Abstract
DNA (cytosine-5)-methyltransferase (DNMT) 1 participates in transcriptional repression of genes by methylation-dependent and -independent mechanisms. Here, DNMT1 is shown to bind p53 and colocalize in the nucleus. DNMT1-mediated methylation is stimulated by p53 in vitro. Upon p53 induction, a reporter construct containing the antiapoptotic gene survivin promoter, which contains a natural p53 binding site, was methylated in WT HCT116 cells but not in DNMT1 null or p53 null cells. Endogenous survivin gene repression involves cooperation between DNMT1 and p53 and is relieved by introduction of DNMT1- or p53-specific small inhibitory RNA. DNMT1 null cells did not exhibit a significant repressive effect for p53 responsive survivin and cdc25C gene expression compared with the parental cells. Normal human fibroblasts also exhibited similar DNMT1- and p53-mediated methylation of the survivin promoter, suggesting cooperation between p53 and DNMT1 in gene silencing.
Keywords: doxorubicin, survivin, small inhibitory RNA
Mammalian cell growth and development is regulated by specific patterns of gene expression, which are controlled by a host of genetic and epigenetic factors. Interaction of these factors with promoter elements can result in transcriptional activation or repression. There are two major epigenetic systems that affect mammalian gene expression at the chromatin level: DNA methylation and histone modification. DNA methylation in the mammalian genome takes place exclusively at the cytosine bases in CpG dinucleotides. Three known classes of catalytically active DNA (cytosine-5)-methyltransferases (DNMTs), DNMT1, DNMT3a, and DNMT3b, have been cloned and characterized (1–3). DNMT1 methylates the daughter strands of newly replicated nuclear DNA to preserve the parental methylation pattern. This enzyme binds to proliferating cell nuclear antigen (PCNA), an auxiliary factor of DNA replication, during S phase (4). The other two enzymes, DNMT3a and DNMT3b, are known as de novo methyltransferases and are believed to participate in establishing the methylation pattern during preimplantation (5). DNA methylation is crucial for mammalian development; neither DNMT1 homozygous knockout mice nor DNMT3b homozygous knockout mice were viable, and DNMT3a knockout mice die 4 weeks after birth (6, 7).
In the past few years, it has become increasingly apparent that aberrant promoter methylation is associated with the loss of gene function and is coordinated with histone deacetylation in mammals (8). Furthermore, DNMTs may enforce gene silencing by recruiting transcriptional repressor protein complexes. DNMT1, as well as DNMT3a and DNMT3b, were found to be associated with histone deacetylase (HDAC) activity in vivo (9, 10), suggesting their involvement in transcriptional repression of chromatin by means of histone deacetylation. Synergy between DNA demethylation and HDAC inhibition in the reexpression of the silenced genes in cancer has been reported (11).
Although the role of DNA methylation and histone modifications in gene expression is well established, the primary signals for specific gene expression mediated by these factors may come from cellular stress or DNA damage. In mammalian cells, a functional p53 tumor suppressor protein responds to a variety of cellular stresses, including DNA damage and aberrantly activated oncogenes, and may induce cell cycle arrest and apoptosis (12, 13). Approximately 1/10th of human gene promoters contain a p53-binding site and therefore may be classified as p53-responsive genes (14). Some are transcriptionally activated; others are transcriptionally repressed by p53. The ratio of up-regulated genes to down-regulated genes was ≈3:2. Affected genes are involved in the cell cycle, angiogenesis, DNA repair and replication, transcription, and apoptosis.
Because the p53 pathway is activated before apoptosis, the switching off of antiapoptotic genes is needed. Activation of p53 leads to down-regulation of survivin, a member of the inhibitor of apoptosis (IAP) family (15). Other IAP family members that are down-regulated by p53 are inhibitors of apoptosis protein 1 (MIHB) and apoptosis protein 2 (MIHC) (16). survivin is overexpressed in embryonic and cancer cells but not in appreciable amounts in most normal cells. The survivin promoter contains two half-sites of the p53 consensus element separated by a three-nucleotide spacer. Chromatin immunoprecipitation (IP) demonstrates that p53 binds to the survivin promoter in vivo (15). Induction of p53 leads to transcriptional and translational repression of survivin in vivo (15). However, detailed mechanisms of repression by p53 have not yet been elucidated. p53 represses genes in several different ways. One of the mechanisms involves an association between p53 and HDACs (17), an interaction mediated by p53 binding to the corepressor protein Sin3a (18). The p53–Sin3a interaction targets HDACs to the promoters of the p53-repressed genes where enzymatic deacetylation of histones on chromatin takes place to create a transcriptionally unfavorable environment (19). A close look at the survivin promoter suggests that it contains a typical CpG island architecture along with the p53-binding site, leading to speculation about the involvement of DNA methylation and p53 in survivin gene regulation. In this report, we have investigated the physical cooperation and functional association between human maintenance DNMT and p53 in response to DNA damage.
Materials and Methods
Cell Culture and Constructs. All cell lines (MCF-7, HEK 293, COS-7, and IMR-90) were obtained from the American Type Culture Collection and were grown as recommended. Parental HCT116 and DNMT and the p53 knockout (DNMT1 null or p53 null) cell lines DNMT1–/– (HCT116 DNMT1–/–, clone 1C1), DNMT1–/–, and DNMT3b–/– (HCT116 DKO) were grown as described in ref. 20. To generate DNA damage, cells were treated for up to 48 h with 0.4 or 1 μM doxorubicin (doxo).
All GST-DNMT constructs are described in ref. 21. Details of the fusion constructs of p53 are available on request. SpII plasmid is described in ref. 15.
IP, GST Pull-Down, and Western Blot Analysis. Antibodies were procured as follows: p53, survivin, and cdc25C from Cell Signaling Technology (Beverly, MA); histone 3 dimethyl lysine 9 (DiMeK9) antibody from Upstate Biotechnology (Lake Placid, NY); DNMT1 and PCNA from New England Biolabs and BD Biosciences; and actin from Sigma. Nuclear extracts were made as described in ref. 22. Co-IP and GST pull-downs were performed as described in ref. 23. Densitometric analysis was performed on Western blots by using nih image 1.59.
DNMT Assay. Methyltransferase assays were carried out by using recombinant human DNMT1 (New England Biolabs) as described in ref. 24. Five hundred nanograms of MCF-7 (breast carcinoma), Micrococcus luteus genomic DNA, and poly(IC) (Sigma) was used for methylation assays with recombinant human p53 expressed in Escherichia coli with a purity of >95%.
Methylation-Sensitive PCR (MSP) Assay. Bisulfite modification of 2 μg of DNA was performed by using the CpGenome DNA Modification Kit (Intergen, Purchase, NY). MSP assay was performed by using the TranSignal Promoter Methylation Detection Kit (Panomics, Redwood City, CA) and AmpliTaq DNA polymerase from Roche Applied Science (Indianapolis).
Plasmid Methylation, Transient Transfections, Luciferase Assay, and Immunocytochemistry. The SpII-Luc (SpII) plasmid was methylated by using M. SssI, M. HpaII, M. MspI, or M. HhaI and cold AdoMet (New England Biolabs). The methylation status was checked by using HpaII, MspI, and HhaI restriction enzymes. Methylated plasmids were purified before transfection. As an internal control, a constant amount of the β-galactosidase reporter pCMVβ (BD Biosciences) also was cotransfected. Cells were transfected with a mixture of DNA and FuGENE 6 (Roche Diagnostics) at a ratio of 1:3 μg/μl. The cells were harvested 48 h posttransfection, and luciferase activity was measured and normalized with β-galactosidase activity. For immunocytochemistry, COS-7 cells were transfected with plasmid and visualized with a Zeiss 200M microscope with a ×63 oil objective lens at 488 nm for GFP-DNMT1, 568 nm for DsRed-p53 fusions, and 460 nm for nuclear staining with Hoechst 33342.
SpII(-p53) plasmid was made by digesting SpII plasmid with the SacII restriction enzyme and religating the plasmid. This plasmid does not contain the p53-binding site.
Methylation Analysis of the survivin Promoter in Genomic DNA and Transfected SpII Plasmid. Adenovirus (Ad) control and Ad expressing p53 (Ad-p53) were obtained from Qbiogene (Carlsbad, CA). Two hours postinfection, HCT116, DNMT1–/–, and p53–/– cells were transfected with SpII. The cells were allowed to recover for 24 h and were treated with 1 μM doxo for 24 h before DNA extraction. The survivin promoter on SpII DNA was amplified by using a survivin promoter-specific 5′ primer (GACCACGGGCAGAGCCACGCGGCG) and a luciferase-specific 3′ primer (CTTTATGTTTTTGGCGTCTTCCA) after digestion with either MspI or HpaII (New England Biolabs) restriction enzymes. The endogenous survivin promoter was amplified by using the same 5′ primer along with the survivin-specific 3′ primer (GCGCCCTGGGCAACCGTCTCCACC). PCR parameters were 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s.
Gene Knockdown Using Gene-Specific Small Inhibitory RNAs (siRNAs). The ShortCut RNAi kit (New England Biolabs) was used to generate 22- to 23-nt-long double-stranded siRNAs. Purified siR-NAs (ShortCut siRNA Mix, New England Biolabs) were transfected into HCT116 cells by using Lipofectamine 2000 (Invitrogen) for 48 h. Transfection used 20 nM siRNAs per 1 × 10e6 cells.
Chromatin IP Assay. HCT116 cells were grown on 150-mm dishes and treated with 1 μM doxo. After 24 h, proteins were cross-linked with DNA by using 1% formaldehyde for 10 min at 37°C. Cells were washed two times with ice-cold PBS, harvested, and lysed with an SDS lysis buffer (Upstate Biotechnology) in the presence of a mixture of protease inhibitors (Sigma). The lysates were sonicated to shear DNA to lengths between 200 and 1,000 bp. After 10-fold dilution of the sonicated cell supernatants in ChIP dilution buffer (Upstate Biotechnology) containing protease inhibitors, 40 μl of protein G coupled to magnetic beads (New England Biolabs) and 100 μg of salmon sperm DNA were added and incubated for 1 h at 4°C with rotation to reduce nonspecific background. IPs were carried out overnight at 4°C with mixing by using 2 μg of DNMT1, p53, HDAC1, DiMeK9 polyclonal, and control GFP monoclonal antibodies. The beads were isolated and washed according to Upstate Biotechnology's instructions. DNA–protein complexes were eluted from the beads with a buffer containing 1% SDS and 0.1 M NaHCO3. The cross-links were reversed by incubating the eluates with NaCl (5 M) for 6 h at 65°C. Proteinase K (Roche) was added for 1 h at 45°C, and the DNA was recovered by phenol/chloroform extraction and ethanol precipitation. Immunoprecipitated DNA was analyzed for the presence of the survivin gene promoter sequence by PCR with proximal (GACCACGGGCAGAGCCACGCGGCG and GCGCCCTGGGCAACCGTCTCCACC) and distal (TCCTGGAACTCGGTTTTGAG and ACCACTTTGGGGCAGAGATG) primer sets by using PCR of 30 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s.
Results
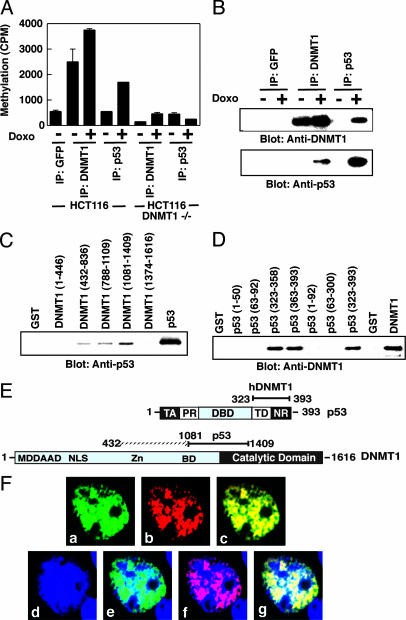
Maintenance DNMT Interacts with p53 in Vivo. To determine whether DNMT1 associates with p53 in vivo, we treated WT and DNMT1–/– HCT116 cells with the DNA damaging agent doxo to activate p53 and then immunoprecipitated protein complexes from the nuclear extract with anti-DNMT1, anti-p53, or control anti-GFP antibodies. Both anti-DNMT1 and anti-p53 antibodies pulled down methyltransferase activity compared with control anti-GFP antibodies in WT HCT116 cells but not in DNMT1–/– cells (Fig. 1A). Immunoprecipitated complexes were further resolved by SDS/PAGE and analyzed by Western blot to identify protein partners. Weak or no interaction between DNMT1 and p53 was observed in uninduced HCT116 cells. In HCT116 cells, a robust p53–DNMT1 complex was observed only after DNA damage by doxo (Fig. 1B). A similar interaction was observed for HEK293 cells (data not shown). To find out whether this result represents direct binding between p53 and DNMT1, GST fusion fragments of DNMT1 covering the whole ORF of the enzyme were used in GST pull-down assays along with purified p53 protein and competitor BSA. Amino acid residues 432–1409 of DNMT1, covering a part of the amino and catalytic region, were bound to p53 (Fig. 1C). A similar binding assay was performed to determine the DNMT1-binding region on p53. GST fusion proteins covering the whole p53 protein were incubated with purified DNMT1 for the pull-down assay. Amino acid residues 323–393 of p53 bound directly to DNMT1 (Fig. 1D). Furthermore, cotransfection of DNMT1 and a p53 mutant (amino acids 1–300) without the DNMT1 binding region did not immunoprecipitate methyltransferase activity (data not shown), confirming that the DNMT1 binding region of p53 is located at the C terminus. The strongest binding of p53 was to amino acids 1081–1409 of DNMT1 (Fig. 1 C and E). To validate the binding in vivo, colocalization experiments were performed. COS-7 cells were transfected with the fusion constructs GFP-DNMT1 and DsRed-p53 WT (Fig. 1F) and visualized by fluorescent microscopy. Green nuclear spots appeared in all of the transfectants (Fig. 1F a), showing DNMT1 in the nucleus, as observed previously (25). Red nuclear spots (Fig. 1Fb) identifying p53 fusions were visible in the nucleus for WT p53 (26). Superimposition of p53 and GFP-DNMT1 resulted in yellow nuclear spots on both the euchromatin and heterochromatin regions (Fig. 1Fc–g), demonstrating colocalization between DNMT1 and WT p53. These results are consistent with a close physical association between p53 and human DNMT1 in vivo.
Fig. 1.
In vivo interaction and analysis of the binding region of DNMT1 and p53. (A) IP of DNMT1 activity in the nuclear extracts of human cells. Antibodies used for IP, along with cell lines, are indicated. GFP antibody was used as a control. (B) Physical interaction between DNMT1 and p53 in nuclear extracts of HCT116 cells. Antibodies used for IP are indicated. Control and doxo-treated cells are indicated by – and +. (C) Mapping of the p53-binding region on DNMT1 by using a GST fusion of DNMT1 and the pull-down procedure described in Materials and Methods. (D) Mapping of the DNMT1 binding region of p53 by using a GST fusion of p53 as in C. Amino acid numbers of the fusions are in parentheses in C and D. (E) A diagram of p53 and DNMT1 showing the binding regions based on C and D. Functional domains of p53 are indicated: TA, transactivation; PR, proline rich; DBD, DNA binding; TD, tetramerization; NR, negative regulation. Methylation DNA-dependent allosteric activation (MDDAAD), bromo domain (BD), and nuclear localization (NLS) regions of DNMT1 are indicated. (F) Colocalization of DNMT1 and p53. Shown are GFP-DNMT1 (green) (a), DsRed-p53 WT (red) (b), GFP-DNMT1 and DsRed-p53 (merged yellow) (c), nucleus (blue) (d), merge nucleus and GFP-DNMT1 (e), merge nucleus and DsRed-p53 WT (f), merge nucleus, GFP-DNMT1, and DsRed-p53 (g).
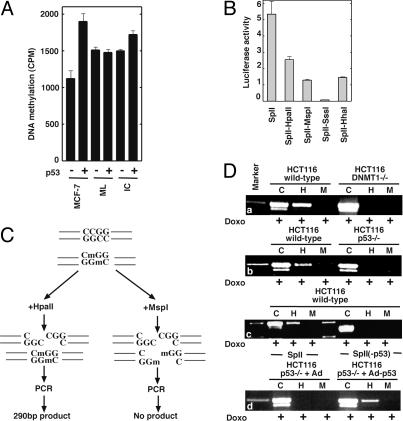
Mechanistic Aspects of p53-Mediated DNA Methylation. DNMTs regulate gene expression by methylation or by recruitment of transcriptional repressors onto the promoter, resulting in gene silencing. Thus, it was important to determine whether p53 binding to DNMT1 has any bearing on methyltransferase activity. Therefore, we incubated various genomic DNA substrates with DNMT1 in the presence or absence of purified p53. A significant increase (≈63%) of methyl group incorporation was observed in the presence of p53 in MCF-7 human genomic DNA compared with a bacterial (M. luteus) DNA substrate (Fig. 2A) that does not contain p53-responsive genes. Thus, DNA methylation stimulation is specific to the human genome, suggesting that the DNMT1 complex with p53 may lead to de novo methylation and gene silencing.
Fig. 2.
DNMT1 and p53 participate in DNA methylation. (A) In vitro determination of DNA methyltransferase activity of DNMT1 in the presence (+) or absence (–) of p53 by using various DNA substrates, MCF-7 genomic DNA, M. luteus (ML), and poly(dI-dC)·poly(dI-dC). (B) In vitro methylation and transcriptional analysis of SpII plasmid. The plasmid was methylated with different prokaryotic methylase, as indicated. (C) A schematic of DNA methylation analysis of the survivin promoter containing SpII plasmid after transfection. Methylated cytosine residues are indicated as “m.” (D) “M” and “H” designate MspI and HpaII restriction enzymes. “C” is undigested DNA control. Molecular weight (Marker) is indicated. The bands shown in the Marker lane are 300 and 200 bp. Cell type and knockout status are indicated at top. Transfection of Ad and Ad-p53 are indicated. Deletion of the p53-binding site on SpII abolished survivin promoter methylation. p53 site deletion resulted in ≈30 bp smaller amplicon in the SpII(-p53) construct, as shown in d.
To validate our hypothesis, we chose the p53-repressive survivin gene promoter reporter construct SpII (15). The survivin gene promoter has a p53-binding site and has several CpG sites that are a target for methylation by DNMT1. To study the role of methylation in survivin promoter regulation, we methylated SpII in vitro, transfected it into HEK 293 cells, and measured luciferase activity. Indeed, luciferase repression correlated with methylated CpG density (Fig. 2B), suggesting that methylation may play a role in survivin gene regulation, perhaps by means of p53 recruitment. To elucidate the role of p53 and DNMT1 in methylation of the survivin promoter, HCT116 WT, HCT116 DNMT1–/–, and HCT116 p53–/– cells were transfected with SpII plasmid containing the survivin promoter and were treated with doxo for p53 induction. The SpII plasmid was reisolated and analyzed for CpG methylation by restriction digestion with MspI or HpaII and PCR of the promoter. If methylation occurs at a CpG within the restriction enzyme recognition sequence CCGG (CmGG, where m is methyl cytosine), then a PCR product of ≈290 bp is expected in samples digested with HpaII, because HpaII restriction is blocked by internal CpG methylation (Fig. 2C). Neither DNMT1–/– nor p53–/– cells were able to methylate the transfected SpII plasmid, as judged from the lack of PCR product [Fig. 2Da and b, lanes H (HpaII) and M (MspI)]. However, SpII transfected in HCT116 WT cells was methylated, as judged from the presence of the ≈290-bp product after HpaII digestion. Furthermore, deletion of the p53-binding site on SpII [SpII(-p53)] disrupted de novo methylation, suggesting that the p53-binding site embedded in the promoter is crucial for promoter methylation in the presence of p53 (Fig. 2Dc, lane H). SpII(-p53) displayed substantial relief of p53-mediated inhibition of luciferase reporter expression compared with SpII (data not shown), as observed previously (15). To confirm the role of p53 in DNA methylation, p53–/– HCT116 cells were infected with Ad or the Ad-p53 gene first and then were transfected with SpII plasmid. Reintroduction of WT p53 in p53–/– HCT116 cells was able to methylate the survivin promoter in the SpII plasmid (Fig. 2Dd, lane H). However, DNMT1–/– cells were not able to methylate the transfected plasmid, despite a high level of p53 expression (data not shown). These experiments demonstrate that p53 is crucial for gene silencing by means of DNA methylation of the survivin promoter in the presence of DNMT1 in SpII.
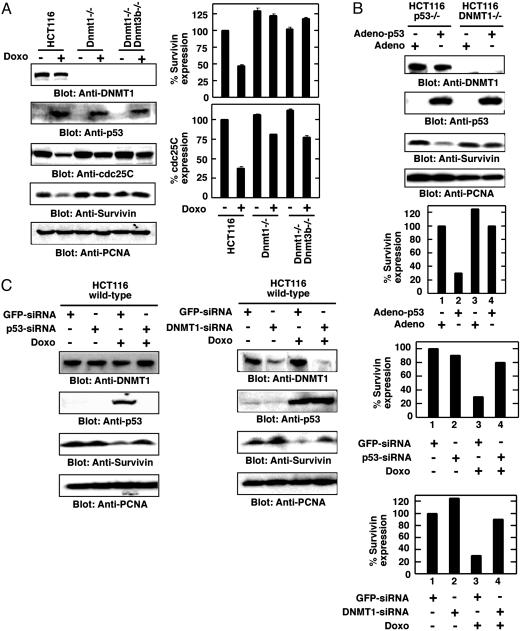
Cooperation Between DNMT1 and p53 for survivin Gene Regulation. To demonstrate interdependence of DNMT1 and p53 in gene regulation, DNMT1, DNMT1 plus DNMT3b knockouts, and parental colorectal cancer HCT116 cells were treated with doxo to induce p53. Cell extracts were examined for p53-responsive gene expression, namely survivin and cdc25C. As anticipated, in HCT116, survivin was down-regulated, as was cdc25C (Fig. 3A). However, both survivin and cdc25C levels remained unchanged or slightly down-regulated in cells lacking DNMT1 or DNMT1 and DNMT3b, strengthening our previous observation of cooperation between p53 and DNMT1 (Fig. 3A Right).
Fig. 3.
DNMT1 and p53 are essential for survivin gene repression. (A) DNMT1 is essential for p53-mediated gene repression in methyltransferase knockout cell lines. Parental HCT116 and knockouts are indicated at top, along with doxo treatment. Note that survivin and cdc25C were down-regulated by p53 induction (doxo treatment), whereas PCNA levels remained unchanged. Densitometric quantitation of survivin and cdc25C expression in MCF-7 cells is shown in Right.(B) Regulation of survivin gene expression in p53 or DNMT1–/– HCT116 cells. Ad constructs are indicated. Densitometric scans of survivin expression are shown at Bottom. (C) Dissection of cooperation between p53 and DNMT1 for survivin gene regulation in colorectal cancer cell lines. Cell lines and knockout status are indicated at top. Transfection of siRNA and doxo treatment is indicated. Survivin expression level was standardized to PCNA level and indicated in Right. Antibodies used are as indicated.
To determine unequivocally the role of p53 and DNMT1 in survivin gene regulation in colorectal cancer cells, we examined the level of DNMT1, p53, survivin, and PCNA expression in cell extracts of HCT116 cells infected with either empty Ad or Ad carrying WT p53. Constitutive expression of WT p53 in HCT116 p53–/– cells but not in HCT116 DNMT1–/– cells led to repression of survivin gene expression (Fig. 3B, lane 2 vs. 4), as observed before with doxo-treated HCT116 or MCF-7 cells (data not shown). Doxo-treated HCT116 p53–/– and p53 mutant T24 cells did not show any changes in survivin gene expression (data not shown), suggesting that p53 is an essential component of survivin gene regulation in the presence of DNMT1.
To confirm the functional association between p53 and DNMT1 in survivin gene regulation, siRNAs (27) for p53 and DNMT1 were used to knock down either induced p53 or endogenous DNMT1 in WT HCT116 cells. In mammalian cells, upon introduction of gene-specific siRNA, specific messages get destroyed, resulting in elimination or reduction of the corresponding protein level (27). This knockdown facilitates functional analysis of a gene product in mammalian cells. We induced p53 in HCT116 cells by doxo treatment and transfected both control and doxo-treated cells with p53-siRNA. Cell extracts were monitored for DNMT1, p53, survivin, and control PCNA expression by using Western blot analysis. Induction of p53 by doxo down-regulated survivin gene expression (Fig. 3C, lane 1 vs. 3), and knockdown of p53 by p53-siRNA relieved this repression (Fig. 3C, lane 2 vs. 4), confirming that p53 is essential for down-regulation of survivin. A similar transfection with DNMT1 siRNA confirmed the participation of DNMT1 in survivin gene regulation in cancer cells. HCT116 cells were either left untreated or treated with doxo to induce p53, followed by DNMT1 siRNA transfection. A Western blot of the cell extract revealed knockdown of endogenous DNMT1 by DNMT1 siRNA (Fig. 3C, anti-DNMT1 blot) without affecting p53 induction (Fig. 3C, anti-p53 blot). As expected, p53 induction in the presence of DNMT1 repressed survivin gene expression in HCT116 cells, as was observed before in MCF-7 cells. Knockdown of DNMT1 in p53-induced cells deregulated survivin gene expression (Fig. 3C, lane 3 vs.4), as observed before in DNMT1 knockout cells (Fig. 3A) or cells depleted for DNMT1 by 5-azacytidine (data not shown). A non-specific control, GFP siRNA, did not show any response (Fig. 3C), confirming the involvement of both DNMT1 and p53 in survivin gene regulation in cancer cells.
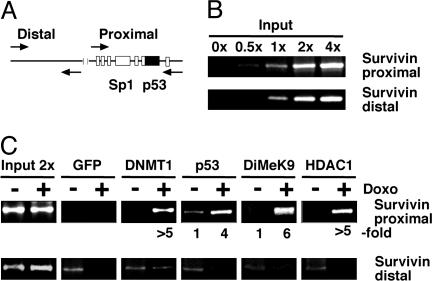
DNMT1 Is Localized on the survivin Promoter upon p53 Induction. We performed chromatin IP assays to demonstrate that p53 recruits DNMT1 onto the survivin promoter. Formaldehyde cross-linked chromatin from control and p53-induced (doxo-treated) cells was immunoprecipitated with antibodies specific for DNMT1, p53, DiMeK9, and HDAC1. In parallel, a nonspecific monoclonal antibody for GFP was included to monitor the specificity of the reaction. Two regions of the survivin promoter were investigated by PCR: a distal region without a p53-binding site and a proximal region close to the transcriptional start site with the p53- and Sp1-binding sites (Fig. 4A). In addition, PCRs were carried out with increasing amounts of input DNA to ensure the linearity of the amplification (Fig. 4B). We were also able to quantitatively immunoprecipitate p53 and DNMT1 by using increasing amounts of specific antibody (data not shown). DNMT1, p53, HDAC1, and lysine 9-methylated histone H3 were found to be associated with the proximal part of the survivin promoter upon doxo-mediated p53 induction, in contrast to the distal part of the survivin promoter that is devoid of a p53-binding site (Fig. 4C). Doxo-mediated binding of p53, DNMT1, and lysine 9-methylated histone H3 on the proximal survivin promoter was severalfold higher compared with the control (background level) or distal region.
Fig. 4.
Presence of p53, DNMT1, HDAC1, and histone H3 dimethyl lysine 9 on the proximal survivin promoter. (A) Relative position of the PCR primers for amplification of the proximal and distal regions of the survivin promoter is shown schematically along with the relative positions of the p53 and Sp1 (open boxes) binding sites. (B) Linearity of PCR amplification by both proximal and distal primer sets, with increasing amount of input DNA as indicated on top. (C) ChIP analysis for the presence of DNMT1, p53, DiMeK9, and HDAC1 on the survivin promoter. Antibodies and doxo-treated samples are indicated on top, and fold induction is indicated on bottom.
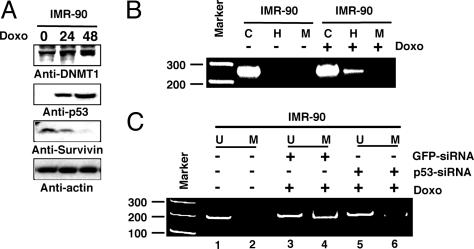
DNMT1 and p53 Cooperate for Gene Regulation in Normal Cells. The mechanisms underlying gene expression in cancer may differ from normal cells because of genetic alterations or mutations. Thus, we examined the interaction of DNMT1 and p53 with each other on survivin gene expression in normal lung fibroblast (IMR-90) cells. Cells were treated with 1 μM doxo to induce p53, and the levels of DNMT1, p53, survivin, and actin were examined 0, 24, and 48 h after treatment. As observed with HCT116 cancer cells, doxo treatment resulted in a time-dependent decrease of survivin expression, with a concordant increase in p53 and similar levels of actin (Fig. 5A). Quantitation of both p53 and survivin showed an inverse correlation between the two (data not shown). Concurrent with p53 stabilization and DNMT1 accumulation, survivin down-regulation occurred.
Fig. 5.
survivin promoter methylation after p53 induction in normal cells. (A) Western blots showing doxo-mediated up- and down-regulation of DNMT1, p53, and survivin using the specific antibodies indicated. (B) “M” and “H” designate MspI and HpaII restriction enzymes. “C” is undigested DNA control. Molecular weight (Marker) is indicated in bp. survivin-specific band shown is ≈240 bp long. (C) Methylation-specific PCR analysis of the survivin promoter after transfection of GFP-siRNA or p53-siRNA with or without doxo treatment. U, unmethylated; M, methylated.
To examine the methylation status of the survivin promoter before and after doxo treatment, genomic DNAs were treated with methylation-sensitive restriction enzyme, followed by PCR (Fig. 5B). The presence of a PCR product in doxo-treated cells demonstrated the presence of CpG methylation in the survivin promoter. To confirm this observation, IMR-90 cells were transfected with GFP- or p53-siRNA, followed by p53 induction with doxo. Genomic DNAs from control and siRNA-transfected cells were subjected to bisulphite modification and methylation-sensitive PCR assay. This assay distinguishes small amounts of methylated sequences from unmethylated sequences by its ability or inability to amplify the DNA by using either unmethylated or methylated sequence-specific primer sets (28). Whereas the control cell DNA did not show any survivin promoter methylation (Fig. 5C, lane 2, absent of survivin band), the GFP-siRNA-transfected doxo-treated DNA did (Fig. 5C, lane 4), demonstrating methylation of the survivin promoter. However, promoter methylation was reduced (Fig. 5C, lane 6) when cells were treated with p53-siRNA, confirming that p53 mediated survivin promoter methylation in the normal cells.
Discussion
Mammalian gene expression is a complex phenomenon involving transcriptional inducers as well as repressors. Some key regulatory proteins, such as p53, act as both transcription factors and repressors. Microarray-based analysis has showed that the number of genes induced or repressed by p53 was nearly equal (16). Cumulative work obtained from different laboratories suggests that p53 is not a stand-alone protein; rather, it participates in a complex network of proteins working in concert (13). A previously unrecognized interaction between DNMT1 and p53 is perhaps the latest addition, but is unlikely to be the last one. We have found that DNMT1, a key factor in transcriptional regulation, is an integral part of the p53 network. As we have demonstrated, the interaction between p53 and DNMT1 is correlated with overall hypermethylation of the survivin promoter, thus contributing to gene repression.
The interaction of human DNMT1 and p53 adds another dimension to p53-mediated gene regulation. Indeed, TROP1 gene methylation is maintained in a WT p53 background but is lost in a p53 mutant (29), presumably affecting DNMT1 interaction. Furthermore, in hepatocellular carcinoma, methylation-mediated inactivation of p14ARF was found to be restricted to WT p53 (30). What we know so far is that DNMT1 preferentially methylates certain types of damaged DNA (31). Our ChIP data suggests that p53, DNMT1, and K9-methylated H3 are in a complex on the survivin promoter after doxo-mediated DNA damage (Fig. 4). We have also observed activation of H3K9 methylation after doxo treatment on human cells (data not shown). Thus, it is possible that both DNA and histone methylation may play a role in survivin repression at DNA damage. Hoffman et al. (15) have also shown p53 binding to the survivin promoter both in vitro and in vivo, supporting our observation. Mirza et al. (32) have contested direct interaction of p53 on the survivin promoter, although they have concluded that the 5′ boundary of survivin promoter repression did include p53-binding sequences and several CpG sites of exon 1. Point mutation on the p53-binding site may have abolished p53 but not the p53–DNMT1 repressor complex binding to CpGs, thus down-regulating the survivin promoter. Based on our results, cooperation between DNMT1 and p53 is essential for survivin gene regulation, either through methylation-dependent or -independent pathways. A plausible model of this repressor complex would require DNA damage-mediated recruitment of HDAC1–DNMT1–p53 complexes to the CpG-rich survivin promoter, a preferred site for DNMT1 binding because of high density of CpGs. Presence of p53 may stabilize the HDAC1–DNMT1–p53 complex on the survivin promoter, either by direct or indirect contacts with DNA by means of methylated histones. HDAC1 may also aid in survivin repression through interaction with p53 by means of the Sin3a repressor complex or by directly binding to DNMT1. HDAC1 binding to DNMT1 through amino acids 686–812 (9) would not affect HDAC1's ability for p53 binding by means of amino acids 1081–1409 (Fig. 4C), thus forming a ternary repressor complex that may mediate survivin repression in both cancer and normal cells. A small increase in DNMT1 in the normal cells in response to p53 accumulation (Fig. 4A), as observed before (33), may have a bearing on hypermethylation that needs further investigation.
Recent reports suggest that cooperation between chromatin protein modification and DNA methylation may be crucial for gene expressions. Bachman et al. (34) have demonstrated that histone modifications and silencing of the p16INK4a gene take place before DNA methylation. Nguyen et al. (35) have shown that K9-methylated H3 is associated with aberrant gene silencing in cancer cells and that this gene silencing is reversed by 5-azacytidine treatments of cancer cells. Thus, histone methylation and DNA methylation, two global epigenetic modifications, appear to be intimately involved in gene regulation. DNMT1 potentially provides a key interaction enzymatically by methylating DNA and recruiting HDAC1 and acting as bridge between p53 and chromatin, reinforcing a repressed chromatin state. This complex may further act to down-regulate other cellular genes after DNA damage, encouraging apoptosis.
Acknowledgments
We thank B. Vogelstein (The Johns Hopkins University, Baltimore) for the DNMT knockout and p53 knockout cell lines, M. Murphy (Fox Chase Cancer Center, Cheltenham, PA) for the gift of the SpII construct, and Y. St-Pierre (University of Quebec, Quebec) for the DsRed vector. We also thank R. J. Roberts, W. Jack, and T. Evans for discussion and D. Comb and New England Biolabs for support and assistance.
Author contributions: P.-O.E. and H.G.C. performed research; P.-O.E. and S.P. analyzed data; S.P. designed research; and S.P. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: DNMT, DNA (cytosine-5)-methyltransferase; PCNA, proliferating cell nuclear antigen; HDAC, histone deacetylase; doxo, doxorubicin; Ad, adenovirus; siRNA, small inhibitory RNA; IP, immunoprecipitation.
References
- 1.Bestor, T., Laudano, A., Mattaliano, R. & Ingram, V. (1988) J. Mol. Biol. 203, 971–983. [DOI] [PubMed] [Google Scholar]
- 2.Yen, R. W., Vertino, P. M., Nelkin, B. D., Yu, J. J., el-Deiry, W., Cumaraswamy, A., Lennon, G. G., Trask, B. J., Celano, P. & Baylin, S. B. (1992) Nucleic Acids Res. 20, 2287–2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okano, M., Xie, S. & Li, E. (1998) Nat. Genet. 19, 219–220. [DOI] [PubMed] [Google Scholar]
- 4.Chuang, L. S., Ian, H. I., Koh, T. W., Ng, H. H., Xu, G. & Li, B. F. (1997) Science 277, 1996–2000. [DOI] [PubMed] [Google Scholar]
- 5.Ramsahoye, B. H., Biniszkiewicz, D., Lyko, F., Clark, V., Bird, A. P. & Jaenisch R. (2000) Proc. Natl. Acad. Sci. USA 97, 5237–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, E., Bestor, T. H. & Jaenisch, R. (1992) Cell 69, 915–926. [DOI] [PubMed] [Google Scholar]
- 7.Okano, M., Bell, D. W., Haber, D. A. & Li, E. (1999) Cell 99, 247–257. [DOI] [PubMed] [Google Scholar]
- 8.Jones, P. A. & Baylin, S. B. (2002) Nat. Rev. Genet. 6, 415–428. [DOI] [PubMed] [Google Scholar]
- 9.Fuks, F., Burgers, W. A., Brehm, A., Hughes-Davies, L. & Kouzarides, T. (2000) Nat. Genet. 24, 88–91. [DOI] [PubMed] [Google Scholar]
- 10.Rountree, M. R., Bachman, K. E. & Baylin, S. B. (2000) Nat. Genet. 25, 269–277. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki, H., Gabrielson, E., Chen, W., Anbazhagan, R., van Engeland, M., Weijenberg, M. P., Herman, J. G. & Baylin, S. B. (2002) Nat. Genet. 31, 141–149. [DOI] [PubMed] [Google Scholar]
- 12.Levine, A. J. (1997) Cell 88, 323–331. [DOI] [PubMed] [Google Scholar]
- 13.Vogelstein, B., Lane, D. & Levine, A. J. (2000) Nature 408, 307–310. [DOI] [PubMed] [Google Scholar]
- 14.Hoh, J., Jin, S., Parrado, T., Edington, J., Levine, A. J. & Ott, J. (2002) Proc. Natl. Acad. Sci. USA 99, 8467–8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman, W. H., Biade S., Zilfou, J. T., Chen, J. & Murphy, M. (2002) J. Biol. Chem. 277, 3247–3257. [DOI] [PubMed] [Google Scholar]
- 16.Kannan, K., Kaminski, N., Rechavi, G., Jakob-Hirsch, J., Amariglio, N. & Givol, D. (2001) Oncogene 20, 3449–3455. [DOI] [PubMed] [Google Scholar]
- 17.Juan, L. J., Shia, W. J., Chen, M. H., Yang, W. M., Seto, E., Lin, Y. S. & Wu, C. W. (2000) J. Biol. Chem. 275, 20436–20443. [DOI] [PubMed] [Google Scholar]
- 18.Zilfou, J. T., Hoffman, W. H., Sank, M., George, D. L. & Murphy, M. (2001) Mol. Cell. Biol. 21, 3974–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy, M., Ahn, J., Walker, K. K., Hoffman, W. H., Evans, R. M., Levine, A. J. & George D. L. (1999) Genes Dev. 13, 2490–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rhee, I., Bachman, K. E., Park, B. H., Jair, K. W., Yen, R. W., Schuebel, K. E., Cui, H., Feinberg, A. P., Lengauer, C., Kinzler, K. W., et al. (2002) Nature 416, 552–556. [DOI] [PubMed] [Google Scholar]
- 21.Pradhan, S. & Kim, G. D. (2002) EMBO J. 21, 779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews, N. C. & Faller, D. V. (1991) Nucleic Acids Res. 19, 2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, G. D., Ni, J., Kelesoglu, N., Roberts, R. J. & Pradhan, S. (2002) EMBO J. 21, 4183–4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pradhan, S., Bacolla, A., Wells, R. D. & Roberts, R. J. (1999) J. Biol. Chem. 274, 33002–33010. [DOI] [PubMed] [Google Scholar]
- 25.Leonhardt, H., Page, A. W., Weier, H. U. & Bestor, T. H. (1992) Cell 71, 865–873. [DOI] [PubMed] [Google Scholar]
- 26.Klibanov, S. A., O'Hagan, H. M., Ljungman, M. (2001) J. Cell Sci. 114, 1867–1873. [DOI] [PubMed] [Google Scholar]
- 27.Denli, A. M. & Hannon, G. J. (2003) Trends Biochem. Sci. 28, 196–201. [DOI] [PubMed] [Google Scholar]
- 28.Herman, J. G., Graff, J. R., Myohanen, S., Nelkin, B. D. & Baylin, S. B. (1996) Proc. Natl. Acad. Sci. USA 93, 9821–9826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nasr, A. F., Nutini, M., Palombo, B., Guerra, E. & Alberti, S. (2003) Oncogene 22, 1668–1677. [DOI] [PubMed] [Google Scholar]
- 30.Tannapfel, A., Busse, C., Weinans, L., Benicke, M., Katalinic, A., Geissler, F., Hauss, J. & Wittekind, C. (2001) Oncogene 20, 7104–7109. [DOI] [PubMed] [Google Scholar]
- 31.Tan, N. W & Li, B. F. (1990) Biochemistry 29, 9234–9240. [DOI] [PubMed] [Google Scholar]
- 32.Mirza, A., McGuirk, M., Hockenberry, T. N., Wu, Q., Ashar, H., Black, S., Wen, S. F., Wang, L., Kirschmeier, P., Bishop, W. R., et al. (2002) Oncogene 21, 2613–2622. [DOI] [PubMed] [Google Scholar]
- 33.Peterson, E. J., Bogler, O. & Taylor, S. M. (2003) Cancer Res. 63, 6579–6582. [PubMed] [Google Scholar]
- 34.Bachman, K. E., Park, B. H., Rhee, I., Rajagopalan, H., Herman, J. G., Baylin, S. B., Kinzler, K. W. & Vogelstein, B. (2003) Cancer Cell 3, 89–95. [DOI] [PubMed] [Google Scholar]
- 35.Nguyen, C. T., Weisenberger, D. J., Velicescu, M., Gonzales, F. A., Lin, J. C., Liang, G. & Jones, P. A. (2002) Cancer Res. 62, 6456–6461. [PubMed] [Google Scholar]