Abstract
Six genes of the Arabidopsis thaliana monosaccharide transporter-like (MST-like) superfamily share significant homology with polyol transporter genes previously identified in plants translocating polyols (mannitol or sorbitol) in their phloem (celery [Apium graveolens], common plantain [Plantago major], or sour cherry [Prunus cerasus]). The physiological role and the functional properties of this group of proteins were unclear in Arabidopsis, which translocates sucrose and small amounts of raffinose rather than polyols. Here, we describe POLYOL TRANSPORTER5 (AtPLT5), the first member of this subgroup of Arabidopsis MST-like transporters. Transient expression of an AtPLT5–green fluorescent protein fusion in plant cells and functional analyses of the AtPLT5 protein in yeast and Xenopus oocytes demonstrate that AtPLT5 is located in the plasma membrane and characterize this protein as a broad-spectrum H+-symporter for linear polyols, such as sorbitol, xylitol, erythritol, or glycerol. Unexpectedly, however, AtPLT5 catalyzes also the transport of the cyclic polyol myo-inositol and of different hexoses and pentoses, including ribose, a sugar that is not transported by any of the previously characterized plant sugar transporters. RT-PCR analyses and AtPLT5 promoter-reporter gene plants revealed that AtPLT5 is most strongly expressed in Arabidopsis roots, but also in the vascular tissue of leaves and in specific floral organs. The potential physiological role of AtPLT5 is discussed.
INTRODUCTION
Linear polyols, such as sorbitol or mannitol, are found in high concentrations in the phloem sap of plants from several families, such as Rosaceae, Apiaceae, or Plantaginaceae (Zimmermann and Ziegler, 1975). After their synthesis in leaves by sugar phosphate reductases and polyol phosphate phosphatases, linear polyols are loaded into the phloem by polyol-H+ symporters that accumulate their substrates to concentrations of several hundred millimolar (Zimmermann and Ziegler, 1975; Lohaus and Fischer, 2002; G. Lohaus, unpublished data). cDNAs encoding these transporters have been cloned recently from celery (Apium graveolens, Apiaceae; Noiraud et al., 2001), from sour cherry (Prunus cerasus, Rosaceae; Gao et al., 2003), and from common plantain (Plantago major, Plantaginaceae; Ramsperger-Gleixner et al., 2004). In celery, the AgMAT1 transporter is discussed to be responsible for the loading of mannitol into the phloem (Noiraud et al., 2001). The same function was also suggested for the sorbitol transporters PmPLT1 and PmPLT2 from common plantain, and in fact these proteins were immunolocalized to phloem companion cells (Ramsperger-Gleixner et al., 2004). By contrast, both sorbitol transporters identified in sour cherry (PcSOT1 and PcSOT2) seem to be responsible for sorbitol import into cherries during the later stages of fruit development. Functional analyses of the encoded proteins from all three species in yeast (Noiraud et al., 2001; Gao et al., 2003; Ramsperger-Gleixner et al., 2004) and of the Plantago transporter after expression in Xenopus oocytes (Ramsperger-Gleixner et al., 2004) showed that irrespective of their physiological substrate, these proteins do catalyze the transport of both mannitol and sorbitol with similar rates.
Arabidopsis thaliana, a member of the Brassicaceae, translocates sucrose in its phloem together with small amounts of raffinose (Haritatos et al., 2000) but no polyols. Nevertheless, Arabidopsis has six genes (At2g16120, At2g16130, At2g18480, At2g20780, At3g18830, and At4g36670) sharing significant homology with the polyol transporter genes mentioned above. The physiological role of these potential sorbitol and/or mannitol transporters was unclear in Arabidopsis, and a physiological role or a substrate specificity different from that in celery, sour cherry, or common plantain seemed reasonable.
Here, we report the isolation of cDNAs for five of these six Arabidopsis polyol transporter-like genes and the detailed characterization of one of the encoded proteins by functional expression of its cDNA in yeast and in Xenopus laevis oocytes. Our data show that in contrast with the previously described polyol transporters from polyol translocating plants, the Arabidopsis homolog AtPLT5 (At3g18830) has a strong preference for sorbitol over mannitol. However, competition analyses revealed that sorbitol transport into AtPLT5-expressing Saccharomyces cerevisiae cells was inhibited by a wide range of other compounds, including polyols with shorter chain lengths, such as xylitol, erythritol, or glycerol, the cyclic polyol myo-inositol or hexoses and pentoses forming pyranose (e.g., glucose and xylose) or furanose rings (e.g., fructose or ribose). Uptake analyses with several of these compounds in AtPLT5-expressing yeast cells and electrophysiological analyses in AtPLT5 cRNA-injected Xenopus oocytes revealed that all tested competitors are also substrates of AtPLT5. The Km values of AtPLT5 were determined for several substrates and were found to be in the millimolar range, characterizing AtPLT5 as a low affinity H+-symporter. The organ-specific expression and the cellular and subcellular localization were determined by RT-PCR analyses in AtPLT5 promoter:β-glucuronidase (GUS) or AtPLT5 promoter:green fluorescent protein (GFP) plants and with anti-AtPLT5 antisera.
RESULTS
Cloning of the AtPLT cDNAs
The open reading frames (ORFs) of the six putative polyol transporter genes from Arabidopsis had been predicted from in silico analyses of the Arabidopsis genome. To prove or disprove these predictions, cDNAs covering the entire ORFs were generated by RT-PCR from whole-plant mRNA and sequenced. Because of their homology to the previously described PmPLT genes from Plantago (Ramsperger-Gleixner et al., 2004), the Arabidopsis genes were named AtPLTs (At2g16120 [AtPLT1], At2g16130 [AtPLT2], At2g18480 [AtPLT3], At2g20780 [AtPLT4], At3g18830 [AtPLT5], and At4g36670 [AtPLT6]).
The obtained cDNA sequences confirmed the predicted and deduced protein sequences of AtPLT1 (511 amino acids), AtPLT2 (511 amino acids), AtPLT3 (508 amino acids), and AtPLT5 (539 amino acids). For AtPLT4, two different ORFs had been predicted, one with 526 amino acids (e.g., NM 127643) and one with 547 amino acids (e.g., AC006234). Our analyses clearly confirmed the shorter ORF encoding the 526–amino acid AtPLT4 protein.
We were not able to isolate a cDNA for AtPLT6. The corresponding gene is predicted to encode the shortest AtPLT protein with only 493 amino acids (e.g., NM 119831). The predicted AtPLT6 intron after 100 bp of the AtPLT6 ORF was confirmed by the sequence of a full-length cDNA obtained during the large-scale cDNA sequencing of clones isolated from a hormone-treated callus (http://www.genoscope.cns.fr; BX827774). However, this clone differs strongly from the genomic AtPLT6 sequence in a downstream region of ∼150 nucleotides and yields the quite likely incorrect protein size of 497 amino acids.
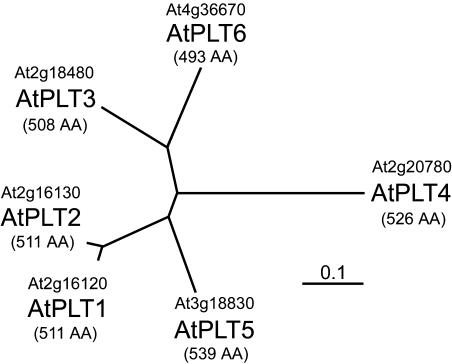
Figure 1 presents a phylogenetic tree based on the experimentally confirmed protein sequences of AtPLT1 to AtPLT5 and on the predicted sequence of AtPLT6 (NM119831). Clearly, AtPLT1 and AtPLT2, which are encoded by adjacent genes on chromosome 2, show the highest degree of sequence conservation (93.5% identity), suggesting that one of these genes formed during a recent duplication event. AtPLT4, the most distant member of the family, shares only ∼60% identical amino acids with all other AtPLTs. AtPLT3, AtPLT5, and AtPLT6 share ∼70 to 80% identity.
Figure 1.
Phylogenetic Tree of the AtPLT Family from Arabidopsis.
The deduced sequences of the six Arabidopsis AtPLTs were aligned with the program ClustalX (Thompson et al., 1997), and an unrooted tree was calculated using TreeViewX software (Page, 1996). The protein names and the MIPS numbers of the corresponding genes are given. The lengths of the proteins (AA, amino acids) were confirmed by sequencing the corresponding cDNAs for AtPLT1 to AtPLT5. The length of the AtPLT6 protein was deduced from the genomic sequence (accessions are given in the text).
Hydropathy analyses of the protein sequences predicted 12 transmembrane helices for all AtPLT proteins (data not shown) and characterized them as a separate group within the Arabidopsis monosaccharide transporter-like (MST-like) superfamily (http://www.arabidopsis.org/info/genefamily/genefamily.html/). The different lengths of the AtPLT proteins result mainly from differences in the N and C termini, with AtPLT5 (539 amino acids) having the longest C terminus, AtPLT4 (526 amino acids) having the longest N terminus, and AtPLT6 (493 amino acids) having both, the shortest N terminus and the shortest C terminus.
The identity values determined for AtPLTs and polyol transporters from other species (AgMAT1, PmPLT1, or PcSOT1) are quite similar (60 to 75%), indicating the high degree of sequence conservation between these proteins from different plant species.
Expression of the AtPLT5 cDNA in S. cerevisiae
To determine the functional properties of this new family of Arabidopsis transporters, we cloned the cDNA of AtPLT5 into the unique EcoRI site of the yeast expression vector NEV-E (Sauer and Stolz, 1994) and used the resulting plasmids harboring cDNA inserts in sense (pYK23) or antisense orientation (pYK24) to transform the yeast strain SEY2102 (Emr et al., 1983). The resulting yeast strains YKY1 (sense AtPLT5) and YKY4 (antisense AtPLT5) were used to study the possible sorbitol and mannitol transport capacity of the recombinant AtPLT5 protein. For optimal expression results, the 5′-flanking regions of the AtPLT5 cDNA were modified. To this end, a 15-bp fragment from the 5′-untranslated region of the high-affinity monosaccharide-H+ symporter gene AtSTP1 (Stadler et al., 1995) was inserted in front of the start ATG. This short sequence (AAGCTTGTAAAAGAA) is known to act as a perfect 5′-untranslated region in yeast and has been used successfully to enhance the expression of otherwise poorly expressed plant cDNAs in yeast (Stadler et al., 1995; Ramsperger-Gleixner et al., 2004).
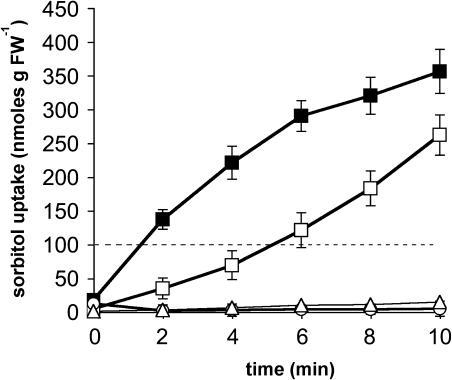
Figure 2 shows that YKY1 cells imported 14C-labeled sorbitol at a high rate, whereas no uptake of sorbitol was seen in YKY4 antisense cells. However, in contrast with all previously described polyol transporters, the AtPLT5 protein did not seem to transport 14C-labeled mannitol (Figure 2). As for other H+-symporters (Sauer et al., 1990; Barth et al., 2003; Ramsperger-Gleixner et al., 2004), we analyzed the uptake of sorbitol in the presence and absence of glucose, a metabolizable carbon source for yeast cells that is expected to enhance proton motive force-dependent transport rates by providing additional energy for the plasma membrane H+-ATPase. In contrast with what has been shown for the polyol transporters PmPLT1 and PmPLT2 from common plantain (Ramsperger-Gleixner et al., 2004), sorbitol transport by AtPLT5 was inhibited by glucose (Figure 2).
Figure 2.
Transport of Sorbitol and Mannitol in YKY1 and YKY4 Cells.
The transport capacity for sorbitol of yeast cells expressing the AtPLT5 cDNA in sense orientation (strain YKY1) was analyzed in the presence (open squares) or absence (closed squares) of 10 mM d-glucose. Open circles show the uptake rates for sorbitol in yeast cells expressing AtPLT5 in antisense orientation (strain YKY4). Mannitol uptake rates in YKY1 cells are shown by open triangles. Values represent the mean of at least three independent transport analyses (±sd). The dashed line indicates the value where the intracellular sorbitol concentration exeeds the extracellular concentration of sorbitol. FW, fresh weight.
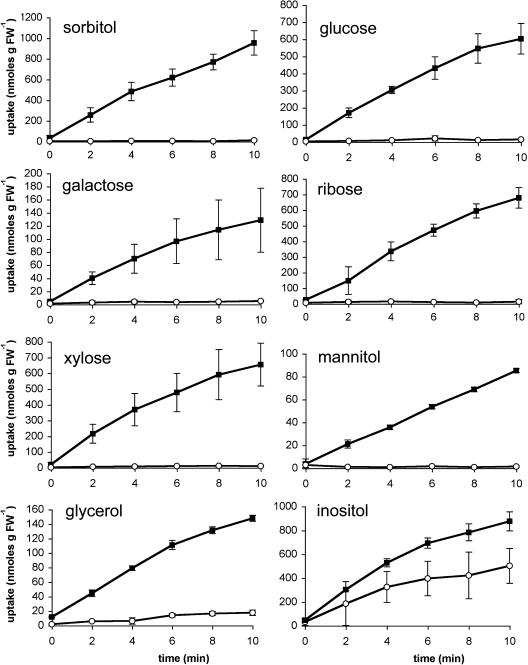
The obvious interpretation for this observed inhibition was that glucose may also be a substrate of AtPLT5. To test this hypothesis, we transformed the yeast strain EBY.VW-4000 with the plasmids pYK23 and pYK24, yielding strains YKY5 (sense AtPLT5) and YKY8 (antisense AtPLT5). Because of multiple gene disruptions, EBY.VW-4000 has no endogenous plasma membrane transporters for d-glucose (Wieczorke et al., 1999) and can therefore be used to analyze the glucose transport capacity of recombinant transporters. Figure 3 shows that AtPLT5 catalyzes the uptake of 14C-labeled glucose in YKY5, whereas no transport of 14C-glucose is seen in the antisense strain YKY8, confirming that both sorbitol and glucose are substrates of AtPLT5. This capacity to transport sugars has not been described for any of the previously published plant polyol transporters.
Figure 3.
Transport of Several Potential Substrates in YKY5 and YKY8 Cells.
The transport capacity of AtPLT5 was analyzed in the hexose transport-deficient yeast line EBY.VW-4000 after transformation with ATPLT5 (strain YKY5) sense or antisense (strain YKY8) constructs. Closed squares show the transport rates of the indicated substrates at initial outside concentrations of 0.1 mM in YKY5 (sense). Open circles show the transport rates of the indicated substrates at initial outside concentrations of 0.1 mM in YKY8 (antisense). Values represent the mean of at least three independent transport analyses (±sd). FW, fresh weight.
For further analyses of the substrate specificity, we studied the uptake of 14C-sorbitol in YKY5 cells in the presence (100-fold excess) or absence of other potential substrates. We tested the inhibitory effect of the disaccharide sucrose, of hexoses and pentoses, of linear polyols with different chain lengths (one to six carbon atoms), and of the cyclic polyol myo-inositol.
Table 1 shows that uptake of 14C-sorbitol was significantly inhibited by many of the tested competitors, suggesting that AtPLT5 may represent a transporter with an unusually wide spectrum of substrates. Unlabeled sorbitol (six carbons), but also linear polyols with five (xylitol), four (erythritol), or three carbons (glycerol) strongly reduced the import of 14C-labeled sorbitol, indicating that AtPLT5 accepts a wide range of linear polyols. Inhibition by a 100-fold excess of mannitol was less pronounced (only ∼35%), and inhibition by a 100-fold excess of dulcitol (galactitol) was only ∼50%. This suggested that despite the negligible rates of 14C-mannitol transport in YKY1 cells (Figure 2), there may be some mannitol uptake by YKY5 cells. No inhibitory effect was seen for the two-carbon polyol glycol and for methanol.
Table 1.
Inhibition of 14C-Sorbitol Uptake by Various Potential Competitors or Inhibitors in YKY5 Cells
| Competitor or Inhibitor | Residual Uptake Rate for 14C-Sorbitol (%) |
|---|---|
| – | 100 |
| Sorbitol | 9.5 ± 1.3 |
| Mannitol | 65.2 ± 4.8 |
| Dulcitol | 49.6 ± 16.5 |
| Xylitol | 3.6 ± 0.8 |
| Erythritol | 11.3 ± 2.6 |
| Glycerol | 37.8 ± 8.2 |
| Glycol | 92.5 ± 3.4 |
| Methanol | 83.4 ± 21.7 |
| Glucose | 52.1 ± 14.6 |
| Arabinose | 4.7 ± 0.6 |
| Xylose | 6.2 ± 1.9 |
| Ribose | 5.7 ± 0.4 |
| myo-inositol | 9.4 ± 0.8 |
| Sucrose | 88.2 ± 21.7 |
| PCMBS | 90.8 ± 12.5 |
| DNP | 45.5 ± 6.9 |
| CCCP | 10.6 ± 5.3 |
Values represent the mean of at least three independent uptake experiments (± sd). Potential competitors (10 mM) or inhibitors (50 μM) were added 30 s before 14C-sorbitol (0.1 mM initial outside concentration). CCCP, carbonyl cyanide-m-chlorophenylhydrazone; DNP, dinitrophenol.
Unexpectedly strong inhibition of 14C-sorbitol uptake was observed also in the presence of a 100-fold excess of myo-inositol, of different pentoses, such as xylose and arabinose, and even of ribose, which forms a furanose ring. No significant inhibition was obtained in the presence of the disaccharide sucrose.
Table 1 also shows the sensitivities of AtPLT5-dependent transport to uncouplers, such as carbonyl cyanide-m-chlorophenylhydrazone and dinitrophenol, and the effect of the SH-group inhibitor p-(chloromercuri)benzene sulfonic acid (PCMBS). Clearly, both uncouplers strongly reduced the transport rates, suggesting that AtPLT5 may catalyze the energy-dependent H+-symport of sorbitol across the yeast plasma membrane. By contrast, PCMBS had no inhibitory effect on AtPLT5-dependent transport (Table 1). This agrees with the results published for most of the polyol transporters from polyol translocating plants (Noiraud et al., 2001; Gao et al., 2003; Ramsperger-Gleixner et al., 2004). PCMBS sensitivity was found only for PmPLT1 (Ramsperger-Gleixner et al., 2004), and in this case, the sensitivity could be attributed to Cys residue Cys61 in this protein. None of the AtPLT proteins have a Cys residue at the corresponding position (in AtPLT5, it is Ile51), which explains the lack of PCMBS sensitivity.
To confirm that the reduced transport rates in the presence of other polyols or sugars (Table 1) result indeed from competition for transport, we analyzed AtPLT5-dependent uptake for selected radiolabeled competitors. In fact, all of the tested monosaccharides (two hexoses and two pentoses) turned out to be transported by AtPLT5 at similar rates (Figure 3). This was an important observation because so far significant transport rates for ribose have not been shown for any of the high-affinity, STP-like monosaccharide transporters from Arabidopsis (Sauer et al., 1990; Büttner and Sauer, 2000).
Figure 3 shows AtPLT5-driven uptake also for glycerol and even for mannitol when analyzed in YKY5 cells. Together with the competition analyses in Table 1, this demonstrates that AtPLT5 can transport linear polyols with chain lengths of three to six carbons. So far, energy-dependent uptake mechanisms for glycerol have not been described for plant cells. The low rates of mannitol transport correlate with its small inhibitory effect on 14C-sorbitol uptake (Table 1), and with the almost undetectable mannitol transport rates seen in Figure 2. Obviously, AtPLT5 is expressed to higher levels in YKY5 cells than in YKY1 cells (see also the difference in sorbitol uptake in Figures 2 [YKY1] and 3 [YKY5]). This difference is attributable to the two yeast strains used for these expression analyses (SEY2102 and EBY.VW-4000).
Unexpectedly, even 14C-inositol was transported with higher rates by YKY5 cells than by YKY8 antisense cells (Figure 3). However, a rather high inherent transport activity for this substrate was also seen in YKY8 control cells.
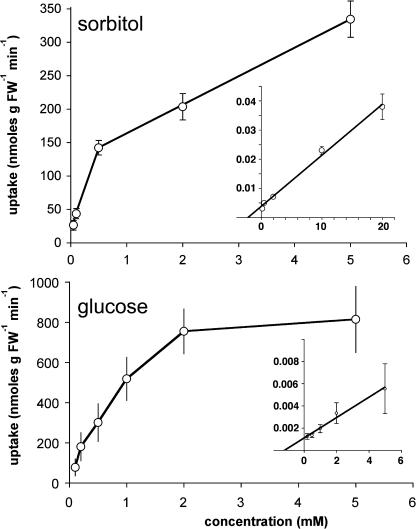
The Km values of recombinant AtPLT5 in yeast were determined for sorbitol (0.5 ± 0.1) and glucose (1.5 ± 0.8) (Figure 4). These values (mean of three analyses) agree with slightly higher sorbitol transport for sorbitol than for glucose (Figure 3). These data also show that the Km value for glucose of AtPLT5 is clearly higher than the Km values previously determined for the members of the high-affinity AtSTP monosaccharide transporters having Km values for glucose between 10 and 150 μM (Sauer et al., 1990; Büttner and Sauer, 2000).
Figure 4.
Km Value of AtPLT5 for Sorbitol Transport and for Glucose Transport in Transgenic Yeast Cells.
Michaelis-Menten diagrams and reciprocal Lineweaver-Burk diagrams (insets) showing the dependence of the sorbitol or glucose transport rates of the extracellular substrate concentrations. All curves represent the data of three independent experiments, and error bars show the standard deviations. FW, fresh weight.
Expression of the AtPLT5 cDNA in X. laevis Oocytes
The fact that sorbitol can be accumulated by AtPLT5-expressing yeast cells to intracellular concentrations exceeding the initial outside concentrations (dashed line in Figure 2) as well as the sensitivity of AtPLT5-driven polyol transport to uncouplers (Table 1) provides indirect evidence for an energy-dependent uptake mechanism. For a direct analysis of its energy dependence and for further analyses of its substrate specificity, AtPLT5 was analyzed in Xenopus oocytes injected with AtPLT5 cRNA (Figure 5).
Figure 5.
Substrate Specificity and Affinities of AtPLT5 in X. laevis Oocytes.
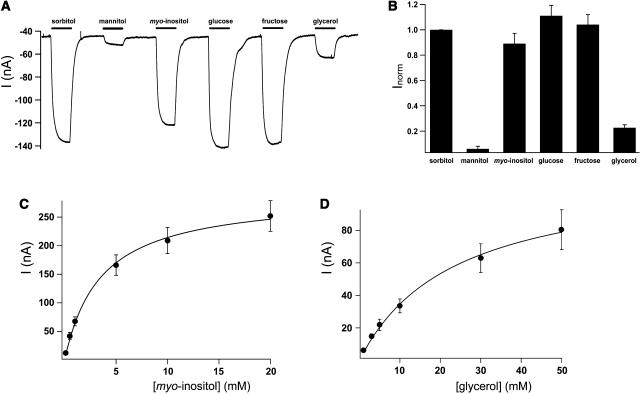
(A) AtPLT5-mediated H+-currents in response to various sugars and sugar alcohols (all 3 mM) were recorded at pH 5.5 and at a membrane potential of −60 mV. Substrates were added for 20 s (black bars on top of current transients).
(B) Normalized AtPLT5-mediated H+-currents (Inorm) gained from three independent experiments as shown in (A). Data were normalized to the currents elicited in response to sorbitol.
(C) Saturation curve for myo-inositol–induced H+-currents (mean of three experiments ± sd). Data were fitted with Michaelis-Menten–type kinetics, revealing a Km value for myo-inositol of 3.5 ± 0.3 mM (at −60 mV and pH 5.5).
(D) Saturation curve for glycerol-induced H+-currents (mean of three experiments ± sd). Data were fitted with a Michaelis-Menten–type kinetics, revealing a Km value for glycerol of 23.4 ± 2.3 mM (at −60 mV and pH 5.5).
Figure 5A shows that similar inward currents were obtained upon perfusion with 3-mM solutions of sorbitol, glucose, fructose, and myo-inositol at an extracellular pH of 5.5. Smaller currents were observed in the presence of 3-mM glycerol and currents almost zero in the presence of mannitol (Figure 5A). This and the normalized values shown in Figure 5B confirm the yeast data for sorbitol and glucose transport (Figure 3) and demonstrate that fructose and inositol also are transported at similar rates. By contrast, and in agreement with the data obtained in the yeast expession system (Figure 3), glycerol is transported at a clearly lower rate (∼20%).
These data confirm that AtPLT5 is also a transporter for myo-inositol and glycerol, two substrates that inhibited the uptake of 14C-sorbitol in yeast (Table 1) but yielded only low transport rates (glycerol in Figure 3) or were difficult to analyze because of a strong inherent transport activity (myo-inositol in Figure 3). The results also confirm that in contrast with all previously described polyol transporters, AtPLT5 does discriminate between sorbitol and mannitol. Finally, the obtained inward currents demonstrate that a positive charge is symported with each of the tested substrates, confirming the interpretation that AtPLT5-driven transport is energy dependent.
Figures 5C and 5D show the Michaelis-Menten kinetics of AtPLT5 for myo-inositol and glycerol in Xenopus oocytes. The Km values were determined at −60 mV and at an extracellular pH of 5.5 and were 3.5 ± 0.3 mM for myo-inositol and 23.4 ± 2.3 mM for glycerol. Additional analyses of the Km values for myo-inositol from 40 to −140 mV revealed that it is voltage (Δψ) dependent and increases with more negative potentials. At −140 mM, the Km of AtPLT5 for myo-inositol is ∼1 mM (data not shown).
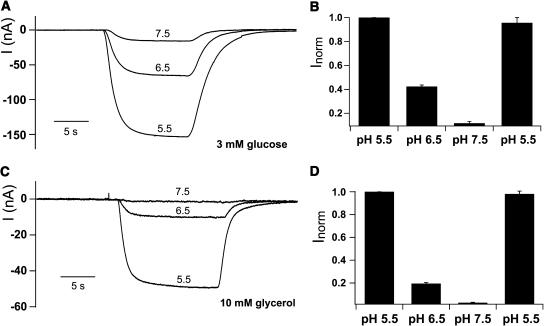
The nature of the cotransported ion was characterized for glucose (Figures 6A and 6B) and glycerol (Figures 6C and 6D). In Na+-free buffer systems, inward currents induced by both substrates increased with decreasing extracellular pH values, indicating that the cotransported ions are protons and characterizing AtPLT5 as an H+-symporter. Figures 6B and 6D show that at extracellular pH values >7, the activity of AtPLT5 is almost zero. An identical pH dependence was obtained for sorbitol uptake in yeast, where transport rates for sorbitol at pH 5.0 and pH 7.0 differed >90% (data not shown).
Figure 6.
Glucose- and Glycerol-Induced AtPLT5 Currents Depend on the H+-Gradient across the Plasma Membrane.
(A) Xenopus oocytes expressing AtPLT5 respond to 3-mM glucose in a pH-dependent manner. Inward H+-currents were elicited by 15-s glucose pulses at pH 7.5, 6.5, or 5.5 at a membrane potential of −60 mV.
(B) Normalized AtPLT5 currents (Inorm) in response to 3-mM glucose recorded at extracellular pH values of 5.5, 6.5, or 7.5. Note the lack of AtPLT5-mediated H+-currents at pH 7.5. The complete reversibility of the pH effects was tested by glucose treatments at pH 5.5 before (first bar) and after (fourth bar) the pH shifts to 6.5 and 7.5 (means of four experiments ± sd; normalized to currents at pH 5.5).
(C) Xenopus oocytes expressing AtPLT5 were challenged as described in (A), but with 10 mM glycerol.
(D) Normalized, AtPLT5-dependent H+-currents (Inorm) in response to 10-mM glycerol at pH 5.5, 6.5, and 7.5 were gained from four independent experiments as shown in (C) (means ± sd, n = 4). The complete reversibility of the pH effects was shown as described in (B).
Transient Expression of an AtPLT5-GFP Construct
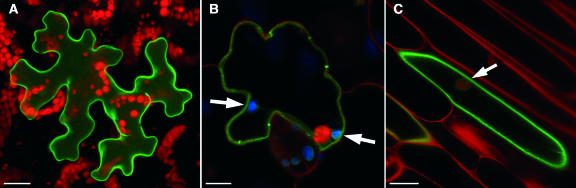
Both in the yeast system and in the Xenopus system, AtPLT5 activity was clearly localized to the plasma membrane. The subcellular localization of AtPLT5 in plant cells was analyzed by transient expression of an AtPLT5 construct that carried an in-frame GFP fusion at its 3′-end. To this end, the plasmid pYK25 that drives expression of this AtPLT5-GFP fusion under the control of an enhanced 35S promoter was used for transient expression in Arabidopsis and onion epidermis cells after transformation with a self-made particle inflow gun.
Analyses of AtPLT5-GFP–expressing Arabidopsis (Figure 7A) and onion (Figure 7B) epidermis cells with a confocal microscope showed that fluorescence resulting from this fusion is confined to the plasma membrane. Thus, AtPLT5 is s plasma membrane–localized H+-symporter.
Figure 7.
Transient Expression of an AtPLT5-GFP Fusion Construct in Arabidopsis and Onion Epidermal Cells.
(A) Confocal image showing green AtPLT5-GFP fluorescence in a cell from the lower epidermis of an Arabidopsis leaf (maximal projection of 10 sections = 20.5 μm). Red staining shows chlorophyll fluorescence of chloroplasts in the mesophyll.
(B) Confocal image (single section) showing green AtPLT5-GFP fluorescence in a cell from the lower epidermis of an Arabidopsis leaf. The leaf was treated with propidium iodide, which results in red staining of nuclei and cell walls. Chloroplasts are shown in blue (false color) and are inside the labeled structure (arrows), which is therefore clearly characterized as the plasma membrane.
(C) Confocal image (single section) showing green AtPLT5-GFP fluorescence in a cell from the inner epidermis of a white onion. The section was stained with propidium iodide as in (B). The arrow demonstrates that AtPLT5-GFP fluoresence is outside the nucleus. This proofs that the labeled structure is the plasma membrane and not the tonoplast.
Bars = 20 μm in (A), 8 μm in (B), and 40 μm in (C).
Analysis of AtPLT5 Expression in AtPLT5 Promoter:GUS and AtPLT5 Promoter:GFP Plants
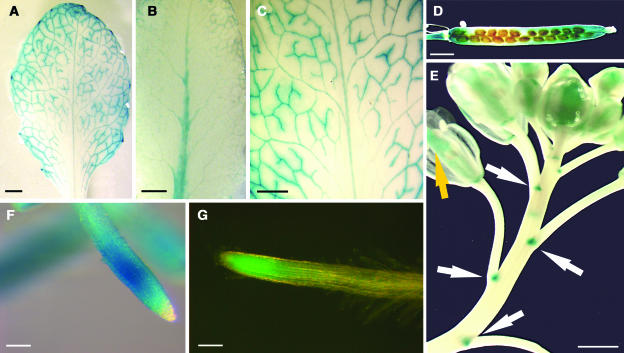
For analysis of the tissue specificity of AtPLT5 expression, we generated and analyzed AtPLT5 promoter:GUS and AtPLT5 promoter:GFP plants. A 1551-bp promoter fragment was used to drive the expression of GUS or GFP in plants that had been selected for BASTA resistance after transformation with the plasmids pYK10 (AtPLT5 promoter:GUS) or pYK13 (AtPLT5 promoter:GFP). We obtained numerous GUS- or GFP-expressing transformants and analyzed 24 independent AtPLT5 promoter:GUS lines and 24 independent AtPLT5 promoter:GFP lines. Figures 8A to 8C show the typical GUS staining found in rosette leaves of AtPLT5 promoter:GUS plants. In all plants analyzed and in most leaves of these plants, GUS staining was detected in or along the vascular tissue. However, typically this staining, which was frequently seen only after a prolonged period of staining, was patchy (Figures 8A and 7C) and often more or less absent from the midrib. In other leaves, however, GUS staining was observed mainly in the midrib (Figure 8B), and sometimes the leaves stayed completely white. Weak GUS staining was also seen along the vascular strands of stems (data not shown), of all sepals (Figure 8E), and of siliques (data not shown). Siliques typically showed an additional, more general staining near both ends (Figure 8D). Distinct AtPLT5 promoter activity was also seen in the axillary regions of all flowers (arrows in Figure 8E). In none of the tested lines was GUS staining seen in seeds, pollen, or petals (data not shown).
Figure 8.
GUS and GFP Reporter Gene Analyses.
(A) GUS-histochemical staining of a rosette-leaf from a AtPLT5-promoter:GUS plant showing patchy GUS-staining in or along the vascular strands.
(B) GUS histochemical staining of a rosette leaf from a AtPLT5-promoter:GUS plant showing significant GUS staining mainly in the midrib.
(C) GUS histochemical staining of a rosette leaf of the patchy type as shown in (A).
(D) GUS histochemical staining of a mature silique showing blue staining mainly at both ends of the silique.
(E) GUS histochemical analysis of a fluorescence showing very weak GUS staining along the vascular strands of the sepals and in the ovary (yellow arrow). GUS staining is also seen in the axillary regions of each individial flower (arrows).
(F) GUS histochemical analysis showing strong GUS staining in a region of ∼300 to 500 μm behind the root tip.
(G) The GFP fluorescence in the tip of an AtPLT5-promoter:GFP root confirms the GUS activity shown in (F).
Bars = 1 mm in (A) to (D), 2 mm in (E), and 0.1 mm in (F) and (G).
In 100% of the plants, the strongest GUS staining was detected over a distance of ∼300 to 500 μm behind the root tip (Figure 8F). Weaker GUS staining was also seen in the upper part of most roots (data not shown). Only this strong GUS activity in the tip regions could was detected in analyses of AtPLT5 promoter:GFP plants (Figure 8G). Leaf- or flower-specific expression has not been found in any of the GFP lines, suggesting that expression in these tissues is much weaker than in roots.
Immunohistochemical Analyses of AtPLT5 Localization
Three anti-AtPLT5 antisera (αAtPLT5-K1, αAtPLT5-K2, and αAtPLT5-M1) were raised against the peptide NH2-CEIGSNKQWKEGDTQSS-COOH that corresponds to amino acids 524 to 539 from the very C terminus of AtPLT5. This sequence is unique to AtPLT5 and is lacking in all other AtPLT proteins with shorter C-terminal sequences.
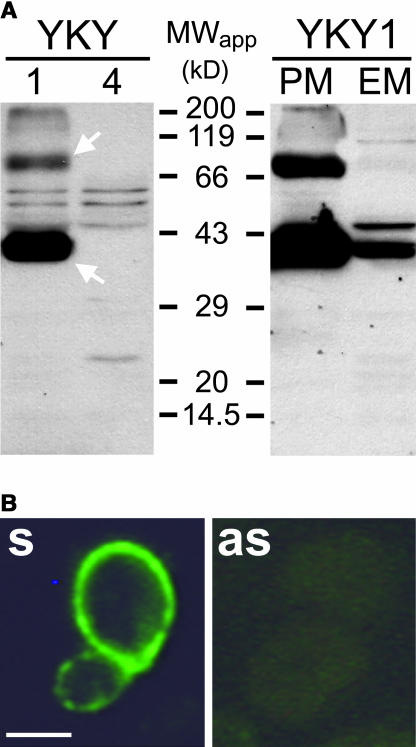
The quality of the obtained sera was tested on detergent extracts from total membranes isolated from YKY1 (sense) and YKY4 (antisense) yeast cells. Figure 9A shows a protein gel blot of these extracts after gel chromatographic separation and incubation with αAtPLT5-K1. A strong signal at an apparent molecular mass of ∼40 kD and a weaker signal at ∼80 kD were detected only in YKY1 cells and were absent in similar extracts from YKY4 controls, indicating that these bands represent the monomeric form (40 kD) and the most likely dimerized form (80 kD) of the AtPLT5 protein. The difference of ∼18 kD between the apparent molecular mass of the monomeric form and the calculated molecular mass of 58.1 kD as well as the appearance of a dimer is typical for highly lipophilic proteins (Beyreuther et al., 1980; Gahrtz et al., 1994; Barth et al., 2003). Similar results were obtained with the other anti-AtPLT5 antisera (data not shown).
Figure 9.
Identification of the AtPLT5 Protein in Transgenic Yeast Cells.
(A) Protein extracts from total membranes (10 μg per lane) from YKY1 (lane 1 on left protein gel blot) and YKY4 cells (lane 4 on left protein gel blot) were separated by gel electrophoresis, transferred to a nitrocellulose filter, and incubated with αAtPLT5-K1 antiserum. AtPLT5 signals at 40 and 80 kD (arrows) are detected only in total membranes of YKY1 cells. The right protein gel blot (10 μg per lane) shows AtPLT5 immunodetection in protein extracts from enriched plasma membranes (PM) and from enriched endomembranes (EM). AtPLT5 signals at 40 and 80 kD are strongly enriched in the plasma membranes. In both protein gel blots, binding of antiserum was visualized with anti-rabbit IgG antiserum conjugated to peroxidase.
(B) Cross sections of yeast cells expressing the AtPLT5 cDNA in sense (s) or antisense orientation (as). Sections were treated with αAtPLT5-K1, decorated with fluorescein isothiocyanate (FITC)–labeled 2nd antibody, and photographed under FITC excitation light. AtPLT5-dependent fluorescence is detected only in the sense strain, where it concentrates at the cell surface, most likely the plasma membrane (see [A]). Bar = 2 μm.
Separation of the yeast total membrane fraction into a plasma membrane–enriched fraction and an endomembrane-enriched fraction (Sauer and Stolz, 2000) localized the majority of the label to the plasma membranes. The small amount of AtPLT5 protein detected in the endomembrane fraction is likely to result from contaminating plasma membranes in this fraction (Figure 9A). This localization was confirmed in immunohistochemical analyses of YKY1 and YKY4 cells, where signals were detected only at the cell surface of the YKY1 sense strain (Figure 9B). No signals were seen in sections of YKY4 antisense cells (Figure 9B).
The antisera were also used in numerous attempts to immunolocalize AtPLT5 in sections of Arabidopsis roots and leaves. Unfortunately, the antisera gave no signals with different embedding and fixation protocols. In view of the specific and strong signals obtained in AtPLT5-expressing yeast cells, we speculate that the amount of AtPLT5 protein in planta is too low for immunohistochemical detection.
Analysis of AtPLT5 Expression by RT-PCR
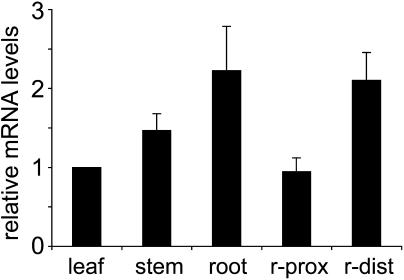
To confirm the tissue-specific expression shown in Figure 8 by an independent technique, we performed light cycler RT-PCR analyses of AtPLT5 mRNA levels in different tissues. Parallel reactions were performed with ACT2 (An et al., 1996) and UBQ10 (Sun and Callis, 1997) standards to minimize potential tissue-specific differences in the expression levels of a single control. Figure 10 gives the data calculated with the ACT2 control; results calculated with the UBG10 are not shown because they were almost identical. The results show only minor differences in AtPLT5 mRNA levels of the analyzed tissues, suggesting that there may be low AtPLT5 expression also in cells and tissues that are not stained in the AtPLT5 promoter:GUS or AtPLT5 promoter:GFP plants. Moreover, these data confirm the higher levels of AtPLT5 expression in the distal regions of the roots, which were also seen in the AtPLT5 promoter:reporter gene plants (Figure 8).
Figure 10.
Real-Time RT-PCRs with Different Arabidopsis Tissues.
Relative AtPLT5 mRNA levels were analyzed in leaves, stems, and roots and in distal (r-dist) and proximal (r-prox) Arabidopsis root preparations. Data were calculated on the basis of AtACT2 control values, normalized for AtPLT5 mRNA levels in leaves, and represent the mean values of four independent analyses (±sd).
Analysis of Mutant Plants Harboring a T-DNA Insertion in the AtPLT5 Gene
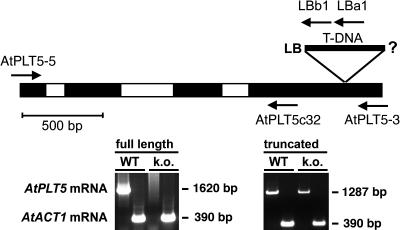
Screening of publically available libraries identified a mutant line (Salk_050162) carrying a T-DNA insertion in the 4th exon of the AtPLT5 gene, 1966 bp after the start ATG (Figure 11). An insertion at this position results in a truncated AtPLT5 mRNA containing only 1374 bp of the AtPLT5 ORF (Figure 11). This mRNA is predicted to encode a protein of only 458 amino acids. In the wild-type AtPLT5 protein, the 81 C-terminal amino acids that are lacking in the truncated mutant protein encode transmembrane helix number 12 and a predicted cytoplasmic C terminus of ∼50 amino acids. The full-length mRNA was no longer present in the mutant line (Figure 11). The truncated AtPLT5 mRNA was, however, identified using primers binding in front of the T-DNA insertion. For the reasons given above, it is not likely that this mRNA encodes a functional protein.
Figure 11.
Identification of a T-DNA Insertion in the AtPLT5 Gene.
The position and orientation of the T-DNA insertion in the AtPLT5 gene of the mutant line Salk_050162 is presented. The gene has three introns (white boxes), and the insertion is located in the 4th exon (black boxes) at a position corresponding to nucleotide 1374 of the AtPLT5 ORF. The orientation of the left border (LB) is indicated, and the opposite end of the insertion has not been characterized. Arrows indicate the position and orientation of primers (AtPLT5-5, AtPLT5-3, AtPLT5c32, LBa1, and LBb1) that were used to discriminate between wild-type and heterozygous or homozygous mutant plants and between full-length and truncated mRNAs. The ethidium bromide–stained agarose gels demonstrate that the 1620-bp full-length mRNA, which is present in wild-type Arabidopsis plants, could no longer be detected in the T-DNA mutant (k.o.) by RT-PCR using the AtPLT5-5 and AtPLT5-3 primers. Under identical conditions, a fragment (390 bp) of the AtACT1 mRNA was amplified in both wild-type and mutant plants. Using the primer combination AtPLT5-5 and AtPLT5c32, the truncated form of the AtPLT5 mRNA (1287 bp) could still be identified in the mutant line.
When the phenotypes of this mutant line and of the corresponding wild-type plants were compared, we observed no differences in the growth phenotype under ambient CO2 concentrations and under the normal growth conditions described in Methods. Growth of mutant and wild-type plants was also compared under salt stress (100 mM NaCl), under drought stress on soil, and finally on Petri plates in the presence or absence of one of the transported substrates (10 and 50 mM sorbitol, inositol, or glycerol; data not shown). Under none of these conditions was a significant difference in plant growth or development observed.
DISCUSSION
This article describes the first member of a new class of plasma membrane–localized H+-symporters from Arabidopsis exhibiting a quite unusual substrate specificity. AtPLT5 represents the first transporter catalyzing the transport of substrates, such as myo-inositol, glycerol, or ribose across this membrane. AtPLT5 represents one of six transporters that form one (AtPLT1 to AtPLT6) of seven subfamilies of the MST-like superfamily in Arabidopsis, which was named after the intensively characterized AtSTP gene family that encodes 14 different plasma membrane–localized monosacharide transporters (Sauer et al., 1990; Büttner and Sauer, 2000).
So far, only members of the STP subfamily of the Arabidopsis MST-like superfamily were characterized by functional expression, although individual members of several other subfamilies have been studied by several groups (At1g08930 [AtERD6], a dehydration-induced gene, Kiyosue et al., 1998; At5g16150 [pGlcT], a putative plastidic transporter, Weber et al., 2000; At5g27350 [AtSFP1], a senescence-induced gene, Quirino et al., 2001). In all of these cases, the functional analysis of the corresponding proteins failed.
Here, a detailed functional characterization of AtPLT5 (At3g18830) after expression of its cDNA in bakers yeast and in Xenopus oocytes is presented. In both expression systems, AtPLT5 was characterized as a polyol/cyclitol/monosaccharide-H+-symporter that is able to catalyze the energy-dependent membrane passage of a wide range of linear polyols (three to six carbon backbone), of cyclic polyols (myo-inositol), and of numerous monosaccharides, including pyranose ring-forming and furanose ring-forming hexoses and pentoses (Figures 3 and 5A).
AtPLT5 Is a Plasma Membrane–Localized Plant Transporter That Mediates Transport of Inositol, Ribose, or Glycerol
Our immunolocalization data in yeast cells (Figure 9B), the accumulation of AtPLT5 protein in yeast plasma membranes (Figure 9A), the activities in yeast and Xenopus plasma membranes (Figures 2, 3, 5, and 6), and finally the localization of an AtPLT5-GFP fusion in Arabidopsis and onion epidermis cells (Figure 7) clearly demonstrate that AtPLT5 is a plasma membrane protein. For several of its identified substrates so far, no transport activity has been described in plant plasma membranes, and for others only uptake via facilitated diffusion systems has been published. For example, the only plant inositol transporters cloned so far were identified in the common ice plant (Mesembryanthemum crystallinum; Mitr1 and Mitr2), and the encoded transporters were shown to be located in the tonoplast (Chauhan et al., 2000). Homologs of the Mitr1 and Mitr2 proteins are present in the Arabidopsis genome (At1g30220, At2g35740, At2g43330, and At4g16480), and like the AtPLT genes, these four genes form a small subgroup within the Arabidopsis MST-like superfamily. To date, none of these four possible Arabidopsis inositol transporters have been functionally characterized, but first localization analyses identified these proteins also in the tonoplast and not in the plasma membrane of Arabidopsis (N. Sauer and S. Schneider, unpublished data).
In animals, a Na+/myo-inositol symporter gene, SMIT1, has been cloned already more than 20 years ago (Kwon et al., 1992). The encoded SMIT1 protein shows significant homology to the animal Na+/glucose symporter SGLT1 (Hediger et al., 1987), but both Na+ symporters do not share significant homology with the plant MST-like genes. Energy-dependent inositol transporter genes (ITR1 and ITR2) that are likely to be H+-symporters have also been cloned from yeast (Nikawa et al., 1991). These proteins have Km values between 0.1 and 0.2 mM and share homology with the Arabidopsis MST-like superfamily.
Glycerol transport activity across plant plasma membranes has so far only been shown for members of the aquaporin family (Weig and Jakob, 2000; Wallace and Roberts, 2004), which perform only facilitated diffusion and are closely related to the Escherichia coli GlpF glycerol permease (Sweet et al., 1990; Maurel et al., 1994; Zardoya et al., 2002). To our knowledge, energy-dependent glyerol transporters of plant plasma membranes or internal membranes have not been described so far. By contrast, genes encoding energy-dependent, plasma membrane–localized transporters for glycerol have been cloned from bakers yeast (GUP1 and GUP2; Oliveira and Lucas, 2004), but the corresponding proteins share no significant similarity with AtPLT5.
Finally, no transporter for ribose has been characterized so far in plant plasma membranes. Although numerous plant monosaccharide transporter genes and cDNAs have been cloned over the last years (Williams et al., 2000), significant amounts of ribose transport have not been shown for any of the analyzed proteins. By contrast, ribose was shown to be excluded from the Chlorella kessleri CkHUP1 monosaccharide transporter and used as a nontransported control substrate (Tanner, 1980). This is quite similar for the different Arabidopsis monosaccharide transporters of the AtSTP family, which transport ribose either not at all or with extremely low rates (Büttner and Sauer, 2000; M. Büttner, unpublished data). Even AtSTP6 (Scholz-Starke et al., 2003), the only Arabidopsis transporter that transports fructose, a furanose ring–forming hexose, at reasonably high rates (50% of the rate of glucose), exhibits only marginal transport rates for ribose (1% of the rate of glucose). This may be explained with binding sites that are optimized for pyranose rings formed by aldohexoses and by several pentoses, such as arabinose or xylose. Fructose or ribose, which form furanose rings in solution, may therefore be poor substrates for STP-type transporters (Büttner and Sauer, 2000; Williams et al., 2000). AtPLT5 does transport ribose, fructose, and glucose at similar rates (Figures 3 and 5A), suggesting little selectivity for the AtPLT5 substrate binding pocket.
A high-affinity transporter for ribose has previously been described in E. coli (Iida et al., 1984), but RbsC, the membrane component of this transporter, has only 10 transmembrane helices (Stewart and Hermodson, 2003) and is not related to the AtPLT transporters of Arabidopsis.
AtPLT5 and the other five members of the AtPLT family share significant homology with known polyol transporters catalyzing the transport of mannitol or sorbitol in polyol translocating plants (Noiraud et al., 2001; Gao et al., 2003; Ramsperger-Gleixner et al., 2004). These previously characterized transporters exhibited drastic differences with respect to their sensitivities to monosaccharides. Whereas both sorbitol transporters from common plantain showed enhanced transport rates in the presence of glucose, which was interpreted with an additional energy supply for an active transport system (Ramsperger-Gleixner et al., 2004), strong inhibition by glucose and other hexoses was seen for the mannitol transporter AgMAT1 from celery (Noiraud et al., 2001) and for the two sorbitol transporters from sour cherry, PcSOT1 and PcSOT2 (Gao et al., 2003). This inhibition was interpreted as an artifact of the yeast expression system or as repression of transporter gene expression by glucose (Noiraud et al., 2001; Gao et al., 2003). In view of our data, it may well be that the observed inhibition of polyol transport by hexoses for AgMAT1, PcSOT1, and PcSOT2 is also because of sugar transport by these proteins.
AtPLT5 Is a H+-Symporter
All plant plasma membrane sugar and polyol transporters of the 12 transmembrane helix-type (STPs, PLTs, and SUCs) were shown to be energy-dependent H+-symporters and to be voltage dependent (Williams et al., 2000; Ramsperger-Gleixner et al., 2004). Uncoupler sensitivity of AtPLT5 in yeast (Table 1) and the intracellular accumulation of sorbitol to concentrations higher than in the external medium (Figure 2) suggested that this may also be the case for this protein. Figure 6 confirms this interpretation, showing that in the absence of Na+ ions, inward currents increased with decreasing pH. The voltage dependence of AtPLT5 was confirmed for myo-inositol transport in the range from 40 to −140 mV (data not shown).
AtPLT5 Is Expressed in Most Plant Tissues
The presented analyses of AtPLT5 promoter:GUS and of AtPLT5 promoter:GFP plants (Figure 8) shows AtPLT5 promoter activity in leaves (preferably along the vascular tissue), in flowers (especially in sepals and siliques), in the root (most strongly in the tip areas), and in the axillary regions of the individual flowers (Figure 8). No AtPLT5 expression was seen in anthers and pollen, in petals, and in ovules (Figure 8E; data not shown). This distribution of AtPLT5 promoter activity correlates well with At3g18830 expression patterns determined in numerous analyses. The Arabidopsis MPSS database found no specific expression of AtPLT5 in only one tissue or organ (http://mpss.udel.edu/at/java.html/), and the result of numerous microarray analyses (http://www.arabidopsis.org/; http://www.cbs.umn.edu/arabidopsis/) was that AtPLT5 is expressed in almost all tissues and organs analyzed, with the exception of pollen and seeds. Moreover, there was a clear difference between the expression in sepals and petals, with AtPLT5 mRNAs being almost absent in petals and found in quite high levels in sepals. All of these data agree with our GUS and GFP analyses.
In our light cycler analyses (Figure 10), we analyzed AtPLT5 mRNA levels in several Arabidopsis tissues and especially the expression in distal and proximal regions of roots. As in the microarray analyses mentioned above, we found only marginal differences between the AtPLT5 expression levels in roots, leaves, and stems. The higher expression levels in the distal parts of the root confirmed the strong GUS activity in Figure 8F.
In summary, our data suggest that AtPLT5 is expressed at rather low levels in most tissues. Our GUS and GFP analyses identify those parts of the plant where expression levels are increased above this basic level. The data are in agreement with microarray analyses.
What Is the Physiological Substrate of AtPLT5?
The broad substrate specificity of the protein on the expression of the gene in multiple tissues and organs and on the unchanged phenotype of Atplt5-k.o. plants (Figure 11) make it difficult to speculate on the physiological role of AtPLT5. Open questions are, for example, (1) which of the identified substrates of AtPLT5 is the main substrate under physiological conditions? (2) Does AtPLT5 transport only one or several of the characterized substrates in planta? (3) Can AtPLT5 transport even other substrates that have not been tested in these analyses? For the last part, the answer is probably yes. We have analyzed only a limited set of monosaccharides and linear or cyclic polyols, and most of these substrates were transported with similar rates (Figures 3 and 5A). It is very likely that other compounds are also accepted as substrates by AtPLT5.
The affinities of AtPLT5 were shown to be 0.5 mM for sorbitol and 1.5 mM for glucose in the yeast system (Figure 4). The affinities for myo-inositol and for glycerol were determined in Xenopus oocytes, and at a constant Δψ of 60 mV they were shown to be 3.5 and 23.4 mM, respectively (Figures 5C and 5D). However, the affinity for myo-inositol increases with more negative potentials and is ∼1 mM at −140 mV (data not shown), which is in the range of the Km values for glucose or sorbitol. This explains why glucose, sorbitol, and myo-inositol are transported with similar rates. It is likely that the affinity for glycerol, which is transported at lower rates (Figures 3 and 5A), shows the same voltage dependence, and one can calculate a Km value of 5 to 7 mM for glycerol in fully energized plasma membranes.
A simple explanation for the function of ATPLT5 might be the retrieval of multiple substrates from the apoplast. All identified substrates are major components of the cellular metabolism and may leak out of the cells. However, more specific physiological functions (e.g., in the cell-to-cell distribution of certain compounds, possibly of different substrates in different tissues) certainly can not be excluded.
METHODS
Strains and Growth Conditions
Arabidopsis thaliana plants were grown in growth chambers on potting soil under a 16-h-light/8-h-dark regime at 22°C and 60% relative humidity or in the greenhouse under ambient conditions. For heterologous expression of AtPLT5 cDNAs in yeast, we used strains SEY2102 (Emr et al., 1983) or EBY.VW-4000 (Wieczorke et al., 1999). The Escherichia coli strain DH5α (Hanahan, 1983) was used for all cloning steps. Transformation of Arabidopsis was performed using Agrobacterium tumefaciens strain GV3101 (Holsters et al., 1980).
cDNA Cloning and Constructs for Expression in Yeast
cDNAs of the six Arabidopsis AtPLT genes were amplified from total RNA isolated from Arabidopsis Columbia-0 with gene-specific primers binding to the very 5′-ends (including the start ATG) or the very 3′-ends (including the stop codon) of the cDNAs. NotI cloning sites were introduced at both ends of AtPLT1 and AtPLT2, and EcoRI cloning sites were introduced at both ends of AtPLT3, AtPLT4, and AtPLT5. The resulting cDNAs were digested with NotI or EcoRI, cloned into the respective sites of the yeast expression vectors NEV-E or NEV-N (Sauer and Stolz, 1994) and sequenced, and the AtPLT5-containing plasmid used for transformation of yeast cells (Gietz et al., 1992). If not otherwise indicated, uptake experiments were performed in 50-mM sodium phosphate buffer, pH 5.0, as described (Sauer et al., 1990).
Heterologous Expression in Xenopus laevis Oocytes
For functional analysis, AtPLT5 cRNA was prepared using the T7 mMESSAGE mMACHINE RNA transcription kit (Ambion, Austin, TX). Oocyte preparation and cRNA injection have been described elsewhere (Becker et al., 1996). In two-electrode voltage-clamp studies, oocytes were perfused with 100-mM KCl-containing solutions, based on Tris/Mes buffers for pH values from 5.5 to 7.5. The standard solution contained 10-mM Tris, pH 5.5, 100-mM KCl, 1-mM CaCl2, and 1-mM MgCl2. Osmolarity was adjusted to 220 mOsmol/kg using sucrose. The content of substrates and the pH values are indicated in the figures and figure legends. Steady state currents (ISS) were recorded with single-pulse protocols to 500-ms test voltages from 40 to −140 mV from a holding potential (VH) of −20 mV. Currents in the absence of substrates were subtracted for leak correction.
AtPLT5 Promoter:GUS and of AtPLT5 Promoter:GFP Constructs and Plant Transformation
A 1551-bp promoter AtPLT5 promoter fragment was PCR amplified from genomic DNA (Arabidopsis Columbia-0) using the primers AtPLT5p-5 (5′-AAAATTCATAAGCTTCATAACAGCGATTGCTCTCG-3′) and AtPLT5p-3 (5′-CATATCGCCATGGTGATAGAGAATGGGGCGAGAGAGA-3′). The fragment was cloned into pGEM-T Easy (Promega, Madison, WI) and sequenced, and the insert was cloned in front of the GFP reporter gene in the vector pGA03 or in front of the GUS reporter gene in the vector pAF6 (pAF6 and pGA03 are pUC19-based plasmids, harboring the GFP [pGA03] or GUS [pAF6] reporter genes). From the resulting plasmids, AtPLT5 promoter:GFP or AtPLT5 promoter:GUS fragments were excised and cloned into pAF16 (Stadler et al., 2005), yielding the plasmids pYK10 (AtPLT5 promoter:GUS) and pYK13 (AtPLT5 promoter:GFP), which were used for transformation of Arabidopsis (Clough and Bent, 1998).
Transient Expression of AtPLT5-GFP
The transient expression plasmid pSO35e was generously provided by M. Büttner (Molecular Plant Physiology, FAU-Erlangen-Nürnberg, Germany). It was made from pAF1, a pUC19-based plasmid (A. Feuerstein and N. Sauer, unpublished data) containing the AtSUC2 promoter (Truernit and Sauer, 1995), the GFP ORF (with a unique NcoI site in the start ATG), and the nopaline synthase terminator. The AtSUC2 promoter of pAF1 was replaced by the 818-bp HindIII/BamHI fragment representing the enhanced 35S promoter from pFF19G (Timmermans et al., 1990).
The AtPLT5 coding sequence was PCR amplified using the primers AtPLT5-NcoI-5 (5′-GACACACCATGGGAATGACAGGTGCCACACCGGAAAA-3′) and AtPLT5-NcoI-3 (5′-GACACACCATGGAACTTTGTGTGTCTCCTT-3′). These primers introduced an NcoI site in front of the AtPLT5 start ATG and a second NcoI site after the last amino acid of the AtPLT5 ORF. This second NcoI site also replaced the stop codon of the original AtPLT5 sequence. The modified AtPLT5 ORF was inserted into the unique NcoI cloning site covering the start ATG of the GFP ORF in the pSO35e plasmid. The correct insertion was confirmed by sequencing, the resulting plasmid was named pYK25.
For transient expression of AtPLT5-GFP from pYK25, 5 mg of surface-sterilized (70% ethanol [v/v]) gold particles (0.3- to 3-μm diameter) were resuspended in 50 μL of water and mixed with 10 μL of pYK25 DNA (1 μg/μL in water). To this mixture, 50 μL 2.5-M CaCl2 and 20 μL of 0.1-M spermidin were added. After the stepwise addition of 100 μL 100% ethanol (−20°C) and of 200 μL 100% ethanol (−20°C), the mixture was kept at −20°C for 30 min and centrifuged for 1 min at 14.000g, and gold particle–attached DNA was resuspended in 50 μL of water (DNA/gold solution).
For bombardment, 9 μL of the DNA/gold solution were pipetted on a filter that was placed into the discharge assembly of a vacuum chamber (0.2 bar). The lower epidermis of Arabidopsis leaves or the inner epidermis of white onion peels (obtained from local markets) were high pressure-sprayed with the DNA/gold solution (using 50-ms helium pulses of 7, 8, or 9 bar). Sprayed leaves and onion peels were incubated for 24 to 48 h on agar plates with MS medium (Sigma-Aldrich, Deisenhofen, Germany) containing 0.3-M mannitol. Fluorescence was detected with a confocal microscope (Leica TCS SP II; Leica Microsystems, Bensheim, Germany).
Immunohistochemical Techniques and Protein Gel Blot Analyses
Peptide-specific antisera against the C-terminal peptide NH2-CEIGSNKQWKEGDTQSS-COOH were generated by Pineda-Antikörper-Service (Berlin, Germany). Yeast cells expressing AtPLT5 in sense or antisense orientation were fixed, embedded, sectioned, and treated with antisera as described (Meyer et al., 2004). Binding of anti-AtPLT5 antibodies to yeast sections was visualized by treatment with anti-rabbit IgG-FITC-isomer 1-conjugate (Sigma-Aldrich). Finally, microscopic slides were mounted in antifading medium (ProLong Antifade kit; Molecular Probes, Leiden, The Netherlands) and viewed under appropriate excitation light.
Protein extracts of different membrane fractions from bakers' yeast were prepared as described (Sauer and Stolz, 2000), separated on SDS polyacrylamide gels (Laemmli, 1970), and transferred to nitrocellulose filters (Dunn, 1986). AtPLT5 protein bands were detected by treatment of the filters with anti-rabbit IgG antiserum-peroxidase conjugate (diluted 1:4000 in blocking buffer) followed by incubation with Lumi-Light protein gel blotting substrate (Roche Diagnostics, Mannheim, Germany).
RT-PCR and Real-Time RT-PCR
Full-length and truncated AtPLT5 mRNAs from wild-type and T-DNA insertion plants were amplified with the primers AtPLT5-5 (5′-GACACAGAATTCAAGCTTGTAAAAGAAATGACAGGTGCCACACCGGAAAA-3′), AtPLT5-3 (5′-GACACAGAATTCCTACGAACTTTGTGTGTCTCCTT-3′), and AtPLT5c32 (5′-CAACCTCAACGGGAATATCTCCG-3′). Real-time RT-PCRs were performed on a RotorGene 2000 (Corbett Research, Sydney, Australia) with the following primers: AtACT2g+846f (5′-ATTCAGATGCCCAGAAGTCTTGTT-3′) and AtACT2g+1295r (5′-GAAACATTTTCTGTGAACGATTCCT-3′) for the actin standard (Arabidopsis ACT2 gene; At3g18780), AtUBQ10g-315f (5′-ACCGTGATCAAGATGCAGATCTTTGT-3′) and AtUBQ10g+163 (5′-TACGGCCATCCTCTAGCTGCTTG-3′) for the ubiquitin standard (Arabidopsis UBQ10 gene; At4g05320), and AtPLT5cW5 (5′-ATCCTCCTTGGTTATGATATAGGAGTGA-3′) and AtPLT5cW3 (5′-GCGATCATGAGAGCATATCCGAC-3′) for AtPLT5.
Epifluorescence and Confocal Microscopy
Images of GFP fluorescence were made with an epifluorescence microscope (Zeiss Axioskop; Carl Zeiss, Jena, Germany) or stereomicroscopes (Zeiss SV11 [Carl Zeiss] or Leica MZFLIII [Leica Microsystems]) with an excitation wavelength of 460 to 500 nm. Emitted fluorescence was monitored at detection wavelengths longer than 510 nm. Confocal microscopy and staining with propidium iodide were performed as described (Stadler et al., 2005).
Acknowledgments
We thank Anja Schillinger and Jennifer Tebart for excellent technical assistance, Christian Lauterbach for helping with the confocal microscopy, and Angelika Wolf for growing the Arabidopsis plants. We are grateful to Andrea Feuerstein and Michael Büttner (both of Molecular Plant Physiology, FAU Erlangen-Nürnberg, Germany) for the pAF1 and pSO35e vectors. This work was supported by a grant of the Deutsche Forschungsgemeinschaft to N.S. (Arabidopsis Functional Genomics Network; Sa 382/13-1).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Norbert Sauer (nsauer@biologie.uni-erlangen.de).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026641.
References
- An, Y.-Q., McDowell, J.M., Huang, S., McKinney, E.C., Chambliss, S., and Meagher, R.B. (1996). Strong, constitutive expression of the Arabidopsis ACT2/ACT8 actin subclass in vegetative tissues. Plant J. 10, 107–121. [DOI] [PubMed] [Google Scholar]
- Barth, I., Meyer, S., and Sauer, N. (2003). PmSUC3: Characterization of a SUT2/SUC3-type sucrose transporter from Plantago major. Plant Cell 15, 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, D., Dreyer, I., Hoth, S., Reid, J.D., Busch, H., Lehnen, M., Palme, K., and Hedrich, R. (1996). Changes in voltage activation, Cs+ sensitivity, and ion permeability in H5 mutants of the plant K+ channel KAT1. Proc. Natl. Acad. Sci. USA 93, 8123–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyreuther, K., Bieseler, B., Ehring, R., Griesser, H.-W., Mieschendahl, M., Müller-Hill, B., and Triesch, I. (1980). Investigation of structure and function of lactose permease of Escherichia coli. Biochem. Soc. Trans. 8, 675–676. [DOI] [PubMed] [Google Scholar]
- Büttner, M., and Sauer, N. (2000). Monosaccharide transporters in plants: Structure, function and physiology. Biochim. Biophys. Acta 1465, 263–274. [DOI] [PubMed] [Google Scholar]
- Chauhan, S., Forsthoefel, N., Ran, Y., Quigley, F., Nelson, D.E., and Bohnert, H.J. (2000). Na+/myo-inositol symporters and Na+/H+-antiport in Mesembryanthemum crystallinum. Plant J. 24, 511–522. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Dunn, S.D. (1986). Effects of the modification of transfer buffer composition on the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal. Biochem. 157, 144–153. [DOI] [PubMed] [Google Scholar]
- Emr, S.D., Schekman, R., Flessel, M.C., and Thorner, J. (1983). An MFα1-SUC2 (α-factor-invertase) gene fusion for study of protein localisation and gene expression in yeast. Proc. Natl. Acad. Sci. USA 80, 7080–7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahrtz, M., Stolz, J., and Sauer, N. (1994). A phloem specific sucrose-H+ symporter from Plantago major L. supports the model of apoplastic phloem loading. Plant J. 6, 697–706. [DOI] [PubMed] [Google Scholar]
- Gao, Z., Maurousset, L., Lemoine, R., Yoo, S.D., Van Nocker, S., and Loescher, W. (2003). Cloning, expression, and characterization of sorbitol transporters from developing sour cherry fruit and leaf sink tissues. Plant Physiol. 131, 1566–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, D., Jean, W.S., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan, D. (1983). Studies on transformation of E. coli with plasmids. J. Mol. Biol. 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Haritatos, E., Ayre, B.G., and Turgeon, R. (2000). Identification of phloem involved in assimilate loading in leaves by the activity of the galactinol synthase promoter. Plant Physiol. 123, 929–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger, M.A., Ikeda, T., Coady, M., Gundersen, C.B., and Wright, E.M. (1987). Expression of size-selected mRNA encoding the intestinal Na+/glucose cotransporter in Xenopus laevis oocytes. Proc. Natl. Acad. Sci. USA 84, 2634–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters, M., Silva, B., Van Vliet, F., Genetello, C., De Block, M., Dhaese, P., Depicker, A., Inze, D., Engler, G., Villarroel, R., Van Montagu, M., and Schell, J. (1980). The functional organization of the nopaline Agrobacterium tumefaciens plasmid pTiC58. Plasmid 3, 212–230. [DOI] [PubMed] [Google Scholar]
- Iida, A., Harayama, S., Iino, T., and Hazelbauer, G.L. (1984). Molecular cloning and characterization of genes required for ribose transport and utilization in Escherichia coli K-12. J. Bacteriol. 158, 674–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue, T., Abe, H., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). ERD6, a cDNA clone for an early dehydration-induced gene of Arabidopsis, encodes a putative sugar transporter. Biochim. Biophys. Acta 1370, 187–191. [DOI] [PubMed] [Google Scholar]
- Kwon, H.M., Yamauchi, A., Uchida, S., Preston, A.S., Garcia-Perez, A., Burg, M.B., and Handler, J.S. (1992). Cloning of the cDNA for a Na+/myo-inositol cotransporter, a hypertonicity stress protein. J. Biol. Chem. 267, 6297–6301. [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lohaus, G., and Fischer, K. (2002). Intracellular and intercellular transport of nitrogen and carbon. In Advances in Photosynthesis and Respiration, Vol. 12, C. Foyer and G. Noctor, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 239–263.
- Maurel, C., Reizer, J., Schroeder, J.I., Chrispeels, M.J., and Saier, M.H., Jr. (1994). Functional characterization of the Escherichia coli glycerol facilitator, GlpF, in Xenopus oocytes. J. Biol. Chem. 269, 11869–11872. [PubMed] [Google Scholar]
- Meyer, S., Lauterbach, C., Niedermeier, M., Barth, I., Sjolund, R.D., and Sauer, N. (2004). Wounding enhances expression of AtSUC3, a sucrose transporter from Arabidopsis sieve elements and sink tissues. Plant Physiol. 134, 684–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikawa, J., Tsukagoshi, Y., and Yamashita, S. (1991). Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J. Biol. Chem. 266, 11184–11191. [PubMed] [Google Scholar]
- Noiraud, N., Maurousset, L., and Lemoine, R. (2001). Identification of a mannitol transporter, AgMaT1, in celery phloem. Plant Cell 13, 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, R., and Lucas, C. (2004). Expression studies of GUP1 and GUP2, genes involved in glycerol active transport in Saccharomyces cerevisiae, using semi-quantitative RT-PCR. Curr. Genet. 46, 140–146. [DOI] [PubMed] [Google Scholar]
- Page, R.D.M. (1996). TREEVIEW: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12, 357–358. [DOI] [PubMed] [Google Scholar]
- Quirino, B.F., Reiter, W.D., and Amasino, R.D. (2001). One of two tandem Arabidopsis genes homologous to monosaccharide transporters is senescence-associated. Plant Mol. Biol. 46, 447–457. [DOI] [PubMed] [Google Scholar]
- Ramsperger-Gleixner, M., Geiger, D., Hedrich, R., and Sauer, N. (2004). Differential expression of sucrose transporter and polyol transporter genes during maturation of common plantain companion cells. Plant Physiol. 134, 147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, N., Friedländer, K., and Gräml-Wicke, U. (1990). Primary structure, genomic organization and heterologous expression of a glucose transporter from Arabidopsis thaliana. EMBO J. 9, 3045–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer, N., and Stolz, J. (1994). SUC1 and SUC2: Two sucrose transporters from Arabidopsis thaliana; expression and characterization in baker's yeast and identification of the histidine tagged protein. Plant J. 6, 67–77. [DOI] [PubMed] [Google Scholar]
- Sauer, N., and Stolz, J. (2000). Expression of foreign transport proteins in yeast. In Practical Approach Series S.A. Baldwin, ed (Oxford: Oxford University Press), pp. 79–105.
- Scholz-Starke, J., Büttner, M., and Sauer, N. (2003). AtSTP6, a new pollen-specific H+-monosaccharide symporter from Arabidopsis thaliana. Plant Physiol. 131, 70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, R., Wolf, K., Hilgarth, C., Tanner, W., and Sauer, N. (1995). Subcellular localization of the inducible Chlorella HUP1 monosaccharide-H+ symporter and cloning of a co-induced galactose-H+ symporter. Plant Physiol. 107, 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler, R., Wright, K.M., Lauterbach, C., Amon, G., Gahrtz, M., Feuerstein, A., Oparka, K.J., and Norbert Sauer, N. (2005). Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. Plant J., in press. [DOI] [PubMed]
- Stewart, J.B., and Hermodson, M.A. (2003). Topology of RbsC, the membrane component of the Escherichia coli ribose transporter. J. Bacteriol. 185, 5234–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, C.-W., and Callis, J. (1997). Independent modulation of Arabidopsis thaliana polyubiquitin mRNAs in different organs and in response to environmental changes. Plant J. 11, 1017–1027. [DOI] [PubMed] [Google Scholar]
- Sweet, G., Gandor, C., Voegele, R., Wittekindt, N., Beuerle, J., Truniger, V., Lin, E.C., and Boos, W. (1990). Glycerol facilitator of Escherichia coli: Cloning of glpF and identification of the glpF product. J. Bacteriol. 172, 424–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, W. (1980). Proton sugar cotransport in lower and higher plants. Ber. Deutsch. Bot. Ges. 93, 167–176. [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmermans, M.C.P., Malig, P., Viera, J., and Messing, J. (1990). The pFF plasmids: Cassettes utilising CaMV sequences for expression of foreign genes in plants. J. Biotechnol. 14, 333–344. [DOI] [PubMed] [Google Scholar]
- Truernit, E., and Sauer, N. (1995). The promoter of the Arabidopsis thaliana SUC2 sucrose-H+ symporter gene directs expression of β-glucuronidase to the phloem: Evidence for phloem loading and unloading by SUC2. Planta 196, 564–570. [DOI] [PubMed] [Google Scholar]
- Wallace, I.S., and Roberts, D.M. (2004). Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Plant Physiol. 135, 1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, A., Servaites, J.C., Geiger, D.R., Kofler, H., Hille, D., Groner, F., Hebbeker, U., and Flügge, U.I. (2000). Identification, purification, and molecular cloning of a putative plastidic glucose translocator. Plant Cell 12, 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weig, A.R., and Jakob, C. (2000). Functional identification of the glycerol permease activity of Arabidopsis thaliana NLM1 and NLM2 proteins by heterologous expression in Saccharomyces cerevisiae. FEBS Lett. 481, 293–298. [DOI] [PubMed] [Google Scholar]
- Wieczorke, R., Krampe, S., Weierstall, T., Freidel, K., Hollenberg, C.P., and Boles, E. (1999). Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464, 123–128. [DOI] [PubMed] [Google Scholar]
- Williams, L.E., Lemoine, R., and Sauer, N. (2000). Sugar transporters in higher plants—A diversity of roles and complex regulation. Trends Plant Sci. 5, 283–290. [DOI] [PubMed] [Google Scholar]
- Zardoya, R., Ding, X., Kitagawa, Y., and Chrispeels, M.J. (2002). Origin of plant glycerol transporters by horizontal gene transfer and functional recruitment. Proc. Natl. Acad. Sci. USA 99, 14893–14896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, M.H., and Ziegler, H. (1975). List of sugars and sugar alcohols in sieve-tube exudates. In Encyclopedia of Plant Physiology, M.H. Zimmermann and J.A. Milburn, eds (Berlin: Springer Verlag), pp. 480–503.