Abstract
Background: MiR-101-3p has been reported to suppress invasion and metastasis in hepatocellular carcinoma (HCC) cells. However, the relevant mechanisms are still unclear. The research seeks to determine systematic value of miR-101-3p in HCC, and comprehensively summarize the predicted target genes as well as their potential function, pathways and networks in HCC. Methods: The miR-101-1 profiles in 353 HCC patients from The Cancer Genome Atlas (TCGA) were analyzed. Meta-analysis was performed to estimate relationship of miR-101 (including precursor and mature miR-101) with clinical features and prognosis in HCC. Further, the promising targets of miR-101-3p were predicted and followed with Gene Ontology (GO), pathway and network analysis. In addition, the functional impact of miR-101-3p was confirmed with in vitro experiments in HCC cells. Results: In TCGA data, low-expression of miR-101-1 might be a diagnostic (AUC: 0.924, 95% CI: 0.894-0.953) and prognostic (HR=1.55) marker for HCC. Down-regulated miR-101-1 also correlated with poor differentiation, advanced TNM stage, lymph node metastasis and high AFP level of HCC. Meta-analysis revealed that miR-101 down-regulation were associated with poor prognosis, high AFP level and advanced TNM stage of HCC. Moreover, 343 hub genes were filtered and miR-101-3p may be involved in intracellular signaling cascade, transcription, metabolism and cell proliferation. Focal adhesion and pathways in cancer were also significantly enriched. In vitro experiments demonstrated that miR-101-3p inhibited proliferation and promoted apoptosis in HCC cells. Conclusions: MiR-101-1 may be a prospective biomarker for diagnosis and prognosis of HCC. Potential targets of miR-101-3p could regulate genesis and development of HCC. The data offers insights into biological significances and promising targets of miR-101-3p for further investigation and potential therapies in HCC.
Keywords: Hepatocellular carcinoma, miR-101-3p, targets, bioinformatics, mechanisms
Introduction
Hepatocellular carcinoma (HCC), the fifth leading carcinoma at diagnosis, was ranked second death cause in male and sixth in female with increasing incidence [1]. Recently, targeted drugs of oncogenic signaling pathways or associated genes have afforded new insights into the treatment of HCC [2]. Despite the advances in diagnosis and treatment, the optimal therapeutic approaches are still limited [3] and the prognosis of HCC remains dismal due to late diagnosis [4]. In order to increase the survival rates of HCC, it is sorely required to search for novel therapeutic targets.
MicroRNAs (miRNAs) are highly conserved small RNAs, which regulate transcription or post-transcription through binding to targeted mRNAs [5]. Extensive researches have demonstrated that miRNAs were involved in various biological processes [6,7]. Recently, accumulating evidences have also suggested that aberrant level of miRNAs was related to proliferation, angiogenesis and metastasis in various human malignancies [8]. Moreover, miRNAs may function as oncogenes or cancer suppressor genes by up or down regulating and influencing different biological functions of target genes [9].
A growing number of studies analyzed the expression of microRNA-101-3p (miR-101-3p, previous name miR-101) and its targeted genes in HCC, and miR-101-3p (the mature miRNA) can be generated from miR-101-1 and miR-101-2 (the precursor miRNA) [8,10]. Researches indicated that miR-101-3p was down-regulated as well as associated with poor prognosis in HCC [10,11], and miR-101-3p noticeably promoted apoptosis and suppressed tumorigenesis, tumor cell migration, invasion and metastasis in HCC cells [12,13]. However, previous researches into levels of miR-101-3p were inconsistent; thus a meta-analysis is required to fully explore the clinical significance of miR-101-3p. As far as we know, some validated targets of miR-101-3p have been documented such as enhancer of zeste homolog 2 (EZH2) [14], RAB GTPase 5A (RAB5A) [15], and dual specificity phosphatase 1 (DUSP1) [16]. However, one single miRNA has hundreds of target genes and the precise molecular mechanisms of miR-101-3p in neoplasia were still not fully understood. Comprehensive analysis of the target genes networks may help deeply understand the effect of miR-101-3p. Therefore, comprehensive and systemic analyses of the clinical value and target proteins of miR-101-3p are necessary to provide novel targets in HCC diagnosis and treatment.
In this study, datasets of miR-101-1 in HCC patients were gained from The Cancer Genome Atlas (TCGA), which included 353 tumors and 47 adjacent non-tumor liver tissues. Integrated meta-analysis quantitatively assessed the role of miR-101 (including precursor and mature miR-101) in HCC. Furthermore, bioinformatics analysis was carried out to predict targets of miR-101-3p and their potential roles by pathway. Gene Ontology (GO) and network analyses were further performed. Finally, the role of miR-101-3p on HepG2 cells was also confirmed with in vitro experiments. The current results explored the comprehensive roles and prospective molecular mechanisms of miR-101-3p, and might provide a potential marker for HCC therapeutic strategy.
Materials and methods
Patients in TCGA database
RNA sequence data were obtained from the TCGA dataset (https://cancergenome.nih.gov/), including 353 HCC patients and 47 adjacent non-cancer liver tissues. In HCC, miR-101-1 level was analyzed, together with its relationship with survival data and other clinical parameters.
Meta-analysis of miR-101
The miR-101 expression levels in HCC patients were obtained from published studies (miR-101-3p) and TCGA database (miR-101-1) up to 30th September, 2016. General information with clinical parameters and survival data was extracted from eligible studies. OR and HR with 95% CI were pooled to evaluate the connection of miR-101 (including precursor and mature miR-101) with HCC quantitatively. Significant heterogeneity was defined when inconsistency index (I2)>50% or P<0.10. Sensitivity analysis and potential publication bias were also assessed. Stata version 12.0 was applied to process all the data.
Prediction of miR-101-3p target genes
Putative genes of dysregulated miR-101-3p were predicted in 13 prediction databases including DIANAmT, mirTarBase, RNA22, miRanda, PICTAR, miRDB, miRWalk, PolymiRTS, PITA, RNAhybrid, Targetscan, Targetminer and TarBase [17]. The target genes which repeated in more than 8 out of 13 programs were retained. Furthermore, validated targets of miR-101-3p were searched in TarBase, mirTarBase and published studies with strong evidences including luciferase reporter assay, WB and RT-PCR.
GO, KEGG and network analysis
To clarify biological functions of the selected target genes, GO enrichment analysis and KEGG analysis were shown by DAVID (https://david.ncifcrf.gov, version 6.7) [17]. The potential functional networks of the selected genes were constructed and visualized with Cytoscape 3.3.0 [18].
Cell proliferation, cycle and apoptosis assays
MiR-101-3p mimics or inhibitor and negative control oligoes (GenePharma, China) with Lipofectamine 2000 (Invitrogen, USA) transfected HepG2 cells. Transfected cells were incubated for 0, 24, 48, 72 and 96 hours individually in 96-well plates. The viability of each group was evaluated by MTT (Beyotime, China) assay [19] and measured absorbance at 490 nm. Cell cycle detection kit was used to detect cell cycle and apoptosis. After 48-hour transfection, cells were collected and processed according to the instruction manual. Then the cell cycle and apoptosis results were obtained from flow cytometry (BECKMAN-COULTER EPICS XL, USA) at 488 nm. The results were assessed by the PI staining technique as previously described [20,21].
Statistical analysis
SPSS20.0 was used to analyze data, which were shown with mean ± standard deviation (SD). The statistical significance between two groups was assessed by ANOVA, and multiple comparisons between groups were analyzed by least significant difference test (LSD-t). Spearman’s rank correlation was employed to analyze the relationship between miR-101-1 and clinical features, and survival data were processed by Kaplan-Meier method. Each experiment was performed in three replications and P<0.05 revealed statistical significance.
Results
Clinical significance of miR-101-1 in HCC with TCGA data
MiR-101-1 level was significantly lower in HCC compared with non-tumor group (Figure 1A). Receiver operating characteristic (ROC) curve was performed to evaluate whether miR-101-1 can be used as a diagnostic marker for HCC. In the cases of HCC compared with non-cancer liver tissues, the area under the curve (AUC) was 0.924 (95% CI: 0.894-0.953) with 89.4% sensitivity and 82.7% specificity (Figure 1B), indicating that low expression level of miR-101-1 may be an ideal marker for HCC diagnosis. In the analysis of miR-101-1 with clinical parameters, low-expression of miR-101-1 was significantly associated with high histological grade, high clinical stage, poor tumor stage, positive lymph node metastasis and high alpha fetal protein (AFP) level of HCC (Table 1). However, no significant differences were noted between miR-101-1 and age, gender, historical risk factor, metastasis stage or liver fibrosis. Moreover, poor prognosis was observed with low expression of miR-101-1 in HCC (HR: 1.55; 95% CI: 1.09-2.22; P=0.015) (Figure 1C).
Figure 1.
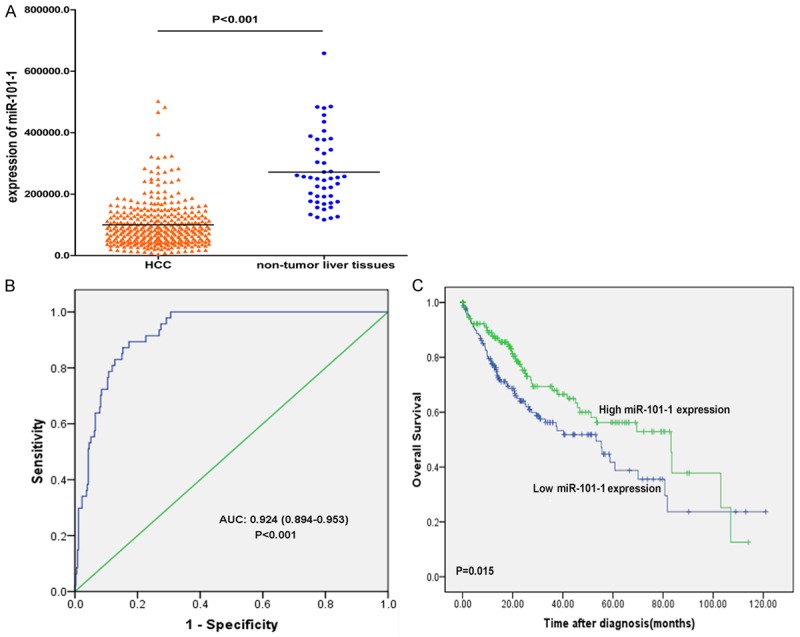
Clinical significance of miR-101-1 in HCC with TCGA data sets. A. MiR-101-1 was obviously downregulated in HCC compared with non-tumor liver tissues; B. ROC curve analysis of miR-101-1 for discriminating HCC from non-tumor liver tissues; C. Kaplan-Meier survival curves showed that lower expression of miR-101-1 with worse survival of HCC patients.
Table 1.
Correlation of miR-101-1 expression with clinicopathological variables in HCC of TCGA data sets
| Clinicopathological features | Cases | MiR-101-1 expression | t | P | ||
|---|---|---|---|---|---|---|
|
| ||||||
| Low | High | |||||
| Age | ≥50 | 287 | 239 | 48 | 1.857 | 0.064 |
| <50 | 66 | 53 | 13 | |||
| Gender | Male | 240 | 199 | 41 | -0.723 | 0.470 |
| Female | 113 | 93 | 20 | |||
| Family history | Yes | 104 | 82 | 22 | 1.791 | 0.074 |
| No | 201 | 170 | 31 | |||
| Historical risk factor | Yes | 242 | 195 | 47 | 1.463 | 0.144 |
| No | 82 | 73 | 9 | |||
| Grade | 3-4 | 129 | 119 | 10 | -3.264 | 0.001 |
| 1-2 | 220 | 170 | 50 | |||
| Stage | III-IV | 86 | 74 | 12 | -2.976 | 0.003 |
| I-II | 245 | 199 | 46 | |||
| Tumor stage | T3-T4 | 89 | 77 | 12 | -2.635 | 0.009 |
| T0-T2 | 262 | 214 | 48 | |||
| Lymph node stage | N1 | 3 | 3 | 0 | -2.615 | 0.009 |
| N0 | 241 | 197 | 44 | |||
| Metastasis stage | M1 | 4 | 3 | 1 | -0.432 | 0.666 |
| M0 | 256 | 211 | 45 | |||
| AFP | ≥400 | 63 | 56 | 7 | -3.379 | 0.001 |
| <400 | 205 | 162 | 43 | |||
| Liver fibrosis | Yes | 135 | 110 | 25 | 1.033 | 0.303 |
| No | 71 | 57 | 14 | |||
| Disease status | Recurred/Progressed | 166 | 131 | 35 | -0.094 | 0.925 |
| Disease free | 139 | 118 | 21 | |||
Annotation: AFP, alpha fetal protein; Historical risk factor including hepatitis B, hepatitis C, alcohol consumption and non-alcoholic fatty liver diseases.
The relationship between miR-101 and clinicopathological parameters or prognosis
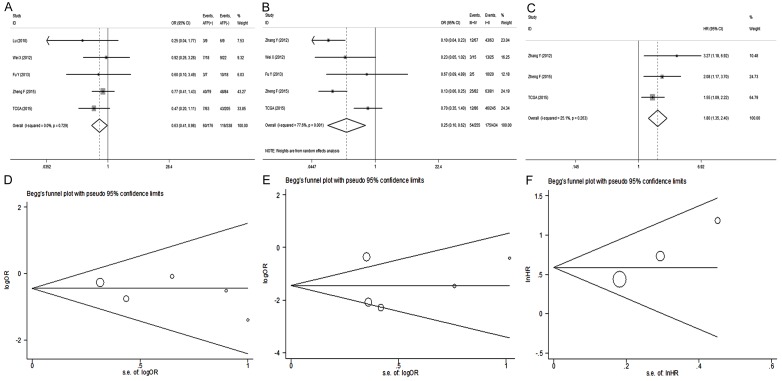
The flow chart of literature selection was shown in Figure 2. Eventually, six papers and TCGA data were selected in this meta-analysis from 2009 to 2015 [11,13,22-25], and general characters were summarized in Table 2. Low expression of miR-101 was significantly associated with the increased level of AFP (OR=0.63; 95% CI: 0.41-0.98; P=0.038) (Figure 3A) and high TNM stage (OR=0.25; 95% CI: 0.10-0.62; P=0.003) (Figure 3B). Other clinicopathological parameters including age, tumor size, grade, HBsAg and cirrhosis were also analyzed, but no statistical significance was observed (Table S1). Furthermore, poor prognosis was observed among HCC patients with down-regulated expression of miR-101 (HR=1.80; 95% CI: 1.35-2.40; P<0.001) without obvious heterogeneity (χ2=2.67; I2=25.1%; P=0.263) (Figure 3C). There was no obvious publication bias estimated with Egger’s and Begg’s test (Figure 3D-F).
Figure 2.
Flow chart of literature selection.
Table 2.
Characteristics of studies and TCGA sets selected for meta-analysis
| First author | Year | Origin of population | Specimen | Case number | Methods | Cut-off | Age | Stage (III) | Histological grade (3) | AFP (+) | HBsAg (+) | Tumor size (>5) | Cirrhosis | Follow-up (months) | Survival analysis | HR statistics | HR (95%CI) (neg) | P |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Li | 2009 | China | Tissue | 20 | qRT-PCR | 2-fold | 47.8 (mean) | N | 10 (3+4) | N | 16 | 12 | 11 | N | N | N | N | N |
| Lu | 2010 | China | Tissue | 18 | qRT-PCR | 0.28 | 47.9±10.3 (mean) | N | 6 | 9 (350) | N | 11 | 7 | N | N | N | N | N |
| Zhang | 2012 | China | Tissue | 130 | qRT-PCR | 0.8 (median) | >50 (44.6%) | 67 (III+IV) | 23 | N | N | N | 86 | 103.2 (median) | OS, DFS | R | 3.27 (1.18-6.92); 2.56 (1.32-5.69) | 0.01; 0.02 |
| Wei | 2012 | China | Tissue | 40 | qRT-PCR | 0.48 | ≥40 (67.5%) | 15 | N | 18 | 25 | 24 | 28 | N | N | N | N | N |
| Fu | 2013 | China | Serum | 25 | qRT-PCR | 10.13 | ≥40 (84.0%) | 5 | N | 7 | 19 | 9 | 18 | N | N | N | N | N |
| Zheng | 2015 | China | Plasma | 163 | qRT-PCR | 2.24 | ≥48.2 (50.9%) | 82 | 40 | 79 (20) | N | 78 | 108 | 63 | OS | R | 2.08 (1.17-3.70) | 0.01 |
| TCGA (miR-101-1) | 2015 | USA | N | 353 | Microarray | 149921 | 59.5 (mean) | 86/331 (III+IV) | 129/349 (3+4) | 63/268 | N | N | N | 9.8 (median) | OS | C | 1.55 (1.09-2.22) | 0.02 |
Annotation: AFP, alpha fetal protein; neg, negative expression of miR-101-3p; N, not given; R, reported; C, calculated; HR, hazard ratio; OS, overall survival; DFS, disease free survival.
Figure 3.
Meta-analysis of miR-101 (including precursor and mature miR-101) expression in HCC patients. A and D. Forest plot and Begg’s funnel plot analysis of miR-101 low expression with AFP level; B and E. Forest plot and Begg’s funnel plot analysis of miR-101 low expression with TNM stage; C and F. Forest plot and Begg’s funnel plot analysis in down-regulated expression of miR-101 with poor prognosis.
Enrichment analysis of candidate targets of miR-101-3p
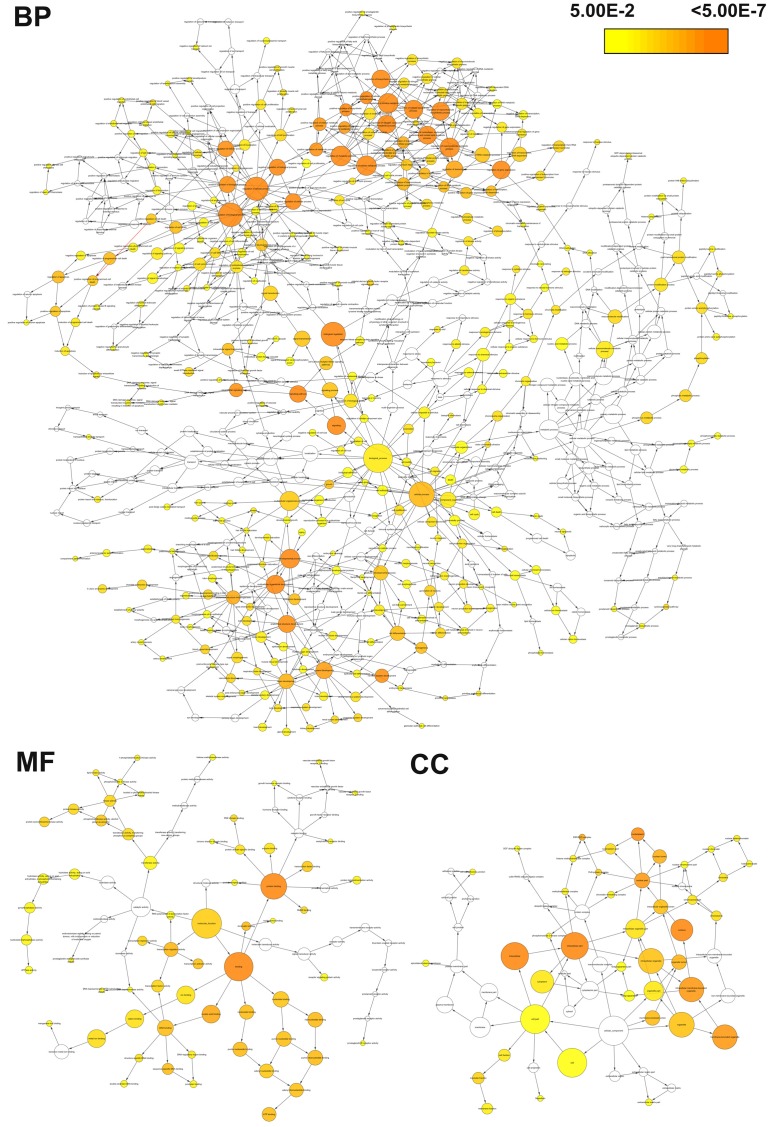
Bioinformatics analysis was performed to gain insight into the functional impact of targeted genes of miR-101-3p. A total of 7924 genes were obtained from predicted databases (Figure 4). Three hundred and forty-three genes were selected including 305 predicted targets with repetition in 8 of 13 programs. Sixty validated genes of miR-101-3p were obtained, among which 16 validated genes were involved in HCC (Table 3). The result of GO analysis was summarized in Table 4 and Figure 5, and the most significantly enriched items for each domain were exhibited according to their p values. The biological process (BP) of correlated genes showed their involvement in intracellular signaling cascade, metabolism, transcription, cell proliferation and biosynthesis processes. The analysis of molecular function (MF) revealed the most correlated functions were chromatin, ATP and transcription factor binding. Moreover, cellular component (CC) was enriched in insoluble fraction and nuclear lumen. Pathways from KEGG were significantly enriched in focal adhesion, colorectal cancer and pathways in cancer (Table 5 and Figure 6). Moreover, MAPK, VEGF and chemokine signaling pathways were also revealed to have statistical significance. The correlation analyses based on the biological significances between targeted genes were further identified in the Cytoscape program and the network relationship was shown in Figure 7.
Figure 4.
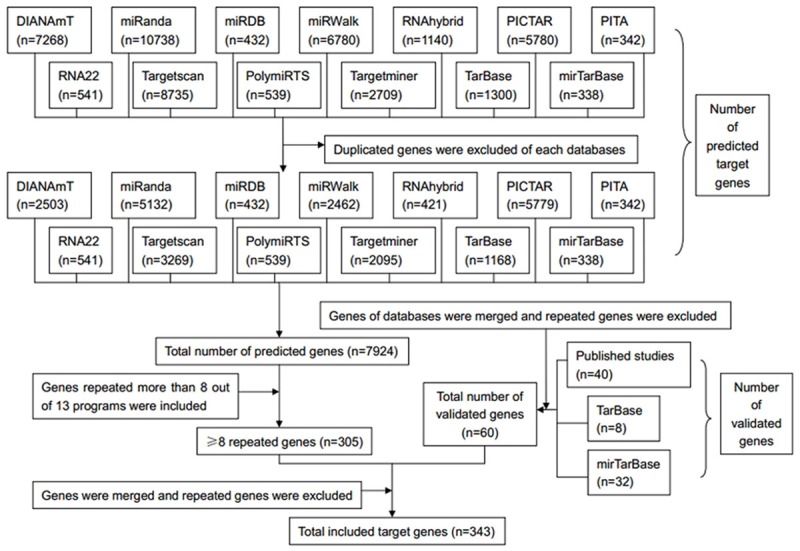
Flow chart of target genes selection.
Table 3.
Validated targets of miR-101-3p in published studies with strong evidences including luciferase reporter assay, western blot and qPCR
| Genes | |||||
|---|---|---|---|---|---|
| All validated | AP1 | DNMT3A | HMGA2 | PIK3CB | RUNX1 |
| ATG4D | DUSP1 | JAK2 | Pim1 | SOCS2 | |
| CDK8 | EED | KLF6 | PRDM16 | SOX9 | |
| c-Met | EP4 | Lin28B | RAB5A | SphK1 | |
| COX2 | EZB1 | MARCH7 | Rac1 | STMN1 | |
| CPEB1 | EZB2 | Mcl-1 | Rap1b | Stmnl | |
| CXCL12 | EZH2 | MITF | RLIP76 | VEGF | |
| CXCR7 | Fos | mTOR | ROCK2 | VEGF-C | |
| Validated in HCC | AP1 | DUSP1 | Mcl-1 | ROCK2 | |
| ATG4D | EED | mTOR | SOX9 | ||
| COX2 | EZH2 | RAB5A | STMN1 | ||
| DNMT3A | Fos | Rap1b | VEGF | ||
Table 4.
GO functional annotation for the most significantly enriched targeted genes of miR-101-3p
| GO ID | GO Term | Count | P Value |
|---|---|---|---|
| Biological process | |||
| GO:0007242 | Intracellular signaling cascade | 54 | 3.27E-07 |
| GO:0006793 | Phosphorus metabolic process | 43 | 3.81E-06 |
| GO:0006796 | Phosphate metabolic process | 43 | 3.81E-06 |
| GO:0045449 | Regulation of transcription | 84 | 1.30E-05 |
| GO:0009891 | Positive regulation of biosynthetic process | 33 | 1.75E-05 |
| GO:0016310 | Phosphorylation | 36 | 2.01E-05 |
| GO:0031328 | Positive regulation of cellular biosynthetic process | 32 | 3.31E-05 |
| GO:0006468 | Protein amino acid phosphorylation | 31 | 4.93E-05 |
| GO:0051173 | Positive regulation of nitrogen compound metabolic process | 30 | 6.58E-05 |
| GO:0010557 | Positive regulation of macromolecule biosynthetic process | 30 | 8.63E-05 |
| GO:0045935 | Positive regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 29 | 9.30E-05 |
| GO:0008283 | Cell proliferation | 23 | 1.04E-04 |
| GO:0045941 | Positive regulation of transcription | 27 | 1.09E-04 |
| GO:0045893 | Positive regulation of transcription, DNA-dependent | 24 | 1.44E-04 |
| GO:0043549 | Regulation of kinase activity | 20 | 1.57E-04 |
| Cellular component | |||
| GO:0005626 | Insoluble fraction | 39 | 5.44E-08 |
| GO:0031981 | Nuclear lumen | 52 | 7.24E-07 |
| GO:0005654 | Nucleoplasm | 37 | 1.57E-06 |
| GO:0005624 | Membrane fraction | 34 | 4.61E-06 |
| GO:0000267 | Cell fraction | 41 | 4.62E-06 |
| GO:0043233 | Organelle lumen | 58 | 5.70E-06 |
| GO:0031974 | Membrane-enclosed lumen | 58 | 1.04E-05 |
| GO:0070013 | Intracellular organelle lumen | 56 | 1.29E-05 |
| GO:0005694 | Chromosome | 22 | 5.92E-05 |
| GO:0044427 | Chromosomal part | 19 | 1.56E-04 |
| Molecular function | |||
| GO:0003682 | Chromatin binding | 15 | 3.44E-06 |
| GO:0005524 | ATP binding | 56 | 1.72E-05 |
| GO:0032555 | Purine ribonucleotide binding | 65 | 2.47E-05 |
| GO:0032553 | Ribonucleotide binding | 65 | 2.47E-05 |
| GO:0032559 | Adenyl ribonucleotide binding | 56 | 2.53E-05 |
| GO:0008134 | Transcription factor binding | 27 | 3.26E-05 |
| GO:0000166 | Nucleotide binding | 75 | 3.28E-05 |
| GO:0017076 | Purine nucleotide binding | 66 | 5.07E-05 |
| GO:0030554 | Adenyl nucleotide binding | 57 | 5.49E-05 |
| GO:0030528 | Transcription regulator activity | 55 | 6.60E-05 |
Annotation: GO, Gene Ontology; Count, number of genes enriched in each GO term.
Figure 5.
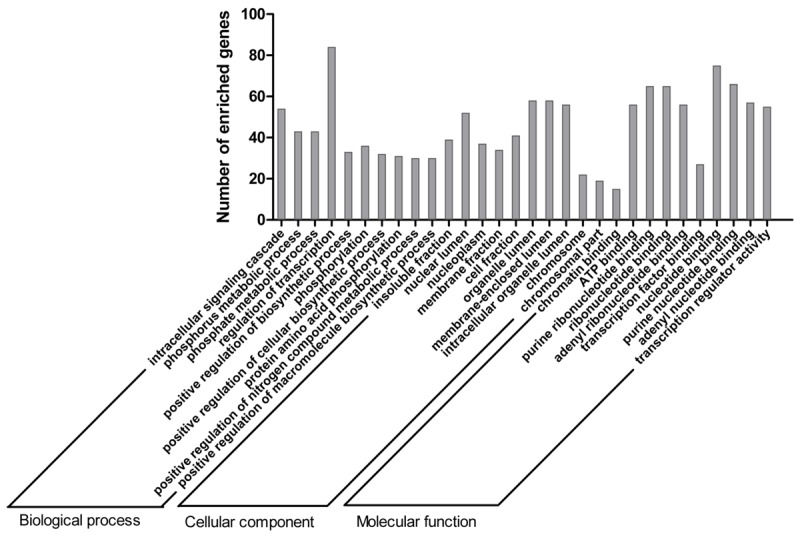
Top 10 GO functional annotation of miR-101-3p targeted genes by DAVID.
Table 5.
KEEG pathways enriched for miR-101-3p targeted genes by DAVID
| KEGG ID | KEGG Term | Count | P Value | Genes |
|---|---|---|---|---|
| hsa04510 | Focal adhesion | 14 | 2.66E-04 | CAV3, PIK3CB, ROCK2, MET, PIP5K1C, PPP1CC, CAPN2, COL5A2, MAPK1, ITGB8, VEGFA, RAC1, PPP1R12A, RAP1B |
| hsa05210 | Colorectal cancer | 9 | 3.42E-04 | MAPK1, FOS, CASP3, PIK3CB, TGFBR1, MET, RAC1, FZD4, FZD6 |
| hsa05200 | Pathways in cancer | 16 | 3.31E-03 | E2F3, PTGS2, PIK3CB, TGFBR1, MITF, MET, FZD4, FZD6, FOS, MAPK1, CASP3, VEGFA, RAC1, HHIP, MTOR, RUNX1 |
| hsa05211 | Renal cell carcinoma | 7 | 3.32E-03 | MAPK1, PIK3CB, MET, VEGFA, GAB1, RAC1, RAP1B |
| hsa04062 | Chemokine signaling pathway | 10 | 1.63E-02 | MAPK1, TIAM2, ROCK2, GNB1, PIK3CB, RAC1, RAP1B, GNG2, JAK2, CXCL12 |
| hsa04930 | Type II diabetes mellitus | 5 | 1.65E-02 | MAPK1, SOCS2, PIK3CB, MTOR, PRKCE |
| hsa05212 | Pancreatic cancer | 6 | 1.72E-02 | MAPK1, E2F3, PIK3CB, TGFBR1, VEGFA, RAC1 |
| hsa04370 | VEGF signaling pathway | 6 | 2.02E-02 | MAPK1, PTGS2, PIK3CB, VEGFA, SPHK1, RAC1 |
| hsa04520 | Adherens junction | 6 | 2.24E-02 | MAPK1, TJP1, NLK, TGFBR1, MET, RAC1 |
| hsa04150 | mTOR signaling pathway | 5 | 2.32E-02 | MAPK1, PIK3CB, VEGFA, PRKAA1, MTOR |
| hsa05221 | Acute myeloid leukemia | 5 | 3.30E-02 | MAPK1, PIK3CB, PIM1, MTOR, RUNX1 |
| hsa04350 | TGF-beta signaling pathway | 6 | 3.56E-02 | MAPK1, ACVR2B, RBL2, ROCK2, TGFBR1, ID4 |
| hsa04310 | Wnt signaling pathway | 8 | 3.90E-02 | VANGL1, ROCK2, PPP2R5A, NLK, RAC1, FBXW11, FZD4, FZD6 |
| hsa04722 | Neurotrophin signaling pathway | 7 | 4.57E-02 | MAPK1, PIK3CB, GAB1, RAC1, SH2B3, RAP1B, KIDINS220 |
| hsa04666 | Fc gamma R-mediated phagocytosis | 6 | 4.90E-02 | MAPK1, PIK3CB, SPHK1, RAC1, PIP5K1C, PRKCE |
Annotation: KEGG, Kyoto Encyclopedia of Genes and Genomes; Count, number of genes enriched in each KEGG term.
Figure 6.
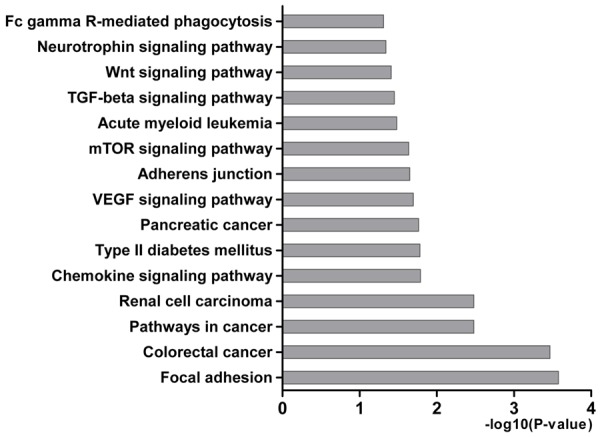
KEEG pathways enriched for miR-101-3p targeted genes by DAVID. The statistical significance level (P-value) was negative 10-based log transformed.
Figure 7.
The constructed networks of miR-101-3p targeted genes by Cytoscape, including biological process (BP), cellular component (CC) and molecular function (MF) networks. Node’s color and size represents the significance of interactions.
MiR-101-3p promoted apoptosis and inhibited proliferation in HepG2 cells
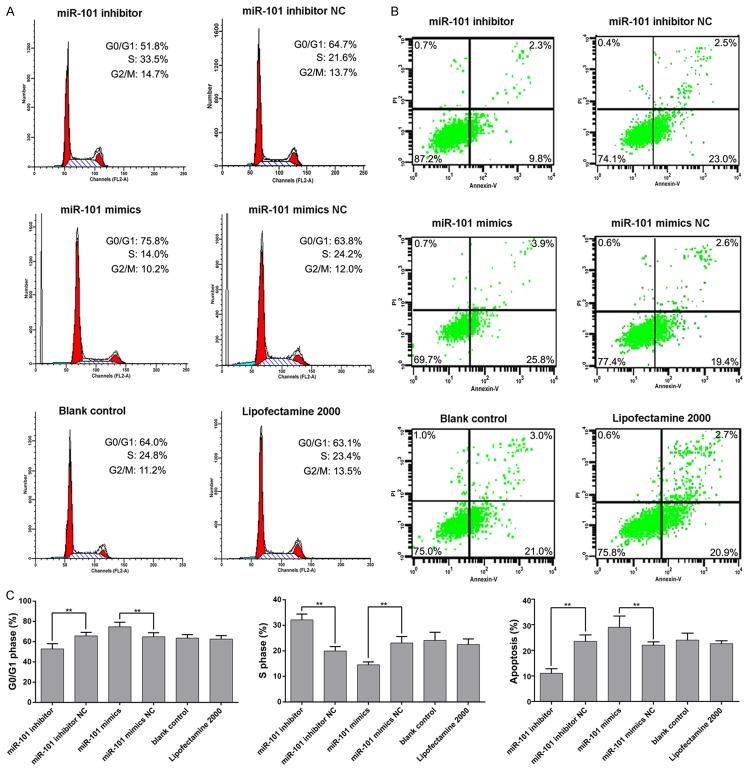
The enforced expression of miR-101-3p significantly inhibited proliferation in mimic transfection group. Obvious proliferation was noted in miR-101-3p inhibitor transfection group compared with control groups. Cells were arrested and on the rise in G0/G1 phase in miR-101-3p mimic group compared with control group, while in inhibitor group, cells increased in S phase with obvious statistical significance (P<0.05, Figure 8A). In cell apoptosis analysis, apoptosis rates rose with mimic contrast with the mock-treated cells especially in the early stage of apoptosis, and opposite result was obtained with inhibitor transfection (P<0.05, Figure 8B).
Figure 8.
Cycle and apoptosis were effected by miR-101-3p in HepG2 cells. A, C. MiR-101-3p mimics and inhibitor effect on cell cycle, cell cycle was arrested in S phase with the effect of miR-101-3p inhibitor and in G0/G1 phase in miR-101-3p mimics transfected group; B, C. MiR-101-3p mimics and inhibitor effect on cell apoptosis, miR-101-3p overexpressed induced apoptosis and opposite result with miR-101-3p inhibitor transfection. Annotation: NC, negative control.
Discussion
Recently, miR-101-3p has been reported to play important roles in tumorigenesis. Decreased miR-101-3p expression was observed in a number of tumors, such as lung, colon, liver, gastric, breast, ovary and prostate cancers [14,26-27]. Comprehensive and systematical analysis of the clinical significance of miR-101-3p in HCC is still limited, and the biological functions as well as mechanisms of anti-tumor are still unexplored.
The data from TCGA showed miR-101-1 quantity was obviously lower in cancer than in non-tumor liver tissues, and miR-101-1 had great diagnostic value of HCC. Down-regulated miR-101-1 was also closely related to poor differentiation, high TNM stage, poor tumor stage, positive lymph node metastasis and high AFP level of HCC. The result indicated that down-regulation of miR-101-1 participated in the progression of HCC. Low expression of miR-101-1 revealed poor overall survival in HCC patients. MiR-101-3p is the majority production of miR-101-1, and the significance of miR-101-1 in TCGA datasets could reflect the importance of miR-101-3p largely. In the research of Zhang et al [23], down-regulated miR-101-3p was an independent prognostic factor, and was frequently detected in advanced tumor grade and clinical stage of HCC tissues. In one recent study of plasma, low-expression of miR-101-3p predicted worse survival rate of HCC patients as well [11].
The clinical value of miR-101 (including precursor miRNA miR-101-1 of TCGA database and mature miRNA miR-101-3p of published studies) was further investigated with a quantitative meta-analysis. This meta-analysis evaluated the prognostic and clinicopathological roles of miR-101 in HCC patients, and this result was likely to show the association of down-regulated miR-101 with poor prognosis, positive AFP expression and higher TNM stage. The result might improve the clinical diagnosis and prognosis of HCC. However, potential bias may exist due to the small number of studies (n=7) and included cases only from Asian and American countries. More investigations with large samples should be performed to clarify the influence of miR-101 in HCC patients.
In recent years, targeted drugs of oncogenic signaling pathways or associated genes have afforded new insights into the treatment of HCC [2], but chemotherapeutic drugs still have limited efficacy [28]. To find more potential targets and detect biological significance as well as potential malignant mechanisms of the altered miR-101-3p expression, systemic bioinformatics analysis was conducted and the signal pathways of genes were evaluated.
In the 343 putative miR-101-3p targets, the most frequent functions were intracellular signaling cascade, transcription, metabolism, cell proliferation, biosynthesis processes, transcriptional activation and chromatin binding, which evidently showed that candidate genes may have a definitive impact on hepatocarcinogenesis. Sixteen validated targets of miR-101-3p in HCC have been reported including activator protein-1 (AP-1), autophagy-related protein 4D (ATG4D), cycloxygenase-2 (COX2), DNA methyltransferase 3A (DNMT3A), DUSP1, embryonic ectoderm development (EED), EZH2, FOS, Mcl-1, mTOR, RAB5A, ras-associated protein-1 b (Rap1b), ROCK2, SOX9, stathmin 1 (STMN1) and VEGF. In recent studies of HCC, transcription factor AP-1 has been verified to participate in the transcription regulation progress mediated by miR-101-3p [10]. MiR-101-3p was shown to play an important role in enhanced apoptosis through the inhibition of autophagy via targeting ATG4D, mTOR, RAB5A and STMN1 in HCC cells [29]. In a latest research of Zheng et al [11], overexpression of miR-101-3p reduced COX2, STMN1 and directly down-regulated ROCK2 in HCC cells. MiR-101-3p was down-regulated and induced aberrant DNA methylation by targeting DNMT3A in hepatitis B virus (HBV) related HCC [24], and miR-101-3p regulated macrophage innate immune responses to LPS via targeting DUSP1 [16]. In the study of Chiang et al [30], miR-101-3p was proved to target EZH2 and EED, acting as a suppressive gene in HCC. Li et al [13] has concluded that miR-101-3p repressed FOS oncogene level and inhibited invasion and migration in HCC cell lines. He et al [31] demonstrated that miR-101-3p enhanced sensitivity of HCC by down-regulating Mcl-1 expression. MiR-101-3p was confirmed to mediate Rap1b level, and was associated with migration and proliferation in HBV-related HCC [32]. Zhang et al [23] indicated that miR-101-3p could target SOX9 and be involved in proliferation and tumorigenicity in HCC cells. Moreover, miR-101-3p, which directly inhibited VEGF-C expression in HCC cells, was assumed to exert a suppressive effect on invasion and migration [33].
A total of 19 pathways were obtained in the analysis of miR-101-3p targets, and most of them have already been proved to be associated with HCC. Previous researches have reported that focal adhesion [34], chemokine signaling pathway [35], type II diabetes mellitus [36], VEGF signaling pathway [37], adherens junction [38], mTOR signaling pathway [39], TGF-beta signaling pathway [40] and Wnt signaling pathway [41] play crucial roles in the genesis and progress of HCC. In addition, the network construction of miR-101-3p-mediated targets showed the similar biological functions with GO analysis by DAVID mentioned above. The BP enriched in intracellular signaling process, transcription, metabolism and biosynthesis processes. The MF focused on the protein or chromatin binding and transcriptional activation. The CC was most correlated with insoluble fraction and nucleus. The highly connected targets are the center of gene regulation and play important roles in biological process and molecular function. The result may lay the foundation for studies on mechanisms of miR-101-3p.
Among the pathways of miR-101-3p targeted genes, focal adhesion kinase (FAK) has been reported to be activated by MET and to promote proliferation in HCC cells [34]. The inhibition of PI3K/Akt/mTOR signaling pathway was shown to arrest cycle and induce apoptosis associated with the expression of mTOR and CXC195 in HCC cells [39,42]. The analysis of other potential pathways revealed that VEGF signaling pathway might influence cell proliferation, cycle and apoptosis by targeting PTGS2 and SPHK1. JAK2, TGFBR1, MAPK1, VEGFA and RAC1 may contribute to cell proliferation and apoptosis, which is related to adherens junction, Fc gamma R-mediated phagocytosis, neurotrophin, chemokine and TGF-beta signaling pathways. GAB1, GNB1, CXCL12 and RAP1B are proliferation related genes and regulated by neurotrophin and chemokine signaling pathways. TGF-beta, Wnt and chemokine signaling pathways probably regulate cell cycle associated with ROCK2, RBL2, FBXW11 and PPP1CC. To further confirm the role of miR-101-3p in HCC, in vitro experiments were performed in HepG2 cells. In this study, enforced miR-101-3p inhibited the proliferation and induced apoptosis, indicating the antitumor feature of miR-101-3p in HCC, which was consistent with previous studies [15,32]. This experimental evidence confirmed the anti-cancer role of miR-101-3p by regulating cell biological processes in HCC.
In conclusion, there is a connection between miR-101-1 and clinical parameters of HCC. Down-regulated miR-101-1 may be a diagnostic marker and related with poor prognosis for HCC patients, and the influence might be induced by proliferation promotion and apoptosis inhibition of HCC cells. MiR-101-3p may perform its biological functions via focal adhesion, pathways in cancer and chemokine signaling pathway by targeting relevant crucial genes. The targeted genes of miR-101-3p are involved in the development progress of HCC and novel targets should be provided for diagnosis and treatment to inhibit tumorigenicity of HCC. The mechanisms of predicted targets have not been fully examined and functional experiments are urgently required for further confirmation.
Acknowledgements
This study was supported by the Fund of National Natural Science Foundation of China (NSFC 81560386) and Guangxi Medical University Training Program for Distinguished Young Scholars (2017). The results published here are partly based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.European Association For The Study of The Liver; European Organisation For Research And Treatment of Cancer. EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Rampone B, Schiavone B, Martino A, Viviano C, Confuorto G. Current management strategy of hepatocellular carcinoma. World J Gastroenterol. 2009;15:3210–3216. doi: 10.3748/wjg.15.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 5.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 6.Sempere LF, Kauppinen S. Handbook of cell signaling. 2nd edition. Oxford, UK: Academic Press; 2009. Translational implications of microRNAs in clinical diagnostics and therapeutics; pp. 2965–2981. [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Xue J, Niu J, Wu J, Wu ZH. MicroRNAs in cancer therapeutic response: friend and foe. World J Clin Oncol. 2014;5:730–743. doi: 10.5306/wjco.v5.i4.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Liu JJ, Lin XJ, Yang XJ, Zhou L, He S, Zhuang SM, Yang J. A novel AP-1/miR-101 regulatory feedback loop and its implication in the migration and invasion of hepatoma cells. Nucleic Acids Res. 2014;42:12041–12051. doi: 10.1093/nar/gku872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng F, Liao YJ, Cai MY, Liu TH, Chen SP, Wu PH, Wu L, Bian XW, Guan XY, Zeng YX, Yuan YF, Kung HF, Xie D. Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet. 2015;11:e1004873. doi: 10.1371/journal.pgen.1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- 13.Li S, Fu H, Wang Y, Tie Y, Xing R, Zhu J, Sun Z, Wei L, Zheng X. MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology. 2009;49:1194–1202. doi: 10.1002/hep.22757. [DOI] [PubMed] [Google Scholar]
- 14.Xu L, Beckebaum S, Iacob S, Wu G, Kaiser GM, Radtke A, Liu C, Kabar I, Schmidt HH, Zhang X, Lu M, Cicinnati VR. MicroRNA-101 inhibits human hepatocellular carcinoma progression through EZH2 downregulation and increased cytostatic drug sensitivity. J Hepatol. 2014;60:590–598. doi: 10.1016/j.jhep.2013.10.028. [DOI] [PubMed] [Google Scholar]
- 15.Sheng Y, Li J, Zou C, Wang S, Cao Y, Zhang J, Huang A, Tang H. Downregulation of miR-101-3p by hepatitis B virus promotes proliferation and migration of hepatocellular carcinoma cells by targeting Rab5a. Arch Virol. 2014;159:2397–2410. doi: 10.1007/s00705-014-2084-5. [DOI] [PubMed] [Google Scholar]
- 16.Wei X, Tang C, Lu X, Liu R, Zhou M, He D, Zheng D, Sun C, Wu Z. MiR-101 targets DUSP1 to regulate the TGF-beta secretion in sorafenib inhibits macrophage-induced growth of hepatocarcinoma. Oncotarget. 2015;6:18389–18405. doi: 10.18632/oncotarget.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He S, Zeng S, Zhou ZW, He ZX, Zhou SF. Hsa-microRNA-181a is a regulator of a number of cancer genes and a biomarker for endometrial carcinoma in patients: a bioinformatic and clinical study and the therapeutic implication. Drug Des Devel Ther. 2015;9:1103–1175. doi: 10.2147/DDDT.S73551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang YM, Dai BL, Zheng L, Zhan YZ, Zhang J, Smith WW, Wang XL, Chen YN, He LC. A novel angiogenesis inhibitor impairs lovo cell survival via targeting against human VEGFR and its signaling pathway of phosphorylation. Cell Death Dis. 2012;3:e406. doi: 10.1038/cddis.2012.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Zhan Y, Zhang D, Dai B, Ma W, Qi J, Liu R, He L. Eupolyphaga sinensis walker displays inhibition on hepatocellular carcinoma through regulating cell growth and metastasis signaling. Sci Rep. 2014;4:5518. doi: 10.1038/srep05518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng L, Wang X, Luo W, Zhan Y, Zhang Y. Brucine, an effective natural compound derived from nux-vomica, induces G1 phase arrest and apoptosis in LoVo cells. Food Chem Toxicol. 2013;58:332–339. doi: 10.1016/j.fct.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Lu HX. Expression of EZH2 gene in hepatocellular carcinoma and to be regulated by miR-101. Guangxi Medical University. 2010:55. [Google Scholar]
- 23.Zhang Y, Guo X, Xiong L, Kong X, Xu Y, Liu C, Zou L, Li Z, Zhao J, Lin N. MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett. 2012;586:4362–4370. doi: 10.1016/j.febslet.2012.10.053. [DOI] [PubMed] [Google Scholar]
- 24.Wei X, Xiang T, Ren G, Tan C, Liu R, Xu X, Wu Z. MiR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell Signal. 2013;25:439–446. doi: 10.1016/j.cellsig.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 25.Fu Y, Wei X, Tang C, Li J, Liu R, Shen A, Wu Z. Circulating microRNA-101 as a potential biomarker for hepatitis B virus-related hepatocellular carcinoma. Oncol Lett. 2013;6:1811–1815. doi: 10.3892/ol.2013.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gui T, Shen K. MiRNA-101: a potential target for tumor therapy. Cancer Epidemiol. 2012;36:537–540. doi: 10.1016/j.canep.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Li JT, Jia LT, Liu NN, Zhu XS, Liu QQ, Wang XL, Yu F, Liu YL, Yang AG, Gao CF. MiRNA-101 inhibits breast cancer growth and metastasis by targeting CX chemokine receptor 7. Oncotarget. 2015;6:30818–30830. doi: 10.18632/oncotarget.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoshida Y, Fuchs BC, Tanabe KK. Prevention of hepatocellular carcinoma: potential targets, experimental models, and clinical challenges. Curr Cancer Drug Targets. 2012;12:1129–1159. [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, An Y, Wang Y, Zhang C, Zhang H, Huang C, Jiang H, Wang X, Li X. MiR-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol Rep. 2013;29:2019–2024. doi: 10.3892/or.2013.2338. [DOI] [PubMed] [Google Scholar]
- 30.Chiang CW, Huang Y, Leong KW, Chen LC, Chen HC, Chen SJ, Chou CK. PKCalpha mediated induction of miR-101 in human hepatoma HepG2 cells. J Biomed Sci. 2010;17:35. doi: 10.1186/1423-0127-17-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He H, Tian W, Chen H, Deng Y. MicroRNA-101 sensitizes hepatocellular carcinoma cells to doxorubicin-induced apoptosis via targeting Mcl-1. Mol Med Rep. 2016;13:1923–1929. doi: 10.3892/mmr.2015.4727. [DOI] [PubMed] [Google Scholar]
- 32.Sheng Y, Ding S, Chen K, Chen J, Wang S, Zou C, Zhang J, Cao Y, Huang A, Tang H. Functional analysis of miR-101-3p and Rap1b involved in hepatitis B virus-related hepatocellular carcinoma pathogenesis. Biochem Cell Biol. 2014;92:152–162. doi: 10.1139/bcb-2013-0128. [DOI] [PubMed] [Google Scholar]
- 33.Liu Z, Wang J, Mao Y, Zou B, Fan X. MicroRNA-101 suppresses migration and invasion via targeting vascular endothelial growth factor-C in hepatocellular carcinoma cells. Oncol Lett. 2016;11:433–438. doi: 10.3892/ol.2015.3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shang N, Arteaga M, Zaidi A, Stauffer J, Cotler SJ, Zeleznik-Le NJ, Zhang J, Qiu W. FAK is required for c-Met/beta-catenin-driven hepatocarcinogenesis. Hepatology. 2015;61:214–226. doi: 10.1002/hep.27402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin Y, Xu SQ, Pan DB, Ye GX, Wu CJ, Wang S, Wang CJ, Jiang JY, Fu J. Silencing of WWP2 inhibits adhesion, invasion, and migration in liver cancer cells. Tumour Biol. 2016;37:6787–6799. doi: 10.1007/s13277-015-4547-z. [DOI] [PubMed] [Google Scholar]
- 36.Jha DK, Mittal A, Gupta SP, Pandeya DR, Sathian B. Association of type II diabetes mellitus with hepatocellular carcinoma occurrence-a case control study from Kathmandu Valley. Asian Pac J Cancer Prev. 2012;13:5097–5099. doi: 10.7314/apjcp.2012.13.10.5097. [DOI] [PubMed] [Google Scholar]
- 37.Ye ZL, Huang Y, Li LF, Zhu HL, Gao HX, Liu H, Lv SQ, Xu ZH, Zheng LN, Liu T, Zhang JL, Jin HJ, Qian QJ. Argonaute 2 promotes angiogenesis via the PTEN/VEGF signaling pathway in human hepatocellular carcinoma. Acta Pharmacol Sin. 2015;36:1237–1245. doi: 10.1038/aps.2015.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson GK, Brimacombe CL, Rowe IA, Reynolds GM, Fletcher NF, Stamataki Z, Bhogal RH, Simoes ML, Ashcroft M, Afford SC, Mitry RR, Dhawan A, Mee CJ, Hubscher SG, Balfe P, McKeating JA. A dual role for hypoxia inducible factor-1alpha in the hepatitis C virus lifecycle and hepatoma migration. J Hepatol. 2012;56:803–809. doi: 10.1016/j.jhep.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen XL, Fu JP, Shi J, Wan P, Cao H, Tang ZM. CXC195 induces apoptosis and endoplastic reticulum stress in human hepatocellular carcinoma cells by inhibiting the PI3K/Akt/mTOR signaling pathway. Mol Med Rep. 2015;12:8229–8236. doi: 10.3892/mmr.2015.4479. [DOI] [PubMed] [Google Scholar]
- 40.Morris SM, Carter KT, Baek JY, Koszarek A, Yeh MM, Knoblaugh SE, Grady WM. TGF-beta signaling alters the pattern of liver tumorigenesis induced by Pten inactivation. Oncogene. 2015;34:3273–3282. doi: 10.1038/onc.2014.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Meng X, Sun X, Liu M, Gao S, Zhao J, Pei F, Yu H. Wnt/beta-catenin signaling pathway may regulate cell cycle and expression of cyclin A and cyclin E protein in hepatocellular carcinoma cells. Cell Cycle. 2009;8:1567–1570. doi: 10.4161/cc.8.10.8489. [DOI] [PubMed] [Google Scholar]
- 42.Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology. 2012;55:1852–1862. doi: 10.1002/hep.25576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.