Abstract
Coronatine (1), a small-molecular virulence factor produced by plant-pathogenic bacteria, promotes bacterial infection by inducing the opening of stomatal pores, the major route of bacterial entry into the plant, via the jasmonate-mediated COI1-JAZ signaling pathway. However, this pathway is also important for multiple plant functions, including defense against wounding by herbivorous insects. Thus, suppression of the COI1-JAZ signaling pathway to block bacterial infection would concomitantly impair plant defense against herbivorous wounding. Here, we report additional, COI1-JAZ-independent, action of 1 in Arabidopsis thaliana guard cells. First, we found that a stereoisomer of 1 regulates the movement of Arabidopsis guard cells without affecting COI1-JAZ signaling. Second, we found using alkyne-tagged Raman imaging (ATRI) that 1 is localized to the endoplasmic reticulum (ER) of living guard cells of Arabidopsis. The use of arc6 mutant lacking chloroplast formation was pivotal to circumvent the issue of autofluorescence during ATRI. These findings indicate that 1 has an ER-related action on Arabidopsis stomata that bypasses the COI1-JAZ signaling module. It may be possible to suppress the action of 1 on stomata without impairing plant defense responses against herbivores.
Short abstract
Bacterial infection of plants causes severe crop losses. A bacterial virulence factor, coronatine (1), facilitates infection by hijacking a plant’s defense pathway. We found an ER-mediated noncanonical pathway of 1-mediated infection in Arabidopsis.
Introduction
Scientists continue to seek novel and effective ways to reduce the impact of bacterial infections upon crop yields. Being able to mitigate plant–pathogen interactions has great potential to improve crop production. The attack of pathogens triggers plant immune responses. A plant hormone, 7-iso-jasmonoyl-l-isoleucine (2, Figure 1a),1 plays important roles in this plant immunity.2−5 The most important physiological role of 2 lies in the activation of induced immunity, which is triggered by the attack of insect herbivores and necrotrophic pathogens as well as tissue injury.62 functions through the coreceptor composed of CORONATINE INSENSITIVE1 (COI1) and JASMONATE ZIM-DOMAIN (JAZ) proteins.7−9 And the COI1-JAZ signaling pathway is considered the only signaling pathway in which 2 is involved.
Figure 1.
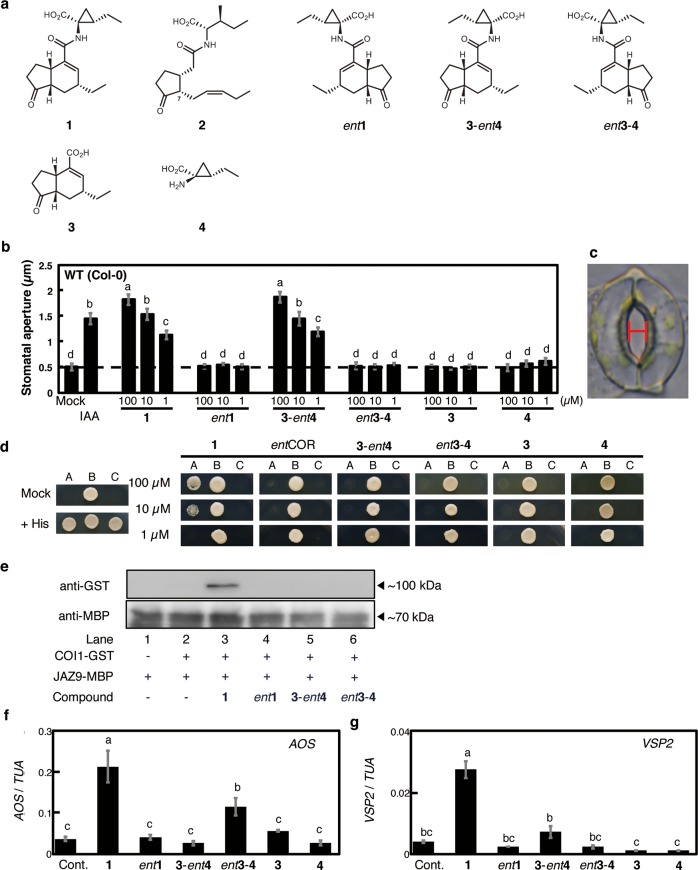
Evaluation of the physiological action of coronatine relative and derivatives. (a) Structures of coronatine (1) relatives and derivatives: 7-epi-jasmonoyl-l-isoleucine (2), coronatine (1), and related compounds (ent1, 3-ent4, ent3-4, 3, and 4). (b) Effect of 1 derivatives on reopening (measured as size of aperture) of Arabidopsis thaliana (Col-0) abaxial stomata by abscisic acid (ABA). Indole-3-acetic acid (IAA), which induces stomatal reopening, was used as a positive control. Dashed line indicates the mean stomatal aperture taken from a control setting, in which Arabidopsis leaf peels with closed stomata were incubated in MES buffer (pH 6.2) containing 2% EtOH. Bars represent mean stomatal aperture and SE (n = 20 stomata). Letters a–d indicate significant differences between the means (ANOVA: P < 0.05). (c) A stoma in the epidermis of Arabidopsis thaliana. The width of the red bar was measured as the stomatal aperture. (d) Yeast two-hybrid (Y2H) assay of compound-induced COI1-JAZ9 coreceptor formation. Lane A: Yeast cells cotransformed with pDEST22-JAZ9 and pDEST32-COI1. Lane B: Yeast cells cotransformed with pDEST22-RalGDS-m1 and pDEST32-Krev1 as a positive control. Lane C: Yeast cells cotransformed with pDEST22-RalGDS-m2 and pDEST32-Krev1 as a negative control. (e) Pull-down assay of purified GST-COI1 (5 nM) with recombinant E. coli expressed MBP-JAZ9 (approximately 40 nM) in the presence of COR derivatives (100 nM). GST-COI1 bound to MBP-JAZ9 were pulled down with amylose resin, and analyzed by immunoblotting. HRP-conjugated anti-GST antibody was used for detection of GST-COI1. Anti-MBP antibody and HRP-conjugated rat IgG antibody was used for showing the amounts of MBP-JAZ9 as the input materials. (f, g) Relative transcript levels of AOS and VSP2 in A. thaliana treated with 1 derivatives. Each relative transcript level was assessed by quantitative RT-PCR. “Cont.” indicates the mean AOS or VSP2 transcript level in a buffer containing 2% EtOH only. Bars represent the mean AOS or VSP2 transcript levels of the test samples (100 μM). Bars represent mean and SE (n = 3). Letters a–c indicate significant differences between the means (ANOVA: P < 0.05).
Coronatine (1, Figure 1a) composed of (+)-coronafacic acid (3, Figure 1a) and (+)-coronamic acid (4, Figure 1a) is a small-molecular phytotoxin produced by Pseudomonas syringae.101 has been considered a structural and biological mimic of 2 and used as a strong agonist of the COI1-JAZ coreceptor.11,12 However, we cannot fully understand the extent to which 1 mimics 2 in plant defense responses, and the unknown targets are expected to be included. For example, 1 suppresses callose deposition to promote infection of P. syringae in a COI1-independent manner.13
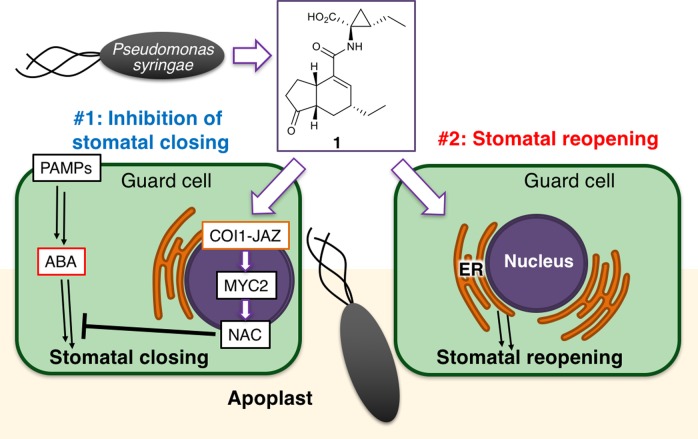
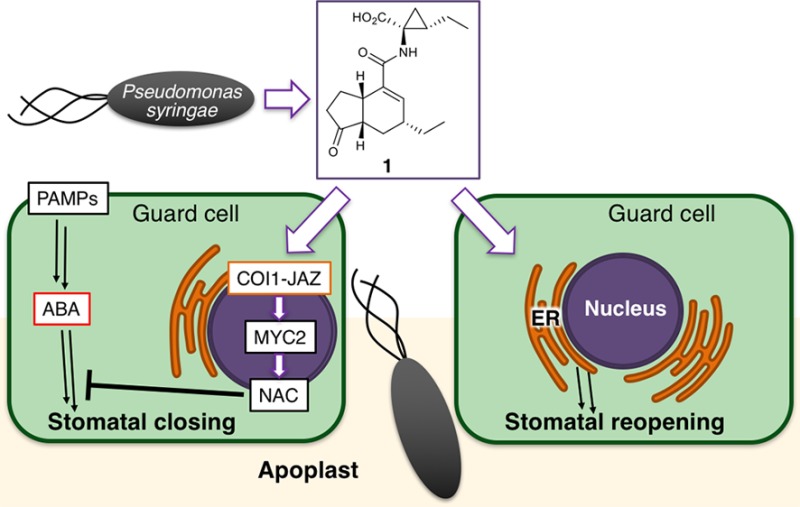
Bacterial virulence factors are molecules produced by bacteria that contribute to their pathogenicity, for instance by facilitating colonization of the host. Previous studies have revealed another role of 1 as a virulence factor in plant–microbe interactions.14 Within a few hours of infection by the bacterium P. syringae, the stomata of Arabidopsis thaliana close (stomatal defense), thus blocking the main route of bacterial entry into the plant apoplast.15P. syringae causes the reopening of closed stomata to counter this stomatal defense, thereby securing the path of bacterial entry into the plant apoplast. Stomatal defense has been considered to be partly under the control of plant hormones, such as abscisic acid (ABA).14,161 has thus been considered to prevent the closure of plant stomata by inhibiting ABA-mediated stomatal closure. Furthermore, this process is thought to take place through the COI1-JAZ-MYC2-NAC signaling cascade (see also Figure 6#1), because 1 fails to inhibit ABA-mediated stomatal closure in both the coi1–3 and anac which is an anac19/anac55/anac72 triple mutant of all three NAC TFs (petunia NAM and Arabidopsis ATAF1, ATAF2, and CUC2 transcription factors).16 NAC TFs are a direct target of MYC TF and function downstream of the COI1-JAZ coreceptor to suppress ABA signaling pathways, leading to the stomatal closure.17 Thus, 1 inhibits stomatal closure through the COI1-JAZ signaling pathway.18 In this way, P. syringae uses 1 to hijack the plant’s own pathway of defense. In principle, inhibition of the COI1-JAZ pathway could therefore improve the stomatal defense of the plant.
Figure 6.
Schematic diagram of the two functions of the bacterial virulence factor, 1, on stomatal guard cells to counter stomatal defense. In Arabidopsis guard cells, 1 produced by Pseudomonas syringae counters stomatal defense through two functions: in #1 (left), 1 inhibits ABA-mediated stomatal closure in response to bacterial PAMPs (pathogen-associated molecular patterns) through the COI1-JAZ-MYC2-NAC signaling cascade, and in #2 (right), it simultaneously promotes the reopening of closed stomata through a COI1-JAZ-independent, ER-related mechanism.
However, inhibiting the COI1-JAZ pathway would present a trade-off between improved stomatal defense and impaired defense against attack of herbivores and necrotrophs, which could impede overall improvement of plant immunity by suppressing the COI1-JAZ pathway. A strategy to suppress the action of 1 on stomata without impairing COI1-JAZ-dependent plant defense responses would therefore be highly desirable.
Here, we report that, in addition to its conventional COI1-JAZ-dependent function, 1 counters stomatal defense through a COI1-JAZ-independent and ER (endoplasmic reticulum)-mediated function. We show this through two independent approaches: the development of a non-COI1-JAZ agonistic 1 derivative with stomatal reopening activity and alkyne-tag in vivo Raman imaging (ATRI)19−21 of 1 in living A. thaliana guard cells. The Raman signal of an alkyne appears in an otherwise silent region where no biomolecules in the cell provide any Raman signal. Especially, the subcellular localization of alkyne-tagged 1 obtained by ATRI provided a pivotal evidence of the COI1-JAZ-independent function. To our knowledge, this is the first successful application of ATRI for visualizing intracellular localization of a small molecule in living plant cells. Our result demonstrated that ATRI is a powerful alternative method for fluorescence imaging because fluorescence-labeled (FL) small molecular ligands such as FL-1 have often lost their bioactivity by the addition of a large fluorescent dye.22
Results and Discussion
Coronatine Triggers the Reopening of Closed Stomata through a COI1-JAZ-Independent Mechanism
Stomatal defense against pathogenic infection can be mimicked by abscisic acid (ABA) treatment because ABA signaling is involved as a key mechanism of stomatal defense.14,16 It has already been shown that 1 can counter both pathogen- and ABA-mediated stomatal closure by inhibiting the ABA signaling cascade through the COI1-JAZ-MYC2-NAC signaling module.16 Then, we evaluated the physiological action of 1 and a series of the stereoisomers against ABA-mediated stomatal closure on the WT (Col-0) and knockout mutants of Arabidopsis.
In a previous study, we prepared optically pure components of 1, 3,22 and 4,23 and then synthesized a stereoisomer (3-ent4; Figure 1a) composed of naturally occurring 3 and enantiomeric 4 ((−)-coronamic acid). We also synthesized the enantiomers of 1 (ent-1) and 3-ent4 (ent3-4; Figure 1a).23 We evaluated the physiological action of these stereoisomers in detail.
Abaxial stomata of 6- to 8-week-old Col-0 A. thaliana were first treated with ABA to induce closure and then treated with 1 or its stereoisomers (ent-1, 3-ent4, ent3-4, 3, and 4). Naturally occurring 1 effectively reopened the closed Col-0 stomata, whereas ent-1, 4, and 3 were ineffective (Figures 1b and 1c). It was surprising that a stereoisomer 3-ent4 was also effective at reopening the stomata of A. thaliana, whereas its enantiomer, ent3-4, was ineffective. Nevertheless, our yeast two-hybrid (Y2H) assays (Figure 1d) and pull-down assay using JAZ9-MBP (Figure 1e) showed that 3-ent4 does not induce COI1-JAZ9 coreceptor formation; nor does it induce expression of COI1-JAZ-regulated JA-biosynthetic gene allene oxide synthase (AOS) and JA-marker gene Vegetative Storage Protein 2 (VSP2) in A. thaliana (Figures 1f–1g). However, weak overlap between transcriptional responses induced by 1 and ent3-4 suggested that ent3-4 maintains some of the biological functions of 1. DNA microarray analyses in Arabidopsis treated by 1, ent3-4, or ent1 demonstrated that 19% of 1-inducible genes (117/605 genes) was overlapped with ent3-4 inducible genes, while as little as 3.8% (23/605 genes) was overlapped with ent1-inducible genes (Figure S1A, Table S1). These results show that the stereochemistry of the 4 moiety in 1 does not affect the function of 1 in stomata and a stereoisomer of 1 (3-ent4) can induce the reopening of closed Arabidopsis stomata against ABA-mediated stomatal closure through a unique mechanism independent of COI1-JAZ.
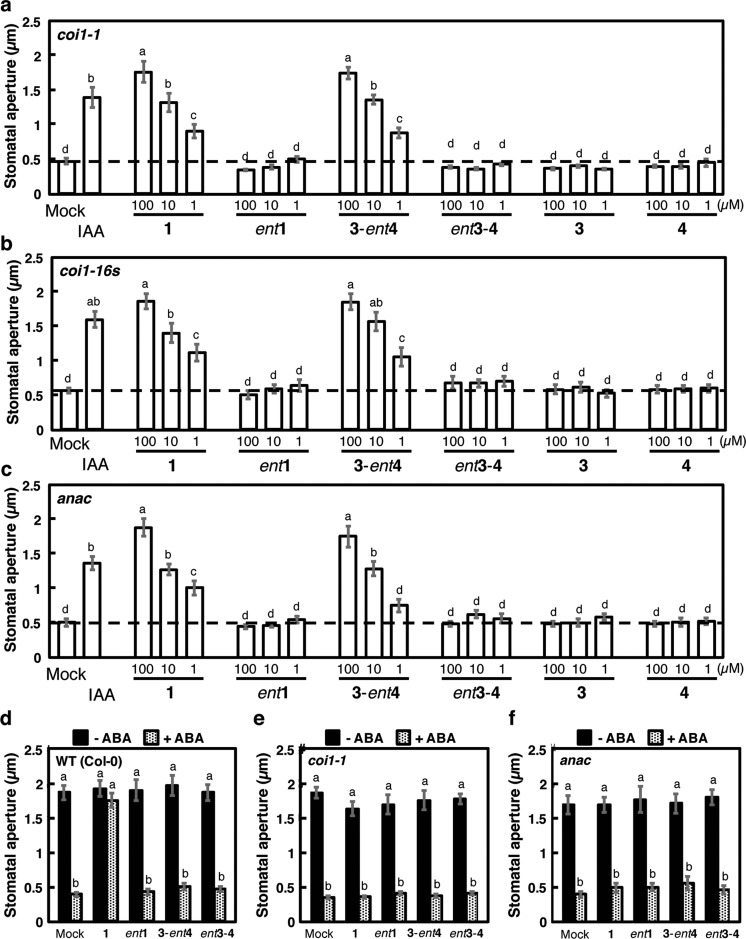
We next examined the COI1-JAZ independent action of 1/3-ent4 against stomatal defense by using knockout mutants of Arabidopsis. All the results on 1 were examined by the comparison of those on 3-ent4 used as a reference. We examined the effect of 1/3-ent4 on the reopening of closed stomata of knockout Arabidopsis mutants, coi1-1,24coi1-16s,25 and anac (Figure 2). The reopening of closed stomata was not affected in all three mutants, either by 1 or by 3-ent4 (Figures 2a, 2b, and 2c). In contrast, different results were obtained in a conventional bioassay used in previous studies.14,16 ABA-induced stomatal closure was inhibited by the coaddition of COR (Figure 2d), and the inhibition was impaired in coi1-1 and anac knockout mutants (Figures 2e and 2f), as reported previously.14,16 However, the ABA-induced stomatal closure was not inhibited by the coaddition of 3-ent4 in wild type plants as well as in both mutants (Figures 2d–2f).
Figure 2.
Physiological evaluation of coronatine and derivatives using knockout mutants of Arabidopsis thaliana. (a–c) Effect of 1 derivatives on reopening of closed stomata of coi1-1 (a), coi1-16s (b), and anac (c) mutants under dark conditions. IAA was used as a positive control. Letters a–d indicate significant differences between the means (ANOVA: P < 0.05). (d–f) Effect of 1 derivatives (30 μM) on stomata of wild type Col-0 (d), coi1-1 (e), and anac (f) with (+ ABA) and without (− ABA) (15 μM) cotreatment. Dashed line indicates mean stomatal aperture under control conditions. Bars represent mean stomatal aperture and SE (n = 20 stomata). Letters indicate significant differences between the means (ANOVA: P < 0.05).
These careful physiological assessments of 1 and the stereoisomer 3-ent4 using knockout mutants of A. thaliana strongly suggest that 1 functions differently in the stomatal reopening after ABA-mediated closure and in the inhibition of ABA-mediated stomatal closure itself, and the reopening of closed stomata mediated by 3-ent4 is independent of the COI1-JAZ-MYC2-NAC signaling cascade (Figures 1b–1f). It is also interesting that the enantiomer ent3-4 was ineffective at stomatal reopening, as this result strongly suggests that molecular recognition of the stereochemistry of the ligand, probably by an unknown receptor protein, is involved in the reopening of closed stomata. Additionally, 1 triggered the reopening of closed stomata in the mutants coi1-1, coi1-16s, and anac even though the COI1-JAZ-MYC2-NAC signaling module is located upstream of the ABA-mediated or SA-related signaling cascades that lead to the inhibition of stomatal closure (Figures 2a–2c).
Our results clearly show that the reopening of closed stomata is regulated by a mechanism that is distinct from the inhibition of ABA/SA-mediated stomatal closure (Figures 1 and 2). This is consistent with the results of our previous in silico docking study22 in which some 1 derivatives were unable to bind the COI1-JAZ coreceptor despite these compounds’ being effective in a stomatal reopening assay.22 We thus hypothesized that 1 counters stomatal defense through two functions: COI1-JAZ-independent reopening of closed stomata and the well-known COI1-JAZ-dependent inhibition of stomatal closure.
Diyne-Tagged 1 and in Vivo Raman Imaging of Subcellular Localization
The subcellular localization of a small molecule can provide important information on its mode of action.26 The nuclear colocalization of COI1 and JAZ9 was observed in A. thaliana and Nicotiana tabacum epidermal cell,27 thus, the information on the subcellular localization of 1 derivative which is effective at stomatal reopening and ineffective as a COI1-JAZ agonist will confirm COI1-JAZ independence of 1-mediated reopening of closed stomata and provide information on the potential mode of action. Our previous attempt by using fluorescein-labeled 1 (FL-1) resulted in failure because the synthetic FL-1 was ineffective at reopening of closed stomata.22 We therefore focused on the in vivo alkyne-tag Raman imaging (ATRI) technique19−21 because the alkyne-tag necessary for ATRI is much smaller than fluorescein and is expected not to affect the bioactivity of original 1. ATRI has been used to examine the subcellular accumulation of alkyne-tagged EdU19 and diyne-tagged coenzyme Q in HeLa cells20 and was recently applied to monitoring the distribution of diyne-tagged sphingomyelin in the lipid raft-mimicking domain of an artificial membrane.21 However, ATRI is a state-of-the-art technique; thus, past reports of ATRI have been confined to proof-of-concept experiments using well-studied examples that have already been verified by other methods. We observed the subcellular localization of 5 in a guard cell, which to our knowledge is the first successful application of ATRI on living plant cells. ATRI using 5 demonstrated a distinct difference in subcellular localization between 1 and the COI1-JAZ module following stomatal reopening.
A 1-based ATRI probe should be effective at stomatal reopening and ineffective as a COI1-JAZ agonist, thus precluding colocalization with the COI1-JAZ machinery in the guard cell. As mentioned for 3-ent4, the 3 moiety in 1 may play an important role in the induction of stomatal reopening, while structural modification of the 4 moiety might not affect the bioactivity of 1. Thus, we designed and synthesized a diyne-tagged 1 (5) consisting of naturally occurring 3 and a diyne-tagged l-amino acid;20 the enantiomer ent-5 was synthesized as a control (Figure 3a, Scheme S1). The use of a methyl ester proved to be suitable for practical use because of its improved bioactivity. Both 5s provided a strong Raman signal at 2260 cm–1 (Figure S2).
Figure 3.
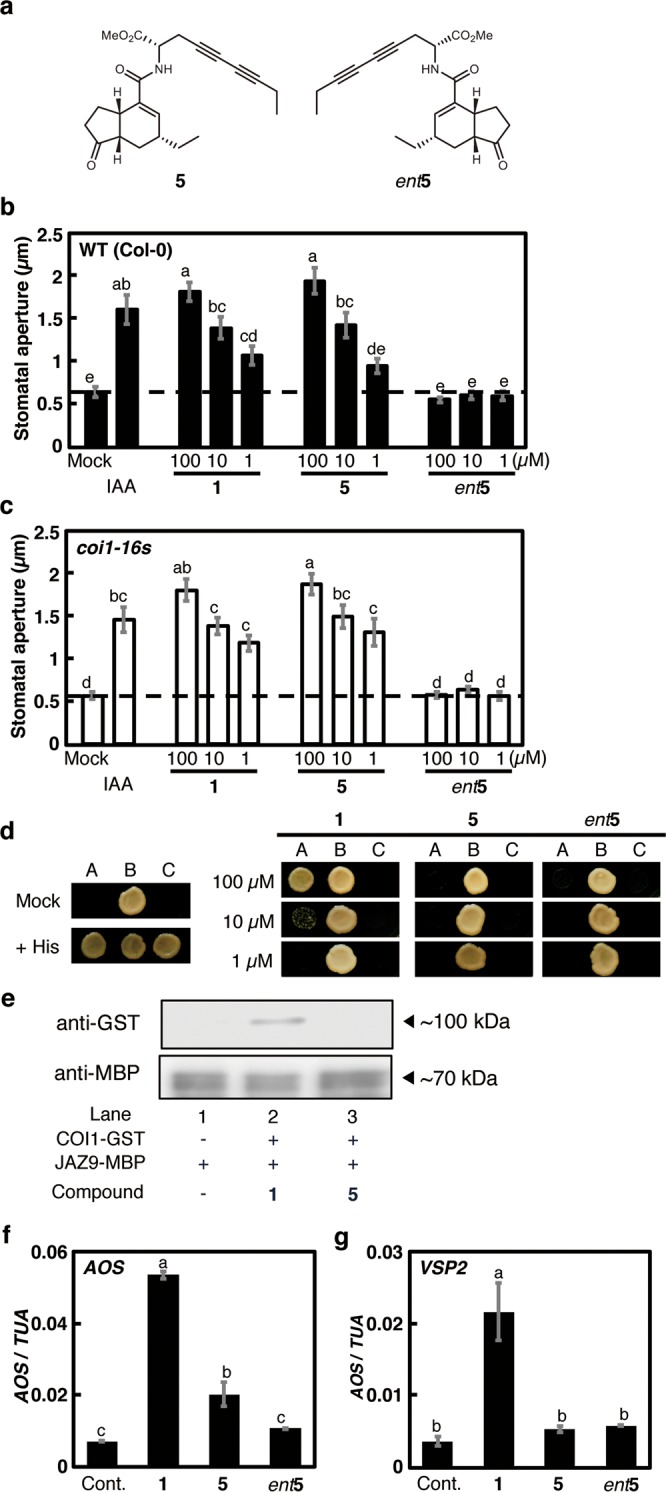
Physiological evaluation of 5s. (a) Structure of 5 and ent5. (b, c) Effect of 1 and 5s on reopening of closed stomata in wild type Col-0 (b) and coi1-16s A. thaliana (c). IAA was used as a positive control. The dashed line indicates mean stomatal aperture of the control without 1 derivatives. Bars represent mean stomatal aperture and SE (n = 20 stomata). Letters a–e indicate significant differences between the means (ANOVA: P < 0.05). (d) Y2H assay of ligand-induced COI1-JAZ9 coreceptor formation. Lane A: Yeast cells cotransformed with pDEST22-JAZ9 and pDEST32-COI1. Lane B: Yeast cells cotransformed with pDEST22-RalGDS-m1 and pDEST32-Krev1 as a positive control. Lane C: Yeast cells cotransformed with pDEST22-RalGDS-m2 and pDEST32-Krev1 as a negative control. (e) Pull-down assay of purified GST-COI1 (5 nM) with recombinant E. coli expressed MBP-JAZ9 (approximately 40 nM) in the presence of diyne 5 (100 nM). All experiments were carried out according to the same procedure as for Figure 1e. (f, g) Relative transcript levels of AOS and VSP2 in A. thaliana treated with 1 and diyne-tagged 1s. Quantitative RT-PCR analyses of relative transcript levels were performed after treatment with each 1 derivative. “Cont.” indicates the mean AOS or VSP2 transcript level under control conditions. Bars represent the mean AOS or VSP2 transcript level of test samples (100 μM). Bars represent mean and SE (n = 3). Letters a–c indicate significant differences between the means (ANOVA: P < 0.05).
Figures 3b–3f show the physiological evaluation of 5 and ent-5, using the same methods as described in Figure 1. 5 was as effective as 1 for stomatal reopening in Col-0, coi1-1, and coi1-16s(25) mutants, whereas ent-5 was ineffective (Figures 3b, 3c, and S4). Moreover, Y2H assays and analysis of gene expressions under the control of COI1-JAZ signaling (AOS and VSP2) suggested that 5 was ineffective as agonists of the COI1-JAZ coreceptor (Figures 3d–3f). These results suggested that 5 is as effective as 1 in triggering the reopening of closed stomata through a COI1-JAZ-independent signaling pathway. Thus, we expected 5 to provide information on the subcellular localization of 1 during COI1-JAZ independent stomatal reopening activity.
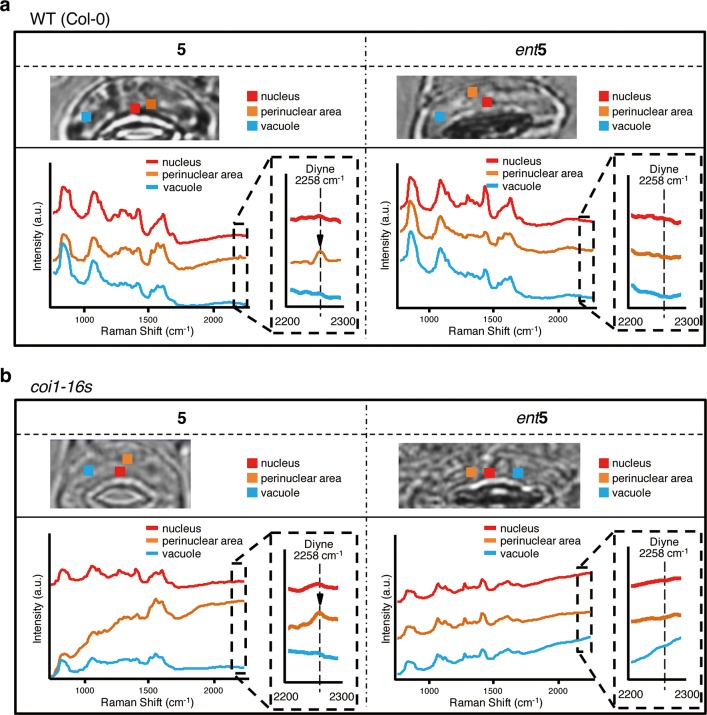
We measured the Raman spectra of subcellular regions of Arabidopsis guard cells, including the nucleus, perinuclear area, vacuole, and dorsal/ventral area of plasma membrane (Figures 4a and 4b). Strong autofluorescence due to the Arabidopsis chloroplast prevents in vivo Raman imaging. Instead, we picked up the Raman signals of 5 (2258 cm–1) in each subcellular region by the comparison of the normalized background signal intensity within the same region. The 5 (2258 cm–1) reopened the closed stomata and then localized around the perinuclear region, but not inside the nucleus, of Col-0 wild type guard cells (Figure 4a). On the other hand, no clear localization was observed using ent-5 (Figure 4a). The localization of 5 was not affected by the presence or absence of the COI1 protein, as shown by its similar localization in the coi1-16s mutant (Figure 4b).
Figure 4.
Raman spectra in each area of stomatal guard cell indicating subcellular localization of 5. Raman spectra in living guard cells of Col-0 (a) and coi1-16s (b) were obtained after 3 h treatment by 100 μM 5 (left)/ent5 (right). The threshold concentration for detecting 5 was ∼100 μM. For details, see Figure S3. Averaged Raman spectra for each subcellular area in the guard cell (1.2 μm × 1.1 μm in size: 3 × 3 pixels) are presented in each color: nucleus area in red, perinuclear area in orange, vacuole area in cyan or blue, plasma membrane (dorsal area) in yellow or green, and plasma membrane (ventral area) in purple. (a) Raman signal of 5 can be found in perinuclear area including ER, whereas no signal was observed in all area for the ent5, and (b) the localization was not affected even in coi1-16s. The light intensity at the sample plane was calculated as 5.8–6.2 mW/μm2 from the ratio of the measured laser power between the sample position and the area of the illumination line. The exposure time for each line was 120 s. Spectra were vertically offset for ease of viewing.
We found that strong autofluorescence due to the Arabidopsis chloroplast precluded in vivo Raman signal-based imaging of 5 in guard cells. To resolve this, we used the arc6 mutant of A. thaliana, which is partly impaired in chloroplast formation,28−30 and in which guard cells without chloroplasts have been reported. As expected, low background autofluorescence from the chloroplast was observed in arc-6 guard cells (Figure S5). The use of arc6 mutant guard cells enabled the in vivo Raman imaging of 5 within the guard cell. Fortunately, 5 remained as effective as 1 for stomatal opening of arc-6 guard cells, whereas ent-5 was ineffective (Figure S6). Figure 5a shows the in vivo Raman imaging of 5 in an arc-6 guard cell. The background noise in this ATRI experiment was very low compared to that using Arabidopsis wild type Col-0 (Figure S7). The clear localization of 5 around the nucleus and plasma membrane (dorsal side) of arc-6 guard cells could be observed, whereas ent-5 showed no specific localization in the same cells (Figures 5a, S8, and S9). In addition, ER-localization of 5 was competitively inhibited by the addition of 1 (Figure S10). We also observed that the fluorescence of ER-marker, ER-Tracker Green, in arc-6 guard cells could be superimposed onto the Raman signal of 5 (Figure 5b).
Figure 5.
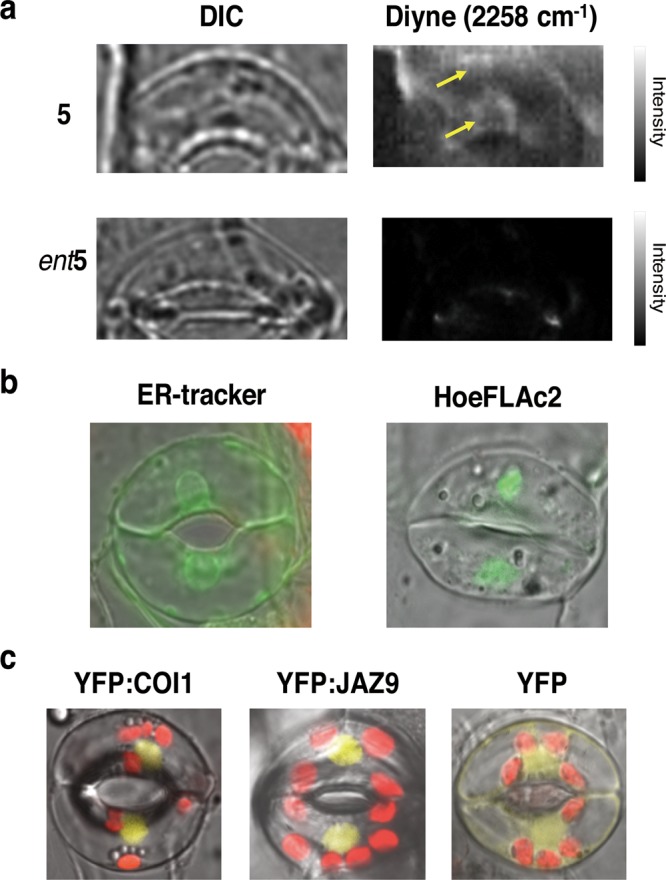
Subcellular localization of 5 and nuclear localization of COI1 and JAZ9 in guard cells. (a) Raman imaging of 5 and ent5 at 100 μM in living guard cells of A. thaliana arc6–1 after 3 h of treatment. 5 localized largely in the perinuclear area including ER, whereas no localization was observed for the ent5. The light intensity at the sample plane was calculated as 6.2 mW/μm2 from the ratio of the measured laser power between the sample position and the illumination line. Exposure time for each line was 120 s. (b) Fluorescence imaging of ER-Tracker (left) and nuclear marker HoeFLAc2 (right). 5 in panel a can be superimposed with ER-tracker, and not with HoeFLAc2. Irradiation at 488 nm, detection at 490–555 nm (green). (c) Both of YFP:COI1 (left, yellow) and YFP:JAZ9 (center, yellow) can be superimposed with HoeFLAc2 in panel b and localized in the nucleus of Arabidopsis guard cell, whereas YFP as a control cannot localize in the nucleus (right, yellow). YFP was detected at 490–555 nm, and chloroplasts were detected at 415–735 nm under irradiation at 488 nm.
The use of the arc-6 mutant was highly successful in avoiding the influence of the strong autofluorescence of chloroplasts in plant cells (Figure 5a). Conventional methods used to circumvent the effects of strong background autofluorescence, such as resonance Raman scattering or the use of longer wavelength irradiation, are ineffective for ATRI in plant cells due to the low Raman signal intensity of diyne. Previous applications of Raman spectroscopy in plant tissues have been restricted to special cases, such as the observation of endogenous polyacetylene in the roots of Daucus carota, which contain no chloroplasts but accumulate a great amount (2.0 g kg–1 FW) of polyacetylene.31 Our strategy using the arc-6 mutant will be a versatile new method for the in vivo application of ATRI in plant cells. While the results presented here were obtained by spontaneous Raman microscopy, the use of newly emerged ATRI coupled with coherent Raman scattering microscopies would offer faster image acquisition, greater detection sensitivity, and less photodamage.32−37 In contrast, the nuclear localization of COI1 and JAZ was clearly observed in P35S-COI1:YFP and P35S-JAZ9:YFP guard cells (Figure 5c). Thus, a distinct difference in localization was confirmed between 5 and the COI1-JAZ coreceptor in Arabidopsis guard cells (Figures 5a and 5c). Our observations strongly suggested that the reopening of closed Arabidopsis stomata by 1 can be linked to the unique localization of 1 in guard cells.
Recently, Higaki et al. reported subcellular microscopic imaging of guard cells using markers for various organelles,38 revealing the typical distribution of organelles in the guard cells of open or closed stomata. In their study, the localization of the ER in the guard cells changed dramatically before and after stomatal opening.38 Our observations of the localization of 5 in arc-6 guard cells seem to correspond well with the distribution of GFP-ER in the guard cells of open stomata. Moreover, the fluorescence pattern of an ER-marker, ER-Tracker Green, in arc-6 guard cells was very similar to the Raman signal of 5 (Figure 5b). It is therefore plausible to conclude that 5 localizes in the ER of arc-6 guard cells. In contrast, the COI1-JAZ machinery was confirmed to localize to the nuclei of guard cells (Figure 5c). This observation corresponds well with the nuclear colocalization of COI1 and JAZ9 in A. thaliana and Nicotiana tabacum epidermal cells, in which the physical interaction between JAZ9 and the TF MYC2 plays a pivotal role.27 The ATRI experiments strongly support the involvement of a COI1-JAZ-independent function of 1 in Arabidopsis guard cells.
The ER thus has an important role in the 1-mediated reopening of stomata. This result provides an important clue for the noncanonical function of 1 in the Arabidopsis guard cell. The swelling of guard cells leading to the stomatal opening is initiated by the activation of H+-ATPase, and subsequent membrane hyperpolarization activates the voltage-dependent inward-rectifying potassium channel KAT-1.39−41 KAT-1 is synthesized and packaged into COPII around the perinuclear region of ER where we observed the Raman signal of 5 and then transported to the plasma membrane through Golgi apparatus.42−46 Ion channel trafficking will strongly affect cellular volume control.41 Thus, our results provide new insight on 1-mediated stomatal reopening that 1 potentially affects the membrane trafficking of KAT-1 in the stomatal guard cell to contribute to the reopening of closed stomata.
Conclusions
In this paper, we described a COI1-JAZ-independent function of 1 on Arabidopsis stomatal guard cells (Figure 6). The bacterial virulence factor, 1, has been considered to inhibit stomatal closure via the ABA-mediated COI1-JAZ signaling pathway. However, AIRI experiments and extensive biological assessments of 1 stereoisomers (ent-1, 3-ent4, ent3-4, 3, and 4) suggested another scenario in which both COI1-JAZ-independent and COI1-JAZ-dependent functions are involved in the 1-mediated breaking of stomatal defense. It is plausible to consider that the functions of 2 (a plant hormone) and 1 (a bacterial virulence factor) in the Arabidopsis guard cell are not identical despite their structural similarity. Only the bacterial virulence factor 1 is involved in the COI1-JAZ-independent reopening of closed stomata.
The discovery of a COI1-JAZ-independent function will provide a possible way to settle a dilemma of plant defense against foreign invaders. Biotrophic bacteria hijack the COI1-JAZ signaling pathway to facilitate infection through the bacterial virulence factor, 1. Inhibition of the COI1-JAZ-independent action of 1 would suppress infection without impairing the COI1-JAZ-dependent plant defense responses against other invaders. A molecular understanding of this novel function could lead to the development of new anti-infection chemicals.
Materials and Methods
All experiments reported in this work were performed three or more times with similar results. In Y2H and in planta assays, we standardize the relative potencies of different ligands used (1 and the stereoisomers, 5/ent5, and 2) before a new set of experiments. Detailed procedures for protein and RNA analyses, assays for stomatal reopening assay, Y2H, Raman and fluorescence imaging, statistical analyses, and chemical syntheses of 5 and ent5 can be found in SI Materials & Methods.
Plant Materials and Growth Conditions
A. thaliana plants were grown on Jiffy-7 peat pellets (Sakata Seed Corporation, Japan) in a Biotron LPH-240SP growth chamber (Nippon Medical & Chemical Instruments Co., Ltd., Japan) at 22 °C under 12 h light (100–118 μmol·m–2·s–1; cool-white fluorescent light)/12 h dark conditions unless stated otherwise. For quantitative PCR analysis, sterilized A. thaliana were grown in 1/2 Murashige & Skoog liquid medium in a Biotron NC-220 growth chamber (Nippon Medical & Chemical Instruments Co., Ltd., Japan) at 22 °C under sterile 16 h light (118 μmol·m–2·s–1; cool-white fluorescent light)/8 h dark conditions.
Stomatal Reopening Assay
The abaxial leaf epidermis of 6- to 8-week-old Arabidopsis plants was peeled and cut to about 2 mm2. For the assay of opening of stomata closed by ABA, the peels were submerged in filtered (with cellulose acetate membrane filter; 25 mm, 0.2 μm, 25CS020AS, ADVANTEC) buffer (25 mM MES (Dojindo, Co., Ltd., Japan)–KOH, pH 6.15, 10 mM KCl) containing 10 μM ABA at 22 °C for 3 h in the light (100–118 μmol·m–2·s–1) to close the stomata. Then the peels with closed stomata were washed and incubated for 3 h with each test sample in buffer (10 mM MES–KOH, pH 6.2, 50 mM KCl) at 22 °C in the dark. For the assay for opening of closed stomata in the dark, the peels were submerged in filtered buffer (10 mM MES–KOH, pH 6.2, 50 mM KCl) at 22 °C for 3 h in the dark to close the stomata. Then the peels with closed stomata were incubated for 3 h with each test sample at 22 °C in the dark. For the competitive inhibition of COR (Sigma-Aldrich Japan Co. Ltd., Japan) and ABA on stomatal aperture, the peels were submerged in filtered buffer (25 mM MES–KOH, pH 6.15, 10 mM KCl) with 15 μM ABA and 30 μM COR or COR derivative at 22 °C for 1 h in the light (100–118 μmol·m–2·s–1). Micrographs were taken under an IX71 microscope (Olympus Corp., Japan) equipped with a DP72 CCD camera (Olympus Corp.). The length of stomatal apertures was measured using ImageJ 1.45S software (http://imagej.net/Welcome).
Quantitative PCR
Seven-day-old plants were incubated in autoclaved 1/2 Murashige & Skoog liquid medium containing 0.5% sucrose and 5 mM MES–KOH (pH 5.8) for 3 h at 22 °C in the dark. Plants were sampled after one more hour of incubation with each filtered 100 μM of test sample at 22 °C in the dark. Total RNA was isolated with an RNeasy Mini Kit (QIAGEN Co., Ltd., Germany). First-strand cDNA was synthesized with ReverTra Ace reverse transcriptase (TOYOBO, Japan) using oligo-dT primers. Quantitative PCR was performed by a StepOnePlus Real-Time PCR System (Life Technologies, USA). Sequences of all primers for quantitative PCR are listed in Table S2. Tubulin-alpha 5 was used as a reference gene.
Y2H Prey-JAZ9 Bait-COI1 Assays
Y2H assays were performed using the ProQuest Two-Hybrid System (Invitrogen, CA, USA). Coding sequences of COI1 and JAZ9 were amplified with the primers shown in Table S3 and cloned into plasmid pENTR/D-TOPO (Invitrogen). Using Gateway technology, COI1 and JAZ9 were respectively transferred to plasmids pDEST32 and pDEST22 and transformed into the MaV203 strain (MATα, leu2-3, 112, trp1-901, his3Δ200, ade2-101, gal4Δ, gal80Δ, SPAL10::URA3, GAL1::lacZ, HIS3UASGAL1::HIS3@LYS2, can1R, cyh2R). Each transformed yeast was grown in 1 mL of SD/–Leu/–Trp medium at 20 °C until the OD600 reached around 0.4 to 0.8. After collection of yeast cells by centrifugation, cells were washed three times with sterile water and the OD600 was adjusted to 0.6. After that, 3 μL of cultures were spotted to SD/–His/–Leu/–Trp medium containing each compound and 1% (v/v) ethanol in each plate. To reduce the number of false-positive colonies, 15 mM 3-amino-1,2,4-triazole was also added to the SD medium. Plates were incubated at 20 °C for 5–6 days to observe the interaction.
Pull-Down Assay for COI1-JAZ Coreceptor Formation
The plasmids of GST-fused COI1 or ASK1 (pFB-GTE-COI1 and pFB-HTB-ASK1) were obtained from Addgene (https://www.addgene.org/). These proteins were coexpressed in insect cells and purified by glutathione affinity chromatography.47 Recombinant MBP-fused JAZ97,22 were expressed in Escherichia coli BL21 cells and purified in amylose resin (New England Biolabs) columns according to the previously reported protocols. In the pull-down experiments, purified COI1-GST (5 nM) with ASK1-GST and coronatine analogues (100 nM) in 500 μL of incubation buffer (50 mM Tris-HCl, pH 7.8, containing 100 mM NaCl, 10% glycerol, 0.1% Tween20, 20 mM 2-mercaptoethanol, 1 μM IP5, EDTA-free complete protease inhibitor cocktail according to the manufacturer’s instructions (Roche)) was added to amylose resin-bound MBP-JAZ9 (25 μL suspension of amylose resin with 40 nM MBP-JAZ9). After 4 h incubation at 4 °C under rotation, the samples were washed with 500 μL of fresh incubation buffer in triplicate. The washed amylose resin was resuspended in 50 μL of SDS–PAGE loading buffer containing maltose (20 mM). Following boiling for 10 min at 60 °C, the samples were loaded on SDS–PAGE and analyzed with Western blotting. The bound COI1-GST were detected using anti-GST HRP conjugate (RPN1236, GE Healthcare).
Statistical Analysis
The data were analyzed by one-way ANOVA followed by a Student–Newman–Keuls post hoc test among all means. Statistical analysis was conducted using CoStat version 6.400 software (CoHort Software, http://www.cohort.com).
Raman Microscopy
Raman spectra and images in Figures 4, 5, S2, S3, S5, S7, S8, and S10 were obtained using laser-scanning Raman microscopy (Raman-11; Nanophoton, Japan) with a 532 nm excitation laser. The beam size was 160 μm × 0.27 μm (X × Y), and the resolution was 0.40 μm × 0.37 μm × >1.0 μm (X × Y × Z) per pixel. The laser beam was focused to a line at the sample and scanned in 1D over a sample to acquire the Raman spectra and image. All data processing was performed using Raman image processing software (Raman Viewer; Nanophoton Corporation, Japan) installed on Raman-11. The detailed conditions for each figure are described in the Supporting Information.
Fluorescence Microscopy
Light micrographs and fluorescent images in Figures 5, S5, and S9 were taken using an IX71 microscope equipped with DP72 CCD camera and WIB filter (Olympus Corp., Japan) or an LSM-710 confocal microscope system (Carl Zeiss, Germany). The detailed conditions for each figure are described in the Supporting Information.
Acknowledgments
Arabidopsis thaliana COI1 mutant (coi1-1 and col1-16s), ANAC mutant (anac) in ecotype Col-0 background were kindly provide by Dr. R. Solano and Dr. A. Chini (National Center for Biotechnology, Madrid, Spain), Dr. Yoshiyuki Murata (Okayama University), and Dr. Xinnian Dong (Duke University), respectively. ARC6 mutant (arc6-1) in ecotype Ws background was kindly provided by Dr. Yasushi Yoshioka (Nagoya University). P35S-YFP, P35S-COI1:YFP, and P35S-YFP:JAZ9 plants in ecotype Col-0 background were kindly provided by Dr. Sheng Yang He (Michigan State University). We thank Prof. Katsumasa Fujita and Dr. Jun Ando (Osaka University) for useful suggestions on ATRI, and Dr. Yoshiyuki Murata and Dr. Shintaro Munemasa (Okayama University) for helpful discussion throughout the manuscript. This work was supported in part by a Grant-in-Aid for Scientific Research (No. 23102012) on Innovative Areas “Chemical Biology of Natural Products” to M.U. from MEXT, Japan; a Grant-in-Aid for Scientific Research (No. 26282207 and No. 17H00885 to M.U.); JSPS A3 Foresight Program to M.U.); and AMED-CREST, AMED, to K.D. and M.S.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.7b00099.
Details of Raman and fluorescence studies, synthesis of diyne probe, microarray data, materials and methods, 1H and 13C NMR spectra, and references (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Fonseca S.; Chini A.; Hamberg M.; Adie B.; Porzel A.; Kramell R.; Miersch O.; Wasternack C.; Solano R. (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 2009, 5 (5), 344–350. 10.1038/nchembio.161. [DOI] [PubMed] [Google Scholar]
- Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100 (4), 681–697. 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C.; Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111 (6), 1021–1058. 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. How Jasmonates Earned their Laurels: Past and Present. J. Plant Growth Regul. 2015, 34 (4), 761–794. 10.1007/s00344-015-9526-5. [DOI] [Google Scholar]
- Wasternack C.; Kombrink E. Jasmonates: structural requirements for lipid-derived signals active in plant stress responses and development. ACS Chem. Biol. 2010, 5 (1), 63–77. 10.1021/cb900269u. [DOI] [PubMed] [Google Scholar]
- Campos M. L.; Kang J. H.; Howe G. A. Jasmonate-triggered plant immunity. J. Chem. Ecol. 2014, 40 (7), 657–675. 10.1007/s10886-014-0468-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A.; Fonseca S.; Fernandez G.; Adie B.; Chico J. M.; Lorenzo O.; Garcia-Casado G.; Lopez-Vidriero I.; Lozano F. M.; Ponce M. R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448 (7154), 666–671. 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Thines B.; Katsir L.; Melotto M.; Niu Y.; Mandaokar A.; Liu G.; Nomura K.; He S. Y.; Howe G. A.; Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 2007, 448 (7154), 661–665. 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- Yan Y.; Stolz S.; Chetelat A.; Reymond P.; Pagni M.; Dubugnon L.; Farmer E. E. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 2007, 19 (8), 2470–2483. 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihara A.; Shiraishi K.; Sato H.; Sakamura S.; Nishiyama K.; Sakai R.; Furusaki A.; Matsumoto T. The structure of coronatine. J. Am. Chem. Soc. 1977, 99 (2), 636–637. 10.1021/ja00444a067. [DOI] [Google Scholar]
- Katsir L.; Schilmiller A. L.; Staswick P. E.; He S. Y.; Howe G. A. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proc. Natl. Acad. Sci. U. S. A. 2008, 105 (19), 7100–7105. 10.1073/pnas.0802332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. Jasmonate passes muster: a receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. 10.1146/annurev.arplant.043008.092007. [DOI] [PubMed] [Google Scholar]
- Geng X.; Cheng J.; Gangadharan A.; Mackey D. The coronatine toxin of Pseudomonas syringae is a multifunctional suppressor of Arabidopsis defense. Plant Cell 2012, 24 (11), 4763–4774. 10.1105/tpc.112.105312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M.; Underwood W.; Koczan J.; Nomura K.; He S. Y. Plant stomata function in innate immunity against bacterial invasion. Cell 2006, 126 (5), 969–980. 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- McLachlan D. H.; Kopischke M.; Robatzek S. Gate control: guard cell regulation by microbial stress. New Phytol. 2014, 203 (4), 1049–1063. 10.1111/nph.12916. [DOI] [PubMed] [Google Scholar]
- Zheng X. Y.; Spivey N. W.; Zeng W.; Liu P. P.; Fu Z. Q.; Klessig D. F.; He S. Y.; Dong X. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 2012, 11 (6), 587–596. 10.1016/j.chom.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montillet J. L.; Hirt H. New checkpoints in stomatal defense. Trends Plant Sci. 2013, 18 (6), 295–297. 10.1016/j.tplants.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Munemasa S.; Oda K.; Watanabe-Sugimoto M.; Nakamura Y.; Shimoishi Y.; Murata Y. The coronatine-insensitive 1 mutation reveals the hormonal signaling interaction between abscisic acid and methyl jasmonate in Arabidopsis guard cells. Specific impairment of ion channel activation and second messenger production. Plant Physiol. 2007, 143 (3), 1398–1407. 10.1104/pp.106.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakoshi H.; Dodo K.; Okada M.; Ando J.; Palonpon A.; Fujita K.; Kawata S.; Sodeoka M. Imaging of EdU, an alkyne-tagged cell proliferation probe, by Raman microscopy. J. Am. Chem. Soc. 2011, 133 (16), 6102–6105. 10.1021/ja108404p. [DOI] [PubMed] [Google Scholar]
- Yamakoshi H.; Dodo K.; Palonpon A.; Ando J.; Fujita K.; Kawata S.; Sodeoka M. Alkyne-Tag Raman Imaging for Visualization of Mobile Small Molecules in Live Cells. J. Am. Chem. Soc. 2012, 134 (51), 20681–20689. 10.1021/ja308529n. [DOI] [PubMed] [Google Scholar]
- Ando J.; Kinoshita M.; Cui J.; Yamakoshi H.; Dodo K.; Fujita K.; Murata M.; Sodeoka M. Sphingomyelin distribution in lipid rafts of artificial monolayer membranes visualized by Raman microscopy. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (15), 4558–4563. 10.1073/pnas.1418088112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egoshi S.; Takaoka Y.; Saito H.; Nukadzuka Y.; Hayashi K.; Ishimaru Y.; Yamakoshi H.; Dodo K.; Sodeoka M.; Ueda M. Dual function of coronatine as a bacterial virulence factor against plants: possible COI1–JAZ-independent role. RSC Adv. 2016, 6 (23), 19404–19412. 10.1039/C5RA20676F. [DOI] [Google Scholar]
- Okada M.; Ito S.; Matsubara A.; Iwakura I.; Egoshi S.; Ueda M. Total syntheses of coronatines by exo-selective Diels-Alder reaction and their biological activities on stomatal opening. Org. Biomol. Chem. 2009, 7 (15), 3065–3073. 10.1039/b905159g. [DOI] [Google Scholar]
- Xie D. X.; Feys B. F.; James S.; Nieto-Rostro M.; Turner J. G. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 1998, 280 (5366), 1091–1094. 10.1126/science.280.5366.1091. [DOI] [PubMed] [Google Scholar]
- Chen J.; Sonobe K.; Ogawa N.; Masuda S.; Nagatani A.; Kobayashi Y.; Ohta H. Inhibition of arabidopsis hypocotyl elongation by jasmonates is enhanced under red light in phytochrome B dependent manner. J. Plant Res. 2013, 126 (1), 161–168. 10.1007/s10265-012-0509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S.; Arita Y.; Honda M.; Iwamoto K.; Matsuyama A.; Shirai A.; Kawasaki H.; Kakeya H.; Kobayashi T.; Matsunaga S.; et al. Marine antifungal theonellamides target 3 beta-hydroxysterol to activate Rho1 signaling. Nat. Chem. Biol. 2010, 6 (7), 519–526. 10.1038/nchembio.387. [DOI] [PubMed] [Google Scholar]
- Withers J.; Yao J.; Mecey C.; Howe G. A.; Melotto M.; He S. Y. Transcription factor-dependent nuclear localization of a transcriptional repressor in jasmonate hormone signaling. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (49), 20148–20153. 10.1073/pnas.1210054109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Asano T.; Fujiwara M. T.; Yoshida S.; Machida Y.; Yoshioka Y. Plant Cells Without Detectable Plastids are Generated in the crumpled leaf Mutant of Arabidopsis thaliana. Plant Cell Physiol. 2009, 50 (5), 956–969. 10.1093/pcp/pcp047. [DOI] [PubMed] [Google Scholar]
- Pyke K. A.; Rutherford S. M.; Robertson E. J.; Leech R. M. arc6, a fertile Arabidopsis mutant with only two mesophyll cell chloroplasts. Plant Physiol. 1994, 106 (3), 1169–1177. 10.1104/pp.106.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson E. J.; Pyke K. A.; Leech R. M. Arc6, an Extreme Chloroplast Division Mutant of Arabidopsis Also Alters Proplastid Proliferation and Morphology in Shoot and Root Apices. J. Cell Sci. 1995, 108, 2937–2944. [DOI] [PubMed] [Google Scholar]
- Roman M.; Dobrowolski J. C.; Baranska M.; Baranski R. Spectroscopic studies on bioactive polyacetylenes and other plant components in wild carrot root. J. Nat. Prod. 2011, 74 (8), 1757–1763. 10.1021/np200265d. [DOI] [PubMed] [Google Scholar]
- Hong S.; Chen T.; Zhu Y.; Li A.; Huang Y.; Chen X. Live-cell stimulated Raman scattering imaging of alkyne-tagged biomolecules. Angew. Chem., Int. Ed. 2014, 53 (23), 5827–5831. 10.1002/anie.201400328. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Paley D. W.; Wei L.; Weisman A. L.; Friesner R. A.; Nuckolls C.; Min W. Multicolor live-cell chemical imaging by isotopically edited alkyne vibrational palette. J. Am. Chem. Soc. 2014, 136 (22), 8027–8033. 10.1021/ja502706q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L.; Hu F.; Shen Y.; Chen Z.; Yu Y.; Lin C. C.; Wang M. C.; Min W. Live-cell imaging of alkyne-tagged small biomolecules by stimulated Raman scattering. Nat. Methods 2014, 11 (4), 410–412. 10.1038/nmeth.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L.; Hu F.; Chen Z.; Shen Y.; Zhang L.; Min W. Live-Cell Bioorthogonal Chemical Imaging: Stimulated Raman Scattering Microscopy of Vibrational Probes. Acc. Chem. Res. 2016, 49 (8), 1494–1502. 10.1021/acs.accounts.6b00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu F.; Lamprecht M. R.; Wei L.; Morrison B.; Min W. Bioorthogonal chemical imaging of metabolic activities in live mammalian hippocampal tissues with stimulated Raman scattering. Sci. Rep. 2016, 6, 39660. 10.1038/srep39660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S.; Matsushita T.; Izuta S.; Katada S.; Ura M.; Ikeda T.; Hayashi G.; Suzuki Y.; Kobayashi K.; Tokunaga K.; et al. Chemically-activatable alkyne-tagged probe for imaging microdomains in lipid bilayer membranes. Sci. Rep. 2017, 7, 41007. 10.1038/srep41007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higaki T.; Kutsuna N.; Hosokawa Y.; Akita K.; Ebine K.; Ueda T.; Kondo N.; Hasezawa S. Statistical organelle dissection of Arabidopsis guard cells using image database LIPS. Sci. Rep. 2012, 2, 405. 10.1038/srep00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. H.; Bohmer M.; Hu H.; Nishimura N.; Schroeder J. I. Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 2010, 61, 561–591. 10.1146/annurev-arplant-042809-112226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L.-M.; Zhao Z.; Assmann S. M. Guard cells: a dynamic signaling model. Curr. Opin. Plant Biol. 2004, 7 (5), 537–546. 10.1016/j.pbi.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Grefen C.; Honsbein A.; Blatt M. R. Ion transport, membrane traffic and cellular volume control. Curr. Opin. Plant Biol. 2011, 14 (3), 332–339. 10.1016/j.pbi.2011.03.017. [DOI] [PubMed] [Google Scholar]
- Sieben C.; Mikosch M.; Brandizzi F.; Homann U. Interaction of the K+-channel KAT1 with the coat protein complex II coat component Sec24 depends on a di-acidic endoplasmic reticulum export motif. Plant J. 2008, 56 (6), 997–1006. 10.1111/j.1365-313X.2008.03658.x. [DOI] [PubMed] [Google Scholar]
- Phillipson B. A.; Pimpl P.; daSilva L. L. P.; Crofts A. J.; Taylor J. P.; Movafeghi A.; Robinson D. G.; Denecke J. Secretory bulk flow of soluble proteins is efficient and COPII dependent. Plant Cell 2001, 13 (9), 2005–2020. 10.2307/3871424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DaSilva L. L. P.; Snapp E. L.; Denecke J.; Lippincott-Schwartz J.; Hawes C.; Brandizzi F. Endoplasmic reticulum export sites and golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 2004, 16 (7), 1753–1771. 10.1105/tpc.022673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M.; Ueda T.; Sato K.; Abe H.; Nagata T.; Nakano A. A dominant negative mutant of Sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 2000, 23 (4), 517–525. 10.1046/j.1365-313x.2000.00823.x. [DOI] [PubMed] [Google Scholar]
- Yang Y. D.; Elamawi R.; Bubeck J.; Pepperkok R.; Ritzenthaler C.; Robinson D. G. Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. Plant Cell 2005, 17 (5), 1513–1531. 10.1105/tpc.104.026757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard L. B.; Tan X.; Mao H.; Withers J.; Ben-Nissan G.; Hinds T. R.; Kobayashi Y.; Hsu F. F.; Sharon M.; Browse J.; et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 2010, 468 (7322), 400–405. 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.