Abstract
Facioscapulohumeral dystrophy (FSHD; OMIM #158900, #158901) is caused by mis-expression of the DUX4 transcription factor in skeletal muscle1. Animal models of FSHD are hampered by incomplete knowledge of the conservation of the DUX4 transcriptional program in other species2–5. Despite divergence of their binding motifs, both mouse Dux and human DUX4 activate genes associated with cleavage-stage embryos, including MERV-L and ERVL-MaLR retrotransposons, in mouse and human muscle cells respectively. When expressed in mouse cells, human DUX4 maintained modest activation of cleavage-stage genes driven by conventional promoters, but did not activate MERV-L-promoted genes. These findings indicate that the ancestral DUX4-factor regulated genes characteristic of cleavage-stage embryos driven by conventional promoters, whereas divergence of the DUX4/Dux homeodomains correlates with retrotransposon specificity. These results provide insight into how species balance conservation of a core transcriptional program with innovation at retrotransposon promoters and provide a basis for animal models that recreate the FSHD transcriptome.
While the transcriptome of human DUX4 expressed in human cells is known6,7, the transcriptome of mouse Dux in mouse cells has been largely unknown8. Both proteins are encoded by retrogenes derived by the retroposition of DUXC mRNA9–11 and both proteins induce apoptosis when expressed in cultured human and mouse muscle cells12,13. Recent studies expressing Dux in human muscle cells12 or DUX4 in mouse cells4,13 showed a partial overlap of regulated genes and a similar consensus binding site12; however, these two proteins have diverged significantly at the sequence level, including their homeodomains. Determination of the degree of similarity in their transcriptional programs might help us understand the rapid evolutionary divergence of Dux and DUX4 and inform murine models of FSHD, a disease which still lacks treatment options.
To compare the Dux transcriptome with the previously published DUX4 transcriptome in FSHD muscle cells, we generated RNA-seq and ChIP-seq datasets for Dux expressed in mouse skeletal muscle cells (see Online Methods). We observed increased expression of 962 genes and decreased expression of 204 genes (Fig. 1a, Supplementary Tables 1–2). In these data, the most upregulated genes were normally expressed in the mouse 2-cell embryo (e.g. Zscan4a-e, Tcstv1/3)14–16, therefore we used gene set enrichment analysis to compare our data to 2-cell-like embryonic stem cells17(GSEA; 2C-like). The top of the Dux transcriptome was significantly enriched for the 2C-like gene signature (258/469 genes in the 2C-like gene signature contributed to the GSEA core enrichment, NES = 12.56, p-value < 0.001; Fig. 1b, Supplementary Table 3, Supplementary Fig. 1). In addition, direct targets of Dux (i.e. genes whose RNA increased expression 4-fold or more and have a ChIP-seq peak within 1kb of the annotated transcriptional start site (TSS)) were enriched in the 2C-like gene signature based on hypergeometric testing (60 direct targets in 2C-like signature/189 total direct targets; 16-fold more direct targets in the 2C-like gene signature than the 3.7 genes expected by chance, p=9.1E-56), including Zscan4a-f, Tcstv1/3, Usp17lb/d, Pramef25 and Zfp352. We further confirmed that robust induction of both Pramef25 and Zscan4c reporter constructs depended on intact Dux binding sites (Supplementary Fig. 2a–b, Supplementary Fig. 3a–b). ChIP-seq peaks at the TSS of each of the five Zscan4-cluster genes supports the hypothesis that Dux directly binds and activates each Zscan4-cluster gene (Supplementary Fig. 3c–h). Although there are two MERV-L elements in the Zscan4 locus, we did not observe RNA-seq reads that spliced from these MERV-Ls to any Zscan4 gene (Supplementary Fig. 3i–j). Importantly, the published 2C-like signature included Dux itself and Dux RNA is expressed in mESC (J. Whiddon, unpublished data). Impartial gene ontology analysis also identified “embryo development” among significantly enriched terms (Supplementary Table 4). Together, these results demonstrated that Dux directly regulates many genes in the 2C-like transcriptome in myoblasts.
Figure 1. Dux and DUX4 activate an early cleavage-stage embryo gene signature in muscle cells of their respective species.
(a) Dux transcriptome in C2C12 mouse muscle cells: red dots are genes affected more than absolute(log2FoldChange)>=2 and adjusted p-value<=0.05. Normalized counts are calculated by DESeq2 (normalized count = read count/size factor, where size-factors are estimated with the median-of-ratios method26). Control samples were un-induced cells of the same cell line.
(b) Gene set enrichment analysis (GSEA): gene set is 2C-like gene signature17, x-axis is log2FoldChange-ranked Dux transcriptome. Enrichment score (ES) increases when a gene in the Dux transcriptome is also in 2C-like gene set and a black vertical bar is drawn in lower panel; ES decreases when a gene isn’t in 2C-like gene set. P-value was empirically determined based on 1,000 permutations of ranked gene lists.
(c) Human DUX4, mouse Dux and canine DUXC homeodomain alignments (%=percent amino acid identity, *=four predicted DNA-contacting residues).
(d) GSEA: gene set is the top 500 most upregulated genes in DUX4-expressing human cells, x-axis is log2FoldChange-ranked Dux transcriptome in mouse cells. This cross-species comparison required limiting both gene set and transcriptome to 1:1 mouse-to-human orthologs. The converse comparison is in Supplementary Figure 4a.
(e) GSEA: gene set is the human orthologs of the mouse 2C-like gene signature, x-axis is log2FoldChange-ranked DUX4 transcriptome in human muscle cells. Both gene set and transcriptome are limited to 1:1 mouse-to-human orthologs. Note: mouse 2C-like gene signature has 469 genes total, 297 of these genes have simple 1:1 mouse-to-human orthology.
Despite considerable sequence divergence in their two DNA-binding homeodomain regions (Fig. 1c), we found that Dux and DUX418 activated orthologous genes in myoblasts of their respective species, including genes in the mouse 2C-like gene signature. For this analysis we only considered genes with simple 1:1 mouse-to-human orthology according to HomoloGene19. GSEA determined that the 500 genes most upregulated by DUX4 were significantly enriched in the genes most upregulated by Dux (NES=8.16, p-value<0.001; Fig. 1d) and vice versa (NES=6.01, p-value<0.001; Supplementary Fig. 4a). GSEA also demonstrated that DUX4 activated the human orthologs of the mouse 2C-like gene signature (NES=2.24, p-value = 0.002, Fig. 1e). It should be noted, however, that these analyses of similarity using the HomoloGene method were conservative. Complex gene families, such as the ZSCAN4, PRAME, THOC4/ALYREF, and USP17 families, were excluded from the HomoloGene dataset because 1:1 orthology cannot be established reliably, but members of each of these families were upregulated in both species. Together, these data demonstrate a strong functional conservation for Dux and DUX4 in the regulation of this 2C-like network in their respective species.
Despite this functional conservation, a de novo motif-finding algorithm20 identified a Dux binding motif in our ChIP-seq data that diverged from the published DUX4 binding motif in the first half of the motif but not the second (Fig. 2a), perhaps reflecting that the four predicted DNA-binding-specificity residues21 are identical between DUX4 and Dux in the second homeodomain but not the first (Fig. 1c). The motif identified in this analysis is similar to the recently published motif for Dux in human muscle cells12, supporting the notion that the Dux binding motif is cell type independent.
Figure 2. Despite binding motif divergence and general transcriptome divergence, DUX4 transcriptome in mouse muscle cells is enriched for the 2C-like gene signature.
(a) Dux and DUX4 binding motifs as derived de novo from ChIP-seq peaks using MEME algorithm. DUX4 ChIP-seq data was previously published6, but re-analyzed using the methods of this study. Note the divergence in the first half of the motif and the conservation of the second half of the motif. E-values listed reflect an estimate of the expected number of motifs, with the given motif’s log likelihood ratio (or higher) and with the same width and site count, that one would find in a similarly sized set of random sequences (where each position in each sequence is independent and letters are chosen according to the background letter frequencies). Histogram to the right shows that 578 peaks out of the 600 used to generate the Dux motif carry a match to the motif and that the motifs are centrally located within each ChIP-seq peak. DUX4 histogram is also shown.
(b) GSEA: gene set is the mouse 2C-like gene signature, x-axis is the log2FoldChange-ranked DUX4 transcriptome in mouse cells. Since the mouse 2C-like gene signature and this DUX4 transcriptome were both identified in mouse cells, neither gene set nor transcriptome was limited to genes with 1:1 mouse-to-human orthology.
Because of the apparent paradox of the functional conservation of Dux and DUX4 transcriptomes and the partial divergence of their binding motifs, we next generated RNA-seq and ChIP-seq datasets for DUX4 in mouse muscle cells to better understand their conservation and divergence (Supplementary Tables 5–6). In this context, DUX4 showed the same binding motif as in human cells (Supplementary Fig. 5a), increased expression of 582 genes and decreased expression of 428 genes (Supplementary Fig. 5b). Although DUX4 regulated many genes that were not orthologous to Dux-regulated genes and overall showed little similarity to the Dux transcriptome (Supplementary Fig. 5c), the genes that were upregulated in both the Dux and DUX4 transcriptomes were enriched for 2C-like genes by hypergeometric testing (p= 1.07e-11) and GSEA showed significant enrichment of the 2C-like gene signature activated by DUX4 in mouse cells (NES = 4.25, p-value<0.001; Fig. 2b; Supplementary Table 7). The activation of this signature, however, was not as robust compared to Dux in mouse cells. For example, Tcstv3 and Zscan4d had log2 fold-changes of only 0.92 and 0.66, respectively, compared to 10.1 and 12.4 by Dux, indicating that the top of the DUX4 transcriptome is enriched for the 2C-like gene signature through moderate induction of many members.
In contrast to the moderate conservation of DUX4’s activation of the conventionally-promoted 2C-like program in mouse cells, activation of 2C-like repetitive elements was specific to Dux. Transcription of certain repetitive elements has been reported in 2C-like mouse ES cells16,22 and we found that Dux, but not DUX4, induced expression of MERV-L elements by 100-fold and pericentromeric satellite DNA by 50-fold (Fig. 3a–c, Supplementary Fig. 6a–c, Supplementary Tables 8–9). ChIP-seq data indicated that MERV-L elements were a direct target of Dux, but not DUX4 (Fig. 3d), and the MERV-L consensus sequence carries a Dux binding site (Supplementary Fig. 6d). Consistently, Dux, but not DUX4, activated a reporter driven by a MERVL element and this activation was lost when we mutated the predicted Dux binding site (Fig. 3e). MERV-L elements have been reported to function as alternative promoters in 2C-embryos16,22, which we observed in Dux-expressing, but not DUX4-expressing, mouse cells using two complementary approaches (Fig. 4a–b, Supplementary Fig. 5d, Supplementary Fig. 7a–c, Supplementary Tables 10–11). These results indicate that DUX4 activated a portion of the 2C-like gene signature in mouse cells, but it did not activate repetitive elements characteristic of the 2C mouse embryo.
Figure 3. Dux, but not DUX4, activates transcription of repetitive elements characteristic of the early embryo in mouse muscle cells.
(a) Expression levels of repeats during Dux expression in mouse cells compared to un-induced cells of the same cell line, broken down by repeat class. For LTR elements broken down by family, see Supplementary Figure 6a–c. Each dot is a repeatName as defined by RepeatMasker. Red color indicates differential expression at absolute(log2-Foldchange)>=1 and adjusted p-value<=0.05. Number in parentheses is log2-FoldChange.
(b) Same as (a) for DUX4-expressing mouse muscle cells compared to un-induced cells of the same cell line.
(c) Same as (a) for DUX4-expressing human muscle cells compared to un-induced cells of the same cell line, data previously published18.
(d) Example of a Dux ChIP-seq peak in MERV-L (MT2-element in RepBase nomenclature). Track height is 200 reads for all tracks. mm10 genome location is chr15:52,742,953–52,744,319.
(e) Luciferase assay comparing the activation of a 2C-active MERV-L element reporter by either Dux, DUX4 or an empty vector. The MERV-L element contains a match to the Dux motif and was mutated as shown in cartoon to the right and the full sequence is in Supplementary Figure 6d. Activation of the mutated MERV-L reporter is also shown.
Data shown are mean fold change over empty vector of 3 cell cultures prepared in parallel for each condition. Error bars are s.e.m. The non-mutated MERV-L reporter activation experiment was repeated on three separate occasions with consistent results. The mutated MERV-L reporter experiment was performed on one occasion.
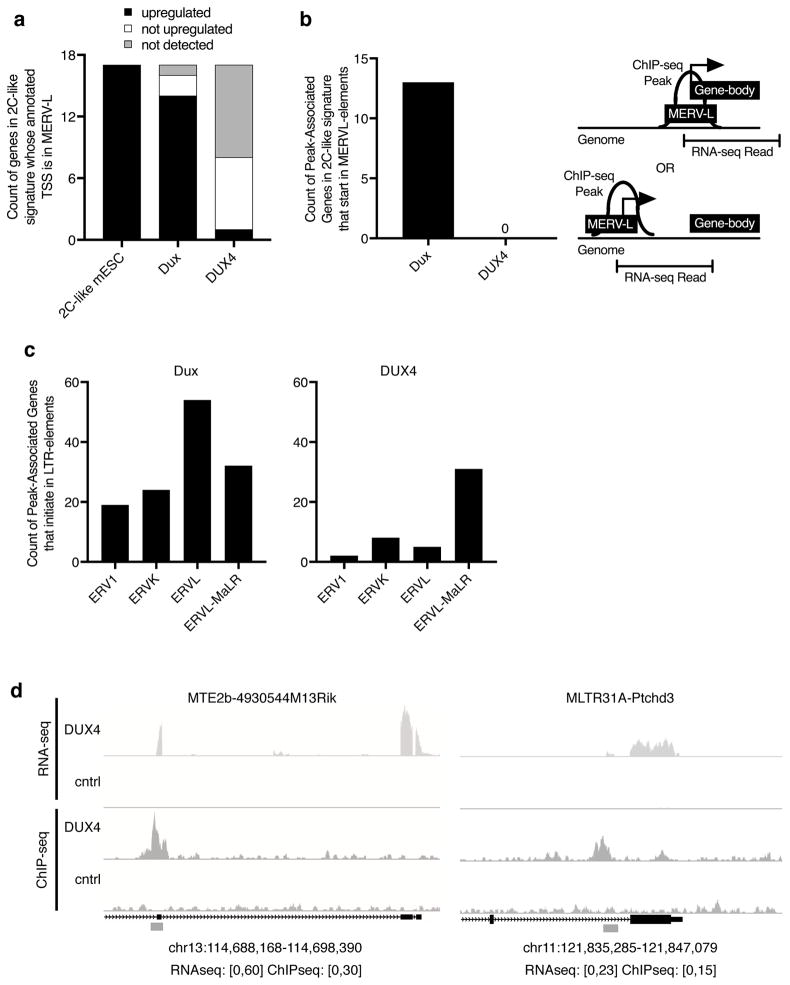
Figure 4. Dux and DUX4 use different types of LTR elements as alternative promoters for protein-coding genes.
(a) Histogram where black bars are counts of genes in the 2C-like signature that are MERV-L-promoted and activated by the indicated factor. White bars are genes detected by RNA-seq, but are not upregulated compared to control samples. Gray bars are genes with no reads by RNAseq. MERV-L promoted genes for this plot were determined by presence of an MT2-type element that overlaps the annotated TSS of a gene in the published 2C-like gene signature17.
(b) Histogram showing the number of genes in the 2C-like signature where the indicated factor bound a MERV-L (MT2-type) element based on ChIP-seq data and there was at least one RNA-seq read that connected the ChIP-seq peak range to an annotated exon in mouse muscle cells, termed “Peak-Associated Genes” (PAGs). Cartoon depiction of PAGs that overlap MERV-Ls is to the right. For two examples of PAGs that start in MERV-L (MT2-type) elements, see Supplementary Figure 7a–b.
(c) LTR-family distribution of PAGs that overlap any LTRs (CHIP-seq peak in an LTR with at least one RNA-seq read that connects the element to an annotated exon). Note that although Dux and DUX4 both have PAGs that start in ERVL-MaLRs, they are predominantly different ERVL-MaLRs (only 1/31 DUX4_PAGs in ERVL-MaLRs was also identified as a Dux_PAG).
(d) Two examples of DUX4 binding an LTR to induce novel transcription. LTR element = gray box. Track height in reads is given in brackets below each browser shot.
Notably, although DUX4 did not bind nor activate MERV-L elements, DUX4 ChIP-seq peaks were 2.6-fold overrepresented in ERVL-MaLR elements in mouse cells (Supplementary Fig. 8a–b) and in at least 30 cases used them as alternative promoters (Fig. 4c). It is important to note, however, that Dux and DUX4 bound to mostly distinct sets of ERVL-MaLR elements with less than 4% of all the bound ERVL-MaLR sites in common and only 1 shared alternative promoter. In some cases, DUX4 binding to an ERVL-MaLR retroelement caused robust expression of the adjacent gene (Fig. 4d), consistent with our previous finding that DUX4 bound ERVL-MaLRs when expressed in human cells and used them as alternative promoters7. That DUX4 bound and activated transcription of specific endogenous retrotransposon elements in the mouse genome that were not activated by Dux, suggests that homeodomain divergence can selectively activate pre-existing subsets of endogenous retrotransposons and induce the expression of adjacent genes.
The above results indicate that Dux and DUX4 have maintained the ability to regulate a set of 2C-like genes in mouse cells despite considerable divergence of their homeodomains; however, conservation does not extend to the retrotransposons activated by each. We used chimeric proteins to identify the regions of Dux and DUX4 responsible for this partial conservation of function (Fig. 5a). The chimera with the Dux homeodomains and the DUX4 carboxy-terminus (MMH) matched the transcriptional activity of Dux (Fig. 5b–d), indicating that the transcriptional divergence between Dux and DUX4 mapped to the region containing the two homeodomains.
Figure 5. Transcriptional divergence between DUX4 and Dux maps to the two DNA-binding homeodomains.
(a) Cartoons of chimeric proteins; MMH is the two Dux homeodomains and the DUX4 C-terminus; MHM is Dux with HD2 from DUX4; HMM is Dux with HD1 from DUX4.
(b–d) RT-qPCR data for 2C-like genes in mouse muscle cells of various classes, defined below. Data shown are mean of 3 separate cell cultures for each condition with s.e.m. error bars. The experiments in (b) and (d) were also repeated on three separate days and showed consistent results. The experiments in (c) were completed on one occasion.
(b) 2C-like genes with MERV-L promoters
(c) 2C-like genes with conventional promoters that are induced by DUX4 and Dux
(d) 2C-like genes with conventional promoters that are induced only by Dux
(e) Cartoons of reciprocal set of chimeric proteins; HHM is the two DUX4 homeodomains and the Dux C-terminus; HMH is DUX4 with HD2 from Dux; MHH is DUX4 with HD1 from Dux.
(f) RT-qPCR data for DUX4-target genes in human rhabdomyosarcoma cells. Data shown are mean of 3 separate cell cultures for each condition with s.e.m. error bars. These experiments were completed on one occasion.
To determine the relative contribution of each homeodomain, we introduced each human homeodomain individually into Dux to create the MHM and HMM chimeras (Fig. 5a). Neither MHM nor HMM activated transcription of MERV-L-promoted genes (Fig. 5b); whereas for 2C-like genes with conventional promoters, the individual DUX4 homeodomains showed different capacities to substitute for the corresponding Dux homeodomain, with MHM consistently showing stronger activation of the target genes compared to HMM (Fig. 5c–d). We confirmed MHM and HMM expression and stability using a reporter assay (Supplementary Fig. 9a). We also performed reciprocal experiments in human cells and again observed the second homeodomains were more equivalent than the first homeodomains (Fig. 5e–f), indicating that the similarity of the second homeodomain was important to maintain the functional conservation of the 2C-like gene signature at conventional promoters.
To further explore the evolutionary conservation of the DUX4-family to activate an early embryo gene signature, we assessed the canine DUXC gene. Both Dux and DUX4 are retroposed copies of an ancestral DUXC mRNA and neither mice nor humans have retained DUXC9–11 (Fig. 1c). When expressed in mouse muscle cells, canine DUXC did not activate MERV-L-promoted genes (Fig. 5b), but did activate transcription of 2C-like genes with conventional promoters (Fig. 5c–d), again indicating that the ancestral DUX4-like gene activated genes characteristic of early cleavage-stage embryos that was independent of retrotransposon-promoted genes.
Our current study shows that Dux and DUX4 activate genes associated with an early 2C-like program when expressed in muscle cells, consistent with a recent study showing Dux and DUX4 regulate the 2C-like program in early embryos23. Despite the divergence of their homeodomains and binding sequences, these factors have maintained the ability to activate the 2C-like gene signature within their own species, but diverged in their ability to activate subsets of retrotransposons, suggesting evolutionary pressure to maintain activation of endogenous genes and a subset of beneficial retrotransposon driven genes, but diverge away from the activation of retrotransposons driving deleterious genes. Genes regulated by all DUX4-family factors likely represent the core ancestral network, while retrotransposon-promoted genes likely contribute species-specific additions. Such comparisons are particularly relevant to FSHD where it remains unclear how to model this disease in non-primate animals. The fact that both DUX4 and Dux expression leads to apoptosis in mouse muscle cells supported the use of DUX4 in mice as a model of FSHD8,24. The cellular toxicity exhibited by cross-species expression might be due to the few classes of genes robustly activated, such as members of the PRAME family, the aggregate action of the larger number of genes moderately activated, such as the 2C/cleavage-stage signature, or the fact that each factor activates classes of retrotransposons and repetitive elements, albeit different classes in different species. Nonetheless, because the pathophysiologic mechanisms of FSHD remain poorly understood, our study suggests that homeodomain divergence might require using Dux to best reproduce the FSHD transcriptional program in murine models of FSHD, although therapies targeting DUX4 RNA or protein would necessarily rely on expression of DUX4. Our study also provides a model for studying genome evolution especially in regards to the critical balance between conservation of a key transcriptional program with the innovation driven by binding to mobile retrotransposon promoters.
Data Availability
The data generated in this publication have been deposited in NCBI’s Gene Expression Omnibus25 and are accessible through GEO series accession number GSE87282 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87282). The RNA-seq for human DUX4 in human myoblasts was previously published and has GEO series accession number GSE85461 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE85461). The ChIP-seq for human DUX4 in human myoblasts was previously published and has GEO series accession number GSE33838 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33838).
Online Methods
General Statistical Methods
Standard statistical tests were used and described for each individual application. Three separate cell cultures for each condition were used for RNA-seq and RT-qPCR as indicated. The ChIP-seq studies were multiple singleton experiments with several antibodies that would IP the same binding domain, as described. No statistical methods were used to predetermine sample size.
Whole genome RNA-sequencing (RNA-seq)
C2C12, mouse myoblasts (ATCC® CRL-1772™), were grown in DMEM (Gibco/Life Technologies) supplemented with 10% fetal bovine serum (Thermo Scientific) and 1% penicillin/streptomycin (Life Technologies). These were obtained from ATCC and passaged without losing the ability to differentiate into myotubes but have not routinely been checked for mycoplasma. We cloned Dux transgene into the pCW57.1 lentiviral vector, a gift from David Root (Addgene plasmid #41393), which has a doxycycline-inducible promoter. Dux and DUX4 transgenes were codon-altered to decrease overall CpG content because this was shown to enhance transgene expression of the inducible DUX4 vector1. To create monoclonal cell lines, we first transduced pCW57.1-Dux into 293T cells (ATCC® CRL-3216™), along with the packaging and envelope plasmids pMD2.G and psPAX2 using lipofectamine 2000 reagent (ThermoFisher). Viral-like-particles containing pCW57.1-DUX4 were a gift from Sean Shadle and were prepared in a similar manner. C2C12 were plated at low density and transduced with lentivirus at a low multiplicity of infection (MOI < 1) in the presence of polybrene. Cells were selected and maintained in 2.6ug/ml puromycin. Individual clones were isolated using cloning cylinders about 7 days after transfection and chosen for analysis based on robust transgene expression following 2ug/ml doxycycline treatment for 36 hours (DUX4, Dux) or 18 hours (MMH).
Three separate cell cultures were prepared for each condition and total RNA was extracted from whole cells using NucleoSpin RNA kit (Macherey-Nagel) following the manufacturer’s instructions. Total RNA integrity was checked using an Agilent 2200 TapeStation (Agilent Technologies, Inc., Santa Clara, CA) and quantified using a Trinean DropSense96 spectrophotometer (Caliper Life Sciences, Hopkinton, MA). RNA-seq libraries were prepared from total RNA using the TruSeq RNA Sample Prep v2 Kit (Illumina, Inc., San Diego, CA, USA) and a Sciclone NGSx Workstation (PerkinElmer, Waltham, MA, USA). Library size distributions were validated using an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA). Additional library quality control, blending of pooled indexed libraries, and cluster optimization were performed using Life Technologies’ Invitrogen Qubit® 2.0 Fluorometer (Life Technologies-Invitrogen, Carlsbad, CA, USA). RNA-seq libraries were pooled (14-plex) and clustered onto two flow cell lanes. Sequencing was performed using an Illumina HiSeq 2500 in “rapid run” mode employing a single-read, 100 base read length (SR100) sequencing strategy. Image analysis and base calling was performed using Illumina’s Real Time Analysis v1.18 software, followed by ‘demultiplexing’ of indexed reads and generation of FASTQ files, using Illumina’s bcl2fastq Conversion Software v1.8.4.
RNA-seq Data Analysis
Reads of low quality were filtered prior to alignment to the reference genome (mm10 assembly) using R (development version 3.4.0) and Bioconductor (3.3.0) to call TopHat2 (version 2.1.0) and Bowtie2 (version 2.2.3). Reads were allowed to map up to 20 locations. Reads overlapping UCSC known genes were counted using the summarizeOverlaps function of the GenomicAlignments package and differential gene expression was determined using DESeq2, which calculated P-values using the Wald test and adjusted P-values for multiple testing using the procedure of Benjamini and Hochberg. DESeq2 estimates variance for each gene using the average expression level across all samples3. Gene Set Enrichment Analysis (GSEA) was performed using the GSEApreranked module of the Broad Institute’s GenePattern4 algorithm. Specifically, we used 1,000 gene list permutations to determine P-value and the classic scoring scheme5. As we only compared to one gene set (from Akiyama et al.6), we did not correct for multiple tests. For GSEA plot interpretation, see Figure 1b legend. For negative control, see Supplementary Figure 1. Gene Ontology analysis (GO) analysis was done using Gene List Analysis tool of the PANTHER Classification System7 (version: 10.0), which calculated P-values using the binomial statistic as described in the PANTHER User Manual. Repeat element analysis was accomplished using an in-house R package named rmskStats (version 0.99.0). It counts reads that fall completely within the RepeatMasker-annotated repeat elements. To account for reads that align to multiple repetitive genome positions, rmskStats adjusts the count of a read to the fraction of the number of reported alignments (the NH column). For example, a read that maps to 5 locations counts as 0.2 read at each location. Using these count results along with the statistical significance calculated by DESeq2, rmskStats then applies hypergeometric tests to infer the enrichment or depletion of families and classes of repeat elements. Reads that support repeats used as alternative promoters or alternative first exons were identified and activation scores were calculated as described previously8, with one difference: we retained reads that linked ChIP-seq peaks to annotated exons regardless of whether they spliced across an intron or not.
Chromatin Immunoprecipitation Coupled to Whole Genome Sequencing (ChIP-seq)
All ChIP-seq experiments were performed using the monoclonal cell lines described in the RNA-seq section above using a doxycycline-inducible system and codon-altered transgenes. To determine DUX4 binding sites in the mouse genome, we compared the DUX4-expresssing cells at 24-hours of induction immunoprecipitated with a 50:50 mixture of the DUX4 antibodies MO488 and MO489 (previously described in Geng et al.9) to DUX4-expressing cells immunoprecipitated with an antibody to an HA-tag, which was not present in these cells. We performed ChIP-seq for Dux using two complementary approaches. First, we immunoprecipitated Dux from Dux-expressing cells at 24-hours of induction using two commercially available Dux antibodies on a Doxycycline-inducible C2C12 clonal cell line prepared as described for RNA-seq (A-19, catalogue number: sc-385089 and S-20, catalogue number: sc-385090, Santa Cruz Biotechnology) and compared to a mock pull-down using an antibody to IgG. Second, we created a monoclonal population of cells with the doxycycline inducible vector expressing a chimeric protein that fused the codon-altered Dux homeodomains with the codon-altered DUX4 carboxy-terminus (MMH). The MMH-chimera maintained the DNA binding domain of Dux and the carboxy-terminal epitopes of DUX4, permitting us to use the same DUX4 antisera to immunoprecipitate the MMH-chimera and DUX4 (Supplementary Fig. 10a). MMH immunoprecipitation was done at 18-hours of induction. We confirmed that the MMH-chimera retained the Dux DNA-binding specificity by comparing the ChIP-seq peaks of the MMH-chimera to those of Dux. Although the Dux antibodies had a lower signal-to-noise ratio, and thus identified fewer peaks, the vast majority of the peaks identified by the Dux-antibody were a subset of the MMH-chimera-identified peaks (Supplementary Fig. 10b). ChIP-seq with one Dux antibody, A-19, found 2,400 peaks, 97.5% of these peaks overlap a peak in the MMH-chimera dataset (51,356 peaks). Similarly, ChIP-seq with a second Dux antibody, S-20, found 628 peaks, 96.7% of these peaks overlap with a peak in the MMH-chimera dataset. Furthermore, the MEME motif prediction algorithm predicted nearly identical motifs from A-19 peaks and MMH-chimera peaks (Supplementary Fig. 10c) and there is a Pearson coefficient of 0.7847 between the MMH-chimera and Dux transcriptomes (Supplementary Fig. 10d). We therefore used the ChIP-seq dataset from the MMH-chimera for all the analyses described in the main text because of the superior signal-to-noise compared to the commercially available antisera to Dux.
Cross-linked ChIP was performed similar to previous reports for other transcription factors10,11. Briefly, ~108 cells were fixed in 1% formaldehyde for 11 minutes, quenched with glycine, lysed, and then sonicated to generate final DNA fragments of 150–600 bp. The soluble chromatin was diluted 1:10 and pre-cleared with protein A:G beads for 2 hours. Remaining chromatin was incubated with primary antibody overnight, then protein A:G beads were added for an additional 2 hours. Beads were washed and then de-crosslinked overnight.
ChIP samples for Dux and DUX4-expressing cells were validated by RT-qPCR and then prepared for sequencing per the Nugen Ovation Ultralow library system protocol with direct read barcodes. ChIP-seq libraries were prepared from immunoprecipitated samples using an Ovation Ultralow Library System kit (NuGEN Technologies., San Carlos, CA, USA). Library size distributions were validated using an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA). Additional library quality control, blending of pooled indexed libraries, and cluster optimization were performed using Life Technologies’ Invitrogen Qubit® 2.0 Fluorometer (Life Technologies-Invitrogen, Carlsbad, CA, USA). ChIP-seq libraries were pooled (12-plex) and clustered onto two flow cell lanes. Sequencing was performed using an Illumina HiSeq 2500 in Rapid Run Mode employing a single-read, 100-base read length (SR100) sequencing strategy. DUX4 ChIP-seq was performed at a separate time from Dux.
ChIP samples for MMH-expressing cells were validated by RT-qPCR and then prepared for sequencing per the NEBnext DNA Library Prep Kit (NEB, E7370L). Adapter ligated DNA was then size selected and purified using AMPure XP beads (Beckman Coulter, A63881). Libraries were quantified, pooled, and sequenced on an Illumina HiSeq 2500 instrument in 125bp, paired-end mode.
ChIP-seq Data Analysis
Image analysis and base calling were performed using Illumina’s Real Time Analysis v1.18 software, followed by ‘demultiplexing’ of indexed reads and generation of FASTQ files, using Illumina’s bcl2fastq Conversion Software v1.8.4. Using R (development version 3.4.0) and Bioconductor (3.3.0), low quality reads that contained one or more N’s in the sequence were filtered out, and the tails were trimmed once 2 to 5 nucleotides had quality encoding less than 4 (phred score 20). Further filtering included eliminating reads with less than 36 nucleotides. The retained reads were then aligned to mm10 using Rsamtools, ShortRead and Rsubread, the Bioconductor version of Subread aligner12. Peak calling was done with MACS213 (macs2 2.1.0.20151222) using DNA from mock pull-down samples as described above for negative controls, only peaks with q-value < 0.01 were considered. MACS2 calculated q-values from p-values using the Benjamini-Hochberg procedure. De novo motif prediction was done with MEME-ChIP 4.11.214–16, based on the top 600 peaks identified by MACS, ranked by q-value, under the expectation of zero or one motif occurrence per sequence and requiring motifs to be between 5–15 nucleotides in length. The Find Individual Motif Occurences (FIMO) component of the MEME-ChIP suite was used to identify good matches to the top predicted motif for DUX4 and Dux genome-wide.
Transient transfection and RT-qPCR
Murine myoblasts (C2C12) cells were cultured according to the description in the RNA-seq section. Human rhabdomyosarcoma cells (RD) were obtained from the American Type Culture Collection (ATCC) and passaged without losing the ability to differentiate into myotubes, but have not been checked for mycoplasma routinely. RD cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% bovine calf serum and 1% penicillin-streptomycin (Gibco). Transient DNA transfections of C2C12 and RD cells were performed using SuperFect (QIAGEN) according to manufacturer specifications. Briefly, 80,000 C2C12 cells were seeded per well of a 6-well plate the day prior to transfection, 2ug DNA/well and 13.4ul SuperFect/well and 250,000 RD cells were seeded per well of a 6-well plate the day prior to transfection, 4ug DNA/well and 8ul SuperFect/well. 24-hours post-transfection, total RNA was extracted from whole cells using NucleoSpin RNA kit (Macherey-Nagel) following the manufacturer’s instructions. One microgram of total RNA was digested with DNAseI (Invitrogen) and then reverse transcribed into first strand cDNA in a 20 uL reaction using SuperScript III (Invitrogen) and oligo(dT) (Invitrogen). cDNA was diluted and used for RT-qPCR with iTaq Universal SYBR Green Supermix (Bio-Rad). Primer efficiency was determined by standard curve and all primer sets used were >90% efficient. Relative expression levels were normalized to the endogenous control locus Timm17b in mouse cells/GAPDH in human cells and empty vector by DeltaDeltaCT. The primers used in this study are listed in Supplementary Table 12.
Transient transfection and dual luciferase assay
Transient DNA transfections of C2C12 cells were performed using SuperFect (QIAGEN) according to manufacturer specifications. Briefly, 16,000 cells were seeded per well of a 24-well plate the day prior to transfection, 1020ng total DNA/well and 5μl SuperFect/well. Cells were co-transfected with a pCS2 expression vector carrying the affector construct indicated (500ng/well), a pCS2 expression vector carrying renilla luciferase (20ng/well) and a pGL3-basic reporter vector (500ng/well) carrying test promoter fragment upstream of the firefly luciferase gene. Cells were lysed 24 hours post-transfection in Passive Lysis Buffer (Promega). Luciferase activities were quantified using reagents from the Dual-Luciferase Reporter Assay System (Promega) following manufacturer’s instructions. Light emission was measured using BioTek Synergy2 luminometer. Luciferase data are given as mean fold change over empty vector ± s.e.m of three separate cell cultures for each condition.
Code Availability
Code that supports the findings of this study are available from the Tapscott Lab’s GitHub (https://github.com/TapscottLab/Dux4FamilyGeneNetwork and https://github.com/TapscottLab/rmskStats).
URLs
bcl2fastq Conversion Software v1.8.4, http://support.illumina.com/downloads/bcl2fastq_conversion_software_184.html; PANTHER User Manual, http://pantherdb.org/help/PANTHER_user_manual.pdf.
Supplementary Material
Acknowledgments
We thank S. Shadle at the Fred Hutchinson Cancer Research Center for making and sharing the pCW57.1-DUX4 virus-like particles used to generate the DUX4-inducible C2C12 clonal cell lines. We thank P. Hendrickson at the Huntsman Cancer Institute for 2C::TdTomato Reporter (Addgene Plasmid #40281), from which we cloned the MERV-L used to create the MERV-L reporter in this study. We thank G. Kardon for critical guidance in the project. The authors would also like to thank members of the research groups of S. Tapscott and H. Malik for helpful discussions and critical reading of the manuscript. This work was supported by: NIH NIAMS R01AR045203 (S.J.T), National Science Foundation Graduate Research Fellowship Program DGE-1256082 (J.L.W), University of Washington Interdisciplinary Training in Genome Sciences T32 HG00035 from the National Human Genome Research Institute (J.L.W), and Friends of FSH Research (J.L.W, S.J.T).
Footnotes
Author Contributions
J.L.W. and S.J.T. conceived and designed the study and wrote the manuscript with the assistance and final approval of all authors. J.L.W. performed and interpreted RNA-seq, ChIP-seq, RT-qPCR and luciferase experiments. A.T.L. performed RT-qPCR and luciferase experiments. CJ.W. performed bioinformatics data analyses. J.W.Z. contributed to the creation and characterization of the MMH-chimera cell line.
Competing Financial Interests
Jennifer Whiddon and Stephen Tapscott have filed a provisional patent application together with Brad Cairns, Edward Grow, Peter Hendrickson based in part on this work (U.S. Provisional Application No. 62/410,078) with the US Patent and Trademark Office titled “Compositions and Methods for Reprogramming Cells and for Somatic Cell Nuclear Transfer Using DUXC Expression”. Authorized by Stephen Tapscott, corresponding author, on behalf of all authors.
References
- 1.Tawil R, van der Maarel SM, Tapscott SJ. Facioscapulohumeral dystrophy: the path to consensus on pathophysiology. Skelet Muscle. 2014;4:12. doi: 10.1186/2044-5040-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lek A, Rahimov F, Jones PL, Kunkel LM. Emerging preclinical animal models for FSHD. Trends Mol Med. 2015;21:295–306. doi: 10.1016/j.molmed.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wallace LM, et al. DUX4, a candidate gene for facioscapulohumeral muscular dystrophy, causes p53-dependent myopathy in vivo. Ann Neurol. 2011;69:540–52. doi: 10.1002/ana.22275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krom YD, et al. Intrinsic epigenetic regulation of the D4Z4 macrosatellite repeat in a transgenic mouse model for FSHD. PLoS Genet. 2013;9:e1003415. doi: 10.1371/journal.pgen.1003415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dandapat A, et al. Dominant lethal pathologies in male mice engineered to contain an X-linked DUX4 transgene. Cell Rep. 2014;8:1484–96. doi: 10.1016/j.celrep.2014.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geng LN, et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young JM, et al. DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet. 2013;9:e1003947. doi: 10.1371/journal.pgen.1003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosnakovski D, Daughters RS, Xu Z, Slack JM, Kyba M. Biphasic myopathic phenotype of mouse DUX, an ORF within conserved FSHD-related repeats. PLoS One. 2009;4:e7003. doi: 10.1371/journal.pone.0007003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leidenroth A, et al. Evolution of DUX gene macrosatellites in placental mammals. Chromosoma. 2012;121:489–97. doi: 10.1007/s00412-012-0380-y. [DOI] [PubMed] [Google Scholar]
- 10.Leidenroth A, Hewitt JE. A family history of DUX4: phylogenetic analysis of DUXA, B, C and Duxbl reveals the ancestral DUX gene. BMC Evol Biol. 2010;10:364. doi: 10.1186/1471-2148-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clapp J, et al. Evolutionary conservation of a coding function for D4Z4, the tandem DNA repeat mutated in facioscapulohumeral muscular dystrophy. Am J Hum Genet. 2007;81:264–79. doi: 10.1086/519311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eidahl JO, et al. Mouse Dux is myotoxic and shares partial functional homology with its human paralog DUX4. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knopp P, et al. DUX4 induces a transcriptome more characteristic of a less-differentiated cell state and inhibits myogenesis. J Cell Sci. 2016;129:3816–3831. doi: 10.1242/jcs.180372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falco G, et al. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–50. doi: 10.1016/j.ydbio.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang W, et al. Zfp206 regulates ES cell gene expression and differentiation. Nucleic Acids Res. 2006;34:4780–90. doi: 10.1093/nar/gkl631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macfarlan TS, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akiyama T, et al. Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 2015;22:307–18. doi: 10.1093/dnares/dsv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jagannathan S, et al. Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coordinators NR. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44:D7–19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noyes MB, et al. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–89. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peaston AE, et al. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 23.Hendrickson PGD, JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, Nix DA, Peterson CM, Tapscott SJ, Carrell DT, Cairns BR. Conserved roles for murine DUX and human DUX4 in activating cleavage stage genes and MERVL/HERVL retrotransposons. Nature Genetics, under consideration. please see companion file.
- 24.Bosnakovski D, et al. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J. 2008;27:2766–79. doi: 10.1038/emboj.2008.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 1.Jagannathan S, et al. Model systems of DUX4 expression recapitulate the transcriptional profile of FSHD cells. Hum Mol Genet. 2016 doi: 10.1093/hmg/ddw271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 5.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akiyama T, et al. Transient bursts of Zscan4 expression are accompanied by the rapid derepression of heterochromatin in mouse embryonic stem cells. DNA Res. 2015;22:307–18. doi: 10.1093/dnares/dsv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mi H, Poudel S, Muruganujan A, Casagrande JT, Thomas PD. PANTHER version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 2016;44:D336–42. doi: 10.1093/nar/gkv1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young JM, et al. DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet. 2013;9:e1003947. doi: 10.1371/journal.pgen.1003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geng LN, et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conerly ML, Yao Z, Zhong JW, Groudine M, Tapscott SJ. Distinct Activities of Myf5 and MyoD Indicate Separate Roles in Skeletal Muscle Lineage Specification and Differentiation. Dev Cell. 2016;36:375–85. doi: 10.1016/j.devcel.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao Y, et al. Genome-wide MyoD binding in skeletal muscle cells: a potential for broad cellular reprogramming. Dev Cell. 2010;18:662–74. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma W, Noble WS, Bailey TL. Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat Protoc. 2014;9:1428–50. doi: 10.1038/nprot.2014.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol. 1994;2:28–36. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this publication have been deposited in NCBI’s Gene Expression Omnibus25 and are accessible through GEO series accession number GSE87282 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE87282). The RNA-seq for human DUX4 in human myoblasts was previously published and has GEO series accession number GSE85461 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE85461). The ChIP-seq for human DUX4 in human myoblasts was previously published and has GEO series accession number GSE33838 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33838).