β-damascenone occurring in a number of fruit and foodstuffs.
It has been suggested that the disruption or alteration in permeability of the thylakoid membranes during the chloroplast to chromoplast transition would facilitate the access of both lipoxygenases and carotenoid cleavage dioxygenases to their substrates, non-esterified fatty acids and carotenoids, respectively (Mathieu et al., 2009). Our data may support this hypothesis, considering that β-ionone showed the strongest correlations with fatty acid derivatives such as (E)-2-heptenal and hexanal (r=0.74 and 0.71, respectively). β-Ionone also shows a notably stronger correlation with geranylacetone than with any other apocarotenoid, which is consistent with both compounds resulting from reactions catalyzed by the same enzyme, as seems to be the case for LeCCD1 enzymes with 9,10 cleavage specificity acting on both cyclic and non-cyclic carotenoid substrates (Simkin et al., 2004).
Two of the volatile compounds located in the fatty acid derivative cluster are 2-ethylfuran and 2-pentylfuran. A metabolic pathway has not yet been elucidated for these compounds but cluster analyses conducted on independent materials of tomato (Tikunov et al., 2005; own unpublished data) and strawberry (Zorrilla-Fontanesi et al., 2012) have repeatedly suggested that both compounds could be derived from fatty acids. This hypothesis is strengthened by the fact that the C9 volatile 2-pentylfuran clustered with C8–C10 fatty acid derivatives in our RIL population, while 2-ethylfuran (a C6 compound) clustered with double-bonded C6 fatty acid derivatives. The latter volatile showed a remarkably strong correlation with (Z)-3-hexenal (r=0.95), suggesting that (Z)-3-hexenal might be postulated as a plausible precursor of 2-ethylfuran.
Identification of volatile QTLs
The 8K SolCap Illumina Infinium SNP tomato array (Sim et al., 2012) containing 7720 SNPs was used to genotype the S. lycopersicum×S. pimpinellifolium RIL population. Of the 7720 markers, 2941 (38%) were monomorphic in the RILs and 538 (7%) could not be reliably genotyped. Therefore, the remaining 4241 markers (around 55% of the SNPs included in the array) were used to generate a genetic linkage map. The markers in the resulting map were assigned to 1704 unique map locations at an average genetic distance of 1.0 cM and a maximum of 8 cM. For QTL analysis, a skeleton map was developed consisting of 297 markers evenly distributed along the tomato genome.
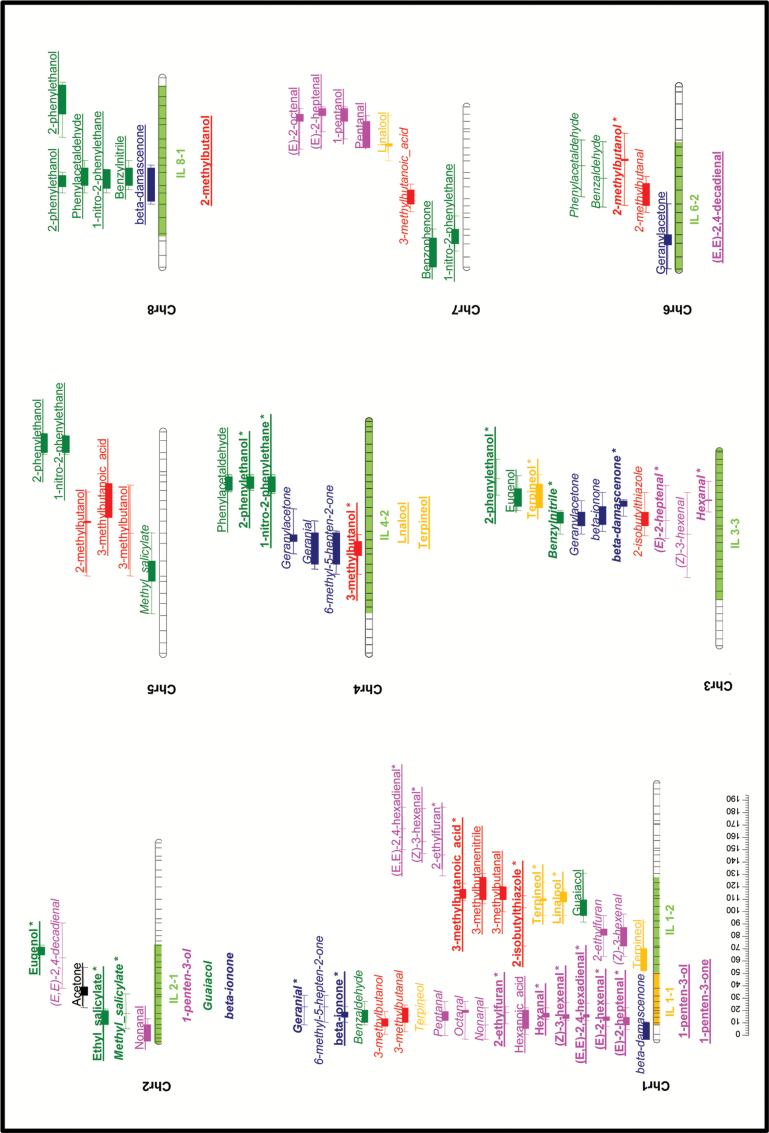
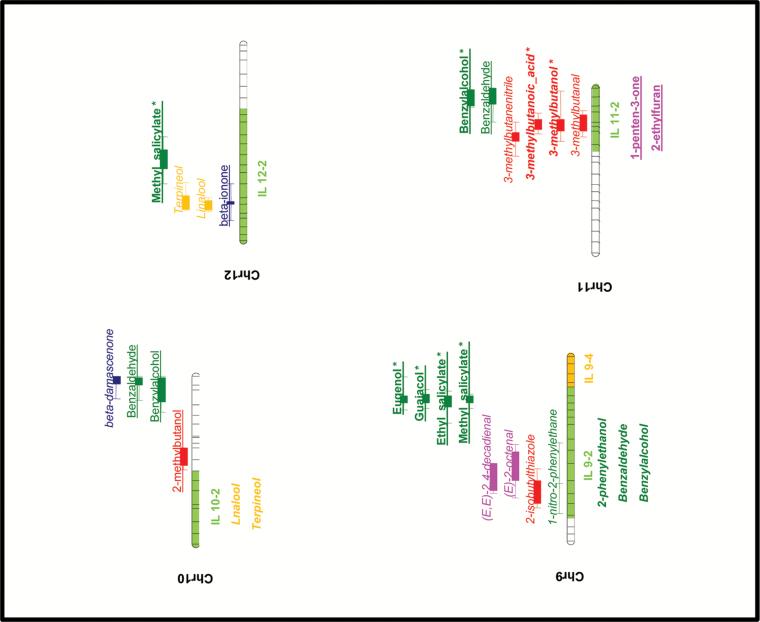
A total of 102 QTLs distributed along all the 12 tomato chromosomes were identified for 39 of the 52 volatile compounds (VOCs) identified in the samples obtained from the 169 RILs (Fig. 4; Supplementary Table S4). Chromosome 1 contained a particularly high number of VOC QTLs—a total of 31 for compounds synthesized via different metabolic pathways. The alleles inherited from S. pimpinellifolium (SP) produced an increase in the levels of volatiles in 42 loci, whereas they produced reduced levels in the remaining 60. Regarding the metabolic pathways, the production of phenolic volatile compounds tended to be increased by S. lycopersicum (SL) alleles, while levels of apocarotenoid volatiles were more likely to be increased by the SP alleles. For the other metabolic pathways, no clear tendency was observed. These results point to S. pimpinellifolium as a species with a potential in terms of breeding for cultivated tomato flavor, as apocarotenoid volatiles have been highlighted as important contributors to liking tomato (Vogel et al., 2010; Tieman et al., 2012).
Fig. 4.
Volatile QTL map obtained from the RIL and IL populations. The metabolic pathways of volatiles are distinguished by using different colors: pink, fatty acid derivatives; green, phenolic compounds; red, branched-chain amino acid-related compounds; orange, terpenoids; blue, apocarotenoids. Loci at which SP alleles induced higher levels of volatiles are marked in italics; those in which SP alleles induced lower levels are marked with plain underlined text. QTLs identified in the RIL population are shown on the right side of each chromosome. The interval corresponds to extreme values of LOD–2 and LOD+2 in the different seasons; the central solid area corresponds to the interval overlapping in the different seasons. QTLs confirmed in the ILs are highlighted in bold and marked with an asterisk. New QTLs identified in the ILs are shown to the left of each chromosome.
Fig. 4.
Continued.
Fatty acid derivatives
QTLs modifying the levels of volatiles derived from fatty acids were identified in five different chromosomes. Note that a region located at the top of chromosome 1 strongly influenced the levels of 10 different volatile compounds. The SP allele decreased levels of six C6 volatiles plus C7 (E)-2-heptenal, while the levels of the non-C6 aliphatic aldehydes pentanal, octanal, and nonanal were increased, indicating the opposite effect of this region on the production of these two groups of compounds, which actually clustered independently and, in most cases (with the exception of pentanal), showed significant negative correlations (Supplementary Table S3). The metabolic pathway of many of these compounds is known, and some key genes involved in their biosynthesis in tomato fruit have been described. Free fatty acids are first oxidized by means of lipoxygenase (LOX) enzymes, and the resulting molecule is subsequently broken down by means of hydroperoxide lyases (HPLs) to produce one non-polar volatile metabolite and one polar non-volatile compound (Rambla et al., 2014). The lipoxygenase gene TomloxC (Chen et al., 2004) has been mapped in this region of chromosome 1 and is a reasonable candidate gene for this major QTL, which in some seasons accounts for >50% of the total variability of some compounds. The HPL gene LeHPL (Matsui et al., 2000) has been mapped at the bottom of the same chromosome in a region where three C6 volatile QTLs were identified. Therefore, this gene may also have an effect on the levels of fatty acid-derived volatiles, although to a much lower extent than TomloxC, as revealed by the R 2 value (Supplementary Table S4).
Phenolic compounds
Over 30 volatile QTLs were identified for phenolic compounds spread across all the tomato chromosomes. Major QTLs accounting for over one-third of the total variability were identified for guaiacol, eugenol, methyl salicylate, and, to a lesser extent, ethyl salicylate at the bottom of chromosome 9 (Supplementary Table S4), in the same region mapped by the NSGT1 glycosyltransferase gene (Tikunov et al., 2013). Many minor QTLs for these compounds were identified at other locations, including one for guaiacol in the same region of chromosome 1 where SlUGT5, a gene involved in the glycosylation of many phenylpropanoids, is located (Louveau et al., 2011). Additionally, QTLs for phenylalanine-derived volatiles benzylnitrile, 1-nitro-2-phenylethane, phenylacetaldehyde, and 2-phenylethanol mapped in the same region of chromosome 8 as LeAADC genes, which have been reported to be involved in their biosynthesis (Tieman et al., 2006a ).
Branched-chain amino acid-related compounds
QTLs for several branched-chain volatiles with chemical structures resembling those of the amino acids leucine or isoleucine have been identified on chromosomes 1, 3, 4, 5, 6, 7, 9, 10, and 11. The biosynthetic pathway of these volatiles has not yet been established in plants, but it has been suggested that these branched-chain amino acids might be their direct precursors (Mathieu et al., 2009). Nevertheless, recent evidence indicates that these volatiles are more likely to be derived from their respective keto acids rather than from the amino acids (Kochevenko et al., 2012). Although QTLs controlling branched-chain volatiles were identified in most of the chromosomes, those for structurally leucine-related volatile compounds (3-methylbutanal, 3-methylbutanol, 3-methylbutanenitrile, 3-methylbutanoic acid, and 2-isobutylthiazole) only mapped in the same position as those for isoleucine-related volatile compounds [2-methylbutanal, 2-methylbutanol, and (E)-2-methyl-2-butenal] on chromosome 5. This indicates that most of the regulation of each group of compounds is independent of group, which is confirmed by the significant, but low, positive correlation observed between the groups.
The only sulfur compound identified in our samples was the branched-chain volatile 2-isobutylthiazole. QTLs for this compound were identified on chromosomes 1, 3, and 9. The 2-isobutylthiazole QTL detected on chromosome 1 co-localized with QTLs for 3-methylbutanol, 3-methylbutanoic acid, and 3-methylbutanenitrile, and the SP alleles reduced the levels of all of them. The other two QTLs identified for 2-isobutylthiazole did not co-localize with loci controlling other volatiles in the same pathway, which points to those regions being likely to harbor the genes responsible for the addition of the sulfur moiety.
Terpenoids
Very little is known about terpenoid biosynthesis in the tomato fruit, and only two of the compounds in this pathway were identified in the samples analyzed. We have detected three and five QTLs that regulate the levels of the terpenic alcohols linalool and terpineol, respectively. These QTLs map on chromosomes 1, 3, 7, and 12. Both compounds provide floral notes, although they are not thought to affect tomato flavor (Buttery and Ling, 1993). SP alleles are associated with reduced levels in all the QTLs identified except for those mapping on chromosome 12. Therefore, these chromosome 12 QTLs would be the target of choice for breeding enhanced levels of these potentially desirable volatiles. The chromosomal regions where a QTL for these compounds maps may contain terpene synthases or other genes involved in their biosynthesis, which could be used in the search for candidate genes for their biosynthetic pathway.
Apocarotenoids
Fourteen QTLs for a set of volatiles derived from carotenoids were identified on chromosomes 1, 3, 4, 6, 8, 10, and 12. A cluster of QTLs controlling the levels of linear apocarotenoids geranylacetone, geranial, and 6-methyl-5-hepten-2-one was identified on chromosome 4. Another cluster of QTLs was identified on chromosome 3 with an effect on both linear and cyclic C13 apocarotenoid volatiles. In both cases, the SP alleles induced increased levels. Interestingly, chromosome 1 harbors a region containing QTLs with the opposite effect on linear compounds 6-methyl-5-hepten-2-one (C8) and geranial (C10) (alleles from SP increasing their levels) and the cyclic C13 compound β-ionone (decreased by SP alleles). It should be noted that carotenoid cleavage dioxygenase 1 genes (LeCCD1A and LeCCD1B), which have been described as participating in the biosynthesis of C13 apocarotenoids (Simkin et al., 2004), also map in this region of chromosome 1.
Up to nine QTLs for the carotenoids lycopene and β-carotene had previously been identified in the same RIL population (Capel et al., 2015). However, apocarotenoid volatile QTLs in general tended not to co-localize with carotenoid QTLs, with the exception of the QTLs for 6-methyl-5-hepten-2-one, geranial, and geranylacetone on chromosome 4 which co-localized with QTLs for both lycopene (the plausible precursor of them all) and β-carotene. This indicates that the main determinants of the production of apocarotenoid volatiles are factors other than substrate accumulation. Therefore, all the other QTLs identified could be due to as yet unknown genes located in these regions implied either directly in the final steps of volatile biosynthesis, or in the accumulation of possible precursors other than the already described lycopene and β-carotene, or in other processes facilitating the interaction between substrates and biosynthetic enzymes.
Introgression and validation of the volatile QTLs in a fresh market tomato genetic background
Twelve SP ILs previously generated from the same cross (Barrantes et al., 2014) carrying single SP introgressions covering 85 of the QTLs identified in the SP RIL population (Fig. 4) were grown at three different locations, and their fruit volatile composition was determined to validate the effect of the QTLs selected.
In contrast to the RIL population, most of the variability observed for the volatile levels in the fruit of these ILs was due to the environment and the genotype, and significant genotype×environment interaction was only observed for a few compounds (Supplementary Table S5). This could be attributed to the high genetic background isogenicity of the ILs, with an average genetic difference between ILs slightly over 10%, whereas between RILs it was ~50%. This reduction of genotypic variability between ILs may also decrease the power of genotype×environment interaction detection.
Thirty-four of the 85 QTLs were confirmed in the 12 ILs, maintaining the direction of the effect on the volatile levels after a single introgression of the S. pimpinellifolium in the fresh market tomato variety ‘Moneymaker’ (Fig. 4). This can be considered indicative of the potential of the QTLs identified for their use in breeding tomato flavor. Nevertheless, the introgression of a genetic region has been described as having a different effect on the volatile profile depending on the genetic background where it is introgressed. Therefore, it seems plausible that the introgression of the same regions of the chromosomes in a different tomato variety might alter different sets of volatiles, as previously observed when introgressing selected regions of cherry tomato in different elite beef tomato varieties (Zanor et al., 2009).
Additionally, 16 new QTLs for volatile compounds corresponding to different metabolic pathways were identified on chromosomes 1, 2, 4, 6, 8, 9, 10, and 11, represented by eight SP ILs (Fig. 4). Some of these new QTLs corresponded to volatiles synthesized in the same metabolic pathway as a VOC QTL previously detected in the RIL population, as in the case of fatty acid derivatives 1-penten-3-ol and 1-penten-3-one in chromosome IL 1-1, which also contained QTLs for another six metabolically related volatiles. This suggests that a gene located in that chromosomal region has a general effect on the whole metabolic pathway, enhancing the accumulation of many different volatiles in the pathway. However, some new QTLs were detected in the ILs unrelated to those mapped in the RIL population. This is the case for the terpenoids linalool and terpineol (in IL 4-2 and IL 10-2), apocarotenoid β-ionone (IL 2-1), branched-chain volatile 2-methylbutanol (IL 8-1), and fatty acid derivatives (E,E)-2,4-decadienal (IL 6-2) and 1-penten-3-one and 2-ethylfuran (IL 11-2). Additionally, we found significant IL effects for some individual compounds at a single location, but other volatiles belonging to the same metabolic pathway changed significantly in the same ILs at other locations. Although these loci were not included in the QTL map presented here, we consider that they correspond to minor genes with an effect on early stages of the metabolic pathway, therefore affecting the levels of a number of compounds downstream. Complete data on volatile levels in the ILs are shown in Fig. 5 and Supplementary Table S6.
Fig. 5.
Heat map showing volatile levels in the ILs. Values correspond to the log2 of the ratio of the levels of each volatile in the ILs in relation to those in S. lycopersicum cv. ‘Moneymaker’, cultivated at each location. Higher levels are shown in red; lower levels in green; identical levels in black. The letter after the IL descriptor indicates where it was cultivated: ‘A’, Alginet; ‘M’, Malaga; ‘O’, Orihuela.
Similarities with QTLs identified in selected ILs developed from S. habrochaites
Two partially overlapping S. habrochaites LA1777 (SH) ILs were selected from the new set of COSII-anchored SH ILs for chromosome 9: ILSH-9C and ILSH-9F (Supplementary Fig. S1). Analyses of their fruit volatile profiles revealed that both produced dramatically reduced levels of guaiacol, methyl salicylate, and eugenol. This is in agreement with the QTLs previously observed in both RIL and IL populations derived from S. pimpinellifolium, and also in near isogenic lines derived from a cherry tomato donor (Zanor et al., 2009). Additionally, ILSH-9F showed significantly reduced levels of (E,E)-2,4-decadienal and 1-nitro-2-phenylethane, for which QTLs were detected in the SP RILs but not in the SP ILs. Finally, QTLs for 2-phenylethanol and benzaldehyde, which were detected in IL 9-2 from ‘Moneymaker’ but had not been identified in the RILs, were also confirmed in ILSH-9F. Surprisingly, no QTLs for volatile compounds had been identified on chromosome 9 when the previously published SH IL/BIL population was analyzed (Mathieu et al., 2009). This might be partially due to the different SH ILs used in the previous study to cover this chromosome 9 region.
Another two non-overlapping ILs—ILSH-11B and ILSH-11C—were selected for chromosome 11 from the new set of COSII-anchored SH ILs (Supplementary Fig. S1). All of the four QTLs for branched-chain volatiles 3-methylbutanol, 3-methylbutanal, 3-methylbutanoic acid, and 3-methylbutanenitrile previously identified in the S. pimpinellifolium RILs were also detected in ILSH-11B. Of these QTLs, only those for 3-methylbutanol and 3-methylbutanoic acid were detected in the SP ILs. Interestingly, QTLs for these compounds were not detected in the previously published SH IL population (Mathieu et al., 2009), but most of them had been described in an IL population developed from S. pennellii (Tieman et al., 2006). ILSH-11C showed significantly higher levels of guaiacol, as described in the earlier SH IL population (Mathieu et al., 2009). However, no QTLs were detected for benzaldehyde and benzyl alcohol, although previous ILs from both SH (Mathieu et al., 2009) and our SP population facilitated the identification of loci for these compounds.
Volatile QTLs identified in tomato show a low degree of overlapping
QTLs for volatile compounds in tomato fruit have been previously identified in a RIL population developed from an intraspecific cross between S. lycopersicum var. cerasiforme inbred line ‘Cervil’ and the S. lycopersicum inbred line ‘Levovil’ (Saliba-Colombani et al., 2001) or in selected ILs generated from the same cross in two inbred line genetic backgrounds (Zanor et al., 2009), in a particular IL (Tadmor et al., 2002), or an IL population derived from S. pennellii in the M82 processing tomato background (Tieman et al., 2006b ), and also in an IL population generated from S. habrochaites in the S. lycopersicum cv. E6203 background (Mathieu et al., 2009). It should be noted that there is a very low degree of overlapping of the QTLs described. Only 26 of the 102 VOC QTLs detected in the SP RIL population had been reported in any of the previous studies, whereas the other 76 QTLs are described here for the first time (Table 1). Moreover, only two of those 26, the QTLs for guaiacol on chromosome 1 (probably corresponding to the SlUGT1 gene) and for phenylacetaldehyde on chromosome 8 (probably due to LeAADC genes), had been reported in more than one previous experiment. This lack of consistent results could be explained in part by the differences in the environmental conditions in each of the experiments. Although environmental conditions explained on average only 3.4% of total variability in this experiment, environmental effects accounted for up to 47% in individual compounds, and this variability would probably be higher when comparing fruits grown in different geographical locations. Additionally, different analytical methods were employed to determine volatile compounds. The precise method used for volatile determination, including both sample preparation and the extraction technique, has a major effect on the compounds that can be detected, as recently stated (Rambla et al., 2015). Therefore, a perfect match in the QTLs identified by different research groups should not be expected even if obtained from the same biological material. Nevertheless, even when some discrepancy exists between the results available, co-localization of different QTLs from metabolically related compounds can be used as a clue to their probable association with the same gene. For example, we identified QTLs for four phenylalanine-derived volatiles: 1-nitro-2-phenylethane, phenylacetaldehyde, 2-phenylethanol, and benzylnitrile, in the same region of chromosome 8 where Saliba-Colombani et al. (2001) only found a QTL for phenylacetaldehyde, and Tadmor et al. (2002) and Tieman et al. (2006b ) described QTLs for phenylacetaldehyde and 2-phenylethanol. In our opinion, the most plausible explanation for these results is that there is a single gene in that region influencing the levels of at least all these four compounds, and the ability to identify a lower or a higher number of volatile QTLs depends largely on the particular analytical method employed by each research group and the environmental conditions in each particular experiment.
Table 1.
Volatile QTLs with possible overlap with previously identified QTLs
|
Saliba-Colombani et al. (2001) Cherry tomato |
Zanor et al. (2009) Cherry tomato |
Tadmor et al. (2002) S. pennellii |
Tieman et al. (2006b) S. pennellii |
Mathieu et al, (2009) S. habrochaites |
|
|---|---|---|---|---|---|
| Chr 1 | Guaiacol | Guaiacol | Guaiacol | ||
| (Z)-3-Hexenal | |||||
| 3-Methylbutanol | |||||
| Pentanal | |||||
| Hexanal | |||||
| Terpineol | |||||
| Chr 2 | Eugenol | ||||
| Chr 4 | 3-Methylbutanol | ||||
| Phenylacetaldehyde | Phenylacetaldehyde | ||||
| 2-Phenylethanol | |||||
| Chr 6 | 2-Methylbutanol | ||||
| Benzaldehyde | |||||
| Chr 8 | Phenylacetaldehyde | Phenylacetaldehyde | Phenylacetaldehyde | ||
| 2-Phenylethanol | 2-Phenylethanol | ||||
| Chr 9 | Guaiacol | Guaiacol | |||
| Eugenol | Eugenol | ||||
| Methyl salicylate | |||||
| Chr 10 | 2-Methylbutanol | ||||
| Benzaldehyde | |||||
| Benzyl alcohol | |||||
| Chr 11 | 3-Methylbutanal | ||||
| 3-Methylbutanol | |||||
| 3-Methylbutanenitrile | |||||
| Benzaldehyde | |||||
| Benzyl alcohol |
Nevertheless, even after considering that part of the lack of redundancy is due to environmental or methodological factors, a substantial part of it can still be claimed to respond to specific genetic variation, as no QTLs for volatiles in the same metabolic pathway had been reported earlier for most of the genetic regions where volatile QTLs have been described here. This is in consonance with the results derived from the comparison between the QTLs reported in ILs derived from S. pennellii (Tieman et al., 2006b ) and S. habrochaites (Mathieu et al., 2009), in which only a minor part of them (10 out of 60) were overlapping, despite the same analytical method having been employed in both studies. Taken all together, this suggests that there is ample genetic variability in wild species of the tomato clade readily available for breeding improved/new fruit flavor and aroma by modulating the levels of volatile compounds.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Schematic overview of tomato chromosomes 9 and 11 showing the map position of the four Solanum habrochaites LA1777 (SH) ILs analyzed.
Fig. S2. Histogram of the distribution of each volatile compound in the RIL population in seasons 1, 2, and 3.
Fig. S3. Hierarchical clusters of volatiles in seasons 2 and 3 independently and an evaluation of the degree of similarity.
Fig. S4. Hierarchical cluster of the RILs according to their volatile profile in seasons 2 and 3 together.
Table S1. Levels of volatile compounds in the RILs with extreme values and in their parental lines..
Table S2. Effect of genotype, environment, and genotype×environment interactions on levels of the volatile compounds in the RILs and statistical significance, as obtained using ANOVA.
Table S3. Correlation between volatiles and significance.
Table S4. Volatile QTL map obtained from the RIL population.
Table S5. Effect of genotype, environment, and genotype×environment interactions on the levels of the volatile compounds in the ILs and statistical significance, as obtained using ANOVA.
Table S6. Levels of volatile compounds in the ILs.
Supplementary Material
Acknowledgements
We thank Erika Moro for valuable help in volatile analysis of the ILs. WB was supported by a fellowship granted by the Universidad de Costa Rica and CSIC-Spain by way of a collaboration agreement between CSIC/UCR. Volatile profiling was performed in the Metabolomic facilities of the IBMCP, CSIC (Spain). This work was supported in part by the Spanish MINECO Project AGL2015-65246-R co-financed by EU FEDER, MINECO Project AGL2011-26957, and the Bilateral agreements of Scientific and Technological Cooperation between the Spanish National Research Council (CSIC) and the Italian National Research Council (CNR). Funding for this project was provided through TRADITOM. TRADITOM has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 634561. Networking activities were supported by COST action Fruit Quality FA 1106.
References
- Alba JM, Montserrat M, Fernández-Muñoz R. 2009. Resistance to the two-spotted spider mite (Tetranychus urticae) by acylsucroses of wild tomato (Solanum pimpinellifolium) trichomes studied in a recombinant inbred line population. Experimental and Applied Acarology 47, 35–47. [DOI] [PubMed] [Google Scholar]
- Abegaz EG, Tandon KS, Scott JW, Baldwin EA, Shewfelt RL. 2004. Partitioning taste from aromatic flavor notes of fresh tomato (Lycopersicon esculentum, Mill) to develop predictive models as a function of volatile and nonvolatile components. Postharvest Biology and Technology 34, 227–235. [Google Scholar]
- Baldwin EA, Goodner K, Plotto A. 2008. Interaction of volatiles, sugars, and acids on perception of tomato aroma and flavor descriptors. Journal of Food Science 73, S294–S307. [DOI] [PubMed] [Google Scholar]
- Barrantes W, Fernandez-del-Carmen A, Lopez-Casado G, Angeles Gonzalez-Sanchez M, Fernandez-Munoz R, Granell A, Monforte AJ. 2014. Highly efficient genomics-assisted development of a library of introgression lines of Solanum pimpinellifolium. Molecular Breeding 34, 1817–1831. [Google Scholar]
- Buttery RG, Ling LC. 1993. Volatile components of tomato fruit and plant-parts—relationship and biogenesis. ACS Symposium Series 525, 23–34. [Google Scholar]
- Capel C, Fernández del Carmen A, Alba JM, et al. 2015. Wide-genome QTL—mapping of fruit quality traits in a tomato RIL population derived from the wild-relative species Solanum pimpinellifolium L. Theoretical and Applied Genetics 128, 2019–2035. [DOI] [PubMed] [Google Scholar]
- Chen G, Hackett R, Walker D, Taylor A, Lin Z, Grierson D. 2004. Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiology 136, 2641–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. 1998. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences, USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowlkes EB, Mallows CL. 1983. A method for comparing 2 hierarchical clusterings. Journal of the American Statistical Association 78, 553–569. [Google Scholar]
- Goulet C, Kamiyoshihara Y, Lam NB, Richard T, Taylor MG, Tieman DM, Klee HJ. 2015. Divergence in the enzymatic activities of a tomato and Solanum pennellii alcohol acyltransferase impacts fruit volatile ester composition. Molecular Plant 8, 153–162. [DOI] [PubMed] [Google Scholar]
- Goulet C, Mageroy MH, Lam NB, Floystad A, Tieman DM, Klee HJ. 2012. Role of an esterase in flavor volatile variation within the tomato clade. Proceedings of the National Academy of Sciences, USA 109, 19009–19014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee HJ, Tieman DM. 2013. Genetic challenges of flavor improvement in tomato. Trends in Genetics 29, 257–262. [DOI] [PubMed] [Google Scholar]
- Kochevenko A, Araújo WL, Maloney GS, Tieman DM, Do PT, Taylor MG, Klee HJ, Fernie AR. 2012. Catabolism of branched chain amino acids supports respiration but not volatile synthesis in tomato fruits. Molecular Plant 5, 366–375. [DOI] [PubMed] [Google Scholar]
- Louveau T, Leitao C, Green S, Hamiaux C, van der Rest B, Dechy-Cabaret O, Atkinson RG, Chervin C. 2011. Predicting the substrate specificity of a glycosyltransferase implicated in the production of phenolic volatiles in tomato fruit. FEBS Journal 278, 390–400. [DOI] [PubMed] [Google Scholar]
- Mageroy MH, Tieman DM, Floystad A, Taylor MG, Klee HJ. 2012. A Solanum lycopersicum catechol-O-methyltransferase involved in synthesis of the flavor molecule guaiacol. The Plant Journal 69, 1043–1051. [DOI] [PubMed] [Google Scholar]
- Mathieu S, Cin VD, Fei Z, Li H, Bliss P, Taylor MG, Klee HJ, Tieman DM. 2009. Flavour compounds in tomato fruits: identification of loci and potential pathways affecting volatile composition. Journal of Experimental Botany 60, 325–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K, Ishii M, Sasaki M, Rabinowitch HD, Ben-Oliel G. 2007. Identification of an allele attributable to formation of cucumber-like flavor in wild tomato species (Solanum pennellii) that was inactivated during domestication. Journal of Agricultural and Food Chemistry 55, 4080–4086. [DOI] [PubMed] [Google Scholar]
- Matsui K, Miyahara C, Wilkinson J, Hiatt B, Knauf V, Kajiwara T. 2000. Fatty acid hydroperoxide lyase in tomato fruits: cloning and properties of a recombinant enzyme expressed in Escherichia coli . Bioscience, Biotechnology, and Biochemistry 64, 1189–1196. [DOI] [PubMed] [Google Scholar]
- Monforte AJ, Tanksley SD. 2000. Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome 43, 803–813. [PubMed] [Google Scholar]
- Orzaez D, Medina A, Torre S, Fernández-Moreno JP, Rambla JL, Fernández-Del-Carmen A, Butelli E, Martin C, Granell A. 2009. A visual reporter system for virus-induced gene silencing in tomato fruit based on anthocyanin accumulation. Plant Physiology 150, 1122–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambla JL, Alfaro C, Medina A, Zarzo M, Primo J, Granell A. 2015. Tomato fruit volatile profiles are highly dependent on sample processing and capturing methods. Metabolomics 11, 1708–1720. [Google Scholar]
- Rambla JL, Tikunov YM, Monforte AJ, Bovy AG, Granell A. 2014. The expanded tomato fruit volatile landscape. Journal of Experimental Botany 65, 4613–4623. [DOI] [PubMed] [Google Scholar]
- Saliba-Colombani V, Causse M, Langlois D, Philouze J, Buret M. 2001. Genetic analysis of organoleptic quality in fresh market tomato. 1. Mapping QTLs for physical and chemical traits. Theoretical and Applied Genetics 102, 259–272. [Google Scholar]
- Salinas M, Capel C, Alba JM, Mora B, Cuartero J, Fernández-Muñoz R, Lozano R, Capel J. 2013. Genetic mapping of two QTL from the wild tomato Solanum pimpinellifolium L. controlling resistance against two-spotted spider mite (Tetranychus urticae Koch). Theoretical and Applied Genetics 126, 83–92. [DOI] [PubMed] [Google Scholar]
- Sefton MA, Skouroumounis GK, Elsey GM, Taylor DK. 2011. Occurrence, sensory impact, formation, and fate of damascenone in grapes, wines, and other foods and beverages. Journal of Agricultural and Food Chemistry 59, 9717–9746. [DOI] [PubMed] [Google Scholar]
- Shen J, Tieman D, Jones JB, Taylor MG, Schmelz E, Huffaker A, Bies D, Chen K, Klee HJ. 2014. A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato. Journal of Experimental Botany 65, 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim SC, Durstewitz G, Plieske J, et al. 2012. Development of a large SNP genotyping array and generation of high-density genetic maps in tomato. PLoS One 7, e40563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simkin AJ, Schwartz SH, Auldridge M, Taylor MG, Klee HJ. 2004. The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles beta-ionone, pseudoionone, and geranylacetone. The Plant Journal 40, 882–892. [DOI] [PubMed] [Google Scholar]
- Skouroumounis GK, Massywestropp RA, Sefton MA, Williams PJ. 1993. beta-Damascenone formation in juices and wines. In: Schreier P, Winterhalter P, eds. Progress in flavour precursor studies: analysis, generation, biotechnology. Proceedings of the International Conference, Würzburg, Germany, September 30–October 2, 1992, 275–278. [Google Scholar]
- Speirs J, Lee E, Holt K, Yong-Duk K, Steele Scott N, Loveys B, Schuch W. 1998. Genetic manipulation of alcohol dehydrogenase levels in ripening tomato fruit affects the balance of some flavor aldehydes and alcohols. Plant Physiology 117, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadmor Y, Fridman E, Gur A, Larkov O, Lastochkin E, Ravid U, Zamir D, Lewinsohn E. 2002. Identification of malodorous, a wild species allele affecting tomato aroma that was selected against during domestication. Journal of Agricultural and Food Chemistry 50, 2005–2009. [DOI] [PubMed] [Google Scholar]
- Tieman D, Bliss P, McIntyre LM, et al. 2012. The chemical interactions underlying tomato flavor preferences. Current Biology 22, 1035–1039. [DOI] [PubMed] [Google Scholar]
- Tieman D, Taylor M, Schauer N, Fernie AR, Hanson AD, Klee HJ. 2006. a Tomato aromatic amino acid decarboxylases participate in synthesis of the flavor volatiles 2-phenylethanol and 2-phenylacetaldehyde. Proceedings of the National Academy of Sciences, USA 103, 8287–8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieman D, Zeigler M, Schmelz E, Taylor MG, Rushing S, Jones JB, Klee HJ. 2010. Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. The Plant Journal 62, 113–123. [DOI] [PubMed] [Google Scholar]
- Tieman DM, Loucas HM, Kim JY, Clark DG, Klee HJ. 2007. Tomato phenylacetaldehyde reductases catalyze the last step in the synthesis of the aroma volatile 2-phenylethanol. Phytochemistry 68, 2660–2669. [DOI] [PubMed] [Google Scholar]
- Tieman DM, Zeigler M, Schmelz EA, Taylor MG, Bliss P, Kirst M, Klee HJ. 2006. b Identification of loci affecting flavour volatile emissions in tomato fruits. Journal of Experimental Botany 57, 887–896. [DOI] [PubMed] [Google Scholar]
- Tikunov Y, Lommen A, de Vos CH, Verhoeven HA, Bino RJ, Hall RD, Bovy AG. 2005. A novel approach for nontargeted data analysis for metabolomics. Large-scale profiling of tomato fruit volatiles. Plant Physiology 139, 1125–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikunov YM, Molthoff J, de Vos RC, et al. 2013. Non-smoky glycosyltransferase1 prevents the release of smoky aroma from tomato fruit. The Plant Cell 25, 3067–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW. 2006. JoinMap® 4. Software for the calculation of genetic linkage maps in experimental populations. Wageningen, The Netherlands: Kyazma BV. [Google Scholar]
- Vogel JT, Tieman DM, Sims CA, Odabasi AZ, Clark DG, Klee HJ. 2010. Carotenoid content impacts flavor acceptability in tomato (Solanum lycopersicum). Journal of the Science of Food and Agriculture 90, 2233–2240. [DOI] [PubMed] [Google Scholar]
- Voorrips RE. 2002. MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of Heredity 93, 77–78. [DOI] [PubMed] [Google Scholar]
- Zanor MI, Rambla JL, Chaïb J, Steppa A, Medina A, Granell A, Fernie AR, Causse M. 2009. Metabolic characterization of loci affecting sensory attributes in tomato allows an assessment of the influence of the levels of primary metabolites and volatile organic contents. Journal of Experimental Botany 60, 2139–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorrilla-Fontanesi Y, Rambla JL, Cabeza A, Medina JJ, Sánchez-Sevilla JF, Valpuesta V, Botella MA, Granell A, Amaya I. 2012. Genetic analysis of strawberry fruit aroma and identification of O-methyltransferase FaOMT as the locus controlling natural variation in mesifurane content. Plant Physiology 159, 851–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.