Highlight
A comprehensive metabolic study of Arabidopsis seeds revealed mQTL hotspots, candidate genes, epistatic interactions, and a potential master regulatory locus of metabolism on the short arm of chromosome IV.
Keywords: Arabidopsis thaliana, gas chromatography–mass spectrometry, metabolic quantitative trait locus, primary metabolism, recombinant inbred line, seed biology.
Abstract
To gain insight into genetic factors controlling seed metabolic composition and its relationship to major seed properties, an Arabidopsis recombinant inbred line (RIL) population, derived from accessions Col-0 and C24, was studied using an MS-based metabolic profiling approach. Relative intensities of 311 polar primary metabolites were used to identify associated genomic loci and to elucidate their interactions by quantitative trait locus (QTL) mapping. A total of 786 metabolic QTLs (mQTLs) were unequally distributed across the genome, forming several hotspots. For the branched-chain amino acid leucine, mQTLs and candidate genes were elucidated in detail. Correlation studies displayed links between metabolite levels, seed protein content, and seed weight. Principal component analysis revealed a clustering of samples, with PC1 mapping to a region on the short arm of chromosome IV. The overlap of this region with mQTL hotspots indicates the presence of a potential master regulatory locus of seed metabolism. As a result of database queries, a series of candidate regulatory genes, including bZIP10, were identified within this region. Depending on the search conditions, metabolic pathway-derived candidate genes for 40–61% of tested mQTLs could be determined, providing an extensive basis for further identification and characterization of hitherto unknown genes causal for natural variation of Arabidopsis seed metabolism.
Introduction
Natural variation is often the result of the contribution of multiple genes, detectable as quantitative trait loci (QTLs), and their interactions (Alonso-Blanco and Koornneef, 2000; Mackay, 2001; Remington and Purugganan, 2003; Koornneef et al., 2004; Shindo et al., 2007). However, it is often poorly understood to what extent the observed variation is attributed to genetic factors (Keurentjes et al., 2006). Linkage and association mapping represent powerful, complementary approaches to connect genetic markers and phenotypic variation (Mackay, 2009). With recent advances in the field of ‘-omics’ sciences, mapping of molecular phenotypes such as transcript (West et al., 2007; Keurentjes et al., 2007) or metabolite levels (Lisec et al., 2008; Toubiana et al., 2012; Alseekh et al., 2015) is increasingly utilized, opening up new possibilities for understanding complex molecular processes.
Recent developments of metabolomic platforms allow parallel and rapid quantification of hundreds to thousands of metabolites, offering new opportunities to study plant metabolomes (Alonso et al., 2015; Soltis and Kliebenstein, 2015). One of the most frequently used metabolic approaches in plant biology is GC-MS (Fiehn, 2008). This technique allows quantification of a broad spectrum of primary metabolites including amino acids, organic acids, and sugars that can be utilized to map metabolic QTLs (mQTLs), as shown in maize (Riedelsheimer et al., 2012), tomato (Schauer et al., 2006, 2008), potato (Carreno-Quintero et al., 2012), and rice (Matsuda et al., 2012; Ying et al., 2012; Li et al., 2016). Based on mQTL mapping, genes encoding enzymes involved in specific biochemical pathways were identified (Saito and Matsuda, 2010; Chan et al., 2011; Angelovici et al., 2013; Li et al., 2014; Strauch et al., 2015; Francisco et al., 2016) and systems for the regulation of metabolic networks elucidated (Fernie and Schauer, 2009; Wen et al., 2015, 2016).
mQTL studies in the model organism Arabidopsis thaliana (Keurentjes et al., 2006, 2008; Lisec et al., 2008, 2009; Rowe et al., 2008) provided the basis to identify genes and polymorphisms causal for natural trait variation (Brotman et al., 2011; Riewe et al., 2016), to characterize hitherto uncharacterized metabolic pathways and networks (Wentzell et al., 2007; Chan et al., 2010; Joseph et al., 2014), and to unravel key regulators of metabolism (Li and Kliebenstein, 2014; Wu et al., 2016). Arabidopsis seed mQTLs were studied for glucosinolates (Kliebenstein et al., 2001), flavonoids (Routaboul et al., 2012), oil content and fatty acids (Hobbs et al., 2004; Sanyal and Randal Linder, 2012), carbohydrates (Calenge et al., 2006), and branched-chain amino acids (BCAAs) (Angelovici et al., 2013). A recent study focused on natural variation for oil, protein, carbon, and nitrogen content by near-infrared spectroscopy (Jasinski et al., 2016). Comprehensive investigations of the genetic basis of the Arabidopsis seed primary metabolome, addressing a broader range of metabolite classes, have rarely been performed (Joosen et al., 2013).
Recombinant inbred line (RIL) populations represent an important genetic resource for the investigation of natural variation in Arabidopsis (Mackay, 2001). A large set of reciprocal RILs was created by crossing the Arabidopsis accessions Col-0 and C24 (Törjék et al., 2006). This RIL population was successfully utilized in several mapping studies, resulting in the identification of genomic regions involved in biomass heterosis at early developmental stages (Meyer et al., 2010), and metabolic and biomass QTLs (Lisec et al., 2008, 2009).
We used this population to address the following questions. (i) Are seed metabolite levels, seed weight, and seed protein content correlated? (ii) Which genomic regions are associated with particular metabolites in mature Arabidopsis seeds and do these loci interact? (iii) Which genomic regions contain genes encoding enzymes involved in pathway reactions related to these metabolites? (iv) Are there common genetic factors affecting multiple metabolites and thus acting as potential master regulators affecting the metabolic composition of mature seeds and what are the most promising candidate regulatory genes?
Materials and methods
Plant material and growing conditions
The mapping population consisted of 393 RILs originating from the reciprocal crosses Col-0×C24 (n=202) and C24×Col-0 (n=191) described by Törjék et al. (2006). F2 plants were propagated by self-pollination using single-seed descent to the F10 generation. Plants were cultivated in two consecutive experiments in a phytotron under controlled conditions [16 h sodium lamp light (250 µmol m−2 s−1); 20 °C; 70% relative humidity/8 h dark; 16 °C; 60% relative humidity] to ensure a constant environment during seed formation. A randomized block design and randomized positions within each block were used. Each line was replicated four times (individual plants). A total of 15–30 seeds per genotype were sown into 6 cm pots filled with Substrate 1 (Klasmann-Deilmann GmbH, Geeste, Germany). Stratification was performed for 3 d at 4 °C in the dark. After 1 week, well-developed and healthy plants were transferred into single pots. The orientation of trays was changed daily; every second day positions of trays in the room were shuffled to minimize position effects. Plants were treated with Novo Nem® F (ÖRE Bio-Protect Biologischer Pflanzenschutz GmbH, Schwentinental, Germany) every second week. Mature seeds were collected using ARACONs (BETATECH, Gent, Belgium), purified, and stored in sealed screw-cap glass vials at 5 °C and 55% relative humidity.
Fractionated metabolite and protein extraction
Twenty seeds of each of three individual plants per RIL were pooled. Polar metabolites and seed proteins were extracted via liquid–liquid extraction, modifying existing protocols (Lisec et al., 2006; Erban et al., 2007; Giavalisco et al., 2011) using a liquid handling system (Biomek® FXP, Beckman Coulter GmbH, Krefeld, Germany). Seed material was deep frozen at –80 °C and homogenized twice for 1 min at 30 Hz using a mixer mill (Retsch GmbH, Haan, Germany). Metabolites were extracted in 0.5 ml of chilled extraction buffer 1 (MTBE:MeOH:H2O; 3:1:0.5 plus internal standard 1:1000) by shaking and ultrasonication for 15 min at 4 °C. After adding 325 μl of extraction buffer 2 (MeOH:H2O; 1:3), samples were centrifuged and two 80 µl aliquots of the upper organic phase were stored for potential further investigation. Subsequently, 150 μl of chloroform were added, followed by shaking and centrifugation for 10 min, leading to a phase inversion. Three 120 μl aliquots of the upper polar phase were transferred into glass vials (CZT Klaus Trott, Kriftel, Germany), dried in a vacuum concentrator, filled with argon, capped, and stored in sealed plastic bags containing silica desiccant at –80 °C. The pellet containing proteins was dried as mentioned above.
Seed protein quantification
Seed protein quantification was performed in 384-well plates using the Bio-Rad Protein Assay (Bio-Rad Laboratories GmbH, Munich, Germany) in three replicates, according to the manufacturer’s instructions. Protein pellets were washed five times with 800 µl of 70% ethanol, dissolved in 800 μl of 60 mM NaOH by shaking for 60 min at 70 °C, and centrifuged for 10 min. The total protein concentration of the supernatant was quantified at 595 nm using an Infinite 200 PRO microplate reader (Tecan Deutschland GmbH, Crailsheim, Germany) and a standard curve of 0–2.5 μg of BSA.
Relative quantification of polar metabolites using GC-MS
Aliquots of the polar phases were in-line derivatized directly prior to injection (Erban et al., 2007) and analyzed using a Gerstel MPS2-XL autosampler (Gerstel, Mühlheim, Germany) and an Agilent 7890 gas chromatograph (Agilent, Santa Clara, CA, USA) coupled to a LECO time-of-flight mass spectrometer (LECO, St. Joseph, MI, USA) (Riewe et al., 2012). Metabolites were identified and assigned using LECO ChromaTOF software including the Statistical Compare package and electron impact spectra library provided by the Golm Metabolome Database (GMD, gmd.mpimp-golm.mpg.de). Extraction of quantitative data was performed using the R-package ‘TargetSearch’ (Cuadros-Inostroza et al., 2009).
Raw data processing and normalization
Metabolite intensities were normalized for seed weight, internal standard (l-alanine-2,3,3,3-d4, 98 atom% D, Isotec Inc., Miamisburg, OH, USA), and individual detector response to correct for potential extraction batch and measurement day effects. Outliers were removed (median ±4× SD), and metabolite data were power transformed to ensure a proximate normal distribution (Box and Cox, 1964). Protein concentrations were batch normalized and seed protein content calculated using the number of seeds, sample, and total buffer volume. The normalized, outlier-corrected, and transformed metabolite data and protein content are summarized in Supplementary Data S1 at JXB online.
Statistical analyses
Statistical analyses were performed in R (R Core Team, 2015). Pearson correlation P-values were multiple testing corrected using false discovery rate (FDR) adjustment (Benjamini and Hochberg, 1995). Principal component analysis (PCA) was performed using the ‘pcaMethods’ package (Stacklies et al., 2007) on centered and Pareto-scaled data.
Molecular markers and linkage map
The RIL population was initially genotyped at the F7 generation with 110 single nucleotide polymorphism (SNP) markers (Törjék et al., 2003, 2006). Further genotyping was performed at the F9 generation using 28 additional simple sequence repeat (SSR) markers (Supplementary Data S2). Markers were derived from previous studies (Loudet et al., 2002; Salathia et al., 2007; Andreuzza et al., 2010; Hou et al., 2010), and partially modified. The marker distributions of the reciprocal subpopulations were compared using a Mantel test (10 000 permutations) of the corresponding similarity matrices obtained by simple matching. The 138 markers were evenly distributed over the five chromosomes, with an average marker distance of 3.4 cM corresponding to ~1 Mbp (Supplementary Data S3).
Analysis of quantitative trait loci
Power-transformed metabolite data, total protein content, and PCA scores were used to map QTLs with the R-package ‘qtl’ (Broman et al., 2003). To check for main effect QTLs, interval mapping and composite interval mapping approaches were performed with Haley–Knott regression (Haley and Knott, 1992). The conditional genotype probabilities were calculated using the ‘calc.genoprob’ function with a step size of 1 cM and an assumed genotyping error probability of 0.0001 with the Kosambi map function. Composite interval mapping was performed using the ‘cim’ function with a pre-defined number of five covariates selected by forward selection and a window size of 20 cM. To correct for type I error rates (false-positive QTLs), a genome-wide logarithm of odds (LOD) score threshold was estimated by 10 000 permutations at alpha 0.05 (Churchill and Doerge, 1994). All QTLs detected were used as the initial QTL model. The ‘stepwiseqtl’ function was used for forward/backward selection of multiple QTL models, with model choice made via a penalized LOD score, with separate penalties on main effects and interactions. Individual ‘heavy’ and ‘light’ penalties were extracted from 3000 ‘scantwo’ permutations with the ‘calc.penalties’ function (Broman and Sen, 2009; Manichaikul et al., 2009). For each trait, the derived multiple QTL model was plugged into the ‘fitqtl’ function to estimate QTL effects and percentages of variance (R2) explained by the individual QTLs from the final simultaneous fit of all QTLs. The 1.5-LOD support intervals (Manichaikul et al., 2006) for each QTL were estimated with the ‘lodint’ function and expanded to the nearest flanking markers.
Candidate gene identification
To identify candidate genes for mQTLs, compound and pathway information of the AraCyc 13.0 database was downloaded from the Plant Metabolic Network (PMN, plantcyc.org). Arabidopsis gene annotation information was obtained from the Arabidopsis Information Portal (ARAPORT 11, araport.org). For each mQTL, a search window was determined according to the 1.5-LOD support interval (Supplementary Data S4). Genes were extracted and tested for either direct association with the corresponding metabolite or indirect association with one of the pathways in which the metabolite is involved. To nominate the most promising candidate genes of the master regulatory locus detected on the short arm of chromosome IV, all 567 genes within the confidence interval of the hotspot on chromosome IV were analyzed for gene ontology (agriGO, bioinfo.cau.edu.cn/agriGO/) and matched against the plant transcription factor database (PlnTFDB 3.0, plntfdb.bio.uni-potsdam.de).
Threshold determination for mQTL hotspots
For each mQTL, the nearest molecular marker to the LOD apex was determined and the numbers of associated mQTLs per marker were added up. The deviation from the random number of co-localizations was calculated by randomly distributing the mQTLs of each metabolite over the 138 marker positions and counting the maximum number of mQTL co-localizations. This procedure was repeated 10 000 times, yielding a distribution of the maximum numbers of mQTLs per marker. The 95% quantile of this distribution corresponded to 15 QTLs. Hence ≥15 mQTLs at the same genome position were regarded as significantly co-localized.
Results and discussion
In the present study, we utilized a previously generated set of reciprocal RILs consisting of 202 Col-0×C24 and 191 C24×Col-0 F10 RILs (Törjék et al., 2006). Pools of 60 seeds from three individuals per genotype were analyzed for their metabolic composition. After exclusion of contaminants and manual data curation, 311 metabolites (64 of known chemical structure) were detected in >85% of all samples. Only these metabolites were taken into further consideration. A comparison of the metabolic composition between the reciprocal subpopulations revealed no statistically significant differences (ANOVA/FDR, P>0.05 for all metabolites). No significant differences in marker distribution between the reciprocal RILs were found (association between marker matrices of 88.3% estimated by Mantel test with 10 000 permutations, P<0.001). Maternal effects, for example due to different cytoplasm, mitochondria, and chloroplasts, were excluded by a two-way ANOVA exemplarily testing for effects of markers, subsets, and marker–subset interactions on leucine abundance (Supplementary Data S5). In addition, a modified cross-validation (Melchinger et al., 2004) was conducted treating the reciprocal RIL subsets individually and including randomly selected subsets in the mapping (Supplementary Data S6). In both cases, the reduction in population size led to a drastic decline of LOD scores and the detection of different combinations of leucine QTLs due to reduced detection power and random sampling effects. Hence the RILs were treated as one population and the corresponding results were used for all subsequent analyses.
Correlation between metabolites, seed protein content, and seed weight
Correlations between levels of metabolites help to gain information about metabolic links (Carreno-Quintero et al., 2012; Hill et al., 2013). Correlated metabolites can be the result of shared metabolic pathways or enzymatic reactions, or indirect regulatory processes affecting different pathways. Pearson correlation/FDR (Supplementary Data S7) analyses resulted in far more positive (97%) than negative (3%) correlations between metabolites (Supplementary Figs S1, S2). At a global level, metabolites were mostly weakly or moderately correlated (0.2<R<0.6). This resembles previous studies in Arabidopsis (Cross et al., 2006; Meyer et al., 2007; Sulpice et al., 2010) and tomato (Schauer et al., 2006). Several significant correlations, including 55 correlations of known metabolites with seed protein content, seven correlations of known metabolites with seed weight content, and hundreds of pairwise correlations between metabolites, were detected. Strong correlations between metabolites of specific sectors of metabolism, including structurally related metabolites, or metabolites from the same biological pathway, were found, as reported previously (Schauer et al., 2006; Lu et al., 2008; Lin et al., 2014). There was a strong correlation between the monosaccharides glucose and fructose (R=0.75). Other high correlations were detected: gentiobiose–erythritol (R=0.91), raffinose–myo-inositol (R=0.85), galactinol–myo-inositol (R=0.59), and raffinose–galactinol (R=0.57). Galactinol, raffinose, and myo-inositol are involved in the reversible reaction producing stachyose (Tanner and Kandler, 1968; Lehle and Tanner, 1973). High correlations between the citric acid cycle metabolites malic acid and citric acid (R=0.79) as well as fumaric acid and malic acid (R=0.55) were detected, supporting the finding that metabolites involved in the same pathway(s), in particular, are highly correlated (Sulpice et al., 2010). The BCAAs valine, leucine, and isoleucine were significantly and highly correlated: valine–isoleucine (R=0.94), leucine–isoleucine (R=0.88), and valine–leucine (R=0.81). This might be due to their structural relationship and largely shared biosynthetic and degradation pathways (Binder, 2010; Angelovici et al., 2013). A dominant role for amino acids in metabolite correlations has previously been described in tomato (Toubiana et al., 2012) and soybean (Lin et al., 2014).
Maturing Arabidopsis seeds mainly accumulate lipids in the form of triacylglycerols (TAGs), and seed storage proteins (SSPs) as storage macromolecules substantially contributing to the weight of mature seeds (Baud et al., 2008). Although primary metabolites serve as feedstock for their synthesis, only low correlations between polar primary metabolites and seed weight were found. Erythritol (R=0.39) and gentiobiose (R=0.36) were the strongest positively correlated, and prephenic acid (R= –0.28) and ornithine (R= –0.26) the strongest negatively correlated metabolites of known structure. As lipids and lipophilic compounds contribute 34.6–46.0% to the seed dry weight (O’Neill et al., 2003), they might be more important than polar primary metabolites.
In total, 276 significant correlations between metabolites and seed protein content were detected, including 55 metabolites of known chemical structure. However, most of these pairs are only moderately or slightly correlated (only 41 with R>0.5). The top four known metabolites correlated with protein content are glutamic acid (R=0.57), glucopyranose (R=0.57), melibiose (R=0.57), and pyroglutamic acid (R=0.56). The high correlation of glutamic acid with seed protein content was expected due to its central role in plant nitrogen transport and protein biosynthesis (Glass et al., 2001). A recent study revealed that the glutamate decarboxylase- (GAD) mediated conversion of glutamate to γ-aminobutyric acid (GABA) during seed development plays an important role in balancing carbon and nitrogen metabolism, and storage reserve accumulation in Arabidopsis seeds, and affects seed protein content (Fait et al., 2011). Glutamic acid is a key compound of cellular metabolism involved in the biosynthesis of many other amino acids. It acts as a substrate for glutamate dehydrogenases and various aminotransferases, providing 2-oxoglutarate for respiration (Forde and Lea, 2007). Moreover, it is a major transport form of nitrogen in plants (Xu et al., 2012). Protein content and seed weight were not found to be significantly correlated.
In summary, moderate to high, mostly positive pairwise correlations were detected, especially for structurally or pathway-related metabolites. Unlike our initial hypothesis, only weak pairwise correlations between individual polar primary metabolites and seed weight could be detected. Lipophilic compounds or combinations of particular metabolites might yield higher correlations, as previously described (Meyer et al., 2007).
Natural variation in primary metabolism and detection of mQTLs and regions associated with seed protein content
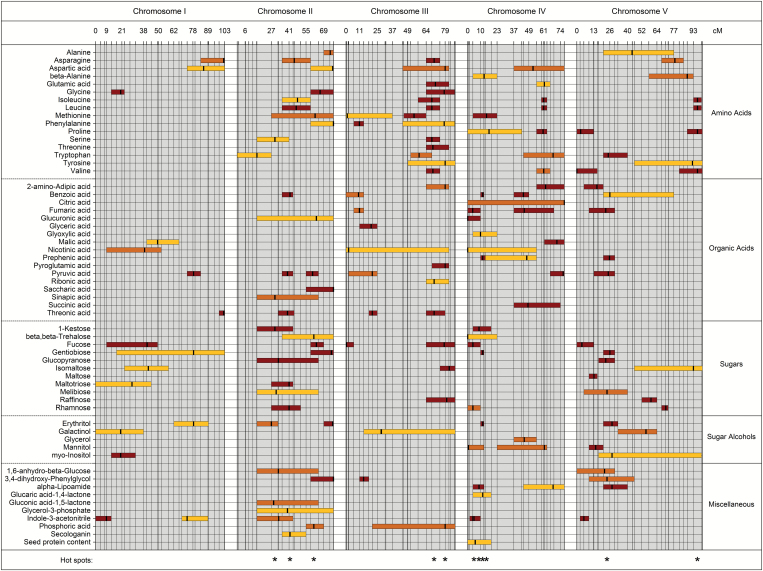
A total of 786 mQTLs affecting the metabolism of mature Arabidopsis seeds were identified by our mapping approach (Supplementary Data S8). For 90% of the 311 analyzed compounds, at least one mQTL was identified, reflecting a remarkably high variation emerging from the cross of two accessions. Lisec et al. (2008) used the same RILs and similar analytical approaches, but investigated leaf material of young vegetatively grown plants and found mQTLs for only 46% of the detected metabolites. Similarly, Rowe et al. (2008) identified mQTLs for 44% of the measured leaf metabolites. In the study presented here, 152 mQTLs were attributed to 58 metabolites of known (Fig. 1) and 634 mQTLs to 222 metabolites of unknown chemical structure. For 72 metabolites, only one QTL was identified. A maximum of seven QTLs was found for unknown mass spectral tags (MSTs) 40, 66, 218, 225, 232, and 245, respectively. Combined main effect QTLs explained up to 88.87% of phenotypic variation of particular metabolites, with explained phenotypic variation ranging from 0.57% to 77.68% for individual QTLs. On average, 7.23% of phenotypic variation is explained by a single QTL, suggesting that for most metabolites multiple unidentified genetic loci may contribute with moderate effects to overall phenotypic variation. In addition, the analysis revealed evidence for 30 epistatic interactions, including four metabolites of known structure: leucine, isoleucine, benzoic acid, and indole-3-acetonitrile (Supplementary Data S9). One QTL associated with seed protein content was detected on chromosome IV with ‘MASC04123’ (4.15 cM) as the nearest marker. The confidence interval for this QTL spanning a region of 18.7 cM on chromosome IV is overlapping with the confidence intervals of 22 QTLs for metabolites of known structure, including proline and methionine, and 146 QTLs for metabolites of unknown structure. However, only a small proportion of phenotypic variation of seed protein content (2.94%) is explained by this QTL.
Fig. 1.
Distribution of mQTLs for metabolites of known chemical structure. Chromosomal locations of significant mQTLs for the 58 metabolites of known chemical structure and the seed protein content are indicated by boxes representing the 1.5-LOD QTL support intervals. Vertical black lines within the boxes indicate the apices of the corresponding LOD curves. The mQTLs are color-coded according to their significance [threshold at alpha of 0.05 (yellow), 0.01 (orange), 0.001 (red)] derived from permutation results of the genome-wide maximum LOD scores. Vertical lines represent marker positions. For a subset, their approximate distance in centiMorgans is indicated. Asterisks at the bottom correspond to the position of identified mQTL hotspots.
The RIL population was previously utilized by Lisec et al. (2008), who quantified 181 leaf metabolites, including 85 compounds of known chemical nature, and identified 157 mQTLs for 84 metabolites (50 of known chemical structure). Comparing this with our findings, a total of 25 known metabolites with mQTLs, predominantly amino acids and organic acids, were detected in both studies. To compare the detected mQTLs, support intervals were extended to the nearest flanking markers to derive physical map positions. This comparison revealed nine mQTLs for seven metabolites with overlapping confidence intervals (Table 1), including a tyrosine mQTL on chromosome V that contains a tyrosine aminotransferase (At5g53970) involved in tocopherol synthesis in Arabidopsis (Riewe et al., 2012). Untargeted LC-MS-based metabolomic approaches and quantitative genetic analysis enabled broad-spectrum molecular dissection of the Arabidopsis leaf metabolite composition of 160 Cvi×Ler RILs (Keurentjes et al., 2006). Mapping of >2000 mass peaks resulted in the identification of mQTLs for ~75% of all mass signals, which is comparable with the detection rate in seeds in the present study. Another study used GC-MS to analyze 210 Bay×Sha RILs (Rowe et al., 2008). They identified a total of 557 metabolites and used them for QTL mapping, leading to the identification of 438 mQTLs for 243 metabolites (36 known metabolites overlapped with those identified in the present study).
Table 1.
Comparison of detected mQTLs in seeds and leaf material
| Metabolite | Chromosome | Support interval | R 2 (%)a | Support interval Lisec et al. (2008) |
R
2 (%)a Lisec et al. (2008) |
|---|---|---|---|---|---|
| Glycine | III | 17.27–23.28 Mbp | 5.42 | 16.24–17.78 Mbp | 8.00 |
| Malic acid | IV | 13.69–18.54 Mbp | 9.85 | 10.67–15.39 Mbp | 4.20 |
| myo-Inositol | I | 3.49–9.36 Mbp | 5.32 | 4.12–6.50 Mbp | 6.50 |
| Raffinose | III | 17.27–23.41 Mbp | 4.30 | 16.24–19.50 Mbp | 4.70 |
| Serine | II | 5.18–10.43 Mbp | 3.30 | 3.00–5.33 Mbp | 5.10 |
| Serine | III | 17.27–19.86 Mbp | 7.54 | 15.17–17.78 Mbp | 6.90 |
| Tyrosine | III | 14.30–23.41 Mbp | 3.36 | 11.77–17.78 Mbp | 4.20 |
| Tyrosine | V | 18.83–26.92 Mbp | 3.50 | 21.92–22.91 Mbp | 9.60 |
a Estimated proportion of the phenotype variance explained by a QTL
By testing all 55 pairwise epistatic interactions between 11 detected metabolite QTL clusters against the average accumulation of 557 metabolites, Rowe et al. (2008) identified 240 metabolites with 1–5 significant epistatic interactions, for a total of 328 significant interactions. Lisec et al. (2008) detected 38 epistatic interactions involving metabolites of known structure. With only four interactions among mQTLs of known metabolites, much fewer incidences of epistasis were observed in the present study on Arabidopsis seed metabolites.
Detection of enzyme-encoding mQTL candidate genes
To identify candidates, genes within the mQTL confidence intervals were extracted and queried for direct or indirect association with the particular metabolite using the AraCyc 13.0 database. Genes encoding enzymes catalyzing reactions that involve the metabolite as substrate or product were considered direct candidates, while genes encoding enzymes that catalyze other reactions within pathways that lead toward the formation of the metabolite or which consume the metabolite were considered indirect candidates. A total of 168 direct candidate genes for 27 of the 52 metabolites of known structure (52%) were found. Expanding search criteria to all pathways in which the metabolite is involved, a total of 765 direct and indirect candidate genes for 33 metabolites (~63%) were determined. These numbers differ from those observed by Lisec et al. (2008) utilizing the same RIL population, but analyzing leaf material. We found a higher percentage of direct candidate genes, which might be attributed to a less conservative calculation of mQTL confidence intervals. In contrast to the 43% increase found by Lisec et al. (2008), the extension of the search criteria to indirect associations increased the percentage of mQTLs with a candidate gene only by 21% in the present study.
Shared mQTLs among branched-chain amino acids, epistatic interactions, and candidate genes
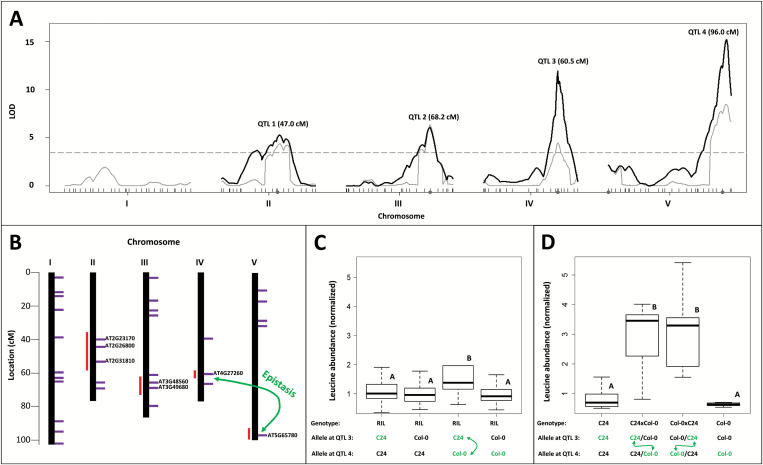
For leucine and isoleucine, shared loci and epistatic interactions between the mQTLs on chromosomes IV (60.5 cM) and V (96.0 cM) were observed (Supplementary Data S8, S9). Furthermore, high correlations were found between leucine, valine, and isoleucine (Supplementary Data S7). These findings prompted us to look at candidate genes within the leucine mQTLs in more detail. Leucine, valine, and isoleucine are characterized by their branched hydrocarbon residues. They form the small group of BCAAs that are critical for protein synthesis and normal plant growth (Yu et al., 2013) and serve as precursors for secondary metabolites (Binder, 2010; Buchanan et al., 2015). Plants synthesize these essential amino acids de novo. Valine and isoleucine are synthesized in two parallel pathways using a single set of four enzymes, whereas the pathway to leucine branches off and requires three additional steps (Binder, 2010). The BCAA biosynthesis in plants occurs in chloroplasts (Diebold, 2002; Binder, 2010), whereas the degradation mostly takes place in mitochondria (Zolman et al., 2001; Beck et al., 2004). Despite the limited mapping resolution provided by RILs, it is possible to identify candidate genes underlying biochemical pathways (Lisec et al., 2008; Brotman et al., 2011). To this end, the four detected mQTLs for leucine (Fig. 2A; Supplementary Data S8) were screened for known and putative pathway genes involved in BCAA metabolism. From three databases, ARAPORT 11 (araport.org), AraCyc 13.0 (PMN, plantcyc.org), and KEGG PATHWAY (www.genome.jp/kegg/pathway.html, last accessed 16 February 2017), genes annotated in leucine biosynthesis and degradation were extracted and their map positions compared with the leucine mQTL confidence intervals. Candidate genes could be identified for all four leucine mQTLs. Three candidate genes: AT2G23170 (GH3.3), AT2G26800 (HML1), and AT2G31810 (AHAS), were associated with the mQTLs on chromosome II. AT2G23170 (GH3.3) encodes an indole-3-acetic acid (IAA)-amido synthetase (Staswick et al., 2005), AT2G26800 a putative hydroxymethylglutaryl-CoA lyase, presumably involved in leucine degradation, and AT2G31810 a small regulatory subunit of the acetolactate synthase (Binder, 2010). The acetolactate synthase is the first common enzyme in the biosynthetic pathways of the BCAAs (Chen et al., 2010) and catalyzes the conversion of two molecules of pyruvate into (S)-2-acetolactate, or one molecule of pyruvate and one molecule of 2-oxobutanoate into 2-aceto-2-hydroxybutyrate (Singh, 1999; Duggleby et al., 2008). Two candidate genes co-localized with the confidence interval of the leucine mQTLs on chromosome III. AT3G48560 (AHAS) encodes the catalytic subunit of the acetolactate synthase (pyruvate decarboxylase). Mutants defective in this gene exhibit increases in all three BCAAs in mature seeds (Lu et al., 2011). AT3G49680 (BCAT3) encodes a BCAA aminotransferase, which is involved in the biosynthesis and degradation of valine, leucine, and isoleucine (Knill et al., 2008). For the confidence interval on chromosome IV, AT4G27260 (GH3.5) could be identified as a direct candidate gene encoding an IAA-amido synthetase, which conjugates various amino acids, including leucine, to IAA. IAA is the prevalent form of auxin, an important phytohormone, affecting many aspects of plant development and plant response to biotic and abiotic stimuli (Woodward and Bartel, 2005). Some of these amino acid modifications can be reversed by amido hydrolases (Davies et al., 1999), suggesting that IAA–amino acid conjugates, such as IAA–leucine, are storage forms of auxin (Staswick et al., 2005). These compounds can be metabolized to contribute to the pool of free auxin, allowing plants to fine-tune their levels of active auxin (Woodward and Bartel, 2005). The confidence interval for the leucine mQTLs on chromosome V harbors AT5G65780 (BCAT5), encoding another member of the BCAA transaminase gene family. Green fluorescent protein tagging localized the protein to the chloroplast, but recent proteomic studies indicated a mitochondrial localization (Diebold, 2002; Taylor, 2004; Zybailov et al., 2008). Although its localization is still controversial and BCAT5 has not yet been characterized in detail, its role in leucine metabolism has been suggested (Schuster et al., 2006; Binder, 2010). A recent genome-wide association study using a diversity panel of 360 Arabidopsis accessions in conjunction with a QTL analysis of a RIL population derived from accessions Bay-0 and Shahdara revealed the unique, catabolic role of the AT1G10070 locus (BCAT2) in BCAA metabolism in Arabidopsis seeds (Angelovici et al., 2013). However, as these accessions are polymorphic for several SNPs and insertion/deletion events, the causative molecular mechanism(s) underlying the phenotypic differences in Bay-0 and Shahdara BCAT2 alleles were not determined. In the present study, no significant association with the BCAT2 locus or any other region on chromosome I was detected. A comparison of the C24 and Col-0 alleles of the BCAT2 locus revealed several polymorphisms (Supplementary Data S10). Only one polymorphism within the coding sequence leading to a glutamic acid to aspartic acid exchange was detected. Both amino acids have comparable properties and are thus unlikely to cause substantial differences of the BCAT2 protein. This absence of functional relevant differences between the C24 and Col-0 alleles could explain the fact that no significant influence of the BCAT2 locus was detected in the C24/Col-0 RIL population. A simplified genetic map with genes involved in leucine biosynthesis and degradation is shown in Fig. 2B. Comparing detected mQTLs for all BCAAs, overlapping confidence intervals were detected. Leucine and isoleucine share all detected mQTLs on chromosomes II, III, IV, and V, including a putative epistatic interaction between mQTLs on chromosomes IV (60.5 cM) and V (96.0 cM). For leucine and valine, mQTLs on chromosomes III, IV, and V share overlapping confidence intervals. For valine there is no mQTL on chromosome II, but an additional mQTL on chromosome V that is not shared by leucine and isoleucine. Its confidence interval harbors one candidate gene (AT5G09300, BCKDH) encoding a putative subunit of the branched-chain keto acid dehydrogenase complex that catalyzes the second step of BCAA degradation (Binder, 2010; Peng et al., 2015).
Fig. 2.
mQTL analysis and candidate gene identification for leucine. (A) LOD profiles were plotted for all five Arabidopsis chromosomes. Gray lines represent LOD profiles calculated with the ‘cim’ function (composite interval mapping). Gray dots indicate selected cofactors. The horizontal dashed gray line corresponds to a CIM alpha threshold of 0.05, estimated by 10 000 permutations. The solid black lines indicate LOD profiles calculated with the ‘stepwiseqtl’ function using a multiple QTL model. The positions of the QTL apices in centiMorgans are given above the curves. (B) A simplified genetic map with known and putative genes involved in leucine biosynthesis and degradation. Purple horizontal lines indicate the locations of genes, directly or indirectly involved in leucine metabolism. Leucine mQTLs were identified on chromosomes II, III, IV, and V. Support intervals are shown as red vertical lines beside the chromosomes. Leucine-related genes, located within the confidence intervals of the mQTLs, are indicated. Identified candidate genes for chromosome II are AT2G23170 (GH3.3), AT2G26800 (HML1), and AT2G31810, for chromosome III AT3G48560 (AHAS) and AT3G49680 (BCAT3), for chromosome IV AT4G27260 (GH3.5), and for chromosome V AT5G65780 (BCAT5). (C) Boxplots of normalized and median divided leucine abundances in seeds of RILs. Samples were subdivided into four groups according to the allelic state at the epistatically interacting loci on chromosomes IV and V. Significant differences between the groups are indicated by upper case letters (ANOVA with post-hoc Tukey HSD, Padj<0.001; number of individuals: nC24/C24=113, nCol-0/C24=20, nC24/Col-0 =82, nCol-0/Col-0=149). (D) Boxplots of normalized and median divided leucine abundances in seeds of parental and reciprocal F1 hybrid plants derived from an independent experiment. Significant differences between the groups are indicated by upper case letters (ANOVA with post-hoc Tukey HSD, Padj<0.05; number of individuals: nC24=7, nC24×Col-0=5, nCol-0×C24=5, nCol-0=5).
To investigate further the influence of the mQTL on chromosomes IV and V and their putative interaction, samples were divided into four groups based on their allelic states and the leucine abundances were plotted (Fig. 2C). The C24 allele at the locus on chromosome V has an increasing effect on the leucine abundance, but only if the Col-0 allele is present at the locus on chromosome IV. The presence of the C24 allele at the locus on chromosome IV leads to no substantial difference in leucine abundance, independent of the allele at the locus on chromosome V. This finding is consistent with the detected epistatic interaction between the mQTLs on chromosomes IV and V and higher leucine abundances in seeds of the reciprocal hybrids compared with their parental accessions (Fig. 2D).
Detection of mQTL hotspots for Arabidopsis seed metabolism
QTL studies in various species identified hotspots (Schauer et al., 2006, 2008; Joosen et al., 2013; Chen et al., 2014; Alseekh et al., 2015; Wen et al., 2015), but their number and position can vary across tissues within a specific population, as found in rice (Gong et al., 2013). There are two potential explanations for these observations. It could be a reflection of an uneven distribution of biosynthetic genes over the genome, or may be a consequence of the occurrence of pleiotropic or regulatory genes of higher hierarchical order controlling multiple metabolic reactions rather than individual metabolic conversions. A study comparing the distribution of metabolic genes in the genome with the mQTL distribution has provided evidence that a large proportion of detected mQTLs, with hitherto unknown metabolic functions, are most probably regulatory genes controlling primary metabolism (Lisec et al., 2008).
The mQTLs detected in this study were not evenly distributed across the Arabidopsis genome. In some regions, mQTLs clustered, whereas other regions were depleted of QTLs. Since 311 metabolic traits and 138 markers were taken into account, stochastic co-localizations of mQTLs are to be expected. The threshold for detection of significant enrichment of mQTLs in certain positions was determined using 10 000 permutations. Markers associated with at least 15 mQTLs were regarded as significant hotspots. In the present study, we found evidence for several mQTL hotspots on chromosomes II, III, IV, and V (Table 2; Supplementary Fig. S3). The smallest number of mQTLs (n=90) and no hotspot were found for chromosome I. On average, substantially fewer mQTLs per marker were detected on chromosome I than expected for a random distribution. For chromosome II, a total of 148 mQTLs and three hotspots were identified. On chromosome III, 143 mQTLs were detected, with evidence for two large hotspots. Chromosome IV shows evidence for 220 mQTLs, and at least four markers were identified as hotspots. Three of them are localized on the short arm of the chromosome and might be regarded as a single hotspot due to their proximity. Their combined median confidence interval is delimited by the markers ‘MASC04123’ and ‘MASC04685’ and extends beyond the short arm of chromosome IV. It spans a region of ~5 Mbp and contains 567 genes. For chromosome V, a total number of 185 mQTLs and the largest hotspot with 94 mQTLs, associated with the marker ‘MASC09209’, were detected. Calculating the median confidence interval over all mQTLs sharing this marker, the hot spot is delimited by the markers ‘MASC09208’ and ‘nga139’, spanning a region of ~6.5 Mbp, and containing 487 genes.
Table 2.
Summary of mQTL hotspots
| Chromosome | Marker | Position (kbp) | Position (cM) | Number of mQTLs |
|---|---|---|---|---|
| II | M2_4269 | 8410.151 | 32.48 | 16 |
| II | MASC02644 | 10 428.938 | 41.29 | 20 |
| II | MASC09222 | 14 375.406 | 58.38 | 34 |
| III | MASC09224 | 18 501.466 | 68.17 | 44 |
| III | MASC02788 | 20 744.711 | 78.77 | 32 |
| IV | MASC04123 | 301.329 | 4.15 | 27 |
| IV | MASC04725 | 1092.491 | 10.21 | 35 |
| IV | MASC05042 | 2188.362 | 12.90 | 44 |
| IV | MASC04685 | 5230.768 | 14.01 | 16 |
| V | MASC09209 | 7717.922 | 26.27 | 94 |
| V | MASC09211 | 25 579.812 | 92.79 | 15 |
Hotspots of mQTLs have been previously reported in Arabidopsis (Keurentjes et al., 2006; Fu et al., 2009; Chan et al., 2010; Joseph et al., 2013). Lisec et al. (2008) identified two mQTL hotspots, on the short arm of chromosome IV and at the bottom of chromosome V, with 16 and 12 mQTLs, respectively. A biomass QTL and multiple mQTLs (Lisec et al., 2008), as well as an early stage biomass heterosis QTL (Meyer et al., 2010) were found in similar positions on the short arm of chromosome IV as the hotspot in the present study. The studies of Rowe et al. (2008) and Joosen et al. (2013) on the Bay×Sha RIL population revealed 11 and 8 mQTL hotspots, respectively. Two major hotspots (AOP on chromosome IV and Elong on chromosome V) may correspond to here reported hotspots on chromosomes IV (10.21 cM and 12.9 cM) and V (26.27 cM). These loci co-localize with known QTLs that determine the transcript accumulation of aliphatic glucosinolate biosynthetic genes and the accumulation and structure of aliphatic glucosinolates (Wentzell et al., 2007). However, another study indicated that expression QTL (eQTL) hotspots may not overlap in different populations (Cubillos et al., 2012).
A region on the short arm of chromosome IV is responsible for the major proportion of metabolic variation
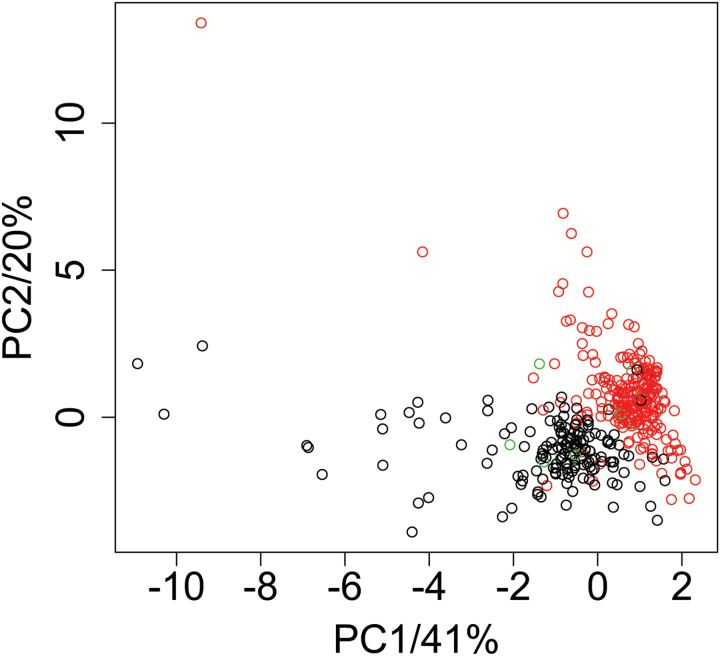
To identify major effects on metabolism, a PCA was performed. The first four principal components explained 41, 20, 7.4, and 5.6% of the variance, respectively. The top loading metabolites of PC1 are unknown MSTs 124, 40, 222, and 105, gentiobiose, and galactinol, and for PC2 unknown MSTs 90, 74, 155, 40, and 85 (Supplementary Data S11). The first two principle components separated the samples into two narrow clusters. Labeling the samples according to extraction batches, measurement time point, or the two RIL subsets did not explain the clustering (Supplementary Data S12). Hence, samples were labeled according to the genotype information sequentially for all 138 markers (Supplementary Data S12). The allelic distribution at marker ‘MASC05042’ on the short arm of chromosome IV (12.90 cM) closely matches the clustering (Fig. 3). All markers in a region of ~23.38 cM, ranging from ‘MASC02820’ to ‘MASC02668’, display a similar pattern, indicating that this region contributes to a large proportion of the overall metabolic variation. To investigate this effect further, PCA scores were included in the QTL analysis to identify genomic regions of high importance affecting multiple correlated metabolites, yielding 13 loci significantly associated with the first four principal components (Supplementary Data S8). Three genomic regions were associated with PC1 responsible for the largest proportion of variance (41%). One QTL was identified on chromosome IV, spanning a region of 25 cM. Another QTL of 14 cM was located on chromosome V. The most prominent QTL for PC1 was identified on the short arm of chromosome IV (12 cM) with ‘MASC05042’ as the nearest marker to the LOD peak. Its confidence interval spans 2.7 cM and includes several mQTLs for PC1 loadings. The PC1 QTL is localized within the mQTL hotspot region on the short arm of chromosome IV that is responsible for the major clustering in the PCA (Table 2; Supplementary Data S12). In previous studies, mQTL hotspots were found at similar positions (Lisec et al., 2008; Rowe et al., 2008; Joosen et al., 2013). The FRIGIDA (FRI; AT4G00650) locus, which encodes a major determinant of Arabidopsis flowering time (Johanson et al., 2000), is located ~8.8 cM (1.9 Mbp) distal to ‘MASC05042’ (marker ‘F6N23ID’ detects a polymorphism within FRIGIDA). The PC1 QTL displays a very sharp peak with a steeply declining LOD profile and the FRIGIDA locus is not included within its confidence interval (Supplementary Fig. S4). Moreover, the top five loadings of PC1 share similar LOD profiles. Considering these facts, it is rather unlikely that allelic variation of FRIGIDA is causal for the observed complex metabolic variation conditioned by the QTLs on the short arm of chromosome IV. In contrast, these findings indicate that another, as yet unknown, master regulatory gene of seed metabolism or a major effector of seed development that has profound consequences on metabolite composition (e.g. through differences in size of certain tissues) is located in the identified region of chromosome IV and is responsible for major parts of the observed phenotypic variation.
Fig. 3.
Principal component analysis of metabolite data. Score plot of the first two principal components PC1 and PC2 explaining 41% and 20% of variance of the data set, respectively. Samples were colored according to the genotype information on chromosome IV/marker: ‘MASC05042’ (12.90 cM). Black, red, and green circles correspond to Col-0, C24, and heterozygous alleles, respectively. Data were normalized, Pareto scaled, and mean centered prior to the calculation of the principal components.
As an entry into the identification of promising candidate regulatory genes for further analyses, all 567 genes within the confidence interval of the hotspot on chromosome IV were analyzed for gene ontology (agriGO, bioinfo.cau.edu.cn/agriGO/; Du et al., 2010) and matched against the plant transcription factor database (PlnTFDB 3.0, plntfdb.bio.uni-potsdam.de; Riaño-Pachón et al., 2007). Several genes were annotated with kinase or phosphatase activity, but annotations gave no direct hints of seed metabolic processes. A total of 38 annotated transcription factors are located within the hotspot (Supplementary Data S13). According to the data accessible in the Arabidopsis eFP Browser 2.0 (www.bar.utoronto.ca/efp2/Arabidopsis/Arabidopsis_eFPBrowser2.html, last accessed 16 February 2017 , Winter et al., 2007), 12 of them display a high relative expression in mature and/or developing Arabidopsis seeds (Table 3). Three of them, AT4G01120, AT4G01280, and AT4G02640, are highly or predominantly expressed in mature (and developing) seeds. A particularly interesting candidate is AT4G02640, which encodes a basic leucine zipper transcription factor, (bZIP10). bZIP10 has been shown to interact with ABI3 (AT3G24650), a central transcriptional regulator of seed maturation (MAT) genes in Arabidopsis, and to activate seed storage protein gene expression synergistically (Lara et al., 2003). Heterodimerization of bZIP53 with bZIP10 significantly enhances DNA binding activity and produces a synergistic increase in target gene activation (Alonso et al., 2009). Furthermore, these bZIP heterodimers interact with ABI3, which further increases MAT gene activation. Comparing the C24 and Col-0 alleles of bZIP10, several polymorphisms (synonymous substitutions) and more interestingly a 7 bp insertion in the putative promotor region of C24 were detected (Supplementary Data S10).
Table 3.
Selection of transcription factor (TF) genes within the mQTL hotspot on chromosome IV expressed in seeds
| AGI locus identifier | TF family | General expression profilea | Seed-specific expression profilea |
|---|---|---|---|
| AT4G00730 | HB | Ubiquitous | Intermediate development |
| AT4G01120 | bZIP | Seed specific | Late development and mature seeds |
| AT4G01250 | WRKY | Preferentially in seeds | Late development |
| AT4G01280 | MYB-related | Seeds and other organs | Mature seeds |
| AT4G01460 | bHLH | Uubiquitous | Intermediate development |
| AT4G01500 | ABI3VP1 | Ubiquitous | Early and intermediate development |
| AT4G01580 | ABI3VP1 | Seeds and other organs | Intermediate development |
| AT4G02020 | SET | Ubiquitous | Early development |
| AT4G02640 | bZIP | ubiquitous | Late development and mature seeds |
| AT4G02670 | C2H2 | Seeds and other organs | Early development |
| AT4G03170 | ABI3VP1 | Seeds and other organs | Late development |
| AT4G04890 | PDF2 | Ubiquitous | Intermediate development |
a According to Arabidopsis eFP Browser 2.0
In conclusion, the results provided by this study substantially enhance our current knowledge about Arabidopsis seed metabolism and natural variation of complex traits. It provides a broad and solid basis for further studies towards broadening the knowledge of factors mediating or regulating plant seed metabolism: detailed investigations can immediately be initiated on the enzyme-encoding candidate genes identified here; metabolic factors, such as transporters or regulators, may be selected via further database searches. Furthermore, upon confirmation and fine mapping of detected mQTLs, for example by the use of introgression lines (ILs), novel factors of hitherto unknown function can be identified. Finally, the evidence provided of a master regulatory locus of seed metabolism on the short arm of chromosome IV and the hints towards a corresponding candidate transcription factor gene are of particular interest as they provide a direct entry into further unraveling of important processes of seed development and maturation.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Histogram of metabolite correlations.
Fig. S2. Correlation matrix.
Fig. S3. Overview of mQTL hotspots.
Fig. S4. LOD profile of PC1 (chromosome IV).
Data S1. Summary of phenotypic data.
Data S2. Additional set of markers.
Data S3. Genetic map.
Data S4. mQTL candidate genes.
Data S5. Summary of the two-way ANOVA.
Data S6. Modified cross-validation of leucine mQTLs.
Data S7. Correlation analysis.
Data S8. Summary of QTL analysis.
Data S9. Summary of epistatic interactions.
Data S10. Comparison of C24 and Col-0 alleles for BCAT2 and bZIP10.
Data S11. Principal component loadings.
Data S12. Principal component plots.
Data S13. Transcription factors within the hotspot on chromosome IV
Supplementary Material
Acknowledgements
We are grateful to Andrea Apelt, Beatrice Knüpfer, and Angelika Flieger for excellent technical assistance. We thank Jahnavi Koppolu and Kristin Langanke who contributed to the phenotypic analysis of the RILs, and Janine Wiebach for bioinformatical support.
References
- Alonso A, Marsal S, Julià A. 2015. Analytical methods in untargeted metabolomics: state of the art in 2015. Frontiers in Bioengineering and Biotechnology 3, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso R, Oñate-Sánchez L, Weltmeier F, Ehlert A, Diaz I, Dietrich K, Vicente-Carbajosa J, Dröge-Laser W. 2009. A pivotal role of the basic leucine zipper transcription factor bZIP53 in the regulation of Arabidopsis seed maturation gene expression based on heterodimerization and protein complex formation. The Plant Cell 21, 1747–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Koornneef M. 2000. Naturally occurring variation in Arabidopsis: an underexploited resource for plant genetics. Trends in Plant Science 5, 22–29. [DOI] [PubMed] [Google Scholar]
- Alseekh S, Tohge T, Wendenberg R, et al. 2015. Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. The Plant Cell 27, 485–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreuzza S, Li J, Guitton AE, Faure JE, Casanova S, Park JS, Choi Y, Chen Z, Berger F. 2010. DNA LIGASE I exerts a maternal effect on seed development in Arabidopsis thaliana. Development 137, 73–81. [DOI] [PubMed] [Google Scholar]
- Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D. 2013. Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. The Plant Cell 25, 4827–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Dubreucq B, Miquel M, Rochat C, Lepiniec L. 2008. Storage reserve accumulation in Arabidopsis: metabolic and developmental control of seed filling. The Arabidopsis Book 6, e0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck HC, Hansen AM, Lauritsen FR. 2004. Catabolism of leucine to branched-chain fatty acids in Staphylococcus xylosus. Journal of Applied Microbiology 96, 1185–1193. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multipletTesting. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300. [Google Scholar]
- Binder S. 2010. Branched-chain amino acid metabolism in Arabidopsis thaliana. The Arabidopsis Book 8, e0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box GEP, Cox DR. 1964. An analysis of transformations. Journal of the Royal Statistical Society. Series B (Methodological) 26, 211–252. [Google Scholar]
- Broman KW, Sen Ś. 2009. A guide to QTL mapping with R/qtl. Dordrecht: Springer. [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19, 889–890. [DOI] [PubMed] [Google Scholar]
- Brotman Y, Riewe D, Lisec J, Meyer RC, Willmitzer L, Altmann T. 2011. Identification of enzymatic and regulatory genes of plant metabolism through QTL analysis in Arabidopsis. Journal of Plant Physiology 168, 1387–1394. [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Gruissem W, Jones RL, eds 2015. Biochemistry & molecular biology of plants. Chichester: Wiley Blackwell. [Google Scholar]
- Calenge F, Saliba-Colombani V, Mahieu S, Loudet O, Daniel-Vedele F, Krapp A. 2006. Natural variation for carbohydrate content in Arabidopsis. Interaction with complex traits dissected by quantitative genetics. Plant Physiology 141, 1630–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno-Quintero N, Acharjee A, Maliepaard C, Bachem CW, Mumm R, Bouwmeester H, Visser RG, Keurentjes JJ. 2012. Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiology 158, 1306–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ. 2011. Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biology 9, e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Hansen BG, Kliebenstein DJ. 2010. The complex genetic architecture of the metabolome. PLoS Genetics 6, e1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Gao Y, Xie W, et al. 2014. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nature Genetics 46, 714–721. [DOI] [PubMed] [Google Scholar]
- Chen H, Saksa K, Zhao F, Qiu J, Xiong L. 2010. Genetic analysis of pathway regulation for enhancing branched-chain amino acid biosynthesis in plants. The Plant Journal 63, 573–583. [DOI] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. 1994. Empirical threshold values for quantitative trait mapping. Genetics 138, 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross JM, von Korff M, Altmann T, Bartzetko L, Sulpice R, Gibon Y, Palacios N, Stitt M. 2006. Variation of enzyme activities and metabolite levels in 24 Arabidopsis accessions growing in carbon-limited conditions. Plant Physiology 142, 1574–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadros-Inostroza A, Caldana C, Redestig H, Kusano M, Lisec J, Peña-Cortés H, Willmitzer L, Hannah MA. 2009. TargetSearch—a Bioconductor package for the efficient preprocessing of GC-MS metabolite profiling data. BMC Bioinformatics 10, 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos FA, Yansouni J, Khalili H, et al. 2012. Expression variation in connected recombinant populations of Arabidopsis thaliana highlights distinct transcriptome architectures. BMC Genomics 13, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B. 1999. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. The Plant Cell 11, 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold R. 2002. The branched-chain amino acid transaminase gene family in Arabidopsis encodes plastid and mitochondrial proteins. Plant Physiology 129, 540–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggleby RG, McCourt JA, Guddat LW. 2008. Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiology and Biochemistry 46, 309–324. [DOI] [PubMed] [Google Scholar]
- Erban A, Schauer N, Fernie AR, Kopka J. 2007. Nonsupervised construction and application of mass spectral and retention time index libraries from time-of-flight gas chromatography–mass spectrometry metabolite profiles. Methods in Molecular Biology 358, 19–38. [DOI] [PubMed] [Google Scholar]
- Fait A, Nesi AN, Angelovici R, et al. 2011. Targeted enhancement of glutamate-to-γ-aminobutyrate conversion in Arabidopsis seeds affects carbon–nitrogen balance and storage reserves in a development-dependent manner. Plant Physiology 157, 1026–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Schauer N. 2009. Metabolomics-assisted breeding: a viable option for crop improvement? Trends in Genetics 25, 39–48. [DOI] [PubMed] [Google Scholar]
- Fiehn O. 2008. Extending the breadth of metabolite profiling by gas chromatography coupled to mass spectrometry. Trends in Analytical Chemistry 27, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Lea PJ. 2007. Glutamate in plants: metabolism, regulation, and signalling. Journal of Experimental Botany 58, 2339–2358. [DOI] [PubMed] [Google Scholar]
- Francisco M, Joseph B, Caligagan H, Li B, Corwin JA, Lin C, Kerwin RE, Burow M, Kliebenstein DJ. 2016. Genome wide association mapping in Arabidopsis thaliana identifies novel genes involved in linking allyl glucosinolate to altered biomass and defense. Frontiers in Plant Science 7, 1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Keurentjes JJ, Bouwmeester H, et al. 2009. System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nature Genetics 41, 166–167. [DOI] [PubMed] [Google Scholar]
- Giavalisco P, Li Y, Matthes A, Eckhardt A, Hubberten HM, Hesse H, Segu S, Hummel J, Köhl K, Willmitzer L. 2011. Elemental formula annotation of polar and lipophilic metabolites using (13) C, (15) N and (34) S isotope labelling, in combination with high-resolution mass spectrometry. The Plant Journal 68, 364–376. [DOI] [PubMed] [Google Scholar]
- Glass ADM, Brito DT, Kaiser BN, et al. 2001. Nitrogen transport in plants, with an emphasis on the regulation of fluxes to match plant demand. Journal of Plant Nutrition and Soil Science 164, 199–207. [Google Scholar]
- Gong L, Chen W, Gao Y, Liu X, Zhang H, Xu C, Yu S, Zhang Q, Luo J. 2013. Genetic analysis of the metabolome exemplified using a rice population. Proceedings of the National Academy of Sciences, USA 110, 20320–20325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haley CS, Knott SA. 1992. A simple regression method for mapping quantitative trait loci in line crosses using flanking markers. Heredity 69, 315–324. [DOI] [PubMed] [Google Scholar]
- Hill CB, Taylor JD, Edwards J, Mather D, Bacic A, Langridge P, Roessner U. 2013. Whole-genome mapping of agronomic and metabolic traits to identify novel quantitative trait loci in bread wheat grown in a water-limited environment. Plant Physiology 162, 1266–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs DH, Flintham JE, Hills MJ. 2004. Genetic control of storage oil synthesis in seeds of Arabidopsis. Plant Physiology 136, 3341–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Li L, Peng Z, et al. 2010. A platform of high-density INDEL/CAPS markers for map-based cloning in Arabidopsis. The Plant Journal 63, 880–888. [DOI] [PubMed] [Google Scholar]
- Jasinski S, Lécureuil A, Durandet M, Bernard-Moulin P, Guerche P. 2016. Arabidopsis seed content QTL mapping using high-throughput phenotyping: the assets of near infrared spectroscopy. Frontiers in Plant Science 7, 1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Joosen RV, Arends D, Li Y, Willems LA, Keurentjes JJ, Ligterink W, Jansen RC, Hilhorst HW. 2013. Identifying genotype-by-environment interactions in the metabolism of germinating arabidopsis seeds using generalized genetical genomics. Plant Physiology 162, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Atwell S, Corwin JA, Li B, Kliebenstein DJ. 2014. Meta-analysis of metabolome QTLs in Arabidopsis: trying to estimate the network size controlling genetic variation of the metabolome. Frontiers in Plant Science 5, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Corwin JA, Züst T, Li B, Iravani M, Schaepman-Strub G, Turnbull LA, Kliebenstein DJ. 2013. Hierarchical nuclear and cytoplasmic genetic architectures for plant growth and defense within Arabidopsis. The Plant Cell 25, 1929–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ, Fu J, de Vos CH, et al. 2006. The genetics of plant metabolism. Nature Genetics 38, 842–849. [DOI] [PubMed] [Google Scholar]
- Keurentjes JJB, Fu J, Terpstra IR, et al. 2007. Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proceedings of the National Academy of Sciences, USA 104, 1708–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ, Sulpice R, Gibon Y, Steinhauser MC, Fu J, Koornneef M, Stitt M, Vreugdenhil D. 2008. Integrative analyses of genetic variation in enzyme activities of primary carbohydrate metabolism reveal distinct modes of regulation in Arabidopsis thaliana. Genome Biology 9, R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Gershenzon J, Mitchell-Olds T. 2001. Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knill T, Schuster J, Reichelt M, Gershenzon J, Binder S. 2008. Arabidopsis branched-chain aminotransferase 3 functions in both amino acid and glucosinolate biosynthesis. Plant Physiology 146, 1028–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D. 2004. Naturally occurring genetic variation in Arabidopsis thaliana. Annual Review of Plant Biology 55, 141–172. [DOI] [PubMed] [Google Scholar]
- Lara P, Oñate-Sánchez L, Abraham Z, Ferrándiz C, Díaz I, Carbonero P, Vicente-Carbajosa J. 2003. Synergistic activation of seed storage protein gene expression in Arabidopsis by ABI3 and two bZIPs related to OPAQUE2. Journal of Biological Chemistry 278, 21003–21011. [DOI] [PubMed] [Google Scholar]
- Lehle L, Tanner W. 1973. The function of myo-inositol in the biosynthesis of raffinose. Purification and characterization of galactinol:sucrose 6-galactosyltransferase from Vicia faba seeds. European Journal of Biochemistry 38, 103–110. [DOI] [PubMed] [Google Scholar]
- Li B, Kliebenstein DJ. 2014. The AT-hook motif-encoding gene METABOLIC NETWORK MODULATOR 1 underlies natural variation in Arabidopsis primary metabolism. Frontiers in Plant Science 5, 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zhang Y, Mohammadi SA, Huai D, Zhou Y, Kliebenstein DJ. 2016. An integrative genetic study of rice metabolism, growth and stochastic variation reveals potential C/N partitioning loci. Scientific Reports 6, 30143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Svedin E, Mo H, Atwell S, Dilkes BP, Chapple C. 2014. Exploiting natural variation of secondary metabolism identifies a gene controlling the glycosylation diversity of dihydroxybenzoic acids in Arabidopsis thaliana. Genetics 198, 1267–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Rao J, Shi J, Hu C, Cheng F, Wilson ZA, Zhang D, Quan S. 2014. Seed metabolomic study reveals significant metabolite variations and correlations among different soybean cultivars. Journal of Integrative Plant Biology 56, 826–836. [DOI] [PubMed] [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, et al. 2008. Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. The Plant Journal 53, 960–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR. 2006. Gas chromatography–mass spectrometry-based metabolite profiling in plants. Nature Protocols 1, 387–396. [DOI] [PubMed] [Google Scholar]
- Lisec J, Steinfath M, Meyer RC, Selbig J, Melchinger AE, Willmitzer L, Altmann T. 2009. Identification of heterotic metabolite QTL in Arabidopsis thaliana RIL and IL populations. The Plant Journal 59, 777–788. [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. 2002. Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theoretical and Applied Genetics 104, 1173–1184. [DOI] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Ajjawi I, et al. 2008. New connections across pathways and cellular processes: industrialized mutant screening reveals novel associations between diverse phenotypes in Arabidopsis. Plant Physiology 146, 1482–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Savage LJ, Larson MD, Wilkerson CG, Last RL. 2011. Chloroplast 2010: a database for large-scale phenotypic screening of Arabidopsis mutants. Plant Physiology 155, 1589–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF. 2001. The genetic architecture of quantitative traits. Annual Review of Genetics 35, 303–339. [DOI] [PubMed] [Google Scholar]
- Mackay TF. 2009. Q&A: Genetic analysis of quantitative traits. Journal of Biology 8, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Dupuis J, Sen S, Broman KW. 2006. Poor performance of bootstrap confidence intervals for the location of a quantitative trait locus. Genetics 174, 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichaikul A, Moon JY, Sen S, Yandell BS, Broman KW. 2009. A model selection approach for the identification of quantitative trait loci in experimental crosses, allowing epistasis. Genetics 181, 1077–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Okazaki Y, Oikawa A, et al. 2012. Dissection of genotype–phenotype associations in rice grains using metabolome quantitative trait loci analysis. The Plant Journal 70, 624–636. [DOI] [PubMed] [Google Scholar]
- Melchinger AE, Utz HF, Schön CC. 2004. QTL analyses of complex traits with cross validation, bootstrapping and other biometric methods. Euphytica 137, 1–11. [Google Scholar]
- Meyer RC, Kusterer B, Lisec J, et al. 2010. QTL analysis of early stage heterosis for biomass in Arabidopsis. Theoretical and Applied Genetics 120, 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, et al. 2007. The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 104, 4759–4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill CM, Gill S, Hobbs D, Morgan C, Bancroft I. 2003. Natural variation for seed oil composition in Arabidopsis thaliana. Phytochemistry 64, 1077–1090. [DOI] [PubMed] [Google Scholar]
- Peng C, Uygun S, Shiu SH, Last RL. 2015. The impact of the branched-chain ketoacid dehydrogenase complex on amino acid homeostasis in Arabidopsis. Plant Physiology 169, 1807–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Remington DL, Purugganan MD. 2003. Candidate genes, quantitative trait loci, and functional trait evolution in plants. International Journal of Plant Sciences 164, S7–S20. [Google Scholar]
- Riaño-Pachón DM, Ruzicic S, Dreyer I, Mueller-Roeber B. 2007. PlnTFDB: an integrative plant transcription factor database. BMC Bioinformatics 8, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsheimer C, Lisec J, Czedik-Eysenberg A, et al. 2012. Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proceedings of the National Academy of Sciences, USA 109, 8872–8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riewe D, Jeon HJ, Lisec J, Heuermann MC, Schmeichel J, Seyfarth M, Meyer RC, Willmitzer L, Altmann T. 2016. A naturally occurring promoter polymorphism of the Arabidopsis FUM2 gene causes expression variation, and is associated with metabolic and growth traits. The Plant Journal 88, 826–838. [DOI] [PubMed] [Google Scholar]
- Riewe D, Koohi M, Lisec J, Pfeiffer M, Lippmann R, Schmeichel J, Willmitzer L, Altmann T. 2012. A tyrosine aminotransferase involved in tocopherol synthesis in Arabidopsis. The Plant Journal 71, 850–859. [DOI] [PubMed] [Google Scholar]
- Routaboul JM, Dubos C, Beck G, Marquis C, Bidzinski P, Loudet O, Lepiniec L. 2012. Metabolite profiling and quantitative genetics of natural variation for flavonoids in Arabidopsis. Journal of Experimental Botany 63, 3749–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ. 2008. Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. The Plant Cell 20, 1199–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Matsuda F. 2010. Metabolomics for functional genomics, systems biology, and biotechnology. Annual Review of Plant Biology 61, 463–489. [DOI] [PubMed] [Google Scholar]
- Salathia N, Lee HN, Sangster TA, et al. 2007. Indel arrays: an affordable alternative for genotyping. The Plant Journal 51, 727–737. [DOI] [PubMed] [Google Scholar]
- Sanyal A, Randal Linder C. 2012. Quantitative trait loci involved in regulating seed oil composition in Arabidopsis thaliana and their evolutionary implications. Theoretical and Applied Genetics 124, 723–738. [DOI] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, Pleban T, Zamir D, Fernie AR. 2008. Mode of inheritance of primary metabolic traits in tomato. The Plant Cell 20, 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, et al. 2006. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nature Biotechnology 24, 447–454. [DOI] [PubMed] [Google Scholar]
- Schuster J, Knill T, Reichelt M, Gershenzon J, Binder S. 2006. Branched-chain aminotransferase4 is part of the chain elongation pathway in the biosynthesis of methionine-derived glucosinolates in Arabidopsis. The Plant Cell 18, 2664–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Bernasconi G, Hardtke CS. 2007. Natural genetic variation in Arabidopsis: tools, traits and prospects for evolutionary ecology. Annals of Botany 99, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, ed 1999. Biosynthesis of valine, leucine and isoleucine. In: Plant amino acids: biochemistry and biotechnology. New York: Dekker, 227–247. [Google Scholar]
- Soltis NE, Kliebenstein DJ. 2015. Natural variation of plant metabolism: genetic mechanisms, interpretive caveats, and evolutionary and mechanistic insights. Plant Physiology 169, 1456–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacklies W, Redestig H, Scholz M, Walther D, Selbig J. 2007. pcaMethods—a bioconductor package providing PCA methods for incomplete data. Bioinformatics 23, 1164–1167. [DOI] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. 2005. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. The Plant Cell 17, 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauch RC, Svedin E, Dilkes B, Chapple C, Li X. 2015. Discovery of a novel amino acid racemase through exploration of natural variation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 112, 11726–11731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulpice R, Trenkamp S, Steinfath M, et al. 2010. Network analysis of enzyme activities and metabolite levels and their relationship to biomass in a large panel of Arabidopsis accessions. The Plant Cell 22, 2872–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner W, Kandler O. 1968. Myo-inositol, a cofactor in the biosynthesis of stachyose. European Journal of Biochemistry 4, 233–239. [DOI] [PubMed] [Google Scholar]
- Taylor NL, Heazlewood JL, Day DA, Millar AH. 2004. Lipoic acid-dependent oxidative catabolism of alpha-keto acids in mitochondria provides evidence for branched-chain amino acid catabolism in Arabidopsis. Plant Physiology 134, 838–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Törjék O, Berger D, Meyer RC, et al. 2003. Establishment of a high-efficiency SNP-based framework marker set for Arabidopsis. The Plant Journal 36, 122–140. [DOI] [PubMed] [Google Scholar]
- Törjék O, Witucka-Wall H, Meyer RC, von Korff M, Kusterer B, Rautengarten C, Altmann T. 2006. Segregation distortion in Arabidopsis C24/Col-0 and Col-0/C24 recombinant inbred line populations is due to reduced fertility caused by epistatic interaction of two loci. Theoretical and Applied Genetics 113, 1551–1561. [DOI] [PubMed] [Google Scholar]
- Toubiana D, Semel Y, Tohge T, et al. 2012. Metabolic profiling of a mapping population exposes new insights in the regulation of seed metabolism and seed, fruit, and plant relations. PLoS Genetics 8, e1002612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Li K, Alseekh S, et al. 2015. Genetic determinants of the network of primary metabolism and their relationships to plant performance in a maize recombinant inbred line population. The Plant Cell 27, 1839–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Liu H, Zhou Y, et al. 2016. Combining quantitative genetics approaches with regulatory network analysis to dissect the complex metabolism of the maize kernel. Plant Physiology 170, 136–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell AM, Rowe HC, Hansen BG, Ticconi C, Halkier BA, Kliebenstein DJ. 2007. Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genetics 3, 1687–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MA, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, Doerge RW, St Clair DA. 2007. Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175, 1441–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. 2007. An ‘Electronic Fluorescent Pictograph’ browser for exploring and analyzing large-scale biological data sets. PLoS One 2, e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward AW, Bartel B. 2005. Auxin: regulation, action, and interaction. Annals of Botany 95, 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Alseekh S, Cuadros-Inostroza Á, et al. 2016. Combined use of genome-wide association data and correlation networks unravels key regulators of primary metabolism in Arabidopsis thaliana. PLoS Genetics 12, e1006363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ. 2012. Plant nitrogen assimilation and use efficiency. Annual Review of Plant Biology 63, 153–182. [DOI] [PubMed] [Google Scholar]
- Ying JZ, Shan JX, Gao JP, Zhu MZ, Shi M, Lin HX. 2012. Identification of quantitative trait loci for lipid metabolism in rice seeds. Molecular Plant 5, 865–875. [DOI] [PubMed] [Google Scholar]
- Yu H, Zhang F, Wang G, Liu Y, Liu D. 2013. Partial deficiency of isoleucine impairs root development and alters transcript levels of the genes involved in branched-chain amino acid and glucosinolate metabolism in Arabidopsis. Journal of Experimental Botany 64, 599–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Monroe-Augustus M, Thompson B, Hawes JW, Krukenberg KA, Matsuda SP, Bartel B. 2001. chy1, an Arabidopsis mutant with impaired beta-oxidation, is defective in a peroxisomal beta-hydroxyisobutyryl-CoA hydrolase. Journal of Biological Chemistry 276, 31037–31046. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. 2008. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3, e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.