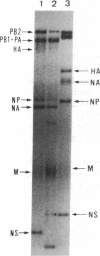
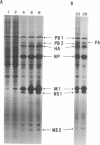
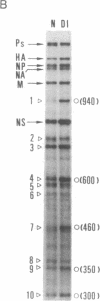
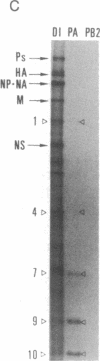
Abstract
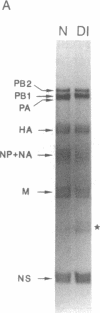
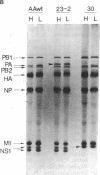
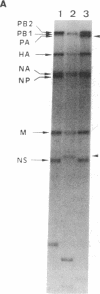
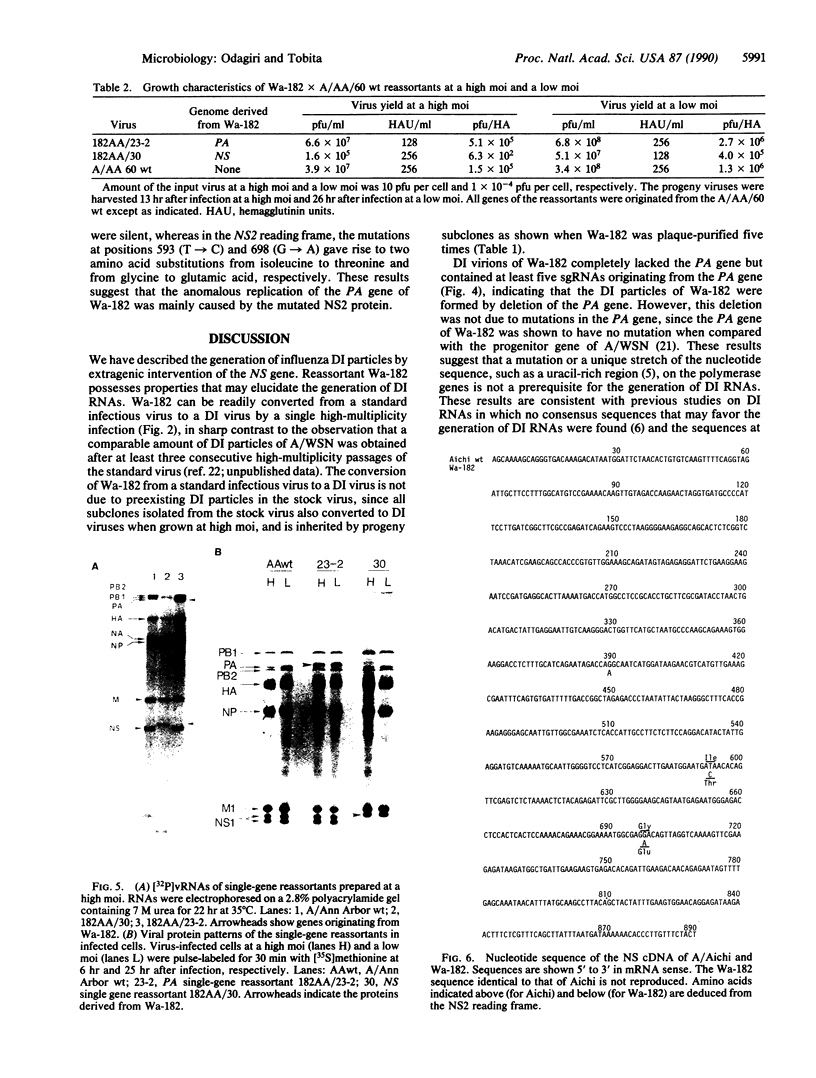
Several consecutive undiluted passages of infectious virus are usually required to obtain defective interfering particles of influenza virus. In contrast, a reassortant (Wa-182) of influenza A/WSN, which we isolated, whose NS gene was replaced with the NS gene of A/Aichi, was readily converted to defective interfering form after only a single high-multiplicity infection. The defective interfering particles of Wa-182 were devoid of the PA gene (RNA segment 3) but possessed several species of subgenomic RNAs of the PA gene origin. Such aberrant replication of the PA gene was shown to be caused by an extragenic effect of the NS gene of Wa-182, because, when the NS gene of Wa-182 was singly transferred to the wild-type A/Ann Arbor/6/60 virus, the recipient showed exactly the same features. Analysis of nucleotide sequence demonstrated that the NS gene of Wa-182 contained three point mutations relative to the wild-type NS gene that resulted in two amino acid substitutions in the nonstructural protein NS2, suggesting that the mutation in NS2 protein affected the normal replication of the PA gene of Wa-182. The results also suggest that the NS2 protein plays an important role in the synthesis of intact genome RNAs.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akkina R. K., Chambers T. M., Nayak D. P. Expression of defective-interfering influenza virus-specific transcripts and polypeptides in infected cells. J Virol. 1984 Aug;51(2):395–403. doi: 10.1128/jvi.51.2.395-403.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaton A. R., Krug R. M. Transcription antitermination during influenza viral template RNA synthesis requires the nucleocapsid protein and the absence of a 5' capped end. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6282–6286. doi: 10.1073/pnas.83.17.6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. Polynucleotide kinase exchange reaction: quantitave assay for restriction endonuclease-generated 5'-phosphoroyl termini in DNA. J Biol Chem. 1977 May 25;252(10):3176–3184. [PubMed] [Google Scholar]
- Crumpton W. M., Dimmock N. J., Minor P. D., Avery R. J. The RNAs of defective-interfering influenza virus. Virology. 1978 Oct 15;90(2):370–373. doi: 10.1016/0042-6822(78)90322-7. [DOI] [PubMed] [Google Scholar]
- Davis A. R., Nayak D. P. Sequence relationships among defective interfering influenza viral RNAs. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3092–3096. doi: 10.1073/pnas.76.7.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S., Winter G. Nucleotide sequences of influenza virus segments 1 and 3 reveal mosaic structure of a small viral RNA segment. Cell. 1982 Feb;28(2):303–313. doi: 10.1016/0092-8674(82)90348-8. [DOI] [PubMed] [Google Scholar]
- Inglis S. C., Barrett T., Brown C. M., Almond J. W. The smallest genome RNA segment of influenza virus contains two genes that may overlap. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3790–3794. doi: 10.1073/pnas.76.8.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda J. M., Davis A. R., Nayak D. P., De B. K. Diversity and generation of defective interfering influenza virus particles. Virology. 1979 May;95(1):48–58. doi: 10.1016/0042-6822(79)90400-8. [DOI] [PubMed] [Google Scholar]
- Janda J. M., Nayak D. P. Defective influenza viral ribonucleoproteins cause interference. J Virol. 1979 Nov;32(2):697–702. doi: 10.1128/jvi.32.2.697-702.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings P. A., Finch J. T., Winter G., Robertson J. S. Does the higher order structure of the influenza virus ribonucleoprotein guide sequence rearrangements in influenza viral RNA? Cell. 1983 Sep;34(2):619–627. doi: 10.1016/0092-8674(83)90394-x. [DOI] [PubMed] [Google Scholar]
- Lamb R. A., Choppin P. W., Chanock R. M., Lai C. J. Mapping of the two overlapping genes for polypeptides NS1 and NS2 on RNA segment 8 of influenza virus genome. Proc Natl Acad Sci U S A. 1980 Apr;77(4):1857–1861. doi: 10.1073/pnas.77.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R. A., Etkind P. R., Choppin P. W. Evidence for a ninth influenza viral polypeptide. Virology. 1978 Nov;91(1):60–78. doi: 10.1016/0042-6822(78)90355-0. [DOI] [PubMed] [Google Scholar]
- Massicot J. G., Murphy B. R., Thierry F., Markoff L., Huang K. Y., Chanock R. M. Temperature-sensitive mutants of influenza virus. Identification of the loci of the two ts lesions in the Udorn-ts-1A2 donor virus and the correlation of the presence of these two ts lesions with a predictable level of attenuation. Virology. 1980 Feb;101(1):242–249. doi: 10.1016/0042-6822(80)90499-7. [DOI] [PubMed] [Google Scholar]
- Murphy B. R., Tolpin M. D., Massicot J. G., Kim H. Y., Parrott R. H., Chanock R. M. Escape of a highly defective influenza A virus mutant from its temperature sensitive phenotype by extragenic suppression and other types of mutation. Ann N Y Acad Sci. 1980;354:172–182. doi: 10.1111/j.1749-6632.1980.tb27966.x. [DOI] [PubMed] [Google Scholar]
- Mücke K., Scholtissek C. Extragenic and intragenic suppression of a transport mutation in the hemagglutinin gene of an influenza A virus as revealed by backcross and sequence determination. Virology. 1987 May;158(1):112–117. doi: 10.1016/0042-6822(87)90243-1. [DOI] [PubMed] [Google Scholar]
- Nayak D. P., Chambers T. M., Akkina R. K. Defective-interfering (DI) RNAs of influenza viruses: origin, structure, expression, and interference. Curr Top Microbiol Immunol. 1985;114:103–151. doi: 10.1007/978-3-642-70227-3_3. [DOI] [PubMed] [Google Scholar]
- Nayak D. P., Sivasubramanian N., Davis A. R., Cortini R., Sung J. Complete sequence analyses show that two defective interfering influenza viral RNAs contain a single internal deletion of a polymerase gene. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2216–2220. doi: 10.1073/pnas.79.7.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Odagiri T., DeBorde D. C., Maassab H. F. Cold-adapted recombinants of influenza A virus in MDCK cells. I. Development and characterization of A/Ann Arbor/6/60 X A/Alaska/6/77 recombinant viruses. Virology. 1982 May;119(1):82–95. doi: 10.1016/0042-6822(82)90067-8. [DOI] [PubMed] [Google Scholar]
- Odagiri T., Tanaka T., Tobita K. Temperature-sensitive defect of influenza A/Ann Arbor/6/60 cold-adapted variant leads to a blockage of matrix polypeptide incorporation into the plasma membrane of the infected cells. Virus Res. 1987 May;7(3):203–218. doi: 10.1016/0168-1702(87)90028-1. [DOI] [PubMed] [Google Scholar]
- Odagiri T., Tobita K. Nucleotide sequence of the PA gene of influenza A/WSN/33(H1N1). Nucleic Acids Res. 1990 Feb 11;18(3):654–654. doi: 10.1093/nar/18.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P., Schulman J. L. Differences in RNA patterns of influenza A viruses. J Virol. 1976 Mar;17(3):876–884. doi: 10.1128/jvi.17.3.876-884.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramig R. F., Fields B. N. Revertants of temperature-sensitive mutants of reovirus: evidence for frequent extragenic suppression. Virology. 1979 Jan 15;92(1):155–167. doi: 10.1016/0042-6822(79)90221-6. [DOI] [PubMed] [Google Scholar]
- Scholtissek C., Spring S. B. Extragenic suppression of temperature-sensitive mutations in RNA segment 8 by replacement of different rna segments with those of other influenza A virus prototype strains. Virology. 1982 Apr 15;118(1):28–34. doi: 10.1016/0042-6822(82)90316-6. [DOI] [PubMed] [Google Scholar]
- Shapiro G. I., Krug R. M. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J Virol. 1988 Jul;62(7):2285–2290. doi: 10.1128/jvi.62.7.2285-2290.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramanian N., Nayak D. P. Defective interfering influenza RNAs of polymerase 3 gene contain single as well as multiple internal deletions. Virology. 1983 Jan 30;124(2):232–237. doi: 10.1016/0042-6822(83)90340-9. [DOI] [PubMed] [Google Scholar]
- VON MAGNUS P. Propagation of the PR8 strain of influenza A virus in chick embryos. III. Properties of the incomplete virus produced in serial passages of undiluted virus. Acta Pathol Microbiol Scand. 1951;29(2):157–181. doi: 10.1111/j.1699-0463.1951.tb00114.x. [DOI] [PubMed] [Google Scholar]
- Wolstenholme A. J., Barrett T., Nichol S. T., Mahy B. W. Influenza virus-specific RNA and protein syntheses in cells infected with temperature-sensitive mutants defective in the genome segment encoding nonstructural proteins. J Virol. 1980 Jul;35(1):1–7. doi: 10.1128/jvi.35.1.1-7.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]