Abstract
The CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated 9) system is widely used as a tool for genome engineering in various organisms. A complex consisting of Cas9 and single guide RNA (sgRNA) induces a DNA double-strand break in a sequence-specific manner, resulting in knockout. Some binary vectors for CRISPR/Cas9 in plants have been reported, but there is a problem with low efficiency. Here, we present a newly developed, highly efficient CRISPR/Cas9 vector for Arabidopsis thaliana, pKAMA-ITACHI Red (pKIR), harboring the RIBOSOMAL PROTEIN S5 A (RPS5A) promoter to drive Cas9. The RPS5A promoter maintains high constitutive expression at all developmental stages starting from the egg cell and including meristematic cells. Even in the T1 generation, pKIR induced null phenotypes in some genes: PHYTOENE DESATURASE 3 (PDS3), AGAMOUS (AG) and DUO POLLEN 1 (DUO1). Mutations induced by pKIR were carried in the germ cell line of the T1 generation. Surprisingly, in some lines, 100% of the T2 plants had the adh1 (ALCOHOL DEHYDROGENASE 1) null phenotype, indicating that pKIR strongly induced heritable mutations. Cas9-free T2 mutant plants were obtained by removing T2 seeds expressing a fluorescent marker in pKIR. Our results suggest that the pKIR system is a powerful molecular tool for genome engineering in Arabidopsis.
Keywords: Arabidopsis thaliana, CRISPR/Cas9, Genome engineering, Knockout, Cas9-free isolation
Introduction
Genome engineering has been rapidly developing and makes it possible to perform specific modification of a target gene. Zinc finger nuclease (ZFN) (Urnov et al. 2010) and transcription activator-like effector nuclease (TALEN) (Cermak et al. 2011) were developed as tools for genome engineering. Both consist of artificial DNA-binding proteins fused to FokI endonuclease. The DNA-binding domain designed to recognize the target DNA sequence binds to the target sequence, and FokI endonuclease cleaves the flanking DNA site. The DNA is cleaved as a double-strand break (DSB), which triggers the DNA repair machinery, generally resulting in a short insertion or deletion via non-homologous end-joining (NHEJ). Since a frameshift in the open reading frame caused by NHEJ disrupts the target gene, a knockout mutant is obtained. ZFN and TALEN have been widely used for genome engineering in various organisms. However, vector construction is not simple because the DNA-binding domain must be customized and tandemly arrayed via several processes to bind to its target DNA sequence. The nucleotide sequences reported herein have been submitted to DDBJ under accession numbers LC189561 (p35S-Cas9), LC189562 (pX2-Cas9), LC189563 (pKIR1.0), LC189564 (pKIR1.1), LC189565 (pKI1.1) and LC189566 (pKI1.1R). Patent: a patent has been applied for on the method to drive Cas9 using the RPS5A promoter for plant genome editing (patent application number: 2016-171590).
Recently, a novel and revolutionary tool for genome engineering, called CRISPR (clustered regularly interspaced short palindromic repeats), has been developed (Jinek et al. 2012, Cong et al. 2013, Mali et al. 2013). The CRISPR/Cas9 (CRISPR-associated 9) system consists of two components, the Cas9 protein and a single guide RNA (sgRNA). Cas9 loading sgRNA scans along DNA to find a target sequence that matches with the 20 bp of the 5′ sequence of sgRNA, with a protospacer adjacent motif (PAM) sequence (NGG), and then the Cas9–sgRNA complex induces a DSB at the target site. Since the DSB site is repaired by NHEJ, as is the case in ZFN and TALEN, we can generate knockout plants using CRISPR/Cas9. CRISPR/Cas9 was reported to be successful in producing gene-modified plants in Arabidopsis thaliana (Feng et al. 2013, Mao et al. 2013, Fauser et al. 2014, Feng et al. 2014, Wang et al. 2015, Osakabe et al. 2016), Marchantia polymorpha L. (Sugano et al. 2014), Oryza sativa (Feng et al. 2013, Miao et al. 2013, Endo et al. 2015, Mikami et al. 2015), Solanum lycopersicum (Brooks et al. 2014) and other plant species. However, there remain some unresolved problems. First, there is still room for improvement in the mutation induction efficiency via the floral dip method, which introduces genes to female reproductive tissue, in A. thaliana (Ye et al. 1999). The efficiency in inflorescences, which contain germ cells, is not high in A. thaliana. This results in low efficiency of transmission to the next generation. Secondly, the isolation of plants without T-DNA for the CRISPR/Cas9 system (referred to as ‘Cas9-free’) requires a PCR experiment as the use of conventional antibiotic selection would result in the death of plants without the T-DNA.
We developed a novel highly efficient CRISPR/Cas9 vector for A. thaliana. To induce inheritable mutations highly efficiently, we tested promoters for Cas9 expression that are constitutively expressed from an initiating cell (an egg cell or a zygote). In Agrobacterium-mediated transformation by floral dipping of A. thaliana, transgenic plants (T1 plants) are produced through transformation of the egg cell formed in the dipped inflorescence (Ye et al. 1999). If Cas9 expression and mutations are induced in the initiating cell or at an earlier developmental stage, the mutations are passed to the daughter cells and then all or most of the cells in the plant, including the meristematic region, will have the induced mutation. Indeed, an egg cell-specific EC1 promoter to express Cas9 was recently used to obtain non-mosaic T1 mutants (Wang et al. 2015). Mosaicism is defined as the presence of cells with different genotypes in a tissue or individual. However, the egg cell-specific Cas9 limits the chance of mutation to a very early embryonic developmental stage. In this study, we used the promoters of RIBOSOMAL PROTEIN S5 A (RPS5A) and WUSCHEL RELATED HOMEOBOX 2 (WOX2). The RPS5A gene is expressed from an early embryonic stage (Weijers et al. 2001) and its promoter is constitutively active, beginning in egg cells (Maruyama et al. 2013). The RPS5A promoter is widely used in imaging analyses to label cells, organelles or structures of interest brightly via expressing fluorescent proteins (Maruyama et al. 2013, Gooh et al. 2015, Liao et al. 2015). Data from imaging analyses show that the RPS5A promoter is capable of supporting high-level gene expression. Continuous Cas9 expression from egg cells achieves mutation induction with high efficiency in the T1 generation. Furthermore, we attempted to select Cas9-free plants non-destructively using a backbone vector, pFAST-R (Shimada et al. 2010), harboring an expression cassette of OLE1–TagRFP (red fluorescent protein) that exhibits red fluorescence in seeds. As reported recently using a similar seed expression system (Gao et al. 2016), a pFAST system that can reveal the existence of transgenes in seeds would provide us with a simple and powerful way to obtain Cas9-free plants.
Results
Vector construction for the CRISPR/Cas9 system in Arabidopsis thaliana
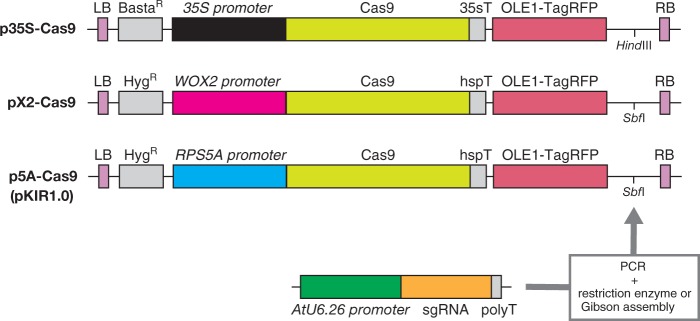
We prepared binary vectors based on the pFAST-R vector series (Shimada et al. 2010) using three promoters to drive human codon-optimized Cas9 expression: Cauliflower mosaic virus 35S (35S), WOX2 and RPS5A. The 35S promoter is a well-known constitutively active promoter. According to previous reports, WOX2 mRNA was detected in the egg cell (Haecker et al. 2004), while fluorescent proteins driven by the WOX2 promoter were observed in the zygote (Ueda et al. 2011, Gooh et al. 2015). The RPS5A promoter begins expression in egg cells (Maruyama et al. 2013, Gooh et al. 2015). The vector carrying 35Sp::Cas9 was designated as p35S-Cas9, WOX2p::Cas9 was designated as pX2-Cas9, and RPS5Ap::Cas9 was designated as p5A-Cas9 (Fig. 1).
Fig. 1.
CRISPR/Cas9 vectors used in this study. Three promoters, 35S, WOX2 and RPS5A, were used to express Cas9. Backbone vectors were from the pFAST-R series and express the red fluorescent protein TagRFP in transgenic seeds. p35S-Cas9 is based on pFAST-R02, while pX2-Cas9 and pKIR1.0 are based on pFAST-R01. An sgRNA cassette amplified by PCR was inserted into the HindIII or SbfI site by restriction enzyme-based cloning or Gibson assembly. LB, left border; BastaR, basta resistance gene; 35sT, 35S terminator; RB, right border; HygR, hygromycin resistance gene; hspT, heat shock protein terminator.
For sgRNA expression, an sgRNA cassette, composed of the AtU6.26 promoter, sgRNA and the Pol III terminator, was also inserted into pFAST-R vectors via restriction enzyme-based cloning or Gibson assembly (Fig. 1). Since pFAST-R vectors harbor OLE1p::OLE1:TagRFP within their T-DNA, the transformants exhibit red fluorescence in their seeds, which helps in transformant selection.
Cas9 driven by the RPS5A promoter induced a knockout phenotype in inflorescences of the T1 generation
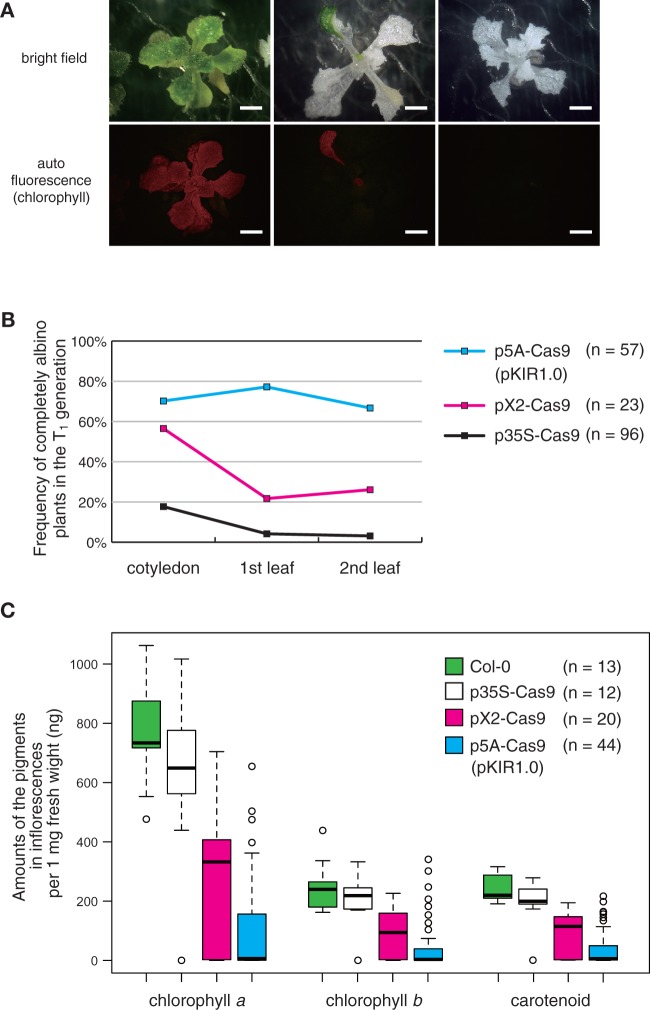
PDS3 knockout.To estimate the knockout efficiency of our vectors, we selected PHYTOENE DESATURASE 3 (PDS3, AT4G14210) as a target gene. The null mutant showed an albino phenotype (Qin et al. 2007), which makes it easy to determine whether both PDS3 alleles are disrupted in the observed tissues. We observed T1 plants transformed by p35S-Cas9 with sgRNA against PDS3 (p35S-Cas9_PDS3). Pairs of leaves in T1 plants were likely to be a mosaic of green and white areas. We judged the pairs of totally white leaves as ‘albino’ in the following observations. In cotyledons, 17.7% of T1 plants displayed the albino phenotype (Fig. 2B). However, as the plants grew, the albino frequency decreased. In the first true leaves, 4.2% had albino leaves and, in the second, 3.1% (Fig. 2B). In contrast, in pX2-Cas9_PDS3 T1 plants, 26.1% had albino second leaves, while the percentage was 66.7% in p5A-Cas9_PDS3 plants (Fig. 2B). Among p5A-Cas9_PDS3 T1 plants, we found three phenotype patterns: green, partially green (partially albino) and completely albino (Fig. 2A).
Fig. 2.
PDS3 gene knockout. (A) T1 plants transformed with pKIR1.0_PDS3. Upper panels are bright field images and lower panels are autofluorescence images. In the left panels, plant leaves are mostly green. In the middle panels, leaves are partially green. In the right panels, leaves are completely albino. Scale bars = 2 mm. (B) The frequency of albino pairs of leaves in T1 plants. The frequency was checked in cotyledons, first leaves and second leaves. (C) Box plots of pigment amounts in the first inflorescence: Chl a and b and carotenoid.
Next, we measured the amounts of three pigments in inflorescence cells. If mutations are detected in the inflorescence, there is an increased probability that the mutation will be inherited by the next generation. To quantify PDS3 knockout efficiency, we measured the amounts of Chl a, Chl b and carotenoid in the inflorescences of T1 plants because the pds3 null mutant has impaired biosynthesis of Chl and carotenoid (Qin et al. 2007). GA3 biosynthesis is disrupted in the pds3 mutant, causing a dwarf phenotype. We collected inflorescences from plants grown on medium with exogenous GA3, which partially rescues the pds3 dwarf phenotype and promotes bolting. All Col-0 (n = 13) and p35S-Cas9_PDS3 (n = 12) plants bolted in this medium. Among the 23 observed pX2-Cas9_PDS3 plants, 20 bolted (87%) and, among the 57 observed p5A-Cas9_PDS3 plants, 44 bolted (77%). Compared with the inflorescences of Col-0 and p35S-Cas9_PDS3, those of pX2-Cas9_PDS3 and p5A-Cas9_PDS3 contained lower amounts of pigment (Fig. 2C). For example, the median amount of Chl a per 1 mg of inflorescence was 730 ng in Col-0, 650 ng in p35S-Cas9, 330 ng in pX2-Cas9_PDS3 and 6.4 ng in p5A-Cas9_PDS3. These results indicate that p5A-Cas9_PDS3 induces mutations more efficiently in both rosette leaves and inflorescences. We designated the p5A-Cas9 vector as pKAMA-ITACHI Red 1.0 (pKIR1.0; Kama-itachi is a mink-like trio of ghosts in Japanese folklore: the first one trips up a target person, the second one injures the person with its nails and the third applies ointment, which is similar to the CRISPR system in terms of scanning, cutting and repairing the target DNA sequence). All subsequent experiments were performed using pKIR.
AGAMOUS knockout.To be transmitted to the next generation, the mutation needs to be harbored in germ cells within the floral meristem. Thus, we examined the knockout level in the floral meristem. We chose AGAMOUS (AG, AT4G18960), which is the gene encoding a transcription factor involved in flower development, as the target gene, and the null mutant has a phenotype of a flower within a flower, called ‘double flower’ (Yanofsky et al. 1990). If the phenotype is observed in pKIR1.0_AG T1 plants, it indicates that both AG alleles in the floral meristem are disrupted. We observed 13 T1 plants, and 12 displayed the double flower phenotype, with one dwarf plant arrested before flowering. Furthermore, in all 12 plants, the phenotype was observed in every flower (Fig. 3). This suggests that both AG alleles in the cells of all floral meristems were mutated by pKIR1.0_AG.
Fig. 3.
AGAMOUS gene knockout. (A) Whole body image of a pKIR1.0_AGAMOUS T1 plant. (B) Enlarged image of the area in the yellow rectangle in (A). (C) A flower exhibiting the ag phenotype. Scale bars = 5 cm (A), 1 cm (B) and 1 mm (C).
pKIR efficiently induced transmittable mutations
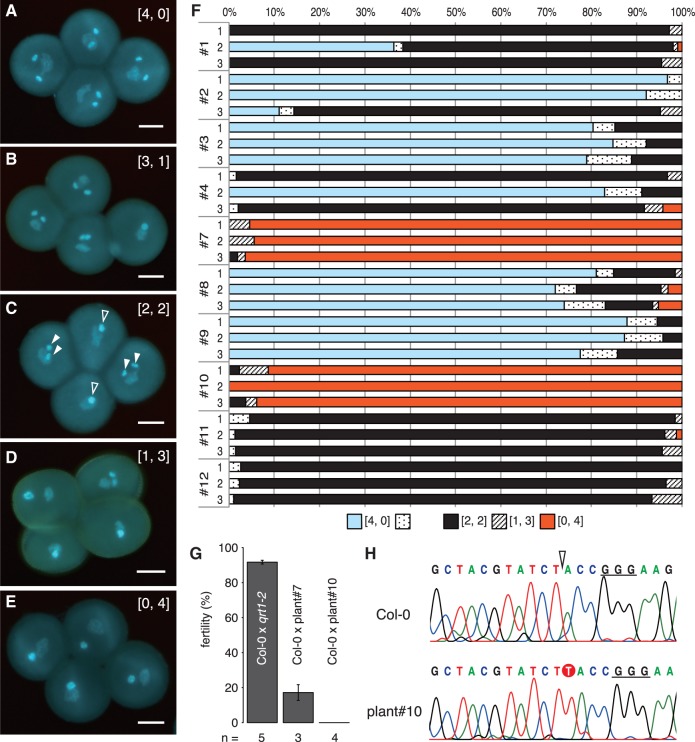
DUO1 knockout.Next, we estimated the knockout efficiency of pKIR1.0 in a male germ cell (sperm cell). If mutations are detected in sperm cells, then mutations may be inherited by the next generation. DUO1 (DUO POLLEN 1, AT3G60460) encodes a male germline-specific R2R3 MYB transcription factor. In the duo1 null pollen grain, cell division of the generative cell is impaired, resulting in a single non-fertile sperm-like cell (Rotman et al. 2005). We prepared pKIR1.0_DUO1 plants in a qrt1-2 mutant background. Since qrt1-2 has a tetrad pollen phenotype (Francis et al. 2006), it helps us to confirm the zygosity of the pollen mother cell. In T1 plants, we found five patterns for the duo1 phenotype in pollen tetrads by 4′,6-diamidino-2-phenylindole (DAPI) staining (Fig. 4A–E). A tetrad displaying two sperm cells in each pollen grain is written as [normal, duo1] = [4, 0]. For example, a tetrad written as [2, 2] displays two pollen grains with two sperm cells and two pollen grains with a single sperm-like cell. A tetrad with a single sperm-like cell is written as [0, 4], and the mother cell is inferred to be DUO1 homozygous or to have biallelic mutations. In T1 plants, we collected three flowers from the first inflorescence and examined the frequency of the duo1 phenotype in the tetrads. There were various frequencies of each phenotype pattern among individuals (Fig. 4F). Some tetrads showed [3, 1] or [1, 3] patterns in T1 plants. One possibility is that DUO1 in such plants is a weak allele. Another is that the mutation is induced after meiosis in the male germline. However, even in qrt1-2, 8.2% of tetrads showed the [3, 1] pattern (n = 7 flowers). These data indicate that a generative cell infrequently fails to undergo cell division. The [3, 1] and [1, 3] patterns in pKIR1.0_DUO1 T1 plants may reflect a failure in cell division rather than mutation induction after meiosis.
Fig. 4.
DUO1 gene knockout. (A–E) Pollen tetrads displaying patterns of the duo1 phenotype. The set numbers in brackets to the upper right of the image indicate the duo1 phenotype pattern in the tetrad. The numbers for normal and duo1 pollen are shown to the right and left, respectively. Scale bars = 10 µm. Filled and open arrowheads in (C) indicate normal sperm cells and duo1 sperm-like cells, respectively. (F) Frequency of the five duo1 phenotype patterns. Three flowers were observed in each pKIR1.0_DUO1 T1 plant. The numbers of observed tetrads ranged from 44 to 151. (G) Fertility of crosses between Col-0 (female) and qrt1-2 (male) or pKIR1.0-DUO1 T1 plants (male). Error bars indicate the SE. ‘n’ indicates the number of observed siliques. (H) DNA sequences of the DUO1 gene from the first cauline leaf in the first inflorescence. The open arrowhead indicates the cleavage site of Cas9 for DUO1 sgRNA. The underlined bases are the PAM sequence of DUO1 sgRNA. The T base inserted by NHEJ is shown as an outline character on a red background.
Notably, plants #7 and #10 had 95.4% and 95.1% [0, 4] tetrads, respectively. Reciprocal crosses of the wild type and these plants showed that plant #7 was slightly fertile (17.2%, n = 3), while #10 was completely infertile (0.0%, n = 4) (Fig. 4G). These data indicate that plant #10 is a duo1 null mutant. Next, we examined the DUO1 sequence of plant #10 using DNA from the first cauline leaf, and found a single T base inserted into the DSB site (Fig. 4H). The cauline leaf of plant #10 was most probably non-mosaic because significantly different peaks for bases were not detected subsequent to the DSB site (Fig. 4H). This result suggests that germ cells, at least in the first inflorescence, were completely null and non-mosaic in DUO1 alleles.
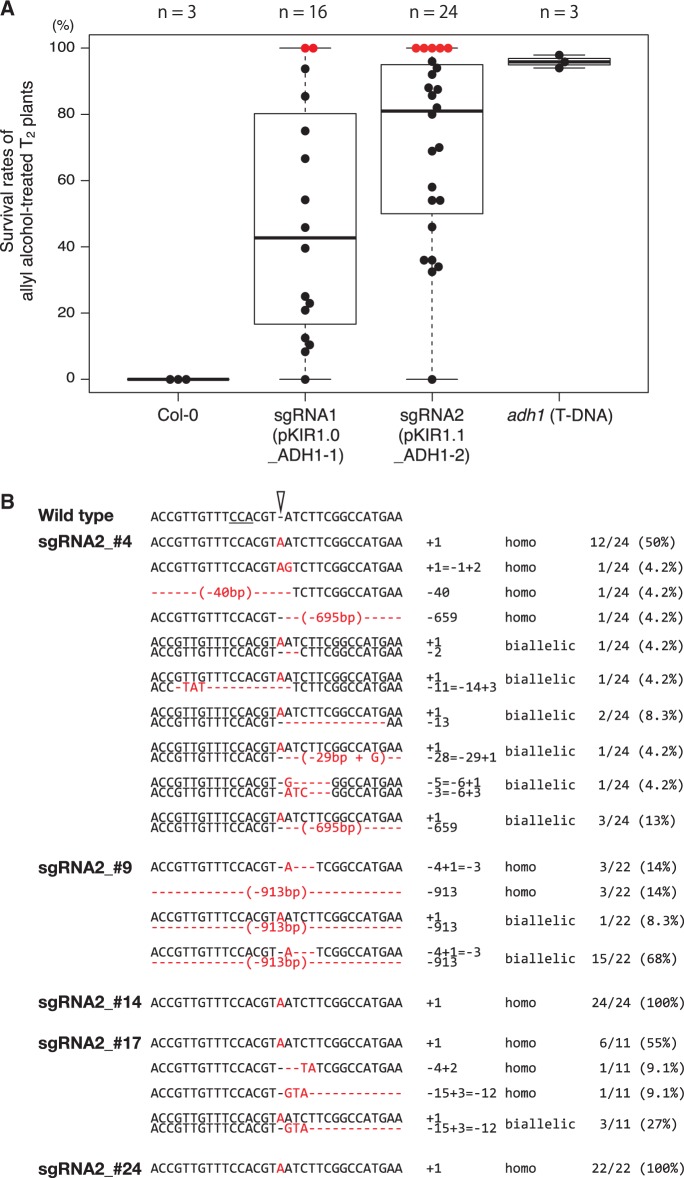
ADH1 knockout.To confirm that mutations by pKIR are efficiently transmitted to the next generation, we examined the transmission efficiency of induced mutations of ADH1 (ALCOHOL DEHYDROGENASE 1, AT1G77120). Since ADH1 encodes an enzyme that converts allyl alcohol into the toxic alcohol, acrolein, wild-type and heterozygous plants are killed and null homozygous or biallelic plants survive in the treatment. We prepared two sgRNAs against different target sequences of ADH1 (pKIR1.0_ADH1-1 and pKIR1.1_ADH1-2; see below for details of pKIR1.1). Dozens of T2 seeds from independent T1 plants were treated with allyl alcohol. Seven days after sowing, we measured the survival rate. The median survival rates of pKIR1.0_ADH1-1 and pKIR1.1_ADH1-2 T2 plants were 43% and 81%, respectively. The rate of Col-0 survival was 0.0% and that of adh1 T-DNA mutants (GABI-Kat 924D03.3) was 96% (Fig. 5A). Surprisingly, we isolated two and five plants in which the T2 seed survival rate was 100% in the pKIR1.0_ADH1-1 and pKIR1.1_ADH1-2 lines, respectively (Fig. 5A). This phenotype of complete resistance against allyl alcohol indicates that ADH1 alleles were already null in all male and female germ lines in the T1 generation.
Fig. 5.
ADH1 gene knockout. (A) Box and bee swarm plots of survival rates in allyl alcohol-treated T2 seeds. Dots indicate individual rates; red dots indicate 100% survival. ‘n’ indicates the number of the experiments in Col-0 and adh1 and that of the independent T2 lines in pKIR1.0_ADH1-1 and pKIR1.1_ADH1-2. (B) Mutation patterns of ADH1 alleles in pKIR1.1_ADH1-2 T2 plants. In the wild-type sequence, the underlined bases are the complementary PAM sequence of the ADH1 sgRNA and the open arrowhead is the cleavage site. The red characters indicate mutated sites.
To fix an induced mutation in and after the T2 generation and isolate a Cas9-free plant, the elimination of the CRISPR/Cas9 system, which consists of RPS5Ap::Cas9 and U6.26p::sgRNA in this study, is necessary in T2 seed selection. Our vector pKIR enabled us to eliminate Cas9 by visualization based on seed fluorescence. Since the T-DNA of pKIR also carries OLE1p::OLE1:TagRFP, which fluoresces red in seeds (Shimada et al. 2010), we could select non-transformants (Cas9-free) or transformants by the absence or presence of red fluorescence, providing an easy and non-destructive strategy for isolating Cas9-free plants. Conventional antibiotic-based selection is unsuited to the isolation of Cas9-free plants because plants without the T-DNA for Cas9 are not able to survive on the selection medium.
We performed Cas9-free plant selection on T2 seeds of pKIR1.1_ADH1-2 plants that showed the 100% survival phenotype (plants #4, #9, #14, #17 and #24) (Fig. 5A). We selected seeds without red fluorescence (Cas9-free seeds) under a fluorescence stereomicroscope and sowed them after allyl alcohol treatment. Next, we collected their leaves and extracted genomic DNA to sequence ADH1 alleles and confirm the absence of Cas9. In T2 plants from lines #4, #9 and #17, we found more than two patterns of alleles and, in those from lines #14 and #24, we found a single pattern (Fig. 5B). These data indicate that all germ cells in T1 plants of lines #14 and #24 had the same mutation. This suggests that the T1 plants were non-mosaic in ADH1 alleles, which may result from mutation induction in an early embryonic stage in the T1 generation. None of the T2 plants harbored the Cas9 expression cassette (Supplementary Fig. S1). Thus, pKIR1.0 enabled the easy and non-destructive isolation of targeted Cas9-free mutants in the T2 generation.
Improvement of pKIR to reduce construction steps
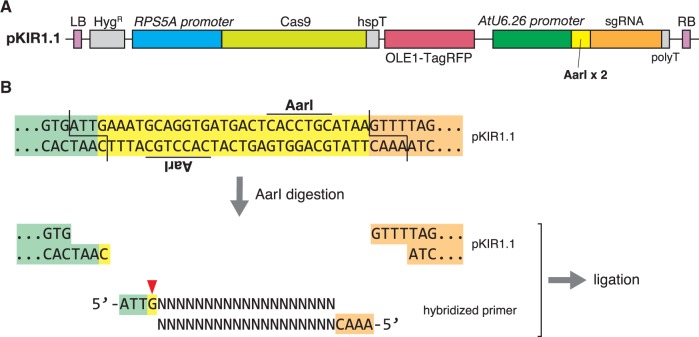
We further improved pKIR1.0 to reduce the number of steps involved in construction. In pKIR1.0 construction, two-step PCR and ligation are required for sgRNA cloning. We developed pKIR1.1, which carries an sgRNA cassette (U6.26p::sgRNA) by default. As shown in Fig. 6, two restriction enzyme sites (AarI) were inserted between the U6.26 promoter and sgRNA scaffold. AarI-digested pKIR1.1 forms four-base overhangs, and a hybridized DNA oligomer with overhangs can be inserted into this site. By annealing two 23 bp primers, we can complete vector construction without PCR. We confirmed that pKIR1.1_PDS3 showed equivalent activity to pKIR1.0_PDS3 (Supplementary Fig. S2).
Fig. 6.
pKIR1.1 construction. (A) For pKIR1.1, U6.26p::2 × AarI:sgRNA was inserted into the SbfI site of pKIR1.0. (B) Flowchart of pKIR1.1-based vector construction. The upper sequence is an enlarged view of the boundary (yellow) between the U6.26 promoter (pale green) and the sgRNA scaffold (orange). This boundary contains an inverted repeat of AarI recognition sites, and AarI digestion generates four-base overhangs. A hybridized primer with overhangs can be inserted into this site as shown. G with a red triangle is the first base for RNA transcription from the U6.26 promoter. G + (N)19 indicates the target sequence.
Discussion
The CRISPR/Cas9 system is a powerful genome engineering tool for modifying target DNA sequences in various organisms. Although CRISPR/Cas9 has been widely used in some plant species, including A. thaliana (Luo et al. 2016), increasing its efficiency to isolate knockout mutants of interest will significantly contribute to the study of plants that show genetic redundancy. In this study, we used three promoters (35S, WOX2 and RPS5A) to express Cas9 in A. thaliana, and found that the RPS5A promoter had the highest efficiency (Fig. 2B). In all targeted genes (PDS3, AG, DUO1 and ADH1), Cas9 driven by RPS5A was sufficient to knock out both alleles in meristematic cells of T1 plants. A binary vector harboring RPS5Ap::Cas9 was constructed and named pKIR, and also contained a TagRFP expression cassette for seeds (OLE1p::OLE1:TagRFP; Shimada et al. 2010). The null phenotype induced by pKIR was observed in germ cells (Fig. 4), and the mutations were efficiently transmitted to the next generation (Fig. 5). Among 24 pKIR1.1_ADH1-2 lines, the median survival rate, denoting an adh1 null rate, was 81%, and five lines showed 100% survival (Fig. 5A).
Various CRISPR/Cas9 systems for A. thaliana have been used for some target genes to examine transmission frequency. For example, Fauser et al. (2014) reported the mutation efficiency of ADH1; the survival rate of T2 plants was 15.3%. Feng et al. (2014) reported the mutation efficiency of several genes. However, neither report mentioned mosaicism. On the other hand, Wang et al. (2015) reported that non-mosaic T1 plants were isolated and the average frequency of T2 plants displaying the mutant phenotype was 24.8%. Although the mutation efficiency depends on the design of the sgRNA and it is difficult to compare the mutation efficiency of different sgRNAs induced by different vectors, our pKIR system produced non-mosaic T1 plants with high efficiency.
The RPS5A promoter, which is used in pKIR for Cas9 expression, is known to begin expression in egg cells (Maruyama et al. 2013) and constitutively maintain expression, even in meristematic cells, which probably contributed to the high efficiency of the mutations reported in this study. In some T2 lines with the 100% allyl alcohol survival phenotype, we found various patterns of mutations (Fig. 5B). These mutations were probably induced independently in different cell lineages rather than successively in the same lineage, based on the mutation sequences. Moreover, the degree of heterogeneity differed between plant lines (Fig. 5B); some lines had a number of mutation patterns, while others had a single one. The single mutation pattern in T2 plants could be due to non-mosaic mutation in the zygote stage of T1 plants. These results suggest that the timing of mutation in apical stem cells was line dependent and occurred anywhere from early developmental stages in zygotes to later developmental stages. Constitutive activity of the RPS5A promoter in meristematic cells may significantly increase the frequency of mutation in stem cells over the entire developmental period.
We used the pFAST system (Shimada et al. 2010) for the easy and non-destructive isolation of Cas9-free plants. Cas9 elimination has two advantages: freedom from off-target mutations and freedom from undesirable mutations of a wild-type allele. With regard to off-target mutations, constitutive Cas9 expression at high levels due to the RPS5A promoter could lead to off-target mutations in non-targeted genes during and after the T2 generation. After removal of the T-DNA for the CRISPR/Cas9 system, there is no possibility of off-target mutation. With regard to undesirable mutations, mosaic mutations in heterozygous T2 mutants can be induced. Mutations within the T-DNA for a rescue experiment to compensate for gene function probably occur as long as the T-DNA for the CRISPR/Cas9 system remains in the genomic DNA. Combination of RPS5Ap::Cas9 and the pFAST system was useful for developing a CRISPR/Cas9 vector.
The efficiency of obtaining mutants of interest depends on various factors. For example, the design of sgRNA significantly influences efficiency (Doench et al. 2014), as shown in the knockout of ADH1 (Fig. 5A). We will continue to improve our pKIR series for better efficiency. For example, to stabilize Cas9 expression and increase the non-mosaic rate, the ADH1 5′-untranslated region may be effective as it is a translational enhancer (Sugio et al. 2008). eSpCas9(1.1), with high specificity for targeted DNA DSBs (Slaymaker et al. 2016), will be useful in reducing off-target mutations. Regarding off-target mutations, a highly efficient CRISPR/Cas9 system may increase the possibility of off-target effects. sgRNA cloning in the pKIR system is easy; the preparation of multiple sgRNAs against a single gene of interest can be done quickly and with little effort. Based on the examination of a few sgRNAs, we avoided mistakenly perceiving the phenotype of an off-target mutation as an on-target mutation (Fig. 5A). A combination of nickase Cas9 and activation-induced cytidine deaminase induces point mutations (Nishida et al. 2016).
The RPS5A promoter was reported to have activity in the division zone of the root meristem (Weijers et al. 2001). This suggests that the pKIR series may induce mutations in the roots of T1 plants. The RPS5A promoter of A. thaliana was active in Torenia fournieri ovular cells, including egg cells (Susaki et al. 2015), and the U6.26 promoter of A. thaliana was active in Nicotiana benthamiana (Jiang et al. 2013). These reports indicate that the RPS5A and U6.26 promoters do not have species specificity and our vectors could potentially be used in other plant species. Taking into consideration the use of RPS5Ap::Cas9 and U6.26p::2 × AarI:sgRNA in binary vectors other than pFAST, we prepared a vector harboring both in pENTR/D-TOPO (pKAMA-ITACHI in pENTR/D-TOPO 1.1, called pKI1.1). This vector, pKI1.1, can be applied to any GATEWAY vectors depending on the selection marker (e.g. antibiotics and seed fluorescence). Users can select vectors from the pKI series for their own purposes, as pKI1.1-derived pKI1.1R_PDS3 showed efficiency equivalent to that of pKIR1.0_PDS3 in the knockout of the PDS3 gene (Supplementary Fig. S3).
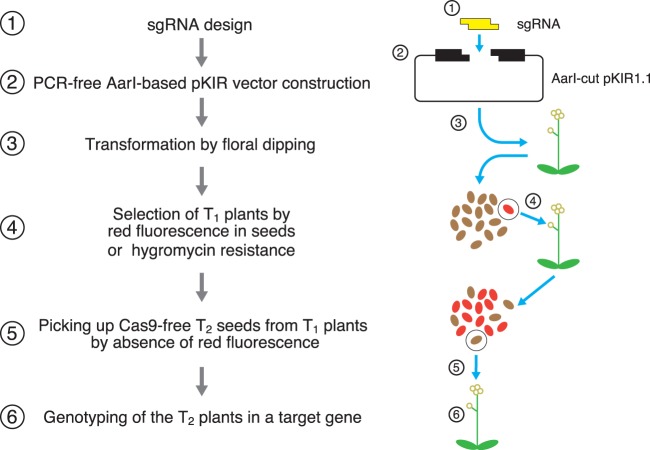
Fig. 7 summarizes the method to obtain Cas9-free knockout mutants by pKIR1.1. pKIR1.1 also contributes to the rapidity of the procedure, with its simple construction through the design of its sgRNA and two 23 bp primers (Fig. 6). Using the six steps shown in Fig. 7, we can now easily obtain knockout mutants of interest. Our pKI vector series based on RPS5A-driven Cas9 provides a useful tool for genome engineering in plants.
Fig. 7.
Flowchart for isolation of Cas9-free target mutants. For construction using pKIR1.1, hybridized 23 bp oligonucleotides containing an sgRNA sequence for the target gene are inserted into AarI-cut pKIR1.1 (Steps 1 and 2; Fig. 6). The pKIR series enables red fluorescence selection of seeds from T1 plants (Steps 3 and 4; hygromycin selection is also available). In T2 selection, seeds without red fluorescence represent Cas9-free plants (Step 5). By genotyping T2 plants, we can obtain a null mutant in the target gene (Step 6).
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana accession Columbia (Col-0) was used for the experiments. For the DUO1 sgRNA experiments, the qrt1-2 (ABRC stock name: CS8846) mutant was used. For the ADH1 sgRNA experiments, the adh1 (GABI-Kat line ID: 924D03.3) mutant was used. Seeds sterilized by PPM (Plant Cell Technology) were sown on 0.3% gellan gum plates containing 2.3 g l–1 Murashige and Skoog salts (Duchefa Biochemie), 1% sucrose, 1 × Gamborg’s vitamin solution (Sigma-Aldrich), 0.05% MES and 300 mg l–1 cefotaxime sodium (pH 5.8). After a few days of incubation at 4°C, they were grown at 22°C under continuous light for about 2 weeks and then transferred to soil or rock wool.
Plasmid construction
All primers in this study are listed in Supplementary Table S1. Cas9 (with a FLAG-tag and nuclear localization signal) was amplified by PCR from pX330-U6-Chimeric_BB-CBh-hSpCas9 (a gift from Feng Zhang, Addgene plasmid # 42230, Cong et al. 2013) and cloned into pENTR/D-TOPO (Thermo Fisher Scientific). The product was called Cas9 in pENTR/D-TOPO.
For the 35Sp::Cas9 construct, Cas9 from pENTR/D-TOPO was transferred into pFAST-R02 (Shimada et al. 2010) by LR reaction. For the WOX2p::Cas9 and RPS5Ap::Cas9 constructs, each promoter was amplified by PCR from MU14 (a gift from Minako Ueda) and DKv39 (a gift from Daisuke Kurihara), respectively, using forward primers with NotI and reverse primers with NcoI, then inserted into the NotI and NcoI sites of Cas9 in pENTR/D-TOPO. HspT (heat shock protein 18.2 terminator, Nagaya et al. 2010) was inserted into the AscI site of these vectors by Gibson assembly. The promoters combined with Cas9:HspT in pENTR/D-TOPO were transferred into pFAST-R01 (Shimada et al. 2010) by LR reaction. These products were pX2-Cas9 and pKIR1.0.
For p35S-Cas9_PDS3, we performed two-step PCR. In the first PCR, U6.26p and sgRNA were amplified separately, and then the products from the first PCR were combined in the second PCR. The second PCR product was inserted into the HindIII site of p35S-Cas9. For pX2-Cas9_PDS3 and pKIR1.0_PDS3, the above second PCR product (U6.26p::PDS3sgRNA) was first subcloned into pUC19 by HindIII, and then U6.26p::PDS3sgRNA was amplified by PCR from the vector and transferred into pX2-Cas9 or pKIR1.0 by Gibson assembly via the SbfI site. For pKIR1.0_AG, pKIR1.0_DUO1 and pKIR1.0_ADH1, all sgRNA cassettes were cloned into pKIR1.0 by two-step PCR and Gibson assembly as for PDS3sgRNA.
Two-step PCR was performed to obtain pKI1.1. The second PCR product (U6.26p::2 × AarI:sgRNA) was inserted into the NotI site of RPS5Ap::Cas9:HspT in pENTR/D-TOPO. To obtain pKIR1.1, the same fragment (U6.26p::2 × AarI:sgRNA) was inserted into the SbfI site of pKIR1.0 via Gibson assembly.
For pKI1.1R, RPS5Ap::Cas9:HspT and U6.26p::2 × AarI:sgRNA from pKI1.1 were transferred into pFAST-R01 (Shimada et al. 2010) by LR reaction. For pKIR1.1_PSD3, pKIR1.1_ADH1-2 and pKI1.1R_PDS3, pKIR1.1 or pKI1.1R was digested by AarI (Thermo Fisher Scientific, #ER1581) with alkaline phosphatase rAPid (Roche, #04898133001). Two primers, consisting of the overhang sequence and the target sequence, were treated with T4 polynucleotide kinase (NEB, #M0201) for phosphorylation and then placed in a thermal cycler with the following settings: 95°C for 5 min, cool down from 95 to 25°C over 14 min (5°C min–1), 25°C for 1 min, and then maintenance at 4°C. The hybridized primer was diluted 250-fold. After the electrophoresis of AarI-digested pKIR1.1 or pKI1.1R in an agarose gel and purification, the diluted hybridized primer was inserted into the digested vector using T4 ligase (NEB, #M0202). The ligation product was treated with Plasmid-Safe ATP-Dependent DNase (epicentre, #E3101K) and then transformed into Escherichia coli.
Plant transformation and selection of transformants
Binary vectors were introduced into Agrobacterium tumefaciens GV3101 by electroporation. Plant transformation was performed using the floral dip method (Clough and Bent 1998). The transformants that displayed red fluorescence in seeds were picked up by a slightly wet toothpick under a stereomicroscope (model SZX16, Olympus) with X-Cite 120LED (Excelitas Technologies).
Pigment measurement
Plants grown on the above-mentioned medium were transferred onto medium with 1.0 mg l–1 GA3 to promote bolting. After bolting, the first inflorescence was collected and its weight was measured. To extract pigments, the inflorescence was transferred into 1 ml of N,N-dimethylformamide and kept at 4°C overnight in the dark. The absorbances at 480, 647 and 664 nm were measured using a DS-11 Spectrophotometer (DeNovix Inc.). The amount of pigment was calculated according to Wellburn (1994).
DAPI staining of pollen tetrads
Flowers were dipped into 100 µl of DAPI staining buffer [1 µg ml–1 DAPI, 0.1% Triton X-100, 1 mM EDTA, 0.1 M sodium phosphate (pH 7.0)] following the method of Park et al. (1998). After brief vortexing and centrifugation, the precipitated pollen tetrads were transferred onto a slide glass. The samples were observed using a BX51N microscope (Olympus) and the staining images were captured by an Axiocam 506 camera (Carl Zeiss).
Allyl alcohol treatment of seeds
Seeds for allyl alcohol treatment were collected from T1 plants with ADH1 sgRNA. The seeds were treated with 20 mM allyl alcohol following the procedure of Jacobs et al. (1988). Sterilization and sowing were performed as described above. To calculate the survival rate, germination was observed 7 d after incubation.
Supplementary data
Supplementary data are available at PCP online.
Funding
This work was supported by the Japan Science and Technology Agency [ERATO project to T.H.]; and the Japan Society for the Promotion of Science [Grant-in-Aid for JSPS Fellows No. 11J06264 to H.T.; Grant-in-Aid for Scientific Research on Innovative Areas Nos. JP16H06465, JP16H06464 and JP16K21727 to T.H.].
Supplementary Material
Acknowledgments
We thank I. Nishimura-Hara for pFAST-R01 and pFAST-R02, F. Zhang for pX330, M. Ueda for MU14, D. Kurihara for DKv39, and D. Maruyama for qrt1-2 seeds. We also thank H. Kakui, S.S. Sugano, H. Takeuchi, Y. Sato and M. Watahiki for their help and advice.
Disclosures
The authors have no conflicts of interest to declare.
Glossary
Abbreviations
- 35S
Cauliflower mosaic virus 35S
- ADH1
ALCOHOL DEHYDROGENASE 1
- AG
AGAMOUS
- CRISPR
clustered regularly interspaced short palindromic repeats
- Cas9
CRISPR-associated 9
- DAPI
4′,6-diamidino-2-phenylindole
- DSB
double-strand break
- DUO1
DUO POLLEN 1
- HspT
heat shock protein 18.2 terminator
- NHEJ
non-homologous end-joining
- PAM
protospacer adjacent motif
- PDS3
PHYTOENE DESATURASE 3
- RFP
red fluorescent protein
- RPS5A
RIBOSOMAL PROTEIN S5 A
- sgRNA
single guide RNA
- TALEN
transcription activator-like effector nuclease
- WOX2
WUSCHEL RELATED HOMEOBOX 2
- ZFN
zinc finger nuclease
The nucleotide sequences reported herein have been submitted to DDBJ under accession numbers LC189561 (p35S-Cas9), LC189562 (pX2-Cas9), LC189563 (pKIR1.0), LC189564 (pKIR1.1), LC189565 (pKI1.1) and LC189566 (pKI1.1R).Patent: a patent has been applied for on 10 the method to drive Cas9 using the RPS5A promoter for plant genome editing (patent application number: 2016-171590)
References
- Brooks C., Nekrasov V., Lippman Z.B., Van Eck J. (2014) Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166: 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cermak T., Doyle E.L., Christian M., Wang L., Zhang Y., Schmidt C., et al. (2011) Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., et al. (2014) Rational design of highly active sgRNAs for CRISPR–Cas9-mediated gene inactivation. Nat. Biotechnol. 32: 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo M., Mikami M., Toki S. (2015) Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 56: 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauser F., Schiml S., Puchta H. (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79: 348–359. [DOI] [PubMed] [Google Scholar]
- Feng Z., Mao Y., Xu N., Zhang B., Wei P., Yang D.-L., et al. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 4632–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Zhang B., Ding W., Liu X., Yang D.-L., Wei P., et al. (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 23: 1229–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis K.E., Lam S.Y., Copenhaver G.P. (2006) Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol. 142: 1004–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Chen J., Dai X., Zhang D., Zhao Y. (2016) An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 171: 1794–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooh K., Ueda M., Aruga K., Park J., Arata H., Higashiyama T., et al. (2015) Live-cell imaging and optical manipulation of Arabidopsis early embryogenesis. Dev. Cell 34: 242–251. [DOI] [PubMed] [Google Scholar]
- Haecker A., Groß-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., et al. (2004) Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 131: 657–668. [DOI] [PubMed] [Google Scholar]
- Jacobs M., Dolferus R., Van den Bossche D. (1988) Isolation and biochemical analysis of ethyl methanesulfonate-induced alcohol dehydrogenase null mutants of Arabidopsis thaliana (L.) Heynh. Biochem. Genet. 26: 105–122. [DOI] [PubMed] [Google Scholar]
- Jiang W., Zhou H., Bi H., Fromm M., Yang B., Weeks D.P. (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao C.-Y., Smet W., Brunoud G., Yoshida S., Vernoux T., Weijers D. (2015) Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 12: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Gilbert B., Ayliffe M. (2016) Applications of CRISPR/Cas9 technology for targeted mutagenesis, gene replacement and stacking of genes in higher plants. Plant Cell Rep. 35: 1439–1450. [DOI] [PubMed] [Google Scholar]
- Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., et al. (2013) RNA-guided human genome engineering via Cas9. Science 339: 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Zhang H., Xu N., Zhang B., Gou F., Zhu J.-K. (2013) Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol. Plant 6: 2008–2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D., Hamamura Y., Takeuchi H., Susaki D., Nishimaki M., Kurihara D., et al. (2013) Independent control by each female gamete prevents the attraction of multiple pollen tubes. Dev. Cell 25: 317–323. [DOI] [PubMed] [Google Scholar]
- Miao J., Guo D., Zhang J., Huang Q., Qin G., Zhang X., et al. (2013) Targeted mutagenesis in rice using CRISPR–Cas system. Cell Res. 23: 1233–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikami M., Toki S., Endo M. (2015) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol. Biol. 88: 561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaya S., Kawamura K., Shinmyo A., Kato K. (2010) The HSP terminator of Arabidopsis thaliana increases gene expression in plant cells. Plant Cell Physiol. 51: 328–332. [DOI] [PubMed] [Google Scholar]
- Nishida K., Arazoe T., Yachie N., Banno S., Kakimoto M., Tabata M., et al. (2016) Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science 353: aaf8729. [DOI] [PubMed] [Google Scholar]
- Osakabe Y., Watanabe T., Sugano S.S., Ueta R., Ishihara R., Shinozaki K., et al. (2016) Optimization of CRISPR/Cas9 genome editing to modify abiotic stress responses in plants. Sci. Rep. 6: 26685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.K., Howden R., Twell D. (1998) The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125: 3789–3799. [DOI] [PubMed] [Google Scholar]
- Qin G., Gu H., Ma L., Peng Y., Deng X.W., Chen Z., et al. (2007) Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res. 17: 471–482. [DOI] [PubMed] [Google Scholar]
- Rotman N., Durbarry A., Wardle A., Yang W.-C., Chaboud A., Faure J.-E., et al. (2005) A novel class of MYB factors controls sperm-cell formation in plants. Curr. Biol. 15: 244–248. [DOI] [PubMed] [Google Scholar]
- Shimada T.L., Shimada T., Hara-Nishimura I. (2010) A rapid and non-destructive screenable marker, FAST, for identifying transformed seeds of Arabidopsis thaliana. Plant J. 61: 519–528. [DOI] [PubMed] [Google Scholar]
- Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. (2016) Rationally engineered Cas9 nucleases with improved specificity. Science 351: 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano S.S., Shirakawa M., Takagi J., Matsuda Y., Shimada T., Hara-Nishimura I., et al. (2014) CRISPR/Cas9-mediated targeted mutagenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 55: 475–481. [DOI] [PubMed] [Google Scholar]
- Sugio T., Satoh J., Matsuura H., Shinmyo A., Kato K. (2008) The 5′-untranslated region of the Oryza sativa alcohol dehydrogenase gene functions as a translational enhancer in monocotyledonous plant cells. J. Biosci. Bioeng. 105: 300–302. [DOI] [PubMed] [Google Scholar]
- Susaki D., Takeuchi H., Tsutsui H., Kurihara D., Higashiyama T. (2015) Live imaging and laser disruption reveal the dynamics and cell–cell communication during Torenia fournieri female gametophyte development. Plant Cell Physiol. 56: 1031–1041. [DOI] [PubMed] [Google Scholar]
- Ueda M., Zhang Z., Laux T. (2011) Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev. Cell 20: 264–270. [DOI] [PubMed] [Google Scholar]
- Urnov F.D., Rebar E.J., Holmes M.C., Zhang H.S., Gregory P.D. (2010) Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11: 636–646. [DOI] [PubMed] [Google Scholar]
- Wang Z.-P., Xing H.-L., Dong L., Zhang H.-Y., Han C.-Y., Wang X.-C., et al. (2015) Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D., Franke-van Dijk M., Vencken R.J., Quint A., Hooykaas P., Offringa R. (2001) An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development 128: 4289–4299. [DOI] [PubMed] [Google Scholar]
- Wellburn A.R. (1994) The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144: 307–313. [Google Scholar]
- Yanofsky M.F., Ma H., Bowman J.L., Drews G.N., Feldmann K.A., Meyerowitz E.M. (1990) The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346: 35–39. [DOI] [PubMed] [Google Scholar]
- Ye G.N., Stone D., Pang S.Z., Creely W., Gonzalez K., Hinchee M. (1999) Arabidopsis ovule is the target for Agrobacterium in planta vacuum infiltration transformation. Plant J. 19: 249–257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.