Abstract
Aims: Bacillithiol (BSH) is utilized as a major thiol-redox buffer in the human pathogen Staphylococcus aureus. Under oxidative stress, BSH forms mixed disulfides with proteins, termed as S-bacillithiolation, which can be reversed by bacilliredoxins (Brx). In eukaryotes, glutaredoxin-fused roGFP2 biosensors have been applied for dynamic live imaging of the glutathione redox potential. Here, we have constructed a genetically encoded bacilliredoxin-fused redox biosensor (Brx-roGFP2) to monitor dynamic changes in the BSH redox potential in S. aureus.
Results: The Brx-roGFP2 biosensor showed a specific and rapid response to low levels of bacillithiol disulfide (BSSB) in vitro that required the active-site Cys of Brx. Dynamic live imaging in two methicillin-resistant S. aureus (MRSA) USA300 and COL strains revealed fast and dynamic responses of the Brx-roGFP2 biosensor under hypochlorite and hydrogen peroxide (H2O2) stress and constitutive oxidation of the probe in different BSH-deficient mutants. Furthermore, we found that the Brx-roGFP2 expression level and the dynamic range are higher in S. aureus COL compared with the USA300 strain. In phagocytosis assays with THP-1 macrophages, the biosensor was 87% oxidized in S. aureus COL. However, no changes in the BSH redox potential were measured after treatment with different antibiotics classes, indicating that antibiotics do not cause oxidative stress in S. aureus.
Conclusion and Innovation: This Brx-roGFP2 biosensor catalyzes specific equilibration between the BSH and roGFP2 redox couples and can be applied for dynamic live imaging of redox changes in S. aureus and other BSH-producing Firmicutes. Antioxid. Redox Signal. 26, 835–848.
Keywords: : Staphylococcus aureus, bacillithiol, bacilliredoxin, redox biosensor, roGFP, oxidative stress
Introduction
Staphylococcus aureus is an opportunistic human pathogen causing not only local skin infections but also life-threatening diseases such as septicemia, endocarditis, and necrotizing pneumoniae (1, 3, 21). The success of the pathogen is mediated by virulence factors and the development of multiple antibiotic-resistant S. aureus strains, such as methicillin-resistant isolates (MRSA) (19). S. aureus has to cope with oxidative stress by reactive oxygen species (ROS), such as hydrogen peroxide (H2O2) and the strong oxidant hypochloric acid by the oxidative burst of macrophages and neutrophils under infection conditions (41).
As defense mechanisms, S. aureus uses various redox-sensing virulence regulators and the thiol-redox buffer bacillithiol (BSH) (5, 13, 20, 28, 30, 32–34). BSH functions in detoxification of ROS, hypochlorite, diamide, methylglyoxal, electrophiles, and antibiotics, such as rifampicin and fosfomycin or heavy metal ions, and protects S. aureus against the oxidative burst by activated macrophages in phagocytosis assays (10, 20, 32, 33). Under hypochlorite stress, BSH forms mixed disulfides with proteins (S-bacillithiolations) as a widespread thiol protection and a redox-switch mechanism that is analogous to S-glutathionylation in eukaryotes (6, 7, 17, 20).
Innovation.
In eukaryotes, glutaredoxin-fused roGFP2 biosensors have been successfully applied for dynamic live imaging of the glutathione redox potential. Here, we have constructed the first genetically encoded bacilliredoxin-fused redox biosensor (Brx-roGFP2) that is specific to measure dynamic changes in the bacillithiol redox potential in the human pathogen Staphylococcus aureus under oxidative stress and infection conditions in vivo. Using this biosensor, we could confirm that different antibiotics do not cause oxidative stress in S. aureus. This Brx-roGFP2 biosensor can be applied to measure redox potential changes across clinical S. aureus isolates and to screen for new redox-active antibiotics to treat methicillin-resistant S. aureus infections.
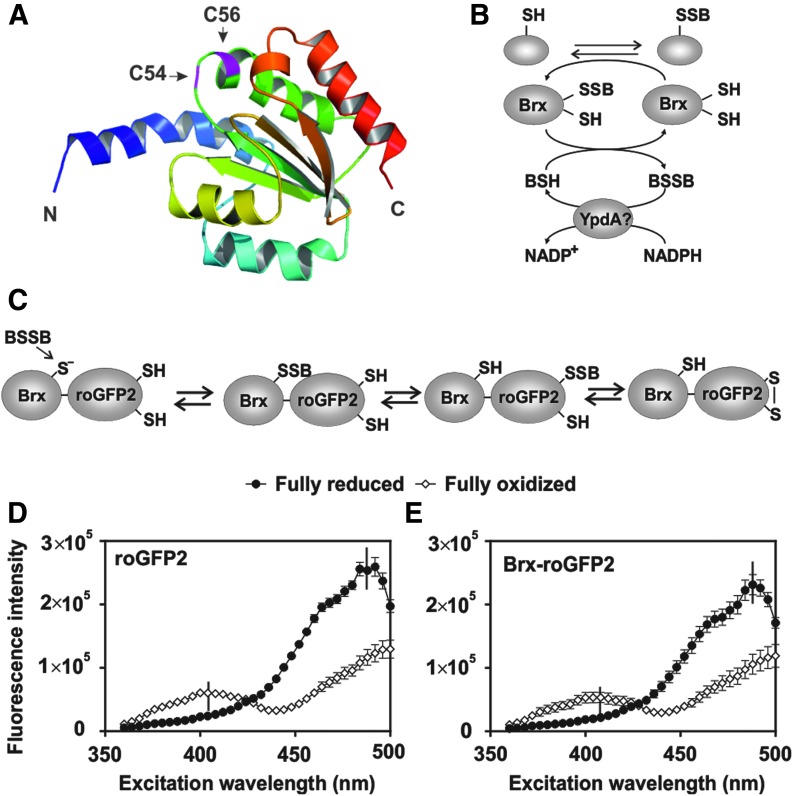
In Bacillus subtilis, two glutaredoxin-like enzymes YphP (BrxA) and YqiW (BrxB) with unusual CGC motifs were characterized as bacilliredoxins (Fig. 1A and Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/ars) that catalyze the reduction of S-bacillithiolated proteins, leading to formation of bacillithiolated Brx (Brx-SSB) as an intermediate of this bacilliredoxin electron pathway (9). Reduction of Brx-SSB requires BSH, resulting in bacillithiol disulfide (BSSB) formation that could be recycled by the putative BSSB reductase YpdA at the expense of nicotinamide adenine dinucleotide phosphate (NADPH) (Fig. 1B) (9, 10, 12, 20).
FIG. 1.
Structure of the bacilliredoxin (Brx) SAUSA300_1321, Brx electron pathway, principle of the Brx-roGFP2 redox pathway, and excitation spectra of Brx-roGFP2 and roGFP2. (A) Bacilliredoxins are Trx-fold proteins of the UPF0403 family with an unusual CGC active-site motif. The structure of Brx (SAUSA300_1321) was generated using the software Phyre2 and PyMol. (B) The S-bacillithiolated proteins are reduced by bacilliredoxins (Brx), leading to Brx-SSB formation. Regeneration of Brx requires BSH and the putative NADPH-dependent BSSB reductase YpdA. (C) In the Brx-roGFP2 fusion, Brx reacts with BSSB, leading to Brx-SSB formation, subsequent transfer of the BSH moiety to the coupled roGFP2, and re-arrangement to the roGFP2 disulfide. The roGFP2 disulfide causes a change of the 405/488 nm excitation ratio. (D, E) Purified roGFP2 and Brx-roGFP2 were fully oxidized and reduced with 5 mM diamide and 10 mM DTT, respectively, and the fluorescence excitation spectra were monitored using the microplate reader (n = 7–9, p < 0.0001 in all samples). In all graphs, mean values are shown, error bars represent the SEM, and p-values are calculated using a Student's unpaired two-tailed t-test by the graph prism software. BSH, bacillithiol; BSSB, bacillithiol disulfide; DTT, dithiothreitol; NADPH, nicotinamide adenine dinucleotide phosphate; SEM, standard error of the mean; Trx, thioredoxin. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
The standard thiol-redox potential of BSH was calculated as E0′(BSSB/BSH) = −221 mV, which is higher than the glutathione redox potential [E0′(GSSG/GSH) = −240 mV] (30, 36). To date, all previous studies have used fluorescent bimane labeling of thiols for quantification of BSH and BSSB levels by high-pressure liquid chromatography under control and stress conditions as an indicator of the changes in the BSH redox potential. According to this method, the BSH/BSSB ratios range from 100:1 to 400:1 in B. subtilis, suggesting that BSH is mostly present in its reduced form (36). Under conditions of S-bacillithiolation provoked by sodium hypochlorite (NaOCl) stress, the level of BSSB increases, indicating a more oxidized BSH redox potential (7). However, the applied methods require disruption of cells and do not allow dynamic measurements of the changes in the BSH redox potential (25, 35).
Thus, recent advances in the design of genetically encoded redox biosensors, such as redox-sensitive green fluorescent proteins (roGFPs), have facilitated the real-time imaging of the cellular redox potential without cell disruption and at high sensitivity under in vivo conditions (11, 24, 35). These roGFP biosensors allow the ratiometric measurements based on two excitation maxima at 405 and 488 nm that change on oxidation (24, 25). RoGFP2 was fused to human glutaredoxin to construct the Grx-roGFP2 biosensor for real-time measurements of dynamic changes in the GSH redox potential (EGSH) in different compartments and different eukaryotic organisms. Grx-roGFP2 detects nanomolar concentrations of GSSG against a backdrop of millimolar GSH within seconds (11, 24).
Recently, roGFP-based biosensors were applied in pathogenic organisms to study EGSH changes under infection conditions and antibiotic treatment, including the malaria parasite Plasmodium falciparum (15) and the Gram-negative bacterium Salmonella Typhimurium (38, 39). In malaria parasites, several antimalarial drugs affected the cellular redox metabolism and showed differential responses of the Grx-roGFP2 biosensor under short- and long-term measurements in vivo (15). In Mycobacterium tuberculosis, an analogous Mrx1-roGFP2 biosensor was developed for dynamic measurements of the mycothiol redox potential (EMSH) in drug-resistant isolates and inside macrophages (2, 24). The Mrx1-roGFP2 biosensor was applied to screen for new ROS-generating anti-TB drugs that affected EMSH (37).
In this study, we have constructed the first bacilliredoxin-fused roGFP biosensor that is highly specific to measure changes of the BSH redox potential in MRSA strains USA300 and COL under oxidative stress and after infection of THP-1 human macrophage-like cells.
Results
Construction of a Brx-roGFP2 biosensor that is highly specific for BSSB
The thioredoxin-fold proteins of the UPF0403 family YphP and YqiW were characterized as bacilliredoxins (BrxA and BrxB) in B. subtilis that function in reduction of S-bacillithiolated proteins and share an unusual CGC active-site motif (Fig. 1A) (9). Both Brx were S-bacillithiolated at their active-site Cys in vivo and in vitro to form Brx-SSB during their catalytic cycle (9). Blast searches identified two bacilliredoxin homologs in S. aureus USA300 (SAUSA300_1321 and SAUSA300_1463). SAUSA300_1321 showed 54% sequence identity with YphP, whereas SAUSA300_1463 shared a stronger sequence identity (68%) with YqiW of B. subtilis (Supplementary Fig. S1). We selected the YphP-homolog SAUSA300_1321 (renamed Brx) for construction of a Brx-roGFP2 fusion protein. S. aureus Brx and Brx Cys-Ala-mutant proteins (BrxA54GC56, BrxC54GA56, and BrxA54GA56) were each fused via the 30aa glycine-serine linker (11) to the N-terminus of roGFP2 to construct the Brx-roGFP2 biosensor variants. Analogous to the reaction of Grx1-roGFP2 and Mrx1-roGFP2 with GSSG or mycothiol disulfide (MSSM) (2, 24), oxidation of the Brx-roGFP2 biosensor should occur specifically by BSSB that targets the active-site Cys54 of Brx to form Brx-SSB. This leads to the transfer of the BSH moiety to the coupled roGFP2, forming S-bacillithiolated roGFP2, which re-arranges to the roGFP2 disulfide and results in ratiometric changes of the excitation maxima at 405 and 488 nm (Fig. 1C).
The His-tagged Brx-roGFP2 protein was expressed in Escherichia coli, purified, and compared with roGFP2 for its ratiometric changes in the fully reduced and oxidized forms using the microplate reader. The thiol-reactive oxidant diamide was used for complete oxidation, and the thiol-reducing compound dithiothreitol (DTT) was applied for complete reduction of the biosensor. Similar to roGFP2, Brx-roGFP2 exhibits two excitation maxima at 405 and 488 nm and responds in a ratiometric manner to 5 mM diamide and 10 mM DTT (2, 24) (Fig. 1D, E). The degree of oxidation (OxD) was calculated according to the fluorescence excitation intensities at 405 and 488 nm of fully oxidized and reduced Brx-roGFP2 probes as previously described (11). For all following measurements, the OxD values of fully reduced and oxidized probes were calibrated as 0 and 1 and the OxD values of the actual measurements were related to these controls.
Furthermore, it was analyzed whether the Brx-roGFP2 response is sensitive to pH changes that could occur during infections. The Brx-roGFP2 probe was diluted into phosphate buffer solutions at different pH values ranging from 5.8 to 8.0 and treated with diamide and DTT (Supplementary Fig. S2). The 405/488 nm excitations ratios were not affected by different pH values, indicating that the probe is insensitive to pH changes.
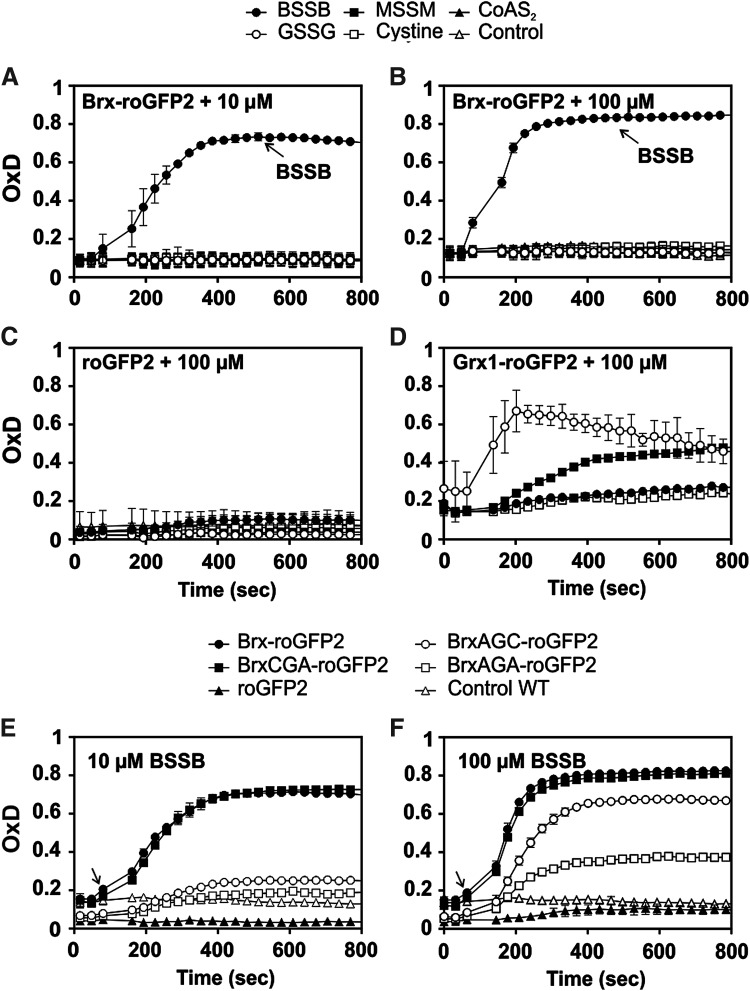
Purified Brx-roGFP2 showed a very fast and specific response to physiological BSSB levels (10–100 μM), but not to other thiol disulfides (GSSG, MSSM, cystine, and CoAS disulfide) (Fig. 2A, B). In contrast, roGFP2 did not respond to 10–100 μM BSSB, confirming that Brx-roGFP2 is specific to detect EBSH changes (Fig. 2C). Furthermore, Grx-roGFP2 was oxidized specifically by 100 μM GSSG, but it was unresponsive to BSSB, indicating that Grx is not specific for the BSH/BSSB redox couple (11) (Fig. 2D).
FIG. 2.
Responses of purified Brx-roGFP2, Grx1-roGFP2, and roGFP2 in vitro to BSSB, GSSG, MSSM, cystine, and CoAS-disulfide. (A, B) Brx-roGFP2 responds specifically to 10 and 100 μM BSSB in vitro but is nonresponsive to GSSG, MSSM, cystine, and CoAS disulfide (n = 3; p < 0.0045 for 10 μM and p < 0.01 for 100 μM in all samples). (C) The roGFP2 probe does not respond to 100 μM thiol disulfides (n = 3; p < 0.0001 for BSSB, GSSG, and MSSM; p = 0.93 for cystine). (D) The Grx1-roGFP2 fusion shows a specific response to 100 μM GSSG, but not to other thiol disulfides (n = 3; p < 0.0033 for all samples). (E, F) The response of Brx-roGFP2 to 10 μM (E) and 100 μM BSSB (F) was compared with the Brx-Cys mutant proteins BrxAGC-roGFP2, BrxCGA-roGFP2, BrxAGA-roGFP2, and roGFP2 (n = 3; p = 0.27 for 10 μM BSSB BrxAGA; p < 0.0001 for all others). Arrows denote the time point of oxidant exposure. The results showed that the Brx-roGFP2 response depends on the active-site Cys54 of Brx. The thiol disulfides were injected into the microplate wells 90 s after the start of measurements, and the biosensor response and OxD were analyzed using the CLARIOstar microplate reader. In all graphs, mean values are shown, error bars represent the SEM, and p-values are calculated using a Student's unpaired two-tailed t-test by the graph prism software. GSSG, oxidized glutathione disulfide; MSSM, mycothiol disulfide; OxD, oxidation degree.
The specificity of Brx for BSSB should be determined by the C54GC56 active-site motif. Hence, the response of Brx-roGFP2 was compared with that of Brx Cys mutant roGFP2 fusions, where the active-site Cys54 and the resolving Cys56 of Brx are each replaced by an alanine (BrxAGC, BrxCGA, and BrxAGA). Brx-roGFP2 and the resolving Brx Cys56 mutant (BrxCGA-roGFP2) showed very fast oxidation by 10–100 μM BSSB, whereas the BrxAGC-roGFP2 and BrxAGA-roGFP2 active-site mutant proteins failed to respond to 10 μM BSSB and showed weaker responses to 100 μM BSSB (Fig. 2E, F). In previous studies, the BSH content was determined as 0.5–1.6 μmol/g raw dry weight in BSH-producing S. aureus strains (36). This equates to 0.5–1.6 mM intracellular BSH, assuming a 50% water content of the cellular biomass as determined for related bacteria (4). The BSH/BSSB ratio was estimated as 1:20–1:40 (28, 32), indicating that the physiological BSSB content should be below 100 μM in S. aureus. Thus, the Brx-roGFP2 biosensor is highly specific to detect physiological levels of 10–100 μM BSSB in vitro.
Response of Brx-roGFP2 in S. aureus MRSA strains COL and USA300 along the growth curve and effect of BSH deficiency on the biosensor response
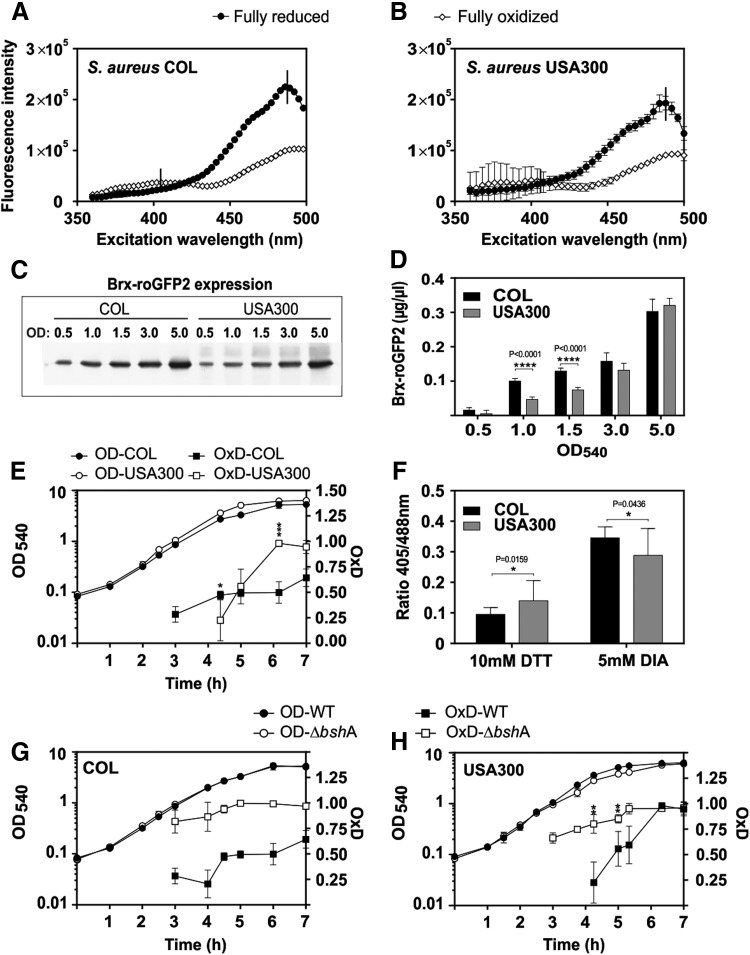
To monitor the changes in EBSH inside S. aureus, Brx-roGFP2 was expressed ectopically from plasmid pRB473 under control of a xylose-inducible promoter in two MRSA isolates COL and USA300. The S. aureus wild-type strains were grown in Luria Bertani (LB) medium overnight with xylose, and the strong roGFP2 fluorescence could be monitored using the microplate reader and fluorescence microscopy. First, we confirmed the ratiometric response of the Brx-roGFP2 biosensor inside S. aureus cells in the fully oxidized and reduced state after treatment with 5 mM diamide and 10 mM DTT, respectively, and monitored the changes at the 405 and 488 nm excitation maxima (Fig. 3A, B). Western blot analyses confirmed that Brx-roGFP2 is expressed as a full-length protein and is not degraded during the growth in S. aureus (Supplementary Fig. S3).
FIG. 3.
Comparison of the Brx-roGFP2 response and expression in S. aureus COL and USA300 wild type and bshA mutants during the growth. (A, B) Ratiometric response of Brx-roGFP2 in S. aureus COL and USA300 after oxidation of cells by 5 mM diamide and reduction by 10 mM DTT (n = 7–8; p < 0.0001 in all samples). (C, D) Expression of Brx-roGFP2 is higher in S. aureus COL compared with USA300 during the log phase (n = 3; p < 0.0001 at OD1–1.5), and (E) the OxD is the most strongly increased during the stationary phase in USA300 (n = 3; p = 0.0009 at 6 h). (F) The dynamic range of Brx-roGFP2 is higher in S. aureus COL compared with USA300 (n = 3; p = 0.0159 DTT; p = 0.0436 diamide). (G, H) The Brx-roGFP2 biosensor is constitutively oxidized in the S. aureus COL and USA300 bshA mutants in comparison to the wild-type strains (n = 3; p < 0.0001 in COL at all time points; p < 0.01 in USA300 at 4–5.5 h). Symbols are defined as follows: nsp > 0.05; *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; and ****p ≤ 0.0001. The OxD was calculated based on 405/488 nm excitation ratios with emission at 510 nm and related to the fully oxidized and reduced controls as described in the Materials and Methods section. In all graphs, mean values are shown, error bars represent the SEM, and p-values are calculated using a Student's unpaired two-tailed t-test by the graph prism software.
Next, the changes of the BSH redox potential were monitored during different stages of growth in LB medium in S. aureus (Fig. 3C–E). In fact, the expression and fluorescence intensity of Brx-roGFP2 varied between the S. aureus COL and USA300 wild-type strains along the growth. Although a significant Brx-roGFP2 fluorescence signal was detected in S. aureus COL already during the exponential growth, the expression and fluorescence of Brx-roGFP2 were much weaker in S. aureus USA300 during the log phase (Fig. 3C, D and Supplementary Fig. S4A, B). In both strains, the OxD of the Brx-roGFP2 increased during the stationary phase, reflecting growth-dependent redox changes (Fig. 3E). Furthermore, we observed that the dynamic range of the 405/488 nm ratios of fully reduced and oxidized Brx-roGFP2 was higher in COL (3.77 ± 0.65) compared with USA300 (2.37 ± 0.98) (Fig. 3F).
However, we confirmed that the level of Brx-roGFP2 did not affect the OxD, since serial dilutions of S. aureus cells that were harvested at an optical density (OD) of 4.0 showed the same OxD level (Supplementary Fig. S4C, D). Based on the OxD of the biosensor, the intracellular BSH redox potential was calculated using the Nernst' equation, ranging from −300 to −270 mV in COL and from −300 to −235 mV in USA300 during exponential growth until transition into the stationary phase (Supplementary Table S5).
We further compared biosensor oxidations between COL and USA300 wild types and isogenic BSH-deficient mutants. The biosensor was fully oxidized in the COL and USA300 bshA mutants, indicating an impaired redox balance and increased oxidative stress in the bshA mutant (Fig. 3G, H). This constitutive biosensor oxidation was also observed in strain RN4220, which is a natural bshC mutant of the S. aureus NCTC8325 lineage (27, 32, 33) (Supplementary Fig. S5). Western blot analyses confirmed that the biosensor is similarly expressed in the COL wild type and bshA mutant (Supplementary Fig. S6).
Response of Brx-roGFP2 in S. aureus COL to oxidative stress
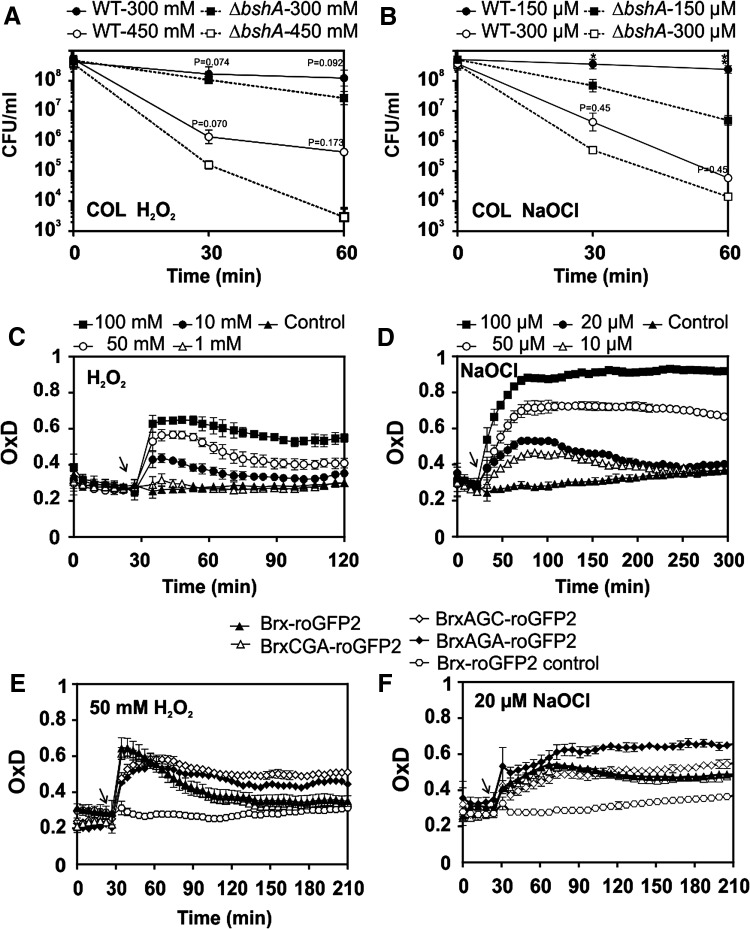
The changes in EBSH were further investigated in S. aureus COL wild type in response to oxidative stress, provoked by H2O2 and NaOCl. Previous studies have shown that NaOCl stress leads to S-bacillithiolation of proteins and a decreased BSH/BSSB redox ratio (7). Since S. aureus is extremely resistant to H2O2 (14, 40), the physiological sub-lethal concentrations of oxidants were determined (Fig. 4A, B). In addition, the role of BSH in the resistance to NaOCl and H2O2 was analyzed in survival assays. The bshA mutant was more sensitive than the wild type to 150 μM NaOCl and 300 mM H2O2, indicating that BSH contributes to oxidative stress resistance.
FIG. 4.
Effect of NaOCl and H2O2 on the survival of S. aureus wild type and bshA mutants and oxidative stress responses of Brx-roGFP2 and Brx Cys-mutant fusions in S. aureus COL. (A, B) The S. aureus COL and USA300 bshA mutants are more sensitive to NaOCl and H2O2 stress compared with the wild type as revealed by survival assays (n = 3; p = 0.07 at 450 mM H2O2; p = 0.0126 at 150 μM NaOCl). (C, D) The Brx-roGFP2 biosensor in S. aureus COL is rapidly and reversibly oxidized by sub-lethal concentrations of 1–50 mM H2O2 and 10–20 μM NaOCl, whereas higher doses result in constitutive biosensor oxidation (n = 4; p = 0.3186 at 1 mM H2O2; p < 0.0001 in all other samples). (E, F) The response of Brx-roGFP2 and BrxCGA-roGFP2 to H2O2 and NaOCl is reversible compared with the active Cys Brx mutant fusions in S. aureus COL, which is not reversible (n = 5; p < 0.0001 in all samples). Arrows denote the time point of oxidant exposure. In all graphs, mean values are shown, error bars represent the SEM, and p-values are calculated using a Student's unpaired two-tailed t-test by the graph prism software. H2O2, hydrogen peroxide; NaOCl, sodium hypochlorite.
The change in EBSH after exposure to 1–100 mM H2O2 and 10–100 μM NaOCl and the time for detoxification of oxidants and recovery of the reduced state were investigated. The biosensor was rapidly and reversibly oxidized by 10–50 mM H2O2, and cells approached a more reduced state within 70 min (Fig. 4C). Remarkably, 100 mM H2O2 did not result in complete oxidation of the biosensor (e.g., OxD = 1), although S. aureus cells were unable to restore their reduced state. Treatment of cells with low doses of 10–20 μM NaOCl resulted in reversible biosensor oxidation and required 200 min for regeneration of the reduced state. The biosensor was fully oxidized by 100 μM NaOCl, but cells were unable to recover (Fig. 4D). Using nonreducing BSH-specific Western blot analysis, we further confirmed that 100 μM NaOCl stress leads to the same increase of S-bacillithiolated proteins in S. aureus COL and COL Brx-roGFP2 strains, indicating that Brx-roGFP2 expression did not affect the S-bacillithiolation pattern (Supplementary Fig. S7).
To analyze the impact of Brx in the biosensor response to the oxidants in vivo, attempts were made to express unfused roGFP2 in S. aureus COL. However, in contrast to the Brx-roGFP2 fusion, expression of unfused roGFP2 failed in S. aureus COL containing the pRB473-roGFP2 plasmid. Thus, we compared the biosensor responses of the various Brx Cys mutant fusions (BrxAGC, BrxCGA, and BrxAGA) with the different oxidants in vivo. The response of Brx-roGFP2 and BrxCGA-roGFP2 to 50 mM H2O2 was fast and reversible, with recovery of the reduced state after 120 min. In contrast, oxidation of the Brx active-site Cys mutant fusions (BrxAGC and BrxAGA) by H2O2 was slower and not fully reversible (Fig. 4E). We further analyzed the responses of the BrxAGC, BrxCGA, and BrxAGA Cys mutant fusions to 20 μM NaOCl. All Brx Cys mutants responded similarly to NaOCl, whereas the BrxAGA double-Cys mutant was unable to recover after 120 min (Fig. 4F). This indicates that a direct response of Brx-roGFP2 to high doses of 50 mM H2O2 and to the strong oxidant NaOCl could, in part, account for the oxidant responses in the absence of the CGC motif of Brx.
To monitor the direct biosensor response to the oxidants, both purified roGFP2 and Brx-roGFP2 proteins were treated with different concentrations of H2O2 and NaOCl (Supplementary Fig. S8). The results showed that both roGFP2 and Brx-roGFP2 respond strongly to 1–10 mM H2O2 and 50–100 μM NaOCl, leading to complete biosensor oxidation. This confirms that the Brx-roGFP2 biosensor could also directly respond to high H2O2 doses and the strong oxidant NaOCl in vivo in the absence of the Brx CGC motif. Another possibility could be that the third conserved Cys of Brx at the C-terminus (Cys144) is S-bacillithiolated by BSSB in vivo in the absence of the CGC motif, leading to subsequent biosensor oxidation.
Confocal laser scanning microscopy of Brx-roGFP2 fluorescence in S. aureus COL wild type and ΔbshA mutant cells
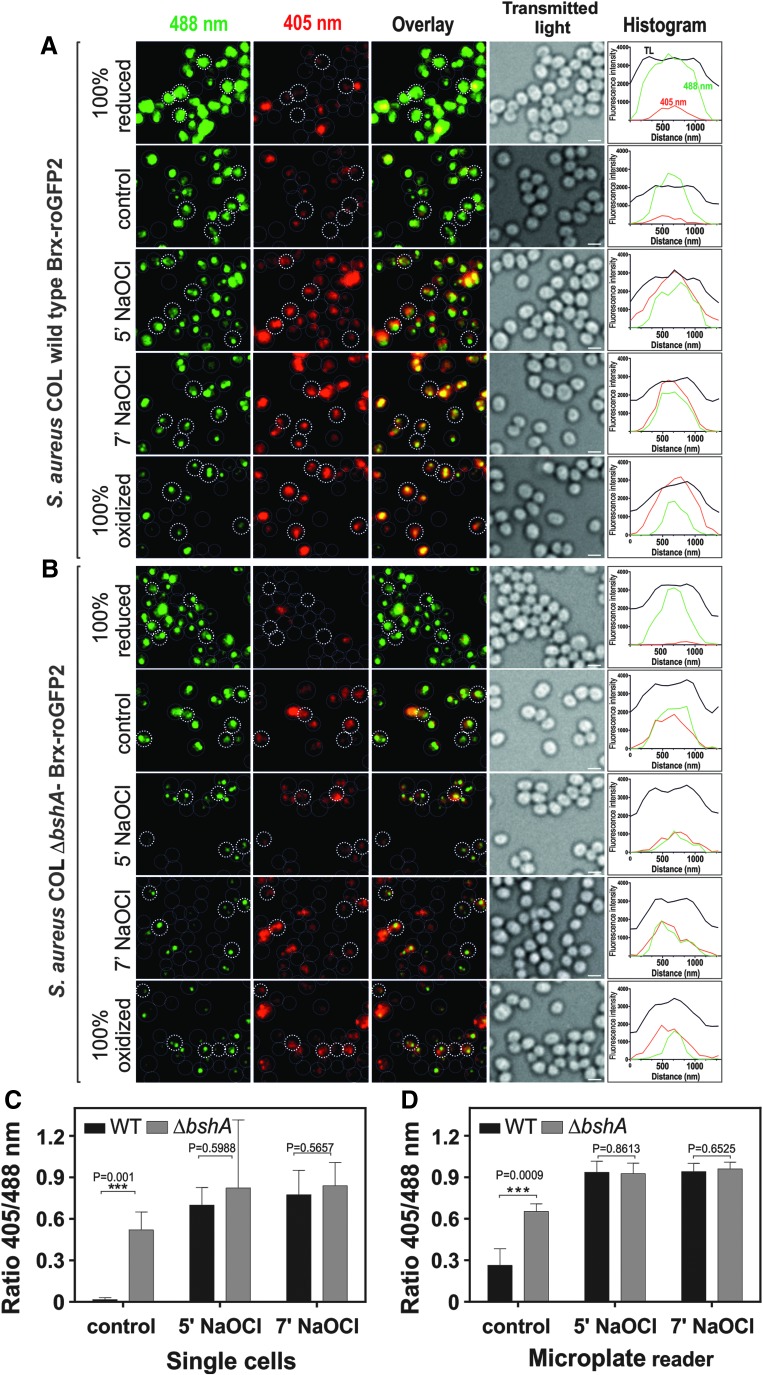
The redox-dependent changes of Brx-roGFP2 fluorescence in S. aureus COL wild type and bshA mutant cells were analyzed using confocal laser scanning microscopy (CLSM) both before and after NaOCl stress. The biosensor fluorescence intensities were measured after excitation at 405 and 488 nm and false-colored in red and green, respectively (Fig. 5). The oxidation state of the biosensor is visualized by an overlay of both red and green images. Confocal imaging showed that Brx-roGFP2 is reduced in nontreated wild-type cells and resembles that of DTT-treated reduced cells with bright fluorescence at the 488 nm excitation wavelength (Fig. 5). NaOCl stress leads to a decreased fluorescence intensity at the 488 nm excitation maximum and a strongly increased fluorescence at the 405 nm excitation maximum, as shown in the histograms. Thus, NaOCl-treated wild-type cells are visualized as red cells in the overlay images similar to the fully oxidized wild-type control. In the S. aureus COL bshA mutant, the Brx-roGFP2 biosensor was fully oxidized under control conditions as shown by the strong fluorescence in the 405 nm (red) channel that resembles the NaOCl-treated sample. The redox states of wild type and bshA mutant cells were calculated from five representative single cells and also using measurements in the microplate reader for comparison (Fig. 5C, D).
FIG. 5.
Live-imaging of S. aureus COL Brx-roGFP2 in the wild type and the bshA mutant during NaOCl stress. (A, B) S. aureus COL Brx-roGFP2 wild type and bshA mutant strains were exposed to 150 μM NaOCl, blocked with NEM, and analyzed using CLSM. Cells treated with 10 mM DTT and 5 mM diamide were used as fully reduced and oxidized controls, respectively. Fluorescence emission was measured at 505–550 nm after excitation at 405 and 488 nm. Fluorescence intensities at the 488 and 405 nm excitation maxima are false-colored in green and red, respectively, and are shown in the overlay images and histograms for single cells. The NaOCl-induced oxidation in the wild type and constitutive oxidation of the biosensor in the ΔbshA mutant are visualized by the overlay images, and the cells are encircled based on the transmitted light image. The histograms show average fluorescence intensities at 405 and 488 nm calculated from five representative single cells. (C) The average 405/488 nm ratios of the wild type and bshA mutant samples were calculated from five representative single cells each for the wild type and bshA mutant that are marked with bold circles (n = 5; p = 0.001 for control WT/bshA). (D) For comparison, the 405/488 nm ratios were also calculated from the same S. aureus samples using the microplate reader (n = 3; p = 0.0009 for control WT/bshA). Symbols are defined as follows: ***p < 0.001. In all graphs, mean values are shown, error bars represent the SEM, and p-values are calculated using a Student's unpaired two-tailed t-test by the graph prism software. CLSM, confocal laser scanning microscopy; NEM, N-ethylmaleimide. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Response of Brx-roGFP2 in S. aureus after internalization by THP-1 macrophages
Next, we measured the changes in EBSH of S. aureus under infection-like conditions during phagocytosis assays with activated THP-1 macrophages. Infection assays were performed with S. aureus COL cells at a multiplicity of infection (MOI) of 25, and fluorescence excitation intensities were monitored at 405 and 488 nm. After 1 h of infection with S. aureus COL, about 70–80% of THP-1 cells showed a green fluorescence. As fully oxidized and reduced controls, infected macrophages were treated with 150 μM NaOCl and 20 mM DTT, respectively, and the mean fluorescence intensity (MFI) at 405 and 488 nm was analyzed using flow cytometry (Supplementary Table S7). The 405/488 nm ratio of the MFI of fully reduced and oxidized THP-1 controls was calibrated to 0% and 100% oxidation and related to the 405/488 nm ratio of the MFI of infected macrophages. In comparison to these fully reduced and oxidized THP-1 controls, the biosensor was 87% oxidized in S. aureus COL during infection after uptake by macrophage-like cells (Supplementary Table S7).
Effects of antibiotics on the Brx-roGFP2 biosensor response in S. aureus COL
We were interested in the changes of the BSH redox potential in response to sub-lethal antibiotics that are commonly used to treat MRSA infections. The aim was to clarify a long debate about the involvement of oxidative stress in the killing mode of antibiotics (16, 18). We have chosen antibiotics with different cellular target sites, including RNA polymerase inhibitors (rifampicin), cell-wall biosynthesis inhibitors (fosfomycin, ampicillin, oxacillin, and vancomycin), aminoglycosides as protein biosynthesis inhibitors that target the ribosome (gentamycin, lincomycin, erythromycin, and linezolid), and fluoroquinolones (ciprofloxacin) that inhibit the DNA gyrase and topoisomerase IV to block DNA replication.
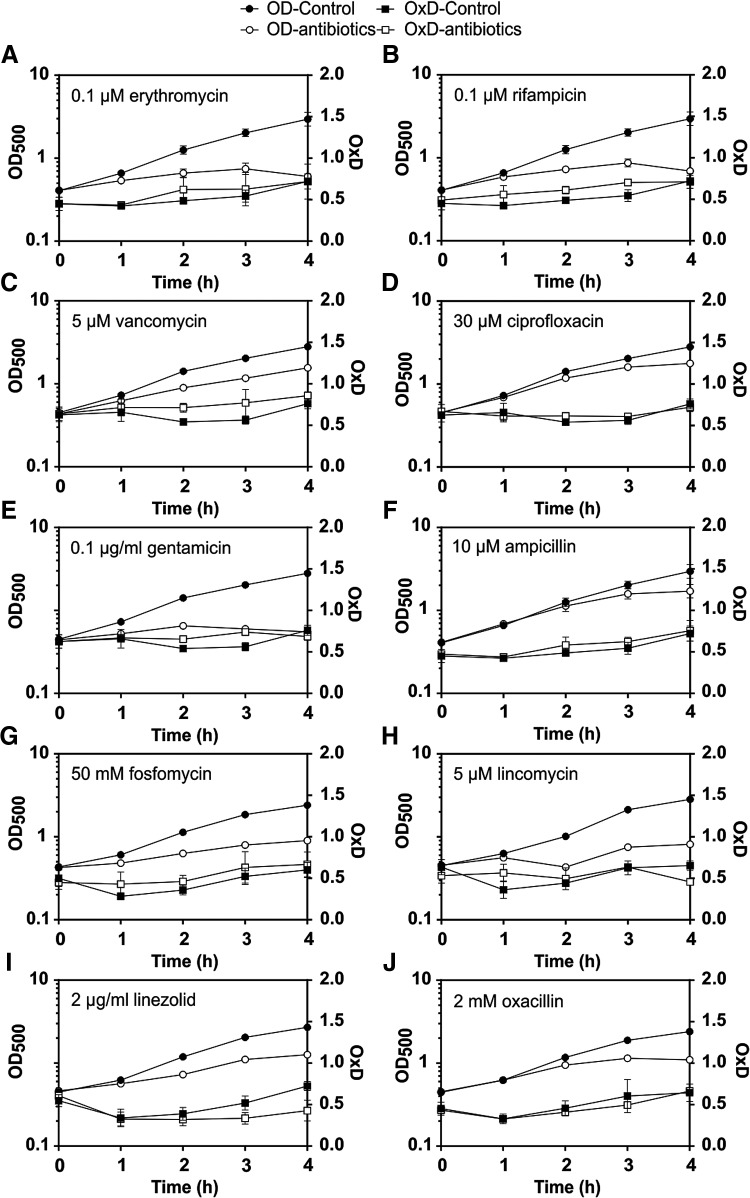
S. aureus COL with Brx-roGFP2 was treated with sub-lethal concentrations of antibiotics and analyzed for its biosensor response. The sub-lethal antibiotics doses that caused a reduced growth rate were determined as 0.1 μM erythromycin, 0.1 μM rifampicin, 5 μM vancomycin, 30 μM ciprofloxacin, 0.5 μg/ml gentamicin, 10 μM ampicillin, 50 mM fosfomycin, 5 μM lincomycin, 2 μg/ml linezolid, and 2 mM oxacillin. The measurement of the Brx-roGFP2 response revealed no increased biosensor oxidation by any of these antibiotics compared with the untreated control (Fig. 6 and Supplementary Table S6). These results document that sub-lethal antibiotics do not cause changes in the BSH redox potential in S. aureus.
FIG. 6.
Growth curves and OxD of S. aureus COL Brx-roGFP2 after exposure to sub-lethal concentrations of antibiotics that decreased the growth rate. S. aureus was exposed to sub-lethal concentrations of antibiotics at an OD500 of 0.5, and the OxD of Brx-roGFP2 was monitored in treated and untreated cells (n = 4; p > 0.05 for OxD control/antibiotics treatment in all samples). The following sub-lethal antibiotics were used: (A) 0.1 μM erythromycin, (B) 0.1 μM rifampicin, (C) 5 μM vancomycin, (D) 30 μM ciprofloxacin, (E) 0.1 μM gentamycin, (F) 10 μM ampicillin, (G) 50 mM fosfomycin, (H) 5 μM lincomycin, (I) 2 μg/ml linezolid, and (J) 2 mM oxacillin. There was no increased oxidation by antibiotics in S. aureus. In all graphs, mean values are shown, error bars represent the SEM, and p-values are calculated using a Student's unpaired two-tailed t-test by the graph prism software.
Discussion
Redox-sensitive GFPs have been recently fused to glutaredoxin and mycoredoxin for dynamic measurements of the intracellular redox potential in eukaryotes and mycobacteria in real time and at high sensitivity and spatiotemporal resolution (35). Here, we coupled the bacilliredoxin (Brx) to roGFP2 to measure the intracellular BSH redox potential in S. aureus cells under infection conditions, ROS, and antibiotics treatments. This Brx-roGFP2 biosensor is highly sensitive and specific for physiological levels of BSSB, whereas unfused roGFP2 and the BrxAGC active-site mutant roGFP2 fusion are much lower responsive to BSSB in vitro.
The role of the active-site Cys of Brx for reduction of S-bacillithiolated OhrR and MetE has been previously shown for B. subtilis BrxA and BrxB (9). The specific reaction of the active site Cys54 with BSSB was verified here for the S. aureus Brx homolog SAUSA300_1321 in the Brx-roGFP2 fusion. Thus, coupling of roGFP2 with the Brx facilitates rapid equilibration of the biosensor with the BSH/BSSB redox pair to selectively measure changes in the BSH redox potential. However, weaker responses of the BrxAGC and BrxAGA active site and double mutants were observed by 100 μM BSSB, which could depend on the third C-terminal Cys144 residue that is also conserved across the UPF0403 family of Brx-homologs (Supplementary Fig. S1).
The Brx-roGFP2 biosensor was applied to monitor the changes in the BSH redox potential inside the archaic hospital-acquired MRSA isolate COL and the community-acquired MRSA strain USA300. In both MRSA strains, BSH is required for survival under oxidative stress and infection-related conditions during phagocytosis with macrophages (32, 33). Here, we confirmed that BSH-deficient mutants of S. aureus COL and USA300 are more sensitive to H2O2 and NaOCl compared with the wild types. We further monitored the perturbations in the BSH redox potential during growth, oxidative challenge, and infection assays with THP-1 macrophages. Increases in EBSH were observed in S. aureus COL and USA300 strains during the stationary phase in LB medium compared with the log phase. However, we confirmed that the expression level of Brx-roGFP2 did not affect the OxD (Supplementary Fig. S4C, D).
The dynamic range of Brx-roGFP2 was lower in USA300 compared with COL, which could depend on the 1.6-fold higher BSH levels in USA300 (32). Differences in the basal level oxidation and dynamic range were also observed between drug-sensitive (3D7) and -resistant (Dd2) P. falciparum parasites (15). In P. falciparum 3D7, the lower basal OxD of the biosensor could be explained by the higher GSH levels. Thus, the higher BSH levels in USA300 could result in a higher reducing capacity and a lower biosensor response to diamide.
We further showed that the Brx-roGFP2 biosensor in S. aureus COL responds rapidly to ROS, such as H2O2 and NaOCl. However, S. aureus is resistant to high levels of 300 mM H2O2 (40). Thus, treatment of S. aureus with 1–10 mM H2O2 resulted in only a slightly increased biosensor oxidation with rapid regeneration of the reduced state. Exposure to 50–100 μM NaOCl stress caused the complete and constitutive oxidation of the biosensor and correlates with the observed S-bacillithiolation of proteins and increased levels of BSSB under NaOCl stress in S. aureus (7). The comparison of the biosensor response of Brx-roGFP2 with that of the Brx Cys mutant fusion revealed a similar response but changes in the recovery after H2O2 and NaOCl stress, which was impaired in the Brx active-site mutants. Thus, the biosensor could also directly respond to the high H2O2 levels and the strong oxidant NaOCl in vivo.
The changes in BSH redox potential were also measured inside S. aureus COL during phagocytosis assays in THP-1 macrophage cell lines. The flow cytometric data showed that the Brx-roGFP2 biosensor was 87% oxidized in S. aureus COL under infection conditions.
The comparison of the biosensor response in S. aureus COL and USA300 wild types and the isogenic bshA mutants as well as in the natural bshC-deficient strain RN4220 revealed a constitutive oxidation of the probe in the absence of BSH. This high biosensor oxidation in BSH-deficient strains is also visualized by confocal imaging at the cellular level. These results are in agreement with the constitutive oxidation of the Grx-roGFP2 biosensor on GSH depletion in Arabidopsis thaliana seeds (23). Thus, our data clearly document the impaired redox balance in the absence of BSH and the major role of BSH in keeping the reduced state of the cytoplasm in S. aureus cells. In addition to BSH depletion, much lower NADPH levels were previously measured in the ΔbshA mutant, which could contribute to the impaired thiol-redox balance (32).
In S. aureus, coenzyme A and Cys were suggested to function as alternative thiol-redox buffers and S. aureus also encodes a CoAS disulfide reductase to keep CoASH in the reduced state (22). However, based on the microscopic and macroscopic pKa values of BSH, the level of the reactive thiolate anion is much higher in BSH compared with CoASH and Cys at physiological pH values (36). Thus, BSH is the only available nucleophilic thiol that reacts with protein disulfides that are formed under oxidative stress in S. aureus (29, 30, 36). Consistent with this notion, neither cystine nor CoAS disulfide were recognized by Brx at physiological concentrations to oxidize the biosensor in vitro.
Previous studies identified the OhrR repressor as redox controlled under organic peroxide and NaOCl stress by S-cysteinylation and S-bacillithiolation (6, 17). BrxA and BrxB were specific for reduction and reactivation of S-bacillithiolated OhrR, but they could not regenerate S-cysteinylated OhrR (9). These results further support our findings about the specificity of Brx of S. aureus for BSSB.
Finally, we studied the changes in the BSH redox potential in S. aureus after treatment with sub-lethal doses of different antibiotics to clarify the role of oxidative stress as a killing mode for antibiotics, a controversial debate among microbiologists (16, 18). However, we could not detect changes in the BSH redox potential after treatment with rifampicin, fosfomycin, ampicillin, oxacillin, vancomycin, aminoglycosides, and fluoroquinolones. Similar to our results, no roGFP2 response was detected by any of these antibiotics in Salmonella (39). However, S. aureus is resistant to 100 mM H2O2 without the killing effect. Thus, S. aureus might be resistant to ROS produced under antibiotics treatment.
In conclusion, we have constructed a novel Brx-roGFP2 biosensor that is highly specific to sense the reduced pool of BSH inside S. aureus cells and that responds to oxidative stress under infection-like conditions inside macrophages. Using this novel tool, we could demonstrate that commonly used antibiotics do not cause oxidative stress when applied to S. aureus and that BSH-deficient mutants have an impaired redox balance and reduced virulence. S. aureus is an important human pathogen with new MRSA strains and other multiple antibiotic-resistant isolates emerging quickly. This novel probe can be applied in drug research to screen for new redox-active antibiotics that affect the BSH redox potential in S. aureus. In addition, the difference in the ROS detoxification capacity and resistance to host defenses can be monitored across emerging MRSA isolates to understand the connection between virulence factor expression, antibiotics resistance, and the BSH redox potential in S. aureus.
Materials and Methods
Bacterial strains, growth conditions, stress, and antibiotics treatments
Bacterial strains, plasmids, and primers are listed in Supplementary Tables S1–S3. For cloning and genetic manipulation, E. coli was cultivated in LB medium. S. aureus strains with the pRB473-XylR-Brx-roGFP2 plasmids were cultivated in LB medium with 1% xylose to ensure constitutive expression of the biosensor. For stress experiments, S. aureus cells were grown in LB until an optical density at 540 nm (OD540) of 1.0 and were transferred to Belitsky minimal medium (BMM) with 1% xylose. The fully reduced control cells were treated with 10 mM DTT and the fully oxidized control was treated with 5 mM diamide for 20 min each, harvested with 10 mM N-ethylmaleimide (NEM) to block the biosensor redox state, and transferred to the microplate wells. The samples for stress exposure were transferred to the microplates, and different oxidants were injected into the wells of microplates. The Brx-roGFP2 biosensor fluorescence emission was measured at 510 nm after excitation at 405 and 488 nm using the CLARIOstar microplate reader (BMG Labtech) as described next for the in vitro measurements. Three biological tests were performed for each stress experiment.
For the survival assays, S. aureus COL wild type and the bshA mutant were treated with NaOCl and H2O2 at an OD500 of 1.0 in BMM and serial dilutions were plated on LB agar plates and counted for colony-forming units. The survival assays were performed in three biological replicates for each strain.
For the determination of the growth-inhibitory and sub-lethal antibiotics concentrations, S. aureus was cultivated in RPMI medium and the antibiotics erythromycin, rifampicin, vancomycin, ciprofloxacin, gentamicin, ampicillin, fosfomycin, lincomycin, linezolid, or oxacillin were added at an OD500 of 0.5 to monitor the reduction in growth as previously described (8). The measurements of the biosensor responses after antibiotics treatment were performed in S. aureus Brx-roGFP2 cells that were grown in RPMI medium and treated with sub-lethal antibiotics doses that reduced the growth rate. Cells were harvested after different times of antibiotics treatment, washed with phosphate-buffered saline (PBS), and blocked with NEM before the microplate reader measurements. Four biological replicates were performed for each antibiotics stress experiment. Sodium hypochlorite, diamide, DTT, H2O2 (35% w/v), and antibiotics (erythromycin, rifampicin, vancomycin, ciprofloxacin, gentamicin, ampicillin, fosfomycin, lincomycin, linezolid, and oxacillin) were purchased from Sigma-Aldrich.
Construction, expression, and purification of Brx-roGFP2 in E. coli
The Grx-roGFP2 containing plasmid pQE60-Grx1-roGFP2 was obtained from Tobias Dick and colleagues (11) and used as a template for construction of the Brx-roGFP2 fusion. The brx gene (SAUSA300_1321) was amplified from chromosomal DNA of S. aureus USA300 by polymerase chain reaction (PCR) using primers SAUSA300-1321yphP-FOR-BamHI-NcoI and SAUSA300-1321yphP-REV-SpeI (Supplementary Table S3), digested with BamHI and SpeI, and inserted into plasmid pQE60-Grx1-roGFP2 that was digested using the same restriction enzymes to generate plasmid pQE60-Brx-roGFP2. The brx-roGFP2 sequence was amplified from plasmid pQE60-Brx-roGFP2 by PCR using primers 1321-roGFP2-FOR-NheI and roGFP2-REV-BamHI, digested with NheI and BamHI, and sub-cloned into pET11b (Novagen) after digestion by the same enzymes to generate plasmid pET11b-Brx-roGFP2.
For construction of the roGFP2 fusions with the Brx-Cys-to-Ala variants, the Cys residues of the C54GC56 active site were replaced by alanine using PCR mutagenesis. For the brxC54A mutant, two first-round PCR reactions were performed using primers SAUSA300-1321yphP-FOR-BamHI-NcoI and SAUSA300-1321-yphP-C56A-REV and primers SAUSA300-1321-yphP-C54A-FOR and SAUSA300-1321yphP-REV-SpeI. For the brxC56A mutant, two first-round PCR reactions were performed using primers SAUSA300-1321yphP-FOR-BamHI-NcoI and SAUSA300-1321-yphP-C56A-REV and primers SAUSA300-1321-yphP-C56A-FOR and SAUSA300-1321yphP-REV-SpeI. The two PCR products of each first-round PCR reaction were hybridized and, subsequently, amplified by a second-round PCR reaction using primers SAUSA300-1321yphP-FOR-BamHI-NcoI and SAUSA300-1321yphP-REV-SpeI. The PCR products from the second-round PCRs were then digested with BamHI and SpeI and inserted into plasmid pQE60-Grx1-roGFP2 that was digested with the same enzymes. Sub-cloning of the Brx-Cys-to-Ala mutant roGFP2 fusions into pET11b was performed as described earlier.
To construct the brxC54A-C56A double mutant, first-round PCR was performed using primers 1321-roGFP2-FOR-NheI and 1321-brx-C54A56A-REV and primers 1321-brx-C54A56A-FOR and SAUSA300-1321yphP-REV-SpeI. Then, the PCR products from first-round PCR reactions were fused by PCR using primers 1321-roGFP2-FOR-NheI and SAUSA300-1321yphP-REV-SpeI. The PCR product from the second-round PCR was then digested with NheI and SpeI and inserted into plasmid pET11b-Brx-roGFP2 that was digested with the same enzymes.
To construct plasmid pET11b-roGFP2, primers roGFP2-FOR-NheI and roGFP2-REV-BamHI were used to amplify roGFP2 from plasmid pQE60-Grx1-roGFP2. The PCR product was digested with NheI and BamHI and inserted into plasmid pET11b that was digested with the same enzymes. The correct sequences of all plasmid inserts were confirmed by PCR amplification and sequencing.
For Brx-roGFP2 expression, E. coli BL21(DE3) plysS containing the pET11-Brx-roGFP2 plasmids was grown in 1 L LB medium and 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added at the exponential phase (OD600 of 0.8) for 16 h at 25°C. Recombinant His-Brx-roGFP2 and the Brx-Cys-to-Ala mutant roGFP2 fusion proteins were purified using PrepEase His-tagged high-yield purification resin (USB) under native conditions according to the manufacturer's instructions (USB). The purified proteins were extensively dialyzed against 10 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 50% glycerol and stored at −80°C.
Microplate reader measurements of Brx-roGFP2 and calculation of OxD and EBSH
To study the Brx-roGFP2 response in vitro, the purified proteins were reduced with 10 mM DTT for 20 min, desalted with Micro-Bio spin columns (Bio-Rad), and diluted to 1 μM in 100 mM potassium phosphate buffer, pH 7.0. Fluorescence excitation spectra of Brx-roGFP2 were analyzed both before and after exposure to the oxidants using the CLARIOstar microplate reader (BMG Labtech) with the Control software version 5.20 R5. Fluorescence excitation spectra were scanned from 360 to 500 nm with a bandwidth of 10 nm, and emission was measured at 510 nm. Gain setting was adjusted for each excitation maximum. The data were analyzed using the MARS software version 3.10 and exported to Excel. Each in vitro measurement was performed in triplicate, as indicated in the figure legend. The OxD of the biosensor was calculated using Equation (1), as previously described (24, 25).
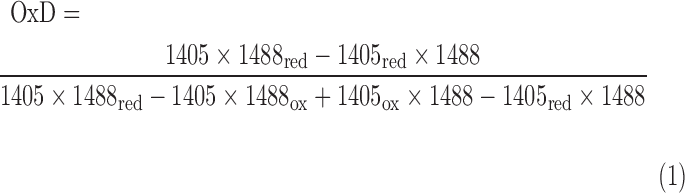 |
The values I405 and I488 are the observed fluorescence excitation intensities at 405 and 488 nm, respectively. The values I405red, I488red, I405ox, and I488ox are the fluorescence excitation intensities at 405 and 488 nm of fully reduced and oxidized probes, respectively.
The biosensor redox potential EroGFP2 was calculated according to the Nernst Equation (2), as previously described (25).
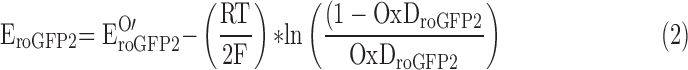 |
Because the biosensor equilibrates with the BSH/BSSB redox couple, EBSH = EroGFP2.
Construction of Brx-roGFP2 fusions in S. aureus
The brx-roGFP2 sequence was amplified with primers SAUSA300-1321-FOR-BamHI-2 and roGFP2-REV-KpnI-3, and the forward primer also includes the Shine-Dalgarno sequence of the brx gene. The PCR product was digested with BamHI and KpnI and inserted into the pRB473-XylR shuttle vector that was digested using the same enzymes to generate pRB473-XylR-Brx-roGFP2. The plasmids were cloned in E. coli DH5α and electroporated into competent cells of S. aureus RN4220. The plasmids were transferred into the S. aureus target strains COL and USA300 by phage transduction using bacteriophage 80 as previously described (26). S. aureus transductants were selected on LB agar with chloramphenicol. The plasmids were isolated and confirmed by PCR and sequencing of the brx-roGFP2 fusion.
Western blot analyses for Brx-roGFP2 expression and BSH-mixed disulfides in S. aureus
S. aureus strains with Brx-roGFP2 were grown in LB with 1% xylose and harvested at different times during the growth. Cells were washed in Tris-buffer (pH 8.0) with 10 mM NEM, disrupted using the ribolyzer and the protein extract was cleared from cell debris by repeated centrifugation. Protein amounts of 25 μg were analyzed by Western blot analysis using mouse-anti-GFP monoclonal antibodies (Cat. No. 12616810949; Tebu Biosciences) as previously described (11). Quantification was performed using the ImageJ software (ver. 1.48, http://imagej.nih.gov) based on a standard curve with purified Brx-roGFP2. The BSH-mixed disulfides were analyzed after exposure of S. aureus cells to 100 μM NaOCl using nonreducing SDS-PAGE and BSH-specific Western blot analysis with polyclonal rabbit BSH-antibodies as previously described (7).
CLSM of S. aureus Brx-roGFP2 strains
S. aureus strains with Brx-roGFP2 were exposed to 150 μM NaOCl, harvested both before and after the stress, blocked with NEM, and analyzed by CLSM using a ZEISS LSM510meta. The microscope was equipped with a 100 × 1.3 M27 EC plan-neofluar oil objective. Fluorescence excitation was performed at 405 and 488 nm, and emission was measured using the 505–550 nm band pass filter. Cells treated with 10 mM DTT and 5 mM diamide were used as fully reduced and oxidized controls, respectively. The argon/2 and 405 nm laser power were set to 20% and 8%, respectively. The smart gain was 948 V. All setting parameters for the CLSM are listed in Supplementary Table S4. The microscope was calibrated with fully oxidized and reduced S. aureus Brx-roGFP2 controls. Quantification of the OxD values was performed from five cells each from each sample, and the experiments were performed in two biological replicates.
Flow cytometry of S. aureus Brx-roGFP2 strains during infection of THP-1 macrophage cell lines
For the infection assays, we used the human monocytic leukemia cell line THP-1, which was purchased from the DSMZ strain collection in Heidelberg (DSMZ-No. ACC-16). The cell line was authenticated by multiplex PCR of minisatellite markers, which revealed a unique DNA profile, and the expression of fusion gene MLL-MLLT3 (MLL-AF9) was confirmed by real-time PCR. Cell cultures were checked for absence of mycoplasma contaminations by PCR on a regular basis. This cell line is not included in the database of commonly misidentified cell lines as maintained by ICLAC. THP-1 cells were cultivated in RPMI-1640 medium with 10% heat-inactivated fetal bovine serum (FBS) and seeded in 60 mm cell culture dishes at a density of 4.5 × 106 cells. Differentiation was induced by 100 nM phorbol 12-myristate 13-acetate (PMA) for 24 h, followed by washing of cells with Hanks' balanced salts solution and the addition of fresh medium without PMA.
After 24 h of incubation, infection assays of THP-1 macrophages with S. aureus were performed as follows. First, S. aureus COL cells expressing Brx-roGFP2 were grown in LB with 1% xylose until OD540 of 0.5 was reached. Bacterial cells were harvested, washed twice, and incubated in RPMI-1640 medium. Infection of THP-1 cells with S. aureus Brx-roGFP2 was performed at an MOI of 25 for 1 h. The infected THP-1 cells were washed twice in PBS buffer, detached with 0.05% trypsin and 0.02% EDTA, centrifuged, and re-suspended in PBS buffer with 1% FBS.
Measurement by flow cytometry was performed on an Attune Acoustic Focusing Cytometer (Life Technologies) with excitation at 405 and 488 nm and emission at 515–545 nm. Ten thousand events were gated, and the MFI was determined with Attune software V2.1.0 or FlowJo V10.07 (Tree Star). For reduced and oxidized controls, infected THP-1 cells were treated with 20 mM DTT and 150 μM NaOCl, respectively. Infection experiments were performed in three biological replicates.
Supplementary Material
Abbreviations Used
- BMM
Belitsky minimal medium
- Brx
bacilliredoxin
- BSH
bacillithiol
- BSSB
bacillithiol disulfide
- CLSM
confocal laser scanning microscopy
- CoASH
coenzyme A
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- FBS
fetal bovine serum
- Grx
glutaredoxin
- GSH
glutathione
- GSSG
oxidized glutathione disulfide
- H2O2
hydrogen peroxide
- IPTG
isopropyl-β-d-thiogalactopyranoside
- LB
Luria Bertani
- MFI
mean fluorescence intensity
- MOI
multiplicity of infection
- Mrx1
mycoredoxin1
- MSH
mycothiol
- MSSM
mycothione or mycothiol disulfide
- NADPH
nicotinamide adenine dinucleotide phosphate
- NaOCl
sodium hypochlorite
- NEM
N-ethylmaleimide
- OD500
optical density at 500 nm
- OxD
oxidation degree
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PMA
phorbol 12-myristate 13-acetate
- roGFP
redox-sensitive green fluorescent protein
- ROS
reactive oxygen species
- SDS-PAGE
sodium dodecyl sulfate–polyacrylamide gel electrophoresis
- SEM
standard error of the mean
- Trx
thioredoxin
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (AN746/4-1) within the SPP1710 on “Thiol-based Redox switches” and by the DFG grants AN746/3-1 and project C1 of the research training group GRK1947 to H.A. The support from the SFB-TR34 (project A8) to M.M. and J.P.-F. is further acknowledged. The authors are grateful to Tobias Dick for providing the plasmid pQE60 with the Grx-roGFP2 fusion and to Ambrose Cheung for the kind gift of the S. aureus bshA mutants in the COL and USA300 backgrounds. They would also like to thank all members of the SPP1710 consortium for stimulating discussions about their Brx-roGFP2 biosensor results.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Archer GL. Staphylococcus aureus: a well-armed pathogen. Clin Infect Dis 26: 1179–1181, 1998 [DOI] [PubMed] [Google Scholar]
- 2.Bhaskar A, Chawla M, Mehta M, Parikh P, Chandra P, Bhave D, Kumar D, Carroll KS, and Singh A. Reengineering redox sensitive GFP to measure mycothiol redox potential of Mycobacterium tuberculosis during infection. PLoS Pathog 10: e1003902, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher HW. and Corey GR. Epidemiology of methicillin-resistant Staphylococcus aureus. Clin Infect Dis 46(Suppl 5): S344–S349, 2008 [DOI] [PubMed] [Google Scholar]
- 4.Bratbak G. and Dundas I. Bacterial dry matter content and biomass estimations. Appl Environ Microbiol 48: 755–757, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen PR, Brugarolas P, and He C. Redox signaling in human pathogens. Antioxid Redox Signal 14: 1107–1118, 2011 [DOI] [PubMed] [Google Scholar]
- 6.Chi BK, Gronau K, Mäder U, Hessling B, Becher D, and Antelmann H. S-bacillithiolation protects against hypochlorite stress in Bacillus subtilis as revealed by transcriptomics and redox proteomics. Mol Cell Proteomics 10: M111.009506, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi BK, Roberts AA, Huyen TT, Bäsell K, Becher D, Albrecht D, Hamilton CJ, and Antelmann H. S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in firmicutes bacteria. Antioxid Redox Signal 18: 1273–1295, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dörries K, Schlueter R, and Lalk M. Impact of antibiotics with various target sites on the metabolome of Staphylococcus aureus. Antimicrob Agents Chemother 58: 7151–7163, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaballa A, Chi BK, Roberts AA, Becher D, Hamilton CJ, Antelmann H, and Helmann JD. Redox regulation in Bacillus subtilis: the bacilliredoxins BrxA(YphP) and BrxB(YqiW) function in de-bacillithiolation of S-bacillithiolated OhrR and MetE. Antioxid Redox Signal 21: 357–367, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaballa A, Newton GL, Antelmann H, Parsonage D, Upton H, Rawat M, Claiborne A, Fahey RC, and Helmann JD. Biosynthesis and functions of bacillithiol, a major low-molecular-weight thiol in Bacilli. Proc Natl Acad Sci U S A 107: 6482–6486, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutscher M, Pauleau AL, Marty L, Brach T, Wabnitz GH, Samstag Y, Meyer AJ, and Dick TP. Real-time imaging of the intracellular glutathione redox potential. Nat Methods 5: 553–559, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Helmann JD. Bacillithiol, a new player in bacterial redox homeostasis. Antioxid Redox Signal 15: 123–133, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hillion M. and Antelmann H. Thiol-based redox switches in prokaryotes. Biol Chem 396: 415–444, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horsburgh MJ, Clements MO, Crossley H, Ingham E, and Foster SJ. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun 69: 3744–3754, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kasozi D, Mohring F, Rahlfs S, Meyer AJ, and Becker K. Real-time imaging of the intracellular glutathione redox potential in the malaria parasite Plasmodium falciparum. PLoS Pathog 9: e1003782, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, and Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 130: 797–810, 2007 [DOI] [PubMed] [Google Scholar]
- 17.Lee JW, Soonsanga S, and Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci U S A 104: 8743–8748, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu Y. and Imlay JA. Cell death from antibiotics without the involvement of reactive oxygen species. Science 339: 1210–1213, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livermore DM. Antibiotic resistance in staphylococci. Int J Antimicrob Agents 16(Suppl 1): S3–S10, 2000 [DOI] [PubMed] [Google Scholar]
- 20.Loi VV, Rossius M, and Antelmann H. Redox regulation by reversible protein S-thiolation in bacteria. Front Microbiol 6: 187, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowy FD. Staphylococcus aureus infections. N Engl J Med 339: 520–532, 1998 [DOI] [PubMed] [Google Scholar]
- 22.Mallett TC, Wallen JR, Karplus PA, Sakai H, Tsukihara T, and Claiborne A. Structure of coenzyme A-disulfide reductase from Staphylococcus aureus at 1.54 A resolution. Biochemistry 45: 11278–11289, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer AJ, Brach T, Marty L, Kreye S, Rouhier N, Jacquot JP, and Hell R. Redox-sensitive GFP in Arabidopsis thaliana is a quantitative biosensor for the redox potential of the cellular glutathione redox buffer. Plant J 52: 973–986, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Meyer AJ. and Dick TP. Fluorescent protein-based redox probes. Antioxid Redox Signal 13: 621–650, 2010 [DOI] [PubMed] [Google Scholar]
- 25.Morgan B, Sobotta MC, and Dick TP. Measuring E(GSH) and H2O2 with roGFP2-based redox probes. Free Radic Biol Med 51: 1943–1951, 2011 [DOI] [PubMed] [Google Scholar]
- 26.Müller M, Reiss S, Schlüter R, Mäder U, Beyer A, Reiss W, Marles-Wright J, Lewis RJ, Pförtner H, Völker U, Riedel K, Hecker M, Engelmann S, and Pane-Farre J. Deletion of membrane-associated Asp23 leads to upregulation of cell wall stress genes in Staphylococcus aureus. Mol Microbiol 93: 1259–1268, 2014 [DOI] [PubMed] [Google Scholar]
- 27.Newton GL, Fahey RC, and Rawat M. Detoxification of toxins by bacillithiol in Staphylococcus aureus. Microbiology 158: 1117–1126, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, Hamilton CJ, Claiborne A, Helmann JD, and Fahey RC. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol 5: 625–627, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perera VR, Newton GL, Parnell JM, Komives EA, and Pogliano K. Purification and characterization of the Staphylococcus aureus bacillithiol transferase BstA. Biochim Biophys Acta 1840: 2851–2861, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perera VR, Newton GL, and Pogliano K. Bacillithiol: a key protective thiol in Staphylococcus aureus. Expert Rev Anti Infect Ther 13: 1089–1107, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.This reference has been deleted.
- 32.Posada AC, Kolar SL, Dusi RG, Francois P, Roberts AA, Hamilton CJ, Liu GY, and Cheung A. Importance of bacillithiol in the oxidative stress response of Staphylococcus aureus. Infect Immun 82: 316–332, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pöther DC, Gierok P, Harms M, Mostertz J, Hochgräfe F, Antelmann H, Hamilton CJ, Borovok I, Lalk M, Aharonowitz Y, and Hecker M. Distribution and infection-related functions of bacillithiol in Staphylococcus aureus. Int J Med Microbiol 303: 114–123, 2013 [DOI] [PubMed] [Google Scholar]
- 34.Rajkarnikar A, Strankman A, Duran S, Vargas D, Roberts AA, Barretto K, Upton H, Hamilton CJ, and Rawat M. Analysis of mutants disrupted in bacillithiol metabolism in Staphylococcus aureus. Biochem Biophys Res Commun 436: 128–133, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwarzländer M, Dick TP, Meye AJ, and Morgan B. Dissecting redox biology using fluorescent protein sensors. Antioxid Redox Signal 24: 680–712, 2016 [DOI] [PubMed] [Google Scholar]
- 36.Sharma SV, Arbach M, Roberts AA, Macdonald CJ, Groom M, and Hamilton CJ. Biophysical features of bacillithiol, the glutathione surrogate of Bacillus subtilis and other firmicutes. Chembiochem 14: 2160–2168, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyagi P, Dharmaraja AT, Bhaskar A, Chakrapani H, and Singh A. Mycobacterium tuberculosis has diminished capacity to counteract redox stress induced by elevated levels of endogenous superoxide. Free Radic Biol Med 84: 344–354, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Heijden J, Bosman ES, Reynolds LA, and Finlay BB. Direct measurement of oxidative and nitrosative stress dynamics in Salmonella inside macrophages. Proc Natl Acad Sci U S A 112: 560–565, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Heijden J, Vogt SL, Reynolds LA, Pena-Diaz J, Tupin A, Aussel L, and Finlay BB. Exploring the redox balance inside gram-negative bacteria with redox-sensitive GFP. Free Radic Biol Med 91: 34–44, 2015 [DOI] [PubMed] [Google Scholar]
- 40.Weber H, Engelmann S, Becher D, and Hecker M. Oxidative stress triggers thiol oxidation in the glyceraldehyde-3-phosphate dehydrogenase of Staphylococcus aureus. Mol Microbiol 52: 133–140, 2004 [DOI] [PubMed] [Google Scholar]
- 41.Winterbourn CC. and Kettle AJ. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid Redox Signal 18: 642–660, 2013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.