Abstract
Network science and engineering provide a flexible and generalizable tool set to describe and manipulate complex systems characterized by heterogeneous interaction patterns among component parts. While classically applied to social systems, these tools have recently proven to be particularly useful in the study of the brain. In this review, we describe the nascent use of these tools to understand human cognition, and we discuss their utility in informing the meaningful and predictable perturbation of cognition in combination with the emerging capabilities of neurofeedback. To blend these disparate strands of research, we build on emerging conceptualizations of how the brain functions (as a complex network) and how we can develop and target interventions or modulations (as a form of network control). We close with an outline of current frontiers that bridge neurofeedback, connectomics, and network control theory to better understand human cognition.
Keywords: graph theory, network neuroscience, neurofeedback, cognition, control theory
The notion that engineering principles are critical for advancing the frontiers of modern neuroscience is not new. Indeed, the marriage of these two disciplines is now commonly known as neuroengineering.1 The purview of this discipline is particularly large, including the use of theoretical, computational, and experimental tools to reveal fundamental principles of neural structure and function across species.2, 3, 4, 5 It also includes efforts to “engineer” the brain in reverse—creating technological systems that perform brain‐like computation—and forward—altering brain structure and modulating brain function in a targeted and theoretically predictable manner.6 Such engineering approaches were recognized in 2013 as key tools for tackling the challenges of mapping the brain.7 In humans specifically, the discipline seeks to reveal the foundations of cognition.
A natural confluence of many of these lines of inquiry lies in brain–machine interfaces (BMIs), which capitalize on emerging instruments for both hardware and software in sensing, signal processing, and machine learning.4, 8, 9 And within the class of all BMIs, one noninvasive tool that receives attention for its potential to inform our understanding of human cognition is neurofeedback.10 Experimental paradigms including neurofeedback begin with the subject receiving sensory or behavioral feedback that is based on the current state of his/her brain activity—as measured by imaging or electrophysiology—in real time.11 The participant then attempts to modulate that activity signal, either increasing it or decreasing it, in response to prompts from the experimenter. With training, many participants are able to learn to modulate the activity in specific brain regions on command.
Historically, neurofeedback training has been developed to assist individuals in the use of BMIs to overcome disabilities, injuries, or mental illness.10, 12, 13, 14, 15, 16, 17, 18 For example, if one could control the activity of the hand motor cortex using mental imagery, that signal could be used to control a robotic hand.4 Or, in the case of mental illness, if one could control a region of the brain whose function is altered by the illness, one might have meaningful relief from undesired symptoms.19 Yet, while clinical applications have remained paramount in the use of these tools over the last decade or more, recent evidence suggests that neurofeedback can in fact also be used to probe the fundamental principles of cognition that explain how the brain relates to behavior.20, 21
The promise of neurofeedback in revealing principles of cognition builds on the fact that—in essence—it is a perturbative approach. Unlike the lesion approaches that were ubiquitous in early studies of neuroanatomy,22 which led to the field of brain mapping,23, 24, 25 neurofeedback enables the investigator to modulate brain activity in a targeted manner by offering the participant a view into the activity of a small volume of neural tissue. As a tool, neurofeedback offers several advantages over stimulation‐based perturbation techniques, such as transcranial magnetic stimulation—subjects can learn to modulate activity on demand without the need for external stimulation hardware, and these learning effects can be sustained over several days.26 Interestingly, such perturbative approaches are in fact the bread and butter of mathematics and physics, where they are used to examine the general structure of the dynamic landscape surrounding a point (see, for example, Refs. 27 and 28). The major benefit of a perturbative approach is that it facilitates generalization of an observation, and by extension the construction of a mechanistic theory. This ability to probe both the canonical form and the broader landscape of a dynamical system has proven fundamentally important in developing mechanistic theories in theoretical physics.
In cognitive neuroscience, the potential to use a perturbative approach like neurofeedback becomes particularly interesting when viewed in light of the emerging field of connectomics.29, 30, 31 Because the brain is not simply a collection of independent units but is instead a complex network of interconnected elements,32, 33 manipulating the activity in one area can have nontrivial effects on other areas—even far from the modulated source.34 Far from the mysteries of quantum mechanics, this action at a distance is a direct consequence of the complex pattern of structural wiring that links brain areas.35, 36, 37 Given this complexity, it is natural to ask, “What distributed network of brain areas is affected when a participant modulates the activity in a single brain region? Can participants learn to modulate two regions at once, or large groups of brain areas? What do the answers to these questions tell us about the distributed computations that support complex cognitive processes? How would we choose the target region for neurofeedback to elicit a specific change in a large‐scale, distributed functional network?”
Answering these questions requires a paradigm shift in our conceptualization of how the brain functions (as a complex network) and how we develop and target interventions or modulations (network control). In this review, we begin by briefly summarizing the use of neurofeedback to probe cognition, and we discuss the insights that these studies have offered into higher level cognitive processes in humans. Next, we highlight both the challenges and potential inherent in acknowledging that altering the activation of a single region can have nontrivial effects throughout the network. To better understand these widespread effects, we briefly describe the principles of network science and the organizational structure currently known to characterize the human connectome. These data lead us into the question of how to perturb the human brain via neurofeedback to move the brain from an initial state to a predictable target state. We describe the utility of network control theory in offering a mathematical framework in which to ground such questions, and we offer an outline of current frontiers that bridge neurofeedback, connectomics, and network control theory to better understand human cognition.
Neurofeedback for cognition: a primer
Like many other quintessentially complex questions in science, understanding how brain function relates to cognition requires a principled empirical approach. Most current efforts implement “open‐loop” forms of inquiry, in which an input stimulus is used to elicit a measurable response in neurophysiology or behavior. At their core, open‐loop methods enable neuroscientists to map the effects of a behavioral perturbation on the observed neural dynamics and, conversely, to map the effects of a neural perturbation on the observed behavior. In the forward direction (perturbing stimuli), one might measure the difference in neural response between different degrees of fearful stimuli. In the reverse direction (perturbing neurophysiology), one might lesion a neural circuit and measure the change in emotional response to those same stimuli. Together, the two types of open‐loop experiments provide a forward mapping of behavior to neural dynamics and a reverse mapping of neural dynamics back to behavior. While these techniques offer particular utility in addressing open questions in cognitive neuroscience, they do not provide a means for addressing how function and behavior change concurrently, as a function of brain state. Indeed, to address this question, one must turn to “closed‐loop” approaches (Fig. 1).
Figure 1.
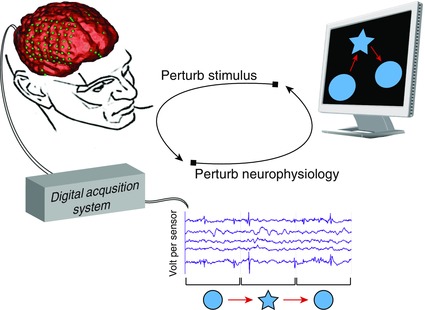
Open‐loop perturbation of brain and behavior. Neuroscience has long exercised an open‐loop approach to probe the characteristics of neural systems. By perturbing behavior and measuring change in neural dynamics or—in reverse—perturbing neural dynamics and measuring change in behavior, this method is used to generate a map between neural dynamics and behavior. For example, consider the following set of experiments investigating the neural basis behind the perception of shapes: to generate a forward mapping between stimulus and neural response, one might measure the change in neural activity of specific brain regions to visual perturbations of object shape; to generate a reverse mapping between neural response and stimulus, one might measure the change in perception of shape due to perturbation of neural activity (perhaps through lesioning or neurostimulation). The forward and reverse mapping are limited in their ability to describe how neural activity and behavior change together on a dynamical continuum. Neurofeedback enables investigators to close the loop around forward and reverse mapping approaches—in real time.
Closing the loop with neurofeedback
Neurofeedback enables closed‐loop scientific inquiry by allowing the subject to directly perturb brain dynamics based on information about his/her current brain state or to indirectly perturb brain dynamics based on feedback about his/her current behavioral state. While both approaches enable the subject to modulate brain activity, they differentially test distinct aspects of cognition by affording flexibility in the design of the feedback signal,11 which we address further below.
To test whether a subject's ability to directly modulate target brain regions improves task performance, investigators can present sensory feedback that scales with the amplitude of brain activity. The use of this technique has revealed that subjects can learn to regulate the activity of subcortical areas, including the amygdala38, 39, 40 and basal ganglia,41 and extended areas of the limbic system, including the insula20, 42 and parahippocampus.43 Interestingly, participants can also learn to modulate the primary sensory motor cortex (primary motor area,44 premotor cortex,45 and supplementary motor area43, 46) as well as higher‐order areas, including the anterior cingulate cortex,20, 42 ventral tegmental area,47 frontal and parietal cognitive control areas,48 anterior midcingulate cortex,43, 46 and frontal cortex.43, 46 Importantly, subjects who learned to successfully modulate regional activity demonstrated improved performance on a variety of cognitive tasks, including tasks eliciting cognitive processes critical for memory,39, 48 mood,38, 49 motor imagery,26, 43, 44, 45, 46, 50 and perception of pain.42 Thus, neurofeedback affords flexibility in targeting distinct functional brain regions associated with different facets of cognition that include sensation, movement, emotion, attention, and learning.
While direct modulation of a neurophysiological signal is a natural place to start, one might also be interested in indirectly perturbing behavioral states and observing concurrent changes in brain activity. Consider examining how the brain dynamically recruits different areas to perform easy versus difficult behavioral tasks—investigators can alter task complexity on the fly based on the amount of activity in the target brain region. To perform this real‐time decoding and mapping of brain activation to behavior, neuroscientists have begun employing machine learning tools that link general linear models with multivariate pattern analysis.11, 51, 52 Briefly, a typical neurofeedback study utilizing these tools would implement the following general process: train a statistical model that discriminates patterns of brain activation in response to different stimuli, present the subject with a stimulus, ask the subject to perform a mental operation based on the stimulus, decode the altered pattern of brain activity using the statistical model, adjust the stimulus on the basis of the new brain state, and have the subject repeat the mental operation. The ability to perturb behavioral state based on underlying brain dynamics has immense utility in revealing functional mechanisms underlying changes in behavior.53 Indeed, recent applications of this technique have yielded critical insights into the mechanisms of attention54, 55 by altering task complexity during decoded states of attentional lapses.56
Learning strategies in neurofeedback training
Exactly how participants learn to regulate regional activity in response to neurofeedback remains incompletely understood. To date, efforts have focused on two complementary strategies. In the first, the participant is provided with cognitive tasks that facilitate the activation of the target region, while in the second, the participant is simply provided with the feedback of regional activation and encouraged to identify his/her own strategy to modulate it. The two techniques have distinct advantages in the study of human cognition.
The earliest work in this field implemented the first approach by providing subjects with instructions to perform mental imagery by imagining the perceptual experience associated with the functional role of the target brain region.21, 42, 50 For example, to activate motor processing areas, one might picture oneself moving a limb, while to activate attention processing areas, one might concentrate on the given task. Evidence suggests that, when given such instructions, subjects display improved modulation of the target brain area within a day of training.26 Put simply, learning to self‐regulate brain activity is akin to learning any other skill.41 When given physiological feedback in addition to a recommended modulation strategy, evidence suggests that subjects display increased performance on related cognitive tasks in comparison to the scenario in which subjects are only given the modulation strategy.42
Nonstrategic training of neurofeedback
Despite the apparent utility of a cognitive modulation strategy, it is also useful to understand to what degree subjects can volitionally modulate brain dynamics. Can subjects simply learn to modulate brain activity when given no pointers as to how to enhance the activity of the target region? Such a capability would be vital in modulating brain activity or connectivity in regions for which we cannot articulate a strategy. These possibilities open up a new realm of scientific investigation that could be particularly helpful in the study of neurological and psychiatric disorders with potentially discordant mappings of functional brain areas57 or in tuning the temporal architecture of brain activity in healthy individuals.58 Studies demonstrate that, even when explicit instructions or strategies are not provided, subjects can search for an effective strategy to self‐regulate functional brain dynamics.26, 38, 47, 57 Such open‐ended experimentation is especially powerful for investigating how subjects employ unique strategies to modulate brain dynamics.38, 40, 57 Indeed, it is of interest to explicitly map individual variability in the chosen control strategy to the subjects' ability to modulate target brain areas. Understanding the relationship between cognitive strategy and effective modulation could inform experimental approaches to fine‐tune the mapping between cognitive processes and functional brain regions (Fig. 2).
Figure 2.
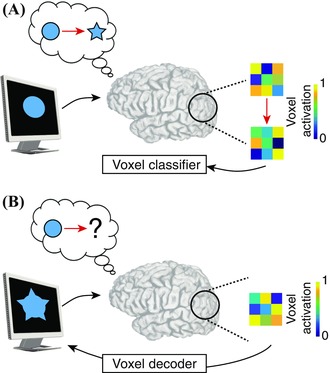
Probing cognitive state with neurofeedback. Indirect perturbation of behavioral state using neurofeedback is a powerful technique to probe the cognitive state space in individuals. Suppose that one wanted to learn the neural basis underlying spatial reasoning ability using a mental manipulation task. (A) The experimenter could present the subject with a circle and a star shape, and ask the subject to observe the stimuli and mentally imagine the circle morphing into a star. Meanwhile, a statistical model could be trained to capture the voxel activation pattern pertaining to the imagined shapes. (B) To probe and quantify the subject's thought process associated with mental manipulation, the experimenter might display the circle to the subject and ask the subject to use his or her mind to manipulate the object into a star. Using the shape‐based model of voxel activation, the experiment could decode the mentally visualized shape from the voxel activation pattern—in real time—and feed the current state of the imagined shape back to the subject. By tracking how subjects explore the cognitive state space while learning how to perform mental operations during a task, investigators could map how individuals use different cognitive strategies to accomplish the task and could further map the distinct neural drivers of these strategies.
Probing cognitive abilities and disabilities
Broadly, direct and indirect neurofeedback are profoundly robust in their ability to modulate brain dynamics. Furthermore, they provide a systematic approach to query functional mechanisms of cognition that are often elusive in open‐loop methods of investigation. By engineering the feedback signal, neurofeedback investigators can flexibly design novel experimental paradigms to study the role of cognition in single brain areas or across multiple brain areas in tandem. This practice has already demonstrated the ability to direct neurofeedback to more focal targets39, 40, 44, 59, 60, 61 or to perturb dynamics across more distributed targets.43, 46, 62 In addition, the difficulty of the neurofeedback training can be titrated to affect a specific change on both attentional circuits and regions devoted to internally directed cognition.63 Particularly exciting applications to cognition include the revelation of associative learning in early visual areas,64 visual perceptual learning induced by decoded functional magnetic resonance imaging (fMRI) neurofeedback,65 and the sensitivity of these learning processes to the subject's state.66, 67
In addition to functioning as a probe for healthy human cognition, neurofeedback can also be used to identify and potentially treat cognitive disabilities. Particularly in populations with impaired mental health, neurofeedback enables the clinical study of mechanisms of dysfunction in neurologic and psychiatric disorders, and it also has the potential to provide noninvasive therapies to minimize the symptoms of such disorders.53 Indeed, soon after its introduction in the late 1960s,12, 13 neurofeedback was popularized in clinical contexts. Studies demonstrated its therapeutic potential in managing epilepsy,14, 15 treating attention‐deficit/hyperactivity disorder,16, 18 and enhancing rehabilitation following stroke.17 Initially, these pioneering studies were limited to electroencephalography (EEG), but with more recent advances in other noninvasive imaging techniques, the tools have been translated to real‐time fMRI.68 Over the last few years, neurofeedback with real‐time fMRI has been used to study schizophrenia,57, 60 depression,39 obesity,61 and addiction.62, 69 While direct neurofeedback is used to improve patients' ability to self‐regulate target brain areas and reduce clinical symptoms,70 indirect neurofeedback is used to study functional dynamics underlying the different cognitive strategies used by healthy subjects and patients to modulate activity in dysfunctional brain regions.57, 71 By using indirect neurofeedback to map cognitive differences between healthy subjects and patients, clinicians may be able to use behaviorally driven approaches to better diagnose individuals with disabilities and mental illness.
Advancing neurofeedback technology
Neurofeedback has yielded critical insights into cognitive function and dysfunction and is now ripe for innovation as neuroscience evolves toward understanding cognition in the broader context of brain networks.33 Indeed, the emerging view of the brain as a network in the mathematical sense has been supported by a growing empirical ability to measure brain activity over a variety of spatial and temporal scales.30, 32 Novel imaging technologies have continued to elucidate a hierarchical organization of the brain,72, 73 ranging from the scale of individual neurons and neuronal populations to that of specialized large‐scale functional areas.74 Bridging the computational units at each scale are complex patterns of structural links facilitating the transmission and processing of information that support cognition and behavior.29 This hierarchical, multiscale network architecture of the brain brings with it unique opportunities for the use of neurofeedback in understanding cognition.
Although prior work demonstrated that neurofeedback is effective in modulating dynamics of (1) individual neurons at submillimeter resolution and fast time scales,75, 76 (2) millimeter‐scale neuronal populations through oscillatory rhythms,58 and (3) macroscale functional domains at slow time scales,59, 68 modulating the way in which brain regions functionally interact is now becoming a tractable frontier. Using statistical models to measure functional connectivity and to construct an effective feedback signal for modulation, several studies demonstrated that subjects can directly perturb functional interactions between brain regions with fMRI‐77, 78, 79, 80, 81 and EEG‐based neurofeedback.82, 83, 84 Moreover, the feedback signal can be adapted to perturb the dynamics of individual connections80, 81 or large groups of connections77, 78, 79 simultaneously.
While neurofeedback can perturb dynamics at different stages and spatial scales of functional processing, equally important is understanding how perturbations can have far‐reaching impact on functional dynamics in non‐targeted brain areas. Several studies identify changes in functional connectivity between the neurofeedback target and other brain regions.26, 47, 60, 61, 85, 86, 87, 88, 89, 90 Other studies demonstrate changes in functional connectivity between pairs of regions completely outside of the area targeted by neurofeedback.26, 85, 87, 88, 89, 91 Moreover, the observed changes in functional connectivity can be complex: upregulating or downregulating target brain regions may increase functional interactions between some brain regions and may decrease functional interactions between other brain regions.87
Taken together, these findings underscore the potential complexity of the effect of neurofeedback on the brain. Yet, to date, little attention has been given to establishing a framework for predicting the impact of neurofeedback on broader network function. Building such a framework is critical for the improvement of feedback paradigms built to identify neurophysiological drivers of cognition and to treat neurological and psychiatric disorders while minimizing side effects. In the following section, we discuss the potential utility of network neuroscience in providing just such a framework.
Network neuroscience
At the confluence of neuroscience, engineering, and physics lies network neuroscience, a burgeoning field that offers a framework for describing brain circuitry at macro‐ and microscales.92 Drawing upon fundamental tools from graph theory,93, 94 network scientists identify important features among elements of a system and measure similarity between these elements—based on these features—to synthesize models that describe an “ecosystem” of interrelated parts.95, 96 These models can be probed and perturbed to understand how an individual element or a group of elements influences the system as a whole.97, 98 This formalism can be used to study how elements of the brain (nodes) structurally or functionally link (edges) to one another and support behavior and cognition in health and disease.99, 100 To construct brain graphs,30 one can measure structural links between brain regions, such as those composed of macroscale white‐matter fibers or microscale synaptic connections. Alternatively, one can measure functional links between brain regions, such as those estimated by similarity in brain dynamics. The pattern of edges between nodes can then be studied mathematically as a graph.30, 32, 33
Mathematical underpinnings of network science
Formally, a graph consists of a set of N nodes and a set of edges .93, 94 To tabulate the strength of edges between network nodes, one can construct an adjacency matrix in which the entry at row i and column j refers to the weight of the edge between node i and node j. Multilayer networks extend the notion of a static graph to a higher‐dimensional graph101 —where nodes or edges change with condition or time, as in the case of dynamic functional brain networks101, 102, 103, 104, 105, 106 —and are represented by an adjacency tensor with N nodes and T layers.107 Using the adjacency matrix formulation, one can compute local, mesoscale, and global statistics to quantify the topological and topographical properties of brain graphs (Fig. 3). On the whole, these graph statistics can provide information about how neural information is represented, processed, and communicated between brain regions.
Figure 3.
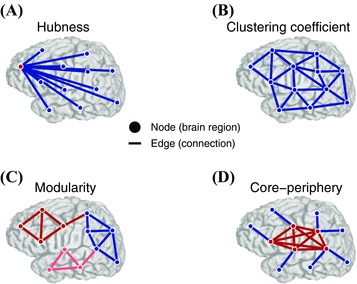
Multiscale topology in brain networks. Brain networks have unique organizing principles at local, meso‐, and global scales that provide information about how neural information is represented, processed, and communicated between brain regions. Brain networks are modeled as a collection of nodes—representing regions of interest with presumably coherent functional responsibilities —and edges—structural connections or functional interactions between brain regions. (A) Node centrality describes the importance of individual nodes in terms of their connectivity relative to other nodes in the network. Nodes with more connections or stronger edges tend to be hubs (red), while nodes with fewer connections tend to be isolated (blue). (B) Clustering coefficient, a measure of connectivity between the neighbors of a node, is another local measure of network topology. Unlike network topologies with strong hubness qualities, as in (A), networks with strong clustering coefficient demonstrate a high density of triangles that is believed to facilitate local information processing. (C) Modularity is a mesoscale topological property that captures communities of nodes that are tightly connected to one another and weakly connected to nodes in other communities. Modular organization underlies a rich functional specialization within individual communities. Here, nodes of different communities are colored red, blue, or pink. (D) Networks with core–periphery structure exhibit a set of tightly connected nodes (core; red) sparsely connected to a set of isolated nodes (periphery; blue). This organization is in stark contrast to the modular organization in (C). The core–periphery architecture is a characteristic of networks that integrate information from isolated regions in a central area. Adapted with permission from Ref. 127.
Local graph measures tell us about the nature of connections from a given node or between neighbors of that node. One simple, yet important, local statistic of a node is centrality—how influential a node is in the context of the broader network.95 Centrality can take many forms, such as degree, the number of edges connected to the node (Fig. 3A), or betweenness, the number of shortest paths between any two nodes that must cross the node in question. Degree centrality has been particularly useful in identifying hubs of neural processing that interact with many different brain regions.97, 108, 109 A second local statistic that is often used to describe brain graphs is the clustering coefficient —the fraction of a node's neighbors that are also connected to one another110 (Fig. 3B). The clustering coefficient describes topological organization thought to underlie local processing of neural information.111, 112
At the mesoscale, one can quantify the tendency of brain regions to form communities —tightly connected groups of nodes that have more connections to one another than to other groups of nodes113, 114 (Fig. 3C). This modular architecture is thought to support the brain's segregation into functionally specialized units.33, 115, 116 Brain graphs can also exhibit core–periphery structure, in which a densely interconnected group of core nodes is connected to a sparsely interconnected group of peripheral nodes117, 118 (Fig. 3D). The core–periphery network structure has been studied for its role in explaining domain‐general versus domain‐specific processing of brain areas,119 in which specialized neural processing in peripheral regions is integrated by a strongly connected core to support higher‐order cognition in learning104, 120 and language.106
Finally, global statistics provide a summary of network topology. For brain networks, the characteristic path length —average shortest path between all node pairs95—is thought to measure how easily neural information can be transferred between brain regions. Brains with shorter path lengths are thought to transfer information more efficiently than brain networks with longer path lengths,121 thereby leading to greater intelligence.122 A similar, dynamical measure of information transfer that has been of recent interest is synchronizability —the ease with which dynamics at each node can synchronize based on the arrangement of edges.123 Synchronizability has also been studied in the context of graph robustness and vulnerability to random and targeted attacks to nodes.124 In brain networks, synchronizability is thought to be maintained between a critical boundary of order and disorder.125
It is important to remember that each statistic is sensitive to network phenomena of a specific scale, and a comprehensive, multiscale quantification of brain network topology requires integration of the output of these measures.120 For instance, local information processing at hub nodes might be projected broadly to mesoscale modules for function‐specific processing that might require globally short path lengths to ultimately integrate and bind the information of each module.100 This framework can be used to query how different elements across multiple scales of network architecture contribute to the processes underlying human cognition.126
Cognitive network neuroscience
Graph theoretic approaches for understanding cognition have become increasingly popular over the past decade, largely due to their utility in describing the interregional relationships between neural processing units elicited by cognitively demanding tasks.105 Application of graph theory to noninvasive brain imaging in humans has yielded critical insight into mechanisms of intelligence, linguistic processing, attention, decision making, learning, memory, and cognitive control.100, 126 Pragmatically, it is useful to separate these insights into those produced by structural brain networks35, 128 and those produced by functional brain networks.97, 125
First, structural brain networks constructed from diffusion‐weighted imaging of white‐matter fiber pathways35, 36 describe the fundamental scaffolding upon which functional brain networks operate.115 Because of their potential role in constraining functional brain dynamics,129, 130 structural brain networks are thought to play important roles in shaping our basic cognitive abilities—such as processing speed, working memory, motor skills, and task switching.126 For example, longitudinal structural imaging has revealed localized changes in network microarchitecture associated with learning new skills,131, 132 and moreover that the strength of structural connections between task‐relevant brain regions predicts individual differences in the rate at which those skills are learned.133 The relevance of structural connectivity for cognition extends to clinical cohorts. For example, patients with anatomical disruptions caused by traumatic brain injury exhibit structural network changes—such as a lengthening of the shortest path length—that are associated with decreased performance on tasks that required switching and inhibition of cognitive resources.134
While structural brain networks reveal important correlates of cognitive ability, a more nuanced dissection of cognitive processing also requires the use of functional imaging methods. Functional brain networks provide a glimpse into network processes (as opposed to structures) that support cognitive function. One recent study highlighted the importance of functional network topology for cognition by tracking longitudinal changes in the behavior of patients with traumatic brain injury; patients with lesions in brain regions that connected to several functional modules exhibited more widespread cognitive deficits compared with patients with lesions in network hubs.135 Patients with traumatic injury also exhibit distributed increases in connectivity during the Stroop task—which requires switching of cognitive resources—that is associated with reduced performance on the task.136 In healthy individuals, differences in global connectivity from specific cognitive control areas predict fluid intelligence.137 Beyond the performance of a single task, evidence suggests that functional brain networks also reconfigure as individuals traverse different cognitive states125 and that the flexibility of this reconfiguration can be used to predict learning in future training sessions.36
Together, these studies underscore the role of network topology and network dynamics as fundamental mechanisms of cognition. Such insights lay the groundwork for exploring how neurofeedback can be used to perturb network properties and thereby more effectively probe drivers of cognition.
Linking neurofeedback to network neuroscience
In the preceeding sections, we explored state‐of‐the‐art capabilities in neurofeedback to perturb brain dynamics, and we introduced a robust framework in network neuroscience for studying the interactions between brain dynamics and the structural networks from which these dynamics originate. Here, we begin to address how the thoughtful integration of neurofeedback and network neuroscience could enhance the study of cognition.
In principle, network neuroscience can be used to identify target brain regions for neurofeedback. Evidence suggests that neurofeedback is differentially effective in distinct brain areas.138 While initial interpretations suggested that this differential effectiveness is due to the inherent differences in the types of cognitive processes that brain regions perform, a second possible explanation is that brain areas are differentially sensitive to neurofeedback based on their connectivity profile. It is intuitively plausible, for example, that regions of the brain that are densely functionally connected with one another (such as the default mode) will be easier to control as a collective—owing to the breadth of potential control mechanisms139 and cognitive strategies—than regions of the brain that are sparsely functionally connected (such as the frontal pole). It will be interesting in the future to determine whether the connectivity profile of a region, or the complexity of the information it processes, is a better predictor of its response to neurofeedback.
Beyond informing our understanding of the impact of neurofeedback on specific brain areas, network neuroscience can also be used to predict the impact of targeted neurofeedback on other brain regions. For example, structural connectivity or resting‐state functional connectivity could pinpoint network hubs, such as cognitive control areas,140, 141, 142 that interact with many other brain areas and serve as a potential target for modulating distributed brain dynamics with neurofeedback. Such a capability could be used to study self‐regulating brain dynamics143 in cognitive control regions and their effective control of other brain areas as humans switch between distinct tasks.142 A similar approach might be employed to investigate whether modulating brain regions that integrate functionally distinct network modules142 could help subjects better learn tasks that require cooperative or competitive interactions between separate cognitive domains140—such as visuomotor interactions in novel skill acquisition.144
More generally, our ability to connect network topology to cognition offers a critical opportunity to model and predict the effects of neurofeedback on cognition. Simultaneously, advancements in neurofeedback can be used to investigate network drivers of cognition: by designing the appropriate feedback signal, neurofeedback can be engineered to modulate not only brain regions but also specific brain networks. This capability opens new doors to test the ability of subjects to modulate specific network properties: Could a subject be trained to modulate the flexibility of a functional module, the participation of a brain region in multiple modules, or even the characteristic path length of the global network? Our ability to modify the feedback signal to accommodate network statistics could have substantial impact on our understanding of how perturbation of network structure affects cognitive ability. Such a capability would also inform the design of novel intervention strategies for patients with neurological disorders or psychiatric disease characterized by disrupted patterns of functional connectivity.
Caveats and future directions
While the prospect of linking neurofeedback and network neuroscience is promising, it is also important to adopt a measured approach in which we acknowledge the pitfalls and limitations inherent in the techniques. First, a note on interpretability: network statistics quantify graph properties from the theoretical perspective of statistical mechanics and information processing, and thus they require a conceptual leap if connected to neurophysiologic phenomena.145, 146 For instance, the relationship between connectivity derived from functional imaging and the amount that two brain regions are in fact communicating 147 remains undetermined. Or consider the shortest path between areas in a brain graph: Does the brain utilize shortest paths?130 Or does it preferentially utilize longer paths or walks?37, 133 Although novel technologies are needed to help bridge these areas of scientific understanding, we remain optimistic that network neuroscience will provide a dynamical systems‐level characterization and understanding of behavioral and cognitive processes.
A second crucial caveat relates to the importance of distinguishing between correlation and causation. A substantial body of work now demonstrates that network measures can be good predictors of cognitive ability based on their degree of correlation to performance metrics. However, these predictions do not equate to causation. In fact, explicitly identifying causation will require a coupling between neurofeedback and network‐based approaches to the study of cognition: by modulating network topology with neurofeedback and evaluating the concurrent change in task performance, we can begin to understand causal effects of network architecture on cognition.
A third and very fundamental consideration relates to the question of whether neurofeedback can be truly targeted to a single region or whether it only ever activates a distributed network. In truth, knowing the degree to which a specific neurofeedback paradigm targets a single region versus many regions remains challenging, and this uncertainty is an important consideration for studies seeking to combine network approaches and neurofeedback training. For example, prior work demonstrates the ability of subjects to regulate brain activity in spatially confined brain regions.59 Specifically, they show that, with training, activation was significantly increased within a predefined target area (somatomotor cortex) relative to the rest of the brain, an observation that suggests that spatial specificity may be achieved with neurofeedback. More recent work confirmed the hypothesis that functional connectivity from the target brain region (ventral tegmental area) to adjoining brain regions is directly caused by volitional activation of the target and not mediated by practice with or without false neurofeedback.47 Thus, they reason that changes in network connectivity are due to the successful perturbation of the target and not associated with the task. This observation, however, still leaves open the question of whether increased functional connectivity occurred in successful perturbations during training or as a result of posttest perturbation. Other work has developed techniques to study the differential effects of perturbation during training and perturbation after training (transfer) on functional connectivity.89 Comparing functional connections stemming from the neurofeedback target, they found more distributed changes during training and specific changes during transfer. These studies present evidence that (1) neurofeedback can activate spatially confined brain regions, (2) target activation and the resulting increase in functional connectivity can be directly related to successful neurofeedback, and (3) methodological innovations are capable of teasing apart changes in functional connectivity due to training perturbations versus transfer perturbations. Nevertheless, it will be important in the future to understand the degree of targeting possible in any given neurofeedback training paradigm, as well as in any given subject, when considering informing such experiments with predictions from network science.
Perhaps most importantly, the possibility of revealing causal relationships between network architecture and cognition with neurofeedback also supports the potential to build computational models or theories of brain network function from first principles.148, 149 Setting aside characterization and description, and even setting aside correlative approaches to link brain network function to observable behavioral variables, models and theories provide mechanistic understanding150 and the ability to generalize inferences to unseen scenarios. What might such a model or theory look like? In the following section, we discuss the potential utility of network control theory in providing just such a model.
Network control theory: a tool to predict impact of modulation
Network control theory is a mathematical modeling framework that addresses the question of how energetic input to a node in a network affects the dynamics of the networked system.151, 152 Stemming from early work in the 1970s on structural controllability,153 the theory is built on two pillars: a model of the system's dynamics and an estimate of the system's network structure.154 In contrast, graph theory stands on only one of these pillars (the network structure) and is devoid of the other (a model of system dynamics). These differences in the nature of the two theories directly affect how they can be used:142, 155 graph theory provides descriptive statistics of a network's organization, while network control theory provides a prediction of how the change in energy at a node will alter the system's dynamics. While graph theory offers tools for characterization, network control theory posits a mechanism for system function.
Traditionally, network control theory stems from the older field of simply control theory, which has been used to inform the control of robotic, technological, and mechanical systems. The simple difference between control theory and network control theory is that network control theory deals with the application of control theory to systems characterized by complex interconnection patterns.151, 152 These patterns significantly affect the control strategies that a system can perform or respond to and further affect the energy required for certain control goals. In the realm of neuroscience, network control theory therefore differs from neural control engineering more generally156 in its explicit treatment of brain network architecture.
A few relevant concepts and tools
In a brain, questions of control can be separated into two types: (1) how does the brain control its own dynamics and (2) how can brain dynamics be controlled via external intervention. When applied to the former question, network control theory can offer insights into cognitive control,157 decision making, and other forms of executive function.142, 158 When applied to the latter, network control theory can inform the use of neural modulation via neurofeedback or brain stimulation,34, 159 the development of cognitive tasks for explicit tuning of brain dynamics, and the understanding of how sensory stimuli impact those dynamics.
To address both types of questions regarding internal and external control, one needs to begin by writing down the two pillars of the theory: a model of the system's dynamics and an estimate of the system's network structure. A simple first step is to use a discrete‐time, time‐invariant, noise‐free model of system dynamics129, 160 and a structural network estimated from diffusion imaging tractography in humans,35 representing the white‐matter pathways crisscrossing cortical and subcortical areas. Of course, these choices are accompanied by model assumptions and caveats associated with the empirical data, both of which need to be acknowledged. (See next section for an explicit treatment of these topics.) Nevertheless, they remain a useful starting point for the application of the theory to human neuroscience.
Within this framework, network control theory offers a few important concepts and tools. The first important concept is that of controllability, which indicates whether a system can be moved from a specified initial state to a specified final state with finite energy and in finite time.151 In initial applications of these algorithms to noninvasive structural neuroimaging data in humans, evidence suggests that the human brain is practically impossible to control from energy injected into a single brain region.142 This result motivates a more nuanced assessment of whether there are particular control strategies that are possible for the brain and whether the system is controllable with a larger number of input sites.
In the engineering literature, common control strategies include (1) average controllability, which describes the ease with which the system can be moved to nearby states on the energy landscape; (2) modal controllability, which describes the ease with which the system can be moved to distant states on the energy landscape; and (3) boundary controllability, which describes the ease with which modules in the network can be coupled or decoupled.152 In applications to human diffusion imaging data, these concepts have proven useful in offering structural explanations for the areas that affect cognitive control142, 158 and the impact of stimulation to the target brain region.34 An even finer account of the brain's dynamics can be obtained by studying the exact transitions from one brain state to another, with assumptions on the need to minimize energy of transitions and minimize the distance that the brain traverses through state space to affect the transition155 (Fig. 4). Applications of these algorithms to human diffusion imaging data have offered an explanation for the anatomical structure of the brain's default mode network155 and a prediction of which regions in the network are most likely to assist in a given state transition.155, 161
Figure 4.
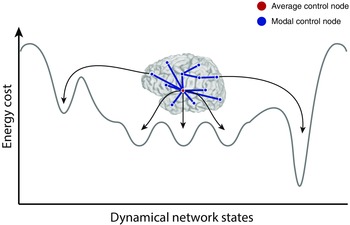
Brain network controllers drive transitions between dynamical states. To accomplish behavioral and cognitive goals, brain networks internally navigate a complex space of dynamical states. Stable brain states may lie in basins of local minimum energy—requiring the brain to expend metabolic energy to move over high‐energy peaks when transitioning from the current state to the next state. Within the space of possible dynamical states, there are easily accessible states and harder‐to‐reach states; in some cases, the accessible states are healthy, while in other cases they may contribute to dysfunction, and similarly for the harder‐to‐reach states. Two commonly observed control strategies in complex systems are average control and modal control. In average control, highly central nodes navigate the brain toward easy‐to‐reach states. In contrast, modal control nodes tend to be isolated brain regions that navigate the brain toward hard‐to‐reach states that may require additional energy expenditure.142 Adapted with permission from Ref. 127.
Limitations, caveats, and extensions of network control theory
Because network control theory is fundamentally a modeling endeavor, it is important to consider model assumptions, model validation, and model extension. First, it is important to acknowledge that the model can only be as accurate as the data used to construct it, and thus one must deal with the inherent limitations associated with constructing the anatomical network from diffusion imaging data.162, 163, 164 Second, when considering a linear model of system dynamics,153 the assumption is that brain network dynamics are linear. In the brain, these linear models have preliminary support in empirically measurable dynamics,129, 160 but further work is needed to delineate the breadth of their applicability. Moreover, it is important to note that, if using this linear approach to model nonlinear dynamics (e.g., those observed in EEG and MEG data), the assumption of linearity nevertheless remains true over short time horizons and in the vicinity of the operating point.165
If one wishes to expand beyond short time horizons and states surrounding the operating point, then one must consider nonlinear models of brain dynamics.166 Which nonlinear models might be relevant while remaining theoretically tractable? This is an open question, but one particularly useful approach might be to use system identification to extract the appropriate nonlinear model from real data.167 Better understanding of control strategies for appropriate nonlinear models of brain dynamics could be particularly important in understanding large‐scale circuit function supporting cognition. For example, synchronization dynamics are thought to play a critical role in facilitating the transfer of information between brain regions at the mesoscale,168 but principles of the brain's endogenous regulation of synchronization remain far from understood. One hypothesis from theoretical physics is that these networks employ a push–pull control strategy in which antagonistic desynchronizing and synchronizing nodes regulate the transfer of information through the network.169 By analyzing the functional network topology of focal and distributed (clinically known as secondarily generalized) seizures, recent work demonstrates that desynchronizing and synchronizing brain regions antagonistically regulate the ability for seizures to synchronize network dynamics.143 Thus, control strategies like push–pull control may be relevant for homeostatic regulation of (nonlinear) synchronization dynamics in the human brain.
Beyond nonlinear extensions, other interesting questions to consider include the possibility that some regions of the state space are inaccessible or pathological,154 that the brain may be underactuated in certain states,170 and that control can be implemented by distributed as well as focal strategies.159 Moreover, although structural controllability theory153 is inherently built on knowledge about the structural connections in a network, it will be interesting in the future to extend these models to statistically estimate the set of either structural or extrasynaptic171 connections that are being utilized at a particular moment in time and how that set might change as humans perform a cognitively effortful task.
Putting it all together: neurofeedback, network neuroscience, and control
What does network control theory add to the conversation between neurofeedback and network neuroscience? Far from being a third wheel, network control theory in fact offers the theoretical framework in which to predict how the activation of a region (or connection or subgraph) by neurofeedback will affect brain network dynamics, moving the brain into a new mental state and thereby altering intrinsic cognitive processes. Thus, in essence, network control theory offers a theoretical backbone on which to begin formulating ideas about the mechanisms of cognition and begin developing theoretically grounded interventions to target their modulation.
To be a bit more concrete, one could use network control theory to simulate the impact of a change in energy at one (or several) regions (or connections) on the subsequent brain network dynamics. Using this approach, one could identify the constellation of regions and connections that—when brought to a specific pattern of activation—would have a predictable effect on brain state (Fig. 5). Then, one could use neurofeedback to produce that pattern of activation, validate the predicted subsequent effect on brain state dynamics, and observe the associated change in cognitive function. In essence, this approach enables a closed loop between theory and experiment, providing a framework in which to develop more general theories of brain function, explain existing empirical data, suggest new directions for empirical research, and inform experimental hypotheses.
Figure 5.
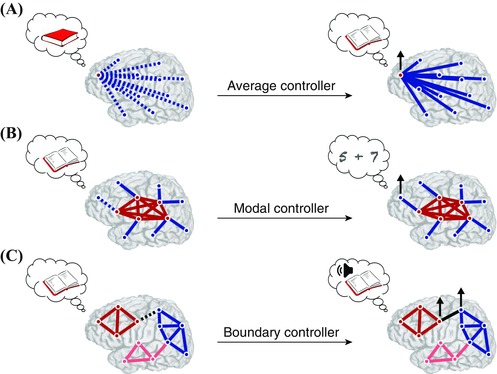
Self‐regulating brain network controllers for cognition. Neurofeedback could be used to teach individuals how to modulate brain activity in important control points that drive changes in dynamical brain state—an experimental tool that would offer tremendous opportunities for studying “cognition dynamics,” or the ability to perform specific tasks based on the current brain state. Furthermore, this approach might be used to train individuals with specific cognitive deficits to better manage their ability to perform certain types of tasks. (A) Suppose that individuals could be trained to upregulate the brain's average controller (red node) to assist in navigating different brain states associated with a specific task, such as opening up and reading a book. (B) If the subject has difficulty with switching between tasks—such as reading and doing math—he/she might be trained to upregulate his/her brain's modal controller (blue node) to switch more efficiently. (C) If the subject has difficulty with comprehension or reading aloud, he/she might be trained to upregulate his/her brain boundary controllers between functional modules associated with language and speech.
Indeed, the confluence of these three disparate disciplines—network control theory, neurofeedback, and network neuroscience—offers much promise in advancing our understanding of human cognition. Both neurofeedback and network neuroscience are well‐developed fields at this point, and they are ripe for integration. Network control theory is the newest field of the three, and its integration will require careful development, characterization, and validation to reach its full promise. Nevertheless, the potential payoff in terms of obtaining a basic, mechanistic understanding of human cognition, seems at this point to be well worth the effort.
Acknowledgments
We thank Andrew C. Murphy for helpful feedback on an earlier version of this manuscript. D.S.B. and A.N.K. would like to acknowledge support from the John D. and Catherine T. MacArthur Foundation, the Alfred P. Sloan Foundation, the Army Research Laboratory and the Army Research Office through Contract Numbers W911NF‐10‐2‐0022 and W911NF‐14‐1‐0679, the National Institute of Health (2‐R01‐DC‐009209‐11, 1R01HD086888‐01, R01‐MH107235, R01‐MH107703, R01MH109520, 1R01NS099348, and R21‐M MH‐106799), the Office of Naval Research, and the National Science Foundation (BCS‐1441502, CAREER PHY‐1554488, BCS‐1631550, and CNS‐1626008). The content is solely the responsibility of the authors and does not necessarily represent the official views of any of the funding agencies.
Competing interests
The authors declare no competing interests.
The copyright line for this article was changed on May 9, 2017 after original online publication.
References
- 1. DiLorenzo D.J. & Bronzino J.D., Eds. 2007. Neuroengineering. CRC Press. [Google Scholar]
- 2. Buettner, H.M. 1995. Neuroengineering in biological and biosynthetic systems. Curr. Opin. Biotechnol. 6: 225–229. [DOI] [PubMed] [Google Scholar]
- 3. Jeong, J.W. et al 2015. Soft materials in neuroengineering for hard problems in neuroscience. Neuron 86: 175–186. [DOI] [PubMed] [Google Scholar]
- 4. Moxon, K.A. & Foffani G.. 2015. Brain–machine interfaces beyond neuroprosthetics. Neuron 86: 55–67. [DOI] [PubMed] [Google Scholar]
- 5. Robinson, J.T. , Jorgolli M. & Park H.. 2013. Nanowire electrodes for high‐density stimulation and measurement of neural circuits. Front. Neural Circuits 7: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson, M.D. et al 2013. Neuromodulation for brain disorders: challenges and opportunities. IEEE Trans. Biomed. Eng. 60: 610–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bin, H. et al 2013. Grand challenges in mapping the human brain: NSF workshop report. IEEE Trans. Biomed. Eng. 60: 2983–2992. [DOI] [PubMed] [Google Scholar]
- 8. Nicolelis, M.A. & Lebedev M.A.. 2009. Principles of neural ensemble physiology underlying the operation of brain–machine interfaces. Nat. Rev. Neurosci. 10: 530–540. [DOI] [PubMed] [Google Scholar]
- 9. Lebedev, M.A. & Nicolelis M.A.. 2006. Brain–machine interfaces: past, present and future. Trends Neurosci. 29: 536–546. [DOI] [PubMed] [Google Scholar]
- 10. Ordikhani‐Seyedlar, M. , Lebedev M.A., Sorensen H.B. & Puthusserypady S.. 2016. Neurofeedback therapy for enhancing visual attention: state‐of‐the‐art and challenges. Front. Neurosci. 10: 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sulzer, J. et al 2013. Real‐time fMRI neurofeedback: progress and challenges. Neuroimage 76: 386–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nowlis, D. & Kamiya J.. 1970. The control of electroencephalographic alpha rhythms through auditory feedback and the associated mental activity. Psychophysiology 6: 476–484. [DOI] [PubMed] [Google Scholar]
- 13. Kamiya, J. 1971. Biofeedback training in voluntary control of EEG alpha rhythms. Calif. Med. 115: 44. [PMC free article] [PubMed] [Google Scholar]
- 14. Seifert, A.R. & Lubar J.F.. 1975. Reduction of epileptic seizures through EEG biofeedback training. Biol. Psychol. 3: 157–184. [DOI] [PubMed] [Google Scholar]
- 15. Lubar, J. & Bahler W.. 1976. Behavioral management of epileptic seizures following EEG biofeedback training of the sensorimotor rhythm. Biofeedback Self Regul. 1: 77–104. [DOI] [PubMed] [Google Scholar]
- 16. Mann, C.A. , Lubar J.F., Zimmerman A.W., et al 1992. Quantitative analysis of EEG in boys with attention‐deficit‐hyperactivity disorder: controlled study with clinical implications. Pediatr. Neurol. 8: 30–36. [DOI] [PubMed] [Google Scholar]
- 17. Rozelle, G. & Budzynski T.. 2011. Neurotherapy for stroke rehabilitation: a single case study. Biofeedback Self Regul. 20: 211–228. [DOI] [PubMed] [Google Scholar]
- 18. Lubar, J. , Swartwood M., Swartwood J. & O'Donnell P.. 1995. Evaluation of the effectiveness of EEG neurofeedback training for ADHD in a clinical setting as measured by changes in T.O.V.A. scores, behavioral ratings, and WISC‐R performance. Biofeedback Self Regul. 20: 83–99. [DOI] [PubMed] [Google Scholar]
- 19. Nicholson, A.A. et al 2017. The neurobiology of emotion regulation in posttraumatic stress disorder: amygdala downregulation via real‐time fMRI neurofeedback. Hum. Brain Mapp. 38: 541–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Weiskopf, N. et al 2003. Physiological self‐regulation of regional brain activity using real‐time functional magnetic resonance imaging (fMRI): methodology and exemplary data. Neuroimage 19: 577–586. [DOI] [PubMed] [Google Scholar]
- 21. DeCharms, R.C. 2007. Reading and controlling human brain activation using real‐time functional magnetic resonance imaging. Trends Cogn. Sci. 11: 473–481. [DOI] [PubMed] [Google Scholar]
- 22. Rorden, C. & Karnath H.‐O.. 2004. Using human brain lesions to infer function: a relic from a past era in the fMRI age? Nat. Rev. Neurosci. 5: 813–819. [DOI] [PubMed] [Google Scholar]
- 23. Martin, N. et al 1992. Imaging techniques for cortical functional localization. Clin. Neurosurg. 38: 132–165. [PubMed] [Google Scholar]
- 24. Sergent, J. 1994. Brain‐imaging studies of cognitive functions. Trends Neurosci. 17: 221–227. [DOI] [PubMed] [Google Scholar]
- 25. Turner, R. 1994. Magnetic resonance imaging of brain function. Ann. Neurol. 35: 637–638. [DOI] [PubMed] [Google Scholar]
- 26. Sepulveda, P. et al 2016. How feedback, motor imagery, and reward influence brain self‐regulation using real‐time fMRI. Hum. Brain Mapp. 37: 3153–3171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kato, T. 1966. Perturbation Theory for Linear Operators. Springer‐Verlag, New York: Springer‐Verlag. [Google Scholar]
- 28. Stewart, G.W. & Sun Ji‐guang. 1990. Matrix Perturbation Theory. Academic Press. [Google Scholar]
- 29. Sporns, O. , Tononi G. & Kotter R.. 2005. The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1: e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bullmore, E.T. & Bassett D.S.. 2011. Brain graphs: graphical models of the human brain connectome. Annu. Rev. Clin. Psychol. 7: 113–140. [DOI] [PubMed] [Google Scholar]
- 31. Matthews, P.M. & Hampshire A.. 2016. Clinical concepts emerging from fMRI functional connectomics. Neuron 91: 511–528. [DOI] [PubMed] [Google Scholar]
- 32. Bassett, D.S. & Bullmore E.. 2006. Small‐world brain networks. Neuroscientist 12: 512–523. [DOI] [PubMed] [Google Scholar]
- 33. Bullmore, E.T. & Sporns O.. 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10: 186–198. [DOI] [PubMed] [Google Scholar]
- 34. Muldoon, S.F. et al 2016. Stimulation‐based control of dynamic brain networks. PLoS Comput. Biol. 12: e1005076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hagmann, P. et al 2008. Mapping the structural core of human cerebral cortex. PLoS Biol. 6: e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bassett, D.S. , Brown J.A., Deshpande V., et al 2011. Conserved and variable architecture of human white matter connectivity. Neuroimage 54: 1262–1279. [DOI] [PubMed] [Google Scholar]
- 37. Crofts, J.J. et al 2011. Network analysis detects changes in the contralesional hemisphere following stroke. Neuroimage 54: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Johnston, S.J. , Boehm S.G., Healy D., et al 2010. Neurofeedback: a promising tool for the self‐regulation of emotion networks. Neuroimage 49: 1066–1072. [DOI] [PubMed] [Google Scholar]
- 39. Young, K.D. et al 2014. Real‐time fMRI neurofeedback training of amygdala activity in patients with major depressive disorder. PLoS One 9: e88785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marxen, M. et al 2016. Amygdala regulation following fMRI‐neurofeedback without instructed strategies. Front. Hum. Neurosci. 10: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Birbaumer, N. , Ruiz S. & Sitaram R.. 2013. Learned regulation of brain metabolism. Trends Cogn. Sci. 17: 295–302. [DOI] [PubMed] [Google Scholar]
- 42. DeCharms, R.C. et al 2005. Control over brain activation and pain learned by using real‐time functional MRI. Proc. Natl. Acad. Sci. U.S.A. 102: 18626–18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Scharnowski, F. et al 2015. Manipulating motor performance and memory through real‐time fMRI neurofeedback. Biol. Psychol. 108: 85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yoo, S.S. , Lee J.H., O'Leary H., et al 2008. Neurofeedback fMRI‐mediated learning and consolidation of regional brain activation during motor imagery. Int. J. Imaging Syst. Technol. 18: 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marins, T.F. et al 2015. Enhancing motor network activity using real‐time functional MRI neurofeedback of left premotor cortex. Front. Behav. Neurosci. 9: 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Auer, T. , Dewiputri W.I., Frahm J. & Schweizer R.. 2016. Higher‐order brain areas associated with real‐time functional MRI neurofeedback training of the somato‐motor cortex. Neuroscience. https://doi.org/10.1016/j.neuroscience.2016.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. MacInnes, J.J. , Dickerson K.C., Chen N. Kuei & Adcock R.A.. 2016. Cognitive neurostimulation: learning to volitionally sustain ventral tegmental area activation. Neuron 89: 1331–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Barnes, J.J. , Nobre A.C., Woolrich M.W., et al 2016. Training working memory in childhood enhances coupling between frontoparietal control network and task‐related regions. J. Neurosci. 36: 9001–9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Harrison, B.J. et al 2008. Modulation of brain resting‐state networks by sad mood induction. PLoS One 3: 1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hwang, H.J. , Kwon K. & Im C.H.. 2009. Neurofeedback‐based motor imagery training for brain–computer interface (BCI). J. Neurosci. Methods 179: 150–156. [DOI] [PubMed] [Google Scholar]
- 51. Norman, K.A. , Polyn S.M., Detre G.J. & Haxby J.V.. 2006. Beyond mind‐reading: multi‐voxel pattern analysis of fMRI data. Trends Cogn. Sci. 10: 424–430. [DOI] [PubMed] [Google Scholar]
- 52. LaConte, S.M. 2011. Decoding fMRI brain states in real‐time. Neuroimage 56: 440–454. [DOI] [PubMed] [Google Scholar]
- 53. Stoeckel, L.E. et al 2014. Optimizing real time fMRI neurofeedback for therapeutic discovery and development. Neuroimage Clin. 5: 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Niazi, A.M. et al 2014. Online decoding of object‐based attention using real‐time fMRI. Eur. J. Neurosci. 39: 319–329. [DOI] [PubMed] [Google Scholar]
- 55. Andersson, P. et al 2011. Real‐time decoding of brain responses to visuospatial attention using 7T fMRI. PLoS One 6: e27638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. DeBettencourt, M.T. , Cohen J.D., Lee R.F., et al 2015. Closed‐loop training of attention with real‐time brain imaging. Nat. Neurosci. 18: 470–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cordes, J.S. et al 2015. Cognitive and neural strategies during control of the anterior cingulate cortex by fMRI neurofeedback in patients with schizophrenia. Front. Behav. Neurosci. 9: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ros, T. , Baars B.J., Lanius R.A. & Vuilleumier P.. 2014. Tuning pathological brain oscillations with neurofeedback: a systems neuroscience framework. Front. Hum. Neurosci. 8: 1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. DeCharms, R.C. et al 2004. Learned regulation of spatially localized brain activation using real‐time fMRI. Neuroimage 21: 436–443. [DOI] [PubMed] [Google Scholar]
- 60. Ruiz, S. et al 2013. Acquired self‐control of insula cortex modulates emotion recognition and brain network connectivity in schizophrenia. Hum. Brain Mapp. 34: 200–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Frank, S. et al 2012. The obese brain athlete: self‐regulation of the anterior insula in adiposity. PLoS One 7: 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Li, X. et al 2014. Volitional reduction of anterior cingulate cortex activity produces decreased cue craving in smoking cessation: a preliminary real‐time fMRI study. Addict. Biol. 18: 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Van De Ville, D. et al 2012. Recovery of the default mode network after demanding neurofeedback training occurs in spatio‐temporally segregated subnetworks. Neuroimage 63: 1775–1781. [DOI] [PubMed] [Google Scholar]
- 64. Amano, K. , Shibata K., Kawato M., et al 2016. Learning to associate orientation with color in early visual areas by associative decoded fMRI neurofeedback. Curr. Biol. 26: 1861–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shibata, K. , Watanabe T., Sasaki Y. & Kawato M.. 2011. Perceptual learning incepted by decoded fMRI neurofeedback without stimulus presentation. Science 334: 1413–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Seitz, A.R. 2013. Cognitive neuroscience: targeting neuroplasticity with neural decoding and biofeedback. Curr. Biol. 23: R210–R212. [DOI] [PubMed] [Google Scholar]
- 67. Scharnowski, F. , Hutton C., Josephs O., et al 2012. Improving visual perception through neurofeedback. J. Neurosci. 32: 17830–17841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cox, R.W. , Jesmanowicz A. & Hyde J.S.. 1995. Real‐time functional magnetic resonance imaging. Magn. Reson. Med. 33: 230–236. [DOI] [PubMed] [Google Scholar]
- 69. Hartwell, K.J. et al 2013. Real‐time fMRI in the treatment of nicotine dependence: a conceptual review and pilot studies. Psychol. Addict. Behav. 27: 501–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sorger, B. , Kamp T., Weiskopf N., et al 2016. When the brain takes ‘BOLD’ steps: real‐time fMRI neurofeedback can further enhance the ability to gradually self‐regulate regional brain activation. Neuroscience. https://doi.org/10.1016/j.neuroscience.2016.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lawrence, J.M. , Hoeft F., Sheau K.E. & Mackey S.C.. 2011. Strategy‐dependent dissociation of the neural correlates involved in pain modulation. Anesthesiology 115: 844–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bullmore, E. et al 2009. Generic aspects of complexity in brain imaging data and other biological systems. Neuroimage 47: 1125–1134. [DOI] [PubMed] [Google Scholar]
- 73. Bassett, D.S. et al 2010. Efficient physical embedding of topologically complex information processing networks in brains and computer circuits. PLoS Comput. Biol. 6: e1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Betzel, R.F. & Bassett D.S.. 2016. Multi‐scale brain networks. Neuroimage. https://doi.org/10.1016/j.neuroimage.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hall, T.M. , Nazarpour K. & Jackson A.. 2014. Real‐time estimation and biofeedback of single‐neuron firing rates using local field potentials. Nat. Commun. 5: 5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Engelhard, B. , Ozeri N., Israel Z., et al 2013. Inducing gamma oscillations and precise spike synchrony by operant conditioning plbibitalic‐via brain–machine interface. Neuron 77: 361–375. [DOI] [PubMed] [Google Scholar]
- 77. Koush, Y. et al 2013. Connectivity‐based neurofeedback: dynamic causal modeling for real‐time fMRI. Neuroimage 81: 422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kim, D.‐Y. , Yoo S.‐S., Tegethoff M., et al 2015. The inclusion of functional connectivity information into fMRI‐based neurofeedback improves its efficacy in the reduction of cigarette cravings. J. Cogn. Neurosci. 27: 1552–1572. [DOI] [PubMed] [Google Scholar]
- 79. Koush, Y. et al 2017. Learning control over emotion networks through connectivity‐based neurofeedback. Cereb. Cortex 27: 1193–1202. [DOI] [PubMed] [Google Scholar]
- 80. Liew, S.‐L. et al 2016. Improving motor corticothalamic communication after stroke using real‐time fMRI connectivity‐based neurofeedback. Neurorehabil. Neural Repair 30: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Megumi, F. , Yamashita A., Kawato M. & Imamizu H.. 2015. Functional MRI neurofeedback training on connectivity between two regions induces long‐lasting changes in intrinsic functional network. Front. Hum. Neurosci. 9: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Coben, R. 2007. Connectivity‐guided neurofeedback for autistic spectrum disorder. Biofeedback 35: 131–135. [DOI] [PubMed] [Google Scholar]
- 83. Ros, T. et al 2013. Mind over chatter: plastic up‐regulation of the fMRI salience network directly after EEG neurofeedback. Neuroimage 65: 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Mottaz, A. et al 2015. Neurofeedback training of alpha‐band coherence enhances motor performance. Clin. Neurophysiol. 126: 1754–1760. [DOI] [PubMed] [Google Scholar]
- 85. Hui, M. , Zhang H., Ge R., et al 2014. Modulation of functional network with real‐time fMRI feedback training of right premotor cortex activity. Neuropsychologia 62: 111–123. [DOI] [PubMed] [Google Scholar]
- 86. Hamilton, J.P. , Glover G.H., Hsu J.J., et al 2011. Modulation of subgenual anterior cingulate cortex activity with real‐time neurofeedback. Hum. Brain Mapp. 32: 22–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Scheinost, D. et al 2013. Orbitofrontal cortex neurofeedback produces lasting changes in contamination anxiety and resting‐state connectivity. Transl. Psychiatry 3: e250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zotev, V. et al 2011. Self‐regulation of amygdala activation using real‐time fMRI neurofeedback. PLoS One 6: e24522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Haller, S. et al 2013. Dynamic reconfiguration of human brain functional networks through neurofeedback. Neuroimage 81: 243–252. [DOI] [PubMed] [Google Scholar]
- 90. Zhang, Q. , Zhang G., Yao L. & Zhao X.. 2015. Impact of real‐time fMRI working memory feedback training on the interactions between three core brain networks. Front. Behav. Neurosci. 9: 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Paret, C. et al 2016. FMRI neurofeedback of amygdala response to aversive stimuli enhances prefrontal–limbic brain connectivity. Neuroimage 125: 182–188. [DOI] [PubMed] [Google Scholar]
- 92. Bassett, D.S. & Sporns O.. 2017. Network neuroscience. Nat. Neurosci. 20: 353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bollobas, B. 1979. Graph Theory: An Introductory Course. New York, Heidelberg, Berlin: Springer‐Verlag. [Google Scholar]
- 94. Bollobas, B. 1985. Random Graphs. Academic Press. [Google Scholar]
- 95. Newman, M.E. 2010. Networks: An Introduction. Oxford University Press. [Google Scholar]
- 96. Newman, M.E.J. 2003. The structure and function of complex networks. SIAM Rev. 45: 167–256. [Google Scholar]
- 97. Achard, S. , Salvador R., Whitcher B., et al 2006. A resilient, low‐frequency, small‐world human brain functional network with highly connected association cortical hubs. J. Neurosci. 26: 63–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Alstott, J. , Breakspear M., Hagmann P., et al 2009. Modeling the impact of lesions in the human brain. PLoS Comput. Biol. 5: e1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Bassett, D.S. & Bullmore E.T.. 2009. Human brain networks in health and disease. Curr. Opin. Neurol. 22: 340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sporns, O. 2014. Contributions and challenges for network models in cognitive neuroscience. Nat. Neurosci. 17: 652–660. [DOI] [PubMed] [Google Scholar]
- 101. Bassett, D.S. et al 2011. Dynamic reconfiguration of human brain networks during learning. Proc. Natl. Acad. Sci. U.S.A. 108: 7641–7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Doron, K.W. , Bassett D.S. & Gazzaniga M.S.. 2012. Dynamic network structure of interhemispheric coordination. Proc. Natl. Acad. Sci. U.S.A. 109: 18661–18668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wymbs, N.F. , Bassett D.S., Mucha P.J., et al 2012. Differential recruitment of the sensorimotor putamen and frontoparietal cortex during motor chunking in humans. Neuron 74: 936–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Bassett, D.S. et al 2013. Task‐based core–periphery organization of human brain dynamics. PLoS Comput. Biol. 9: e1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Mattar, M.G. , Cole M.W., Thompson‐Schill S.L. & Bassett D.S.. 2015. A functional cartography of cognitive systems. PLoS Comput. Biol. 11: e1004533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Chai, L.R. , Mattar M.G., Blank I.A., et al 2016. Functional network dynamics of the language system. Cereb. Cortex 26: 4148‐4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Muldoon, S.F. & Bassett D.S.. 2016. Network and multilayer network approaches to understanding human brain dynamics. Philos. Sci. 83: 710–720. [Google Scholar]
- 108. Sporns, O. , Honey C.J. & Kotter R.. 2007. Identification and classification of hubs in brain networks. PLoS One 2: e1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Zalesky, A. et al 2010. Whole‐brain anatomical networks: does the choice of nodes matter? Neuroimage 50: 970–983. [DOI] [PubMed] [Google Scholar]
- 110. Watts, D.J. & Strogatz S.H.. 1998. Collective dynamics of ‘small‐world’ networks. Nature 393: 440–442. [DOI] [PubMed] [Google Scholar]
- 111. Scholtens, L.H. , Schmidt R., de Reus M.A. & van den Heuvel M.P.. 2014. Linking macroscale graph analytical organization to microscale neuroarchitectonics in the macaque connectome. J. Neurosci. 34: 12192–12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Kitzbichler, M.G. , Henson R.N.A., Smith M.L., et al 2011. Cognitive effort drives workspace configuration of human brain functional networks. J. Neurosci. 31: 8259–8270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Porter, M.A. , Onnela J.‐P. & Mucha P.J.. 2009. Communities in networks. Am. Math. Soc. 56: 1082–1097, 1164–1166, 2009. [Google Scholar]
- 114. Fortunato, S. 2010. Community detection in graphs. Phys. Rep. 486: 75–174. [Google Scholar]
- 115. Mišić, B. et al 2016. Network‐level structure–function relationships in human neocortex. Cereb. Cortex 26: 3285–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sporns, O. & Betzel R.F.. 2016. networks Modular brain. Annu. Rev. Psychol. 67: 613–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Borgatti, S.P. & Everett M.G.. 2000. structures Models of core/periphery. Soc. Netw. 21: 375–395. [Google Scholar]
- 118. Everett, M.G. & Borgatti S.P.. 1999. Peripheries of cohesive subsets. Soc. Netw. 21: 397–407. [Google Scholar]
- 119. Fedorenko, E. & Thompson‐Schill S.L.. 2014. Reworking the language network. Trends Cogn. Sci. 18: 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Bassett, D.S. et al 2013. Robust detection of dynamic community structure in networks. Chaos 23: 013142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Achard, S. & Bullmore E.. 2007. Efficiency and cost of economical brain functional networks. PLoS Comput. Biol. 3: e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Li, Y. et al 2009. Brain anatomical network and intelligence. PLoS Comput. Biol. 5: e1000395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Arenas, A. , Díaz‐Guilera A., Kurths J., et al 2008. Synchronization in complex networks. Phys. Rep. 469: 93–153. [Google Scholar]
- 124. Wang, X.F. & Chen G.. 2002. Synchronization in scale‐free dynamical networks: robustness and fragility. IEEE Trans. Circuits Syst. I Fundam. Theory Appl. 49: 54–62. [Google Scholar]
- 125. Bassett, D.S. , Meyer‐Lindenberg A., Achard S., et al 2006. Adaptive reconfiguration of fractal small‐world human brain functional networks. Proc. Natl. Acad. Sci. U.S.A. 103: 19518–19523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Medaglia, J.D. , Lynall M.‐E. & Bassett D.S.. 2015. Cognitive network neuroscience. J. Cogn. Neurosci. 27: 1471–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Bassett, D.S. , Khambhati A.N. & Grafton S.T.. 2016. Emerging frontiers of neuroengineering: a network science of brain connectivity. Annu. Rev. Biomed. Eng. arXiv:1612.08059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Bassett, D.S. et al 2008. Hierarchical organization of human cortical networks in health and schizophrenia. J. Neurosci. 28: 9239–9248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Honey, C.J. et al 2009. Predicting human resting‐state functional connectivity from structural connectivity. Proc. Natl. Acad. Sci. U.S.A. 106: 2035–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Goni, J. et al 2014. Resting‐brain functional connectivity predicted by analytic measures of network communication. Proc. Natl. Acad. Sci. U.S.A. 111: 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Scholz, J. , Klein M.C., Behrens T.E. & Johansen‐Berg H.. 2009. Training induces changes in white‐matter architecture. Nat. Neurosci. 12: 1370–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Schlegel, A.A. , Rudelson J.J. & Tse P.U.. 2012. White matter structure changes as adults learn a second language. J. Cogn. Neurosci. 24: 1664–1670. [DOI] [PubMed] [Google Scholar]
- 133. Kahn, A.E. et al 2016. Structural pathways supporting swift acquisition of new visuomotor skills. Cereb. Cortex 27: 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Caeyenberghs, K. et al 2014. Altered structural networks and executive deficits in traumatic brain injury patients. Brain Struct. Funct. 219: 193–209. [DOI] [PubMed] [Google Scholar]
- 135. Warren, D.E. et al 2014. Network measures predict neuropsychological outcome after brain injury. Proc. Natl. Acad. Sci. U.S.A. 111: 14247–14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Caeyenberghs, K. et al 2012. Graph analysis of functional brain networks for cognitive control of action in traumatic brain injury. Brain 135: 1293–1307. [DOI] [PubMed] [Google Scholar]
- 137. Cole, M.W. et al 2012. Global connectivity of prefrontal cortex predicts cognitive control and intelligence. J. Neurosci. 32: 8988–8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Harmelech, T. , Friedman D. & Malach R.. 2015. Differential magnetic resonance neurofeedback modulations across extrinsic (visual) and intrinsic (default‐mode) nodes of the human cortex. J. Neurosci. 35: 2588–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Gu, S. et al 2015. Emergence of system roles in normative neurodevelopment. Proc. Natl. Acad. Sci. U.S.A. 112: 13681–13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Fornito, A. , Harrison B.J., Zalesky A. & Simons J.S.. 2012. Competitive and cooperative dynamics of large‐scale brain functional networks supporting recollection. Proc. Natl. Acad. Sci. U.S.A. 109: 12788–12793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Cole, M.W. et al 2013. Multi‐task connectivity reveals flexible hubs for adaptive task control. Nat. Neurosci. 16: 1348–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Gu, S. et al 2015. Controllability of structural brain networks. Nat. Commun. 6: 8414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Khambhati, A.N. , Davis K.A., Lucas T.H., et al 2016. Virtual cortical resection reveals push–pull network control preceding seizure evolution. Neuron 91: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Bassett, D.S. , Yang M., Wymbs N.F. & Grafton S.T.. 2015. Learning‐induced autonomy of sensorimotor systems. Nat. Neurosci. 18: 744–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Bassett, D.S. & Lynall M.‐E.. 2013. Network methods to characterize brain structure and function. In The Cognitive Neurosciences: The Biology of the Mind, 5th Ed. Gazzaniga M.S., Ivry R.B. & Mangun G.R., Eds: 1–27. Cambridge, MA: MIT Press.
- 146. Rubinov, M. & Bassett D.S.. 2011. Emerging evidence of connectomic abnormalities in schizophrenia. J. Neurosci. 31: 6263–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Papo, D. , Zanin M., Pineda‐Pardo J.A., et al 2014. Functional brain networks: great expectations, hard times and the big leap forward. Philos. Trans. R. Soc. B Biol. Sci. 369: 20130525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Valiant, L.G. 2014. What must a global theory of cortex explain? Curr. Opin. Neurobiol. 25: 15–19. [DOI] [PubMed] [Google Scholar]
- 149. Valiant, L.G. 2006. A quantitative theory of neural computation. Biol. Cybern. 95: 205–211. [DOI] [PubMed] [Google Scholar]
- 150. Craver, C.F. 2005. Beyond reduction: mechanisms, multifield integration and the unity of neuroscience. Stud. Hist. Philos. Biol. Biomed. Sci. 36: 373–395. [DOI] [PubMed] [Google Scholar]
- 151. Liu, Y.‐Y. , Slotine J.‐J. & Barabasi A.‐L.. 2011. Controllability of complex networks. Nature 473: 167–173. [DOI] [PubMed] [Google Scholar]
- 152. Pasqualetti, F. , Zampieri S. & Bullo F.. 2014. Controllability metrics and algorithms for complex networks. IEEE Trans. Control Netw. Syst. 1: 40–52. [Google Scholar]
- 153. Kailath, T. 1980. Linear Systems. Prentice‐Hall. [Google Scholar]
- 154. Motter, A.E. 2015. Networkcontrology. Chaos 25: 097621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Betzel, R.F. , Gu S., Medaglia J.D., et al 2016. Optimally controlling the human connectome: the role of network topology. Sci. Rep. 6:30770. https://doi.org/10.1038/srep30770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Schiff, S.J. 2011. Neural Control Engineering: The Emerging Intersection between Control Theory and Neuroscience. MIT Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. MacDonald, A.W. , Cohen J.D., Stenger V. & Carter C.S.. 2000. Dissociating the role of the dorsolateral prefrontal and anterior cingulate cortex in cognitive control. Science 288: 1835–1838. [DOI] [PubMed] [Google Scholar]
- 158. Medaglia, J.D. et al 2016. Cognitive control in the controllable connectome. arXiv:1606.09185. [DOI] [PMC free article] [PubMed]
- 159. Ching, S. , Brown E.N. & Kramer M.A.. 2012. Distributed control in a mean‐field cortical network model: implications for seizure suppression. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 86: 021920. [DOI] [PubMed] [Google Scholar]
- 160. Galan, R.F. , Ermentrout G.B. & Urban N.N.. 2008. Optimal time scale for spike‐time reliability: theory, simulations, and experiments. J. Neurophysiol. 99: 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Gu, S. et al 2017. Optimal trajectories of brain state transitions. Neuroimage 148: 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Jbabdi, S. & Johansen H.‐Berg. 2011. Tractography: where do we go from here? Brain Connect. 1: 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Pestilli, F. , Yeatman J.D., Rokem A., et al 2014. Evaluation and statistical inference for human connectomes. Nat. Methods 11: 1058–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Dell'Acqua, F. & Catani M.. 2012. Structural human brain networks: hot topics in diffusion tractography. Curr. Opin. Neurol. 25: 375–383. [DOI] [PubMed] [Google Scholar]
- 165. Leith, D.J. & Leithead W.E.. 2000. Survey of gain‐scheduling analysis and design. Int.J. Control 73: 1001–1025. [Google Scholar]
- 166. Cornelius, S.P. , Kath W.L. & Motter A.E.. 2013. Realistic control of network dynamics. Nat. Commun. 4: 1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Lepage, K.Q. , Ching S. & Kramer M.A.. 2013. Inferring evoked brain connectivity through adaptive perturbation. J. Comput. Neurosci. 34: 303–318. [DOI] [PubMed] [Google Scholar]
- 168. Singer, W. 1999. Neuronal synchrony: a versatile code for the definition of relations? Neuron 24: 49–65, 111–125. [DOI] [PubMed] [Google Scholar]
- 169. He, Z. , Wang X., Zhang G.‐Y. & Zhan M.. 2014. Control for a synchronization–desynchronization switch. Phys. Rev. E 90: 012909. [DOI] [PubMed] [Google Scholar]
- 170. Ching, S. & Ritt J.T.. 2013. Control strategies for underactuated neural ensembles driven by optogenetic stimulation. Front. Neural Circuits 7: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Bentley, B. et al 2016. The multilayer connectome of Caenorhabditis elegans . PLoS Comput. Biol. 12: e1005283. [DOI] [PMC free article] [PubMed] [Google Scholar]


