ABSTRACT
Influenza A H15 viruses are members of a subgroup (H7-H10-H15) of group 2 hemagglutinin (HA) subtypes that include H7N9 and H10N8 viruses that were isolated from humans during 2013. The isolation of avian H15 viruses is, however, quite rare and, until recently, geographically restricted to wild shorebirds and waterfowl in Australia. The HAs of H15 viruses contain an insertion in the 150-loop (loop beginning at position 150) of the receptor-binding site common to this subgroup and a unique insertion in the 260-loop compared to any other subtype. Here, we show that the H15 HA has a high preference for avian receptor analogs by glycan array analyses. The H15 HA crystal structure reveals that it is structurally closest to H7N9 HA, but the head domain of the H15 trimer is wider than all other HAs due to a tilt and opening of the HA1 subunits of the head domain. The extended 150-loop of the H15 HA retains the conserved conformation as in H7 and H10 HAs. Furthermore, the elongated 260-loop increases the exposed HA surface and can contribute to antigenic variation in H15 HAs. Since avian-origin H15 HA viruses have been shown to cause enhanced disease in mammalian models, further characterization and immune surveillance of H15 viruses are warranted.
IMPORTANCE In the last 2 decades, an apparent increase has been reported for cases of human infection by emerging avian influenza A virus subtypes, including H7N9 and H10N8 viruses isolated during 2013. H15 is the other member of the subgroup of influenza A virus group 2 hemagglutinins (HAs) that also include H7 and H10. H15 viruses have been restricted to Australia, but recent isolation of H15 viruses in western Siberia suggests that they could be spread more globally via the avian flyways that converge and emanate from this region. Here we report on characterization of the three-dimensional structure and receptor specificity of the H15 hemagglutinin, revealing distinct features and specificities that can aid in global surveillance of such viruses for potential spread and emerging threat to the human population.
KEYWORDS: influenza virus, H15 subtype, receptor binding, glycan arrays, X-ray crystallography
INTRODUCTION
Seasonal migration between breeding and wintering grounds is a common strategy for many species of birds to find better food sources and optimal environments for breeding. These birds migrate along flyways (normally north to south and back) that traverse different countries and continents. As an unwanted consequence, migrating birds become effective carriers of different kinds of viruses, parasites, and bacteria (1). Shorebirds and wild waterfowl are considered the natural reservoir of most avian influenza A viruses (IAV) (2). The IAV are classified according to serology and from the antigenic properties of their two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). Sixteen HA (H1-16) and nine NA (N1-9) subtypes of influenza A viruses have been identified in birds, giving 144 possible combinations (3, 4). HA plays a pivotal role in the initiation of virus infection by binding to terminal sialic acids (N-acetylneuraminic acid [NeuAc]) of glycan receptors on host cells followed by pH-dependent fusion of the viral envelope with the host cell membrane in the acidic environment of endosomal compartments (5). On the other hand, the NA protein, as a receptor-destroying enzyme, contributes to the release of the progeny viruses from the host cell. The HA influenza A virus subtypes can be phylogenetically separated into two groups, and each group can be further divided into subgroups (Fig. 1A) (4, 6, 7). Beside birds, IAV can circulate in a variety of mammals, including humans, pigs, horses, mink, felines, and marine mammals. Recently, two new subtypes of influenza A viruses (H17N10 and H18N11) were identified in bats (8–12).
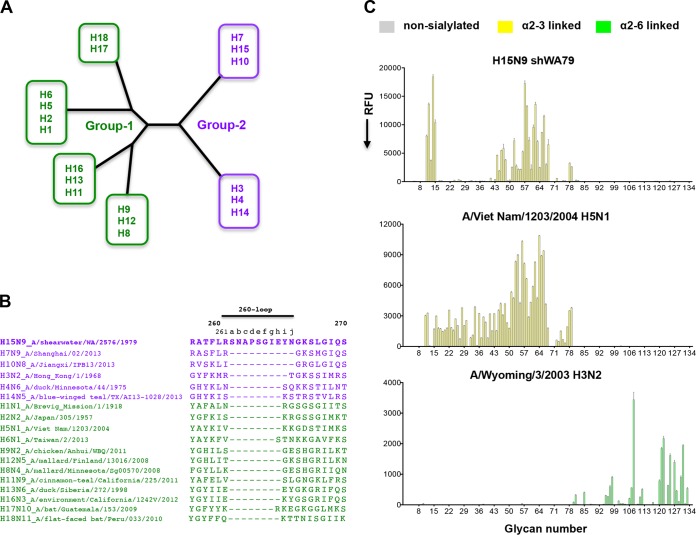
FIG 1.
The amino acid sequence of the elongated 260-loop and the receptor binding specificity of the H15N9 shWA79 HA. (A) Schematic representation of the phylogenetic tree of influenza A virus HAs. The HAs are divided into group 1 and group 2, each of which can be further subdivided into subgroups. (B) Alignment of the HA 260-loop sequences of representative influenza viruses from all influenza A virus HA subtypes. The alignment indicates a marked elongation of the 260-loop of H15 HA compared to other HAs. The sequences of the group 1 and group 2 HAs are colored green and purple, respectively. (C) Glycan microarray analysis of the receptor binding specificity of recombinant H15N9 shWA79 HA indicates avian-type specificity and preferential binding of short, sulfated linear glycans (glycan numbers 11 to 15) and long, branched, O-linked and N-linked glycans. The A/Viet Nam/1203/2004 H5N1 and the A/Wyoming/3/2003 H3N2 HAs were used as controls for α2-3- and α2-6-linked sialoside binding, respectively. The mean signal (in relative fluorescence units [RFU]) and standard error were calculated from six independent replicates on the array. α2-3-linked sialosides are shown by yellow bars (glycans 11 to 79 on the x axis), and α2-6-linked sialosides are shown by green bars (glycans 80 to 135). Glycans 1 to 10 are nonsialylated control glycans and shown in gray. Glycans imprinted on the array are listed in Table S1 in the supplemental material.
Adaptation of influenza A viruses to circulate in a new host requires a switch in receptor specificity of the HA toward the specific sialic acid receptors that are expressed on the target host cells. The HAs of avian viruses preferentially bind sialoside receptors that contain sialic acid linked by an α2-3 linkage to galactose (avian-type receptors), whereas viruses that infect and transmit in humans prefer α2-6-linked sialic acids (human-type receptors) (13, 14).
Influenza viruses can be divided into two phylogenetic geographic clades, the Americas and Eurasia, which also includes the Australian subclade (15, 16). These two geographic areas are connected through western Siberia that serves as a junction for four major avian flyways and thereby converge and intermingle the bird populations from Europe, Africa, Asia, Oceania, and North America. Thus, western Siberia is a hub for reassortment of influenza viruses from different lineages and hosts. Isolation and identification of H15 viruses are quite rare, with only nine sequences of H15 HAs out of more than 50,000 influenza A virus sequences in the influenza virus databases (NCBI Influenza Virus Resource and the Global Initiative on Sharing All Influenza Data [GISAID]). Until recently, the isolation of H15 viruses in birds was exclusive to Australia with only seven reported between 1979 and 1983. However, during a relatively recent surveillance of avian influenza viruses in western Siberia (2008) and Ukraine (2010), two new H15 viruses were isolated (17, 18), demonstrating some spread of H15 viruses outside of Australia. Since western Siberia is the major junction of avian flyways, the new isolates suggest that H15 viruses could potentially spread to the rest of the world.
Three cases of human infections by new subtypes of avian-origin IAV (H7N9, H6N1, and H10N8) were reported during 2013 (19–25), two of which, H7N9 and H10H8, are members of the same group 2 HA subgroup that includes the H15 HA viruses (H7-H10-H15 subgroup) (Fig. 1A). Since the first case of human infection by the H7N9 virus was reported in March 2013, 1,223 laboratory-confirmed cases have now been filed with a high mortality rate of ∼31% (as of 14 February 2017, http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en). For H10N8, two of the three cases reported were lethal. However, no human infection by an H15 HA virus has so far been reported.
These emerging subtypes appear to be part of an increase in avian IAV that have caused sporadic human infections in the past 2 decades, indicating a potential major threat to human health worldwide. A recent study compared the pathogenicity and virulence properties of a set of chimeric viruses expressing 13 different contemporary avian influenza virus HA subtypes to the pathogenicity and virulence of an H1N1 virus that contains mutations that facilitate viral replication in mammalian cells. Viruses with avian H1, H6, H7, H10, and H15 HAs caused severe disease in mice and damage to human lung cells (26). This study further demonstrates the potential of avian-origin viruses of the H7-H10-H15 subgroup to be a severe health risk for the human population.
Here, we report the crystal structure and receptor specificity of an H15 HA. The glycan array results reveal that the H15 HA has a high preference for avian receptor analogs, especially sulfated and long-branched glycans, and that mutagenesis of the key residues that switch receptor specificity in the receptor-binding sites (RBS) of H1, H2 and H3 pandemic viruses does not change receptor specificity in H15 HA. The H15 HA is structurally closest to H7N9 HA and retains the conformation of the extended 150-loop (loop beginning at position 150) that is conserved in H7 and H10 HAs. Although similar to H7 and other group 2 HAs, the HA1 subunits of the head domain of the H15 HA are tilted related to the stem domain, thereby acquiring a more open conformation for the head domain of the H15 trimer compared to all other HAs. In addition, an extended 260-loop that is specific to H15 HAs widens and extends the exposed surface of the HA and can contribute to antigenic variation in H15 HAs.
RESULTS
Binding specificity of shWA79 H15N9 HA.
Isolation of H15 viruses is quite rare with only nine sequences of H15 HAs available in the influenza virus sequence databases. Alignment of these H15 HA sequences illustrates high conservation with viruses isolated in Australia and Russia (see Fig. S1A in supplemental material). Influenza A virus HAs are phylogenetically divided into two groups (groups 1 and 2) based on antigenic properties and can be further classified into subgroups (6, 7, 10); the H15 HAs are within the H7-H10-H15 subgroup of group 2 HAs (Fig. 1A). Sequence alignment of the A/shearwater/Western Australia/2576/1979 (shWA79) H15N9 HA with other group 2 HAs (human pandemic H3N2 and recent H7N9 and H10N8 strains) reveal that the H15 HA contains the same insertion of two amino acids in the 150-loop that is a feature of the H7-H10-H15 HA subgroup (Fig. S1B). In addition, the 260-loop of H15 HAs contains a unique insertion of 7 to 10 residues compared to all other subtypes (Fig. 1B) and nine residues compared to H3 HAs, which are also used for reference numbering (Fig. S1B).
The receptor-binding site (RBS) of avian IAV contain four key highly conserved residues that have been associated with conferring avian-type specificity: Glu190, Gly225, Gln226, and Gly228 (H3 numbering). The H15 HAs also contain these RBS residues (Fig. S1A). To directly assess the receptor binding properties of the shWA79-H15N9 HA, we used an expanded custom sialoside glycan array that is comprised of diverse α2-3 sialosides (numbers 11 to 79) and α2-6 sialosides (numbers 80 to 135) (Table S1) (27). The H15 HA has a strong preference for α2-3 sialylated glycans with no detectable binding to α2-6 sialosides (Fig. 1C). More specifically, the H15 HA binds preferentially to short, sulfated, linear glycans (glycan numbers 11 to 15) and to long, branched, O-linked and N-linked glycans terminating in α2-3-linked sialic acids (glycan numbers 44 to 48 and 51 to 68).
Structural characterization of shWA79 H15N9 HA.
To investigate the structural features and properties of the H15 HA, we determined its crystal structure at 3.1-Å resolution (Table 1) with two HA protomers in the asymmetric unit. Overall, its structure is similar to those of other influenza A virus HA trimer structures (Fig. 2A). Seven N-linked glycosylation sites per HA protomer are predicted (28) (N22, N38, N92, and N165 in HA1 and N82, N154, and N158 in HA2), and interpretable electron density was observed at HA1 Asn38 and HA2 Asn158, with more limited density (that was not modeled) at HA1 Asn92 and HA2 Asn154 (Fig. 2A).
TABLE 1.
Data collection and refinement statistics for H15 HA apo and receptor analog complex
| Parameter | Value fora: |
|
|---|---|---|
| Apo | 3'-SLN | |
| Data collection statistics | ||
| Beamline | APS-23 | APS-23 |
| Wavelength (Å) | 1.0332 | 1.0332 |
| Space group | P3 | P3 |
| Unit cell (Å) | a = b = 115.9; c = 128.4 | a = b = 116.6; c = 129.1 |
| Resolution (Å) | 50.00–3.10 (3.15–3.10) | 50.00–2.75 (2.80–2.75) |
| No. of observations | 143,348 | 132,482 |
| No. of unique reflections | 34,747 (1,596) | 49,813 (2,489) |
| Completeness (%) | 98.7 (90.7) | 97.4 (99.3) |
| I/σ(I) | 9.1 (1.7) | 10.8 (1.7) |
| CC1/2b | 0.94 (0.89) | 0.87 (0.52) |
| Rsymc | 0.18 (0.44) | 0.14 (0.99) |
| Rpimd | 0.08 (0.23) | 0.08 (0.58) |
| Redundancy | 4.1 (3.1) | 2.7 (2.6) |
| Refinement statistics | ||
| Resolution (Å) | 50.00–3.10 (3.15–3.10) | 50.00–2.75 (2.80–2.75) |
| No. of reflections | 34,651 | 49,791 |
| Rcryste/Rfreef | 0.221/0.263 | 0.222/0.275 |
| No. of atoms | ||
| Protein | 7,788 | 7,736 |
| Carbohydrate | 84 | 162 |
| Water | 105 | |
| Wilson B (Å2) | 46 | 43 |
| Avg B value (Å2) | ||
| Overall | 48 | 46 |
| Water | 44 | |
| Ligand | 72 | |
| RMSDg from ideal geometry | ||
| Bond length (Å) | 0.014 | 0.014 |
| Bond angle (°) | 2.2 | 1.9 |
| Ramachandran plot (%)h | ||
| Favored | 96.8 | 95.5 |
| Outliers | 0.2 | 0.2 |
| PDB ID | 5TG8 | 5TG9 |
Values in parentheses are the outer shell statistics.
CC1/2 is the Pearson correlation coefficient between two random half data sets.
Rsym = Σhkl Σi | Ihkl,i − 〈Ihkl〉 |/Σhkl Σi Ihkl,i, where Ihkl,i is the scaled intensity of the ith measurement of reflection h, k, l, and 〈Ihkl〉 is the average intensity for that reflection.
Rpim = Σhkl [1/(n − 1)]1/2 Σi | Ihkl,i − 〈Ihkl〉 |/Σhkl Σi Ihkl,i, where n is the redundancy.
Rcryst = Σhkl | Fo − Fc |/Σhkl | Fo | × 100, where Fo and Fc are the observed and calculated structure factors, respectively.
Rfree was calculated as for Rcryst, but on a test set of 5% of the data excluded from refinement.
RMSD, root mean square deviation.
Calculated using MolProbity (62).
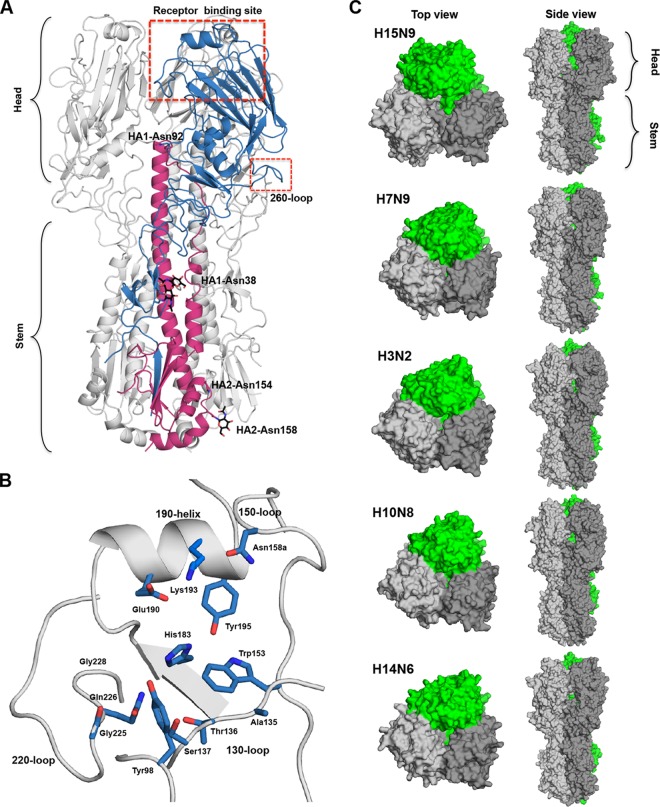
FIG 2.
Crystal structure of the H15N9 shWA79 HA. (A) Schematic cartoon representation of the H15N9 shWA79 HA trimer. One protomer is colored in blue and magenta for the HA1 and the HA2 subunits. The RBS and the 260-loop are marked with red rectangles. N-linked glycans that could be modeled in the electron density maps and the corresponding asparagine are shown as sticks and labeled in black. (B) Schematic representation of the RBS of the H15 HA. The key residues for receptor binding are shown as sticks and colored in blue. (C) The wider head domain of the H15 HA trimer is shown in the top and side view. Surface representations of the group 2 H15, H7, H3, H10, and H14 HA trimers indicate that the H15 head domain subunits tilt more outwards relative to the stem region compared to other subtypes.
Superposition of the H15 HA onto other avian HAs and to pandemic HAs (H1, H2, and H3) confirms that H15 is structurally closest to H7 HA (C-α root mean square deviation [RMSD] of 1.1 Å compared to 1.3 to 1.7 Å for other group 2 HAs and 2.5 to 2.7 Å for representative group 1 HAs). Similar results were also obtained from superposition of the HA1, HA2, and RBS subdomains (Table 2). Like other influenza A virus HAs, the H15 RBS contains the absolutely conserved Tyr98, Trp153, His183, and Tyr195 on the floor of the binding pocket (5) as well as the other key structural elements that include the 130-loop, 150-loop, 190-helix, and 220-loop (Fig. 2B) that delineate the sides and ends of the RBS.
TABLE 2.
Comparison of H15 HA with group 1 and other group 2 subtypes
| HA strain | PDB code | RMSD values of C-α atoms (Å)a |
RBS subdomainb | Alignment score (%)c | ||
|---|---|---|---|---|---|---|
| Monomer | HA1 | HA2 | ||||
| H3N2_A/Hong_Kong/1/1968 | 4FNK | 1.3 (462) | 1.4 (301) | 0.6 (160) | 0.9 (126) | 47 |
| H14N6_A/Mallard/Gurjev/244/1982 | 3EYK | 1.4 (484) | 1.3 (313) | 0.6 (168) | 0.9 (130) | 49 |
| H7N9_A/Shanghai/02/2013 | 4N5J | 1.1 (486) | 0.5 (316) | 0.4 (168) | 0.4 (149) | 79 |
| H10N8_A/Jiangxi/IPB13a/2013 | 4XQ5 | 1.7 (486) | 0.7 (316) | 0.6 (168) | 0.6 (145) | 65 |
| H1N1_A/California/04/2009 | 3AL4 | 2.5 (428) | 1.9 (266) | 1.0 (127) | 0.6 (109) | 40 |
| H2N2_A/Japan/305/1957 | 3KU5 | 2.7 (424) | 1.8 (264) | 1.0 (140) | 0.6 (100) | 41 |
| H5N1_A/Vietnam/4/2003 | 2FK0 | 2.8 (415) | 2.0 (244) | 0.9 (136) | 0.7 (109) | 41 |
The numbers in parentheses are the numbers of residues included in the comparison.
The receptor-binding subdomain (RBS subdomain) is defined as residues 117 to 265 of the HA1 subunit (H3 numbering). The number in parentheses are the number of residues.
Calculated for the HA sequence (Fig. S1) using Clustal W2 (ENBL-EBI) (63).
Despite the high structural similarity between the H15 and H7 protomers, the superposition of the H15 and H7 HA trimer indicates that the head domain of H15 is wider, adopting a more open conformation (Fig. 2C). Similar results were obtained by superposition of the H15 trimer on other group 2 HAs (Fig. 2C). To investigate the potential cause of this change in configuration, we calculated the center of mass of the HA1 domain, the head domain (residues 45 to 295 of the HA1 subunit), and the RBS subdomain (residues 117 to 265 of the HA1 subunit) of the group 2 HAs and then measured the angle between the stem domain and the center of mass of the head domains (by measuring the angle between C-α of conserved HA2 stem residues E86 and Q125 to the calculated center of mass [Table 3]). The results show a decrease in the angle of the H15 HA in comparison to other group 2 HAs that splay apart the head domain (Table 3). Measuring the distances between the center of mass of the head domain of one protomer to the center of mass of another protomer in the HA trimer indicated a significant increase in the distance only for the H15 HA (Table 3), suggesting that the unique, more open conformation of the H15 trimer is due to tilt of the head domain relative to the stem region, thereby resulting in a widening of the trimer apex.
TABLE 3.
Increased width of the head domain of H15 HAa
| HA strain | PDB code | HA1 |
Head domain |
RBS subdomain |
|||
|---|---|---|---|---|---|---|---|
| Angleb (°) | Distancec (Å) | Angle (°) | Distance (Å) | Angle (°) | Distance (Å) | ||
| H15N9 | 5TG8 | 112.1 | 36.6 | 121.7 | 40.8 | 131.0 | 39.2 |
| H3N2 | 4FNK | 114.7 | 32.3 | 126.7 | 34.9 | 135.0 | 31.4 |
| H14N6 | 3EYK | 116.3 | 32.6 | 128.1 | 35.0 | 136.2 | 31.5 |
| H7N9 | 4N5J | 115.3 | 32.7 | 127.0 | 35.3 | 136.0 | 31.8 |
| H10N8 | 4XQ5 | 118.9 | 32.7 | 128.3 | 35.4 | 137.4 | 32.5 |
The center of mass was calculated by Pymol using the “center of mass” script (https://pymolwiki.org/index.php/Center_of_mass). H15N9 HA is shown in boldface type.
The angle between the stem region and center of mass was calculated between the C-α atom of the conserved HA2 residues E86 and Q125 to the center of mass of the HA1 domain, head domain (HA1 residues 45 to 295), or RBS subdomain (HA1 residues 117 to 260).
The distance between the center of mass of the HA1 domain, head domain, or RBS subdomain on one protomer of the HA trimer to that of the adjacent protomer.
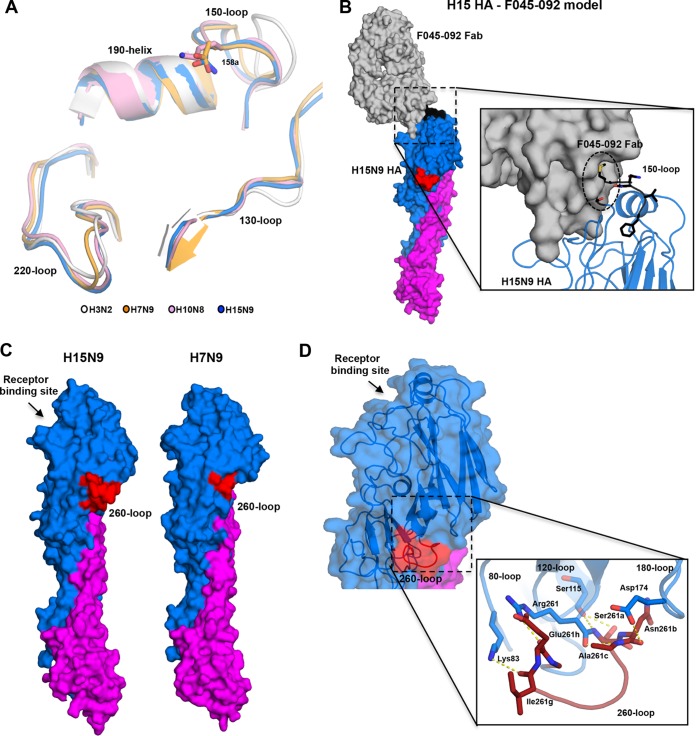
The RBS subdomain superposition confirms that the extended 150-loop of H15 HA is highly similar in conformation to those of H7 and H10 HAs and folds over toward the RBS (Fig. 3A). This extended 150-loop can perturb the binding of natural antibodies that broadly neutralize seasonal strains as illustrated for antibodies C05 and F045-092 (29, 30) (Fig. 3B). The 260-loop contains a unique extension of nine residues compared to H3 and other HAs (Fig. 1B, 2A, 3C, and S1B). In the H15 structure, this large insert can be fully modeled as an extended loop that stretches toward the base of the HA head (Fig. 2A) and broadens the head domain (Fig. 3C). The H15 260-loop conformation is stabilized by polar interactions with the 80-loop, 120-loop, and 170-loop (Fig. 3D).
FIG 3.
The elongated 150- and 260-loops of the H15 HA. (A) The conformation of the 150-loop of the group 2 HAs. Superposition of the RBS subdomains of the H15N9 shWA79 HA (blue), A/Jiangxi/IPB13/2013 H10N8 HA (pink) (PDB entry 4XQ5), A/Shanghai/02/2013 H7N9 HA (orange) (PDB entry 4N5J), and pandemic H3N2 HA (gray) (PDB entry 4NFK). The conserved structural elements of the HA RBS (130-loop, 150-loop, 190-helix, and 220-loop) are labeled, and the side chains of H15 Asn158a, H10 Lys158a and H7 Asp158a are shown as sticks. (B) A model of the complex between H15 HA (blue and purple) and F045-092 Fab that targets the RBS (gray) indicates potential steric clashes between the extended 150-loop (black) and the variable region of the F045-092 Fab. The model was constructed by superposition of H15 HA and A/Victoria/3/1975 (H3N2) HA1 in complex with Fab045-092 (PDB entry 4O58) (30). (C) Conformation of the extended 260-loop of H15N9 shWA79 HAs. Surface representation of one HA protomer of the H15N9 shWA79 shows the extended 260-loop of H15 HAs (left). The HA protomer of the A/Shanghai/02/2013 H7N9 HA (right) is shown for comparison. The HA1 and HA2 subunits are colored blue and magenta, respectively, and the 260-loop is shown in red. (D) The elongated 260-loop of shWA79 H15N9 extends toward the base of the head domain and creates polar interactions with the 80-loop, 120-loop, and 180-loop. The HA head domain is colored blue, the stem domain is magenta, and the elongated 260-loop is red. The residues that appear to stabilize the conformation of the 260-loop are shown as sticks.
The increase in HA accessible surface as a result of the elongated 260-loop may vary the antigenicity and perhaps even immunogenicity of H15 HAs compared to other subtypes (Fig. 3C). Modeling the binding of broadly neutralizing antibodies that recognize different epitopes in the HAs shows that the extended 260-loop would not be expected to perturb recognition and binding of broadly neutralizing antibodies to the HA stem (e.g., CR8020 and CR9114) (31, 32) or to antibodies that bind to the apex of the HA head (e.g., 2D1) (33) (Fig. 4). In contrast, the elongated 260-loop would be expected to affect which types of antibodies can recognize and be elicited to the central and lower part of the HA head (e.g., such as H5M9-like antibodies) (34) (Fig. 4).
FIG 4.
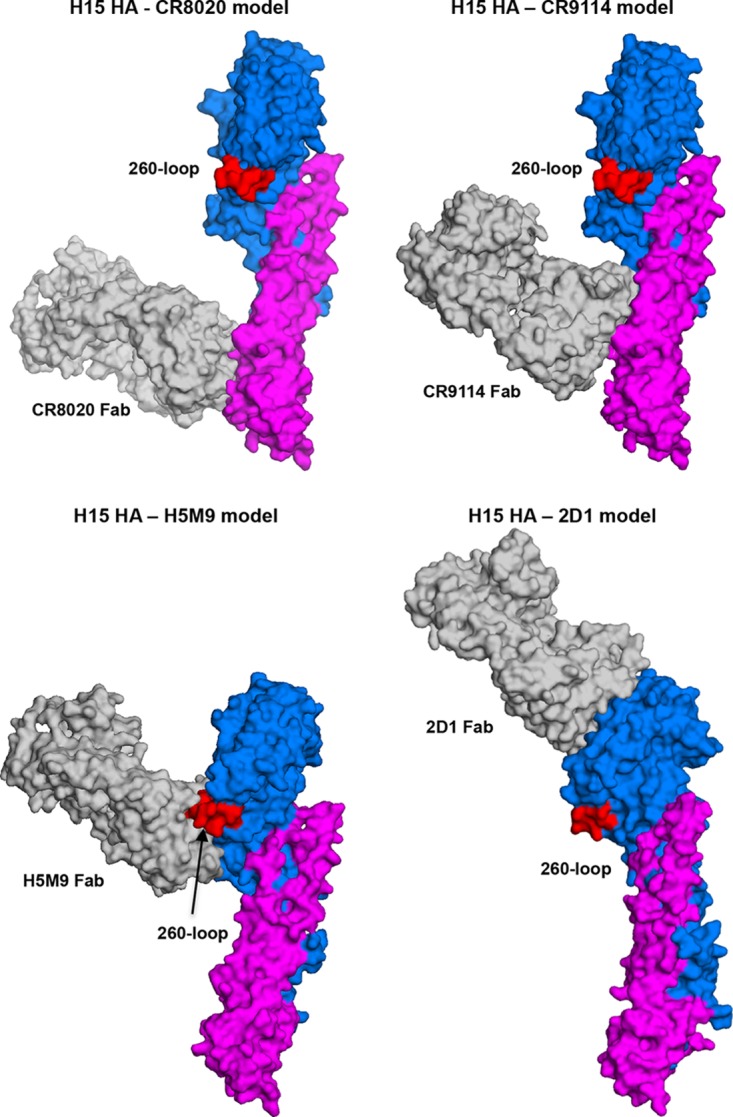
Models of complexes between H15 HA and CR8020, CR9114, H5M9, and 2D1 Fabs (gray) indicate that the elongated 260-loop (red) of the H15 HA can affect binding of antibodies to the lower part of the HA head (H5M9), but not to the HA stem (CR8020 and CR9114) or upper part of the HA head (2D1). For stem binding antibodies, the H15 HA2 chain was superimposed onto the HA2 chain of A/Hong_Kong/1/1968 (H3N2) (for CR8020, PDB entry 3SDY) (31) and A/Viet Nam/1203/2004 (H5N1) (for CR9114, PDB entry 4FQI) (32). For head binding antibodies, the H15 HA1 chain was superimposed onto the HA1 chain of A/Viet Nam/1203/2004 (H5N1) (for H5MP, PDB entry 4MHH) (34) and A/Brevig Mission/1/1918 (H1N1) (for 2D1, PDB entry 3LZF) (33).
Structural characterization of shWA79 H15N9 HA in complex with an avian receptor analog.
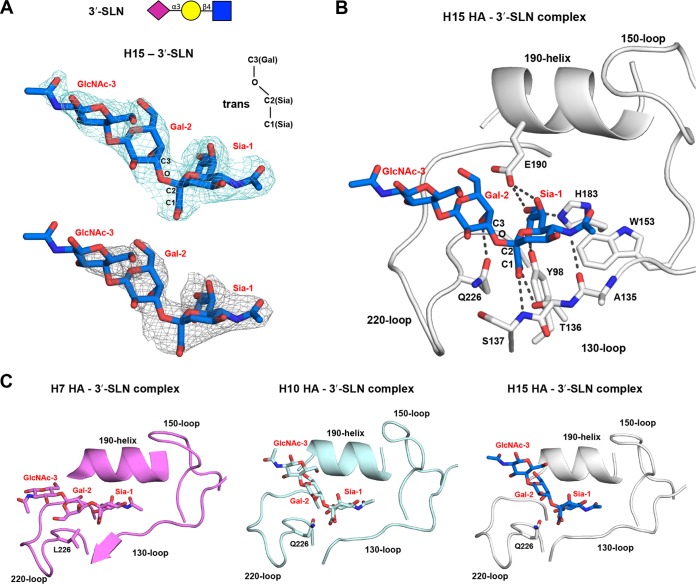
The crystal structure of H15 HA in complex with avian receptor analog 3′-SLN (NeuAcα2-3Galβ1-4GlcNAc) was determined at 2.8-Å resolution (Table 1), and interpretable electron density was observed for all three sugar rings (Fig. 5). The 3′-SLN binds in a trans conformation (with regard to the C-1Sia–C-2Sia–O–C-3Gal bond where C-1Sia is the C-1 for sialic acid), where the second and third sugars of the 3′-SLN exit the RBS between the 220-loop and the 190-helix. Similar to other HA subtypes, Sia-1 makes the usual conserved hydrogen bonds with Tyr98 and His183 and with the 130-loop, 190-helix, and 220-loop (Fig. 5B), and hydrophobic interactions with Trp153. In addition, Gal-2 hydrogen bonds with the polar side chain of Gln226, which supports the trans conformation of avian receptors as previously reported for other avian HAs (35–38). Superposition H15 HA with equivalent complexes of avian H7 and H10 HAs (PDB identifier [ID] 4BSD and 4XQU) (39, 40) reveals that the avian analog in the H15 complex is more similar to the H10 complex than to the H7 complex, probably due to the Q226L substitution in the RBS of H7N9 (Fig. 5C).
FIG 5.
Crystal structure of the H15N9 shWA79 HA in complex with an avian receptor analog. (A) Electron density maps (2Fo − Fc; top) and unbiased 2Fo-Fc electron density map (bottom) of avian receptor analog 3′-SLN in the crystal structure of the H15N9 shWA79 HA contoured at a 1σ level. The glycan structure of the 3′-SLN analog is represented above. Abbreviations: Sia, sialic acid; Gal, galactose; GlcNAc, N-acetylglucosamine. (B) Representation of the interactions between 3′-SLN and the H15 HA RBS. The RBS conserved structural elements are labeled and shown as cartoons. Selected residues and the receptor analog are labeled and shown as sticks. (C) The avian analog 3′-SLN in crystal structures with H10 and H15 HAs adopts a similar trans conformation, but in the H7 3′-SLN complex, it binds in a cis conformation (with regard to the C-1Sia–C-2Sia–O–C-3Gal bond). The H7 HA contains Leu at position 226, while the H10 and H15 HAs have Gln. The superposition was performed on the sialic acid of the sialoside.
Potential for H15 HA to switch its receptor specificity.
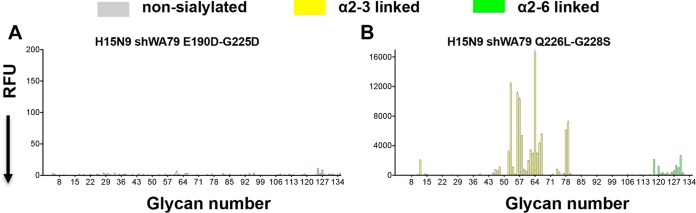
Previous studies on the H1N1, H2N2, and H3N2 pandemic influenza viruses have shown that only two substitutions of four key RBS residues were required to switch the receptor specificity from the avian type to the human type. For H1N1 subtypes (1918 and 2009 pandemic strains), E190D and G225D substitutions were required (41–43), but for H2N2 and H3N2 subtypes (1957 and 1968 pandemics), a different pair of substitutions, Q226L and G228S, was responsible for the specificity switch (41, 44). To examine which if any of these mutations influence the receptor specificity of the H15 HA, we mutated and expressed the H15N9E190D-G225D and the H15N9Q226L-G228S double mutants and interrogated the receptor specificity on the custom sialoside glycan array. The H15N9E190D-G225D double mutant lost binding to avian analogs without gaining significant ability to bind human analogs (Fig. 6A). In contrast, for the H15N9Q226L-G228S double mutant, binding of avian analogs was generally maintained. However, binding of the H15N9Q226L-G228S double mutant to short sulfated linear glycans (glycan numbers 11 to 15) was largely lost, and very weak binding toward long, branched, human receptor analogs was observed (glycan numbers 118 to 130) (Fig. 6B).
FIG 6.
Glycan microarray assay of the receptor binding specificities of the H15 HA RBS mutants. The receptor binding specificity of recombinant HAs of the H15N9E190D-G225D (A) and the H15N9Q226L-G228S (B) double mutants were examined using the custom sialoside microarray. Mutations in the RBS of the H15 HA that are associated with a switch in receptor binding specificity of pandemic H1/H2/H3 viruses did not switch receptor binding specificity of the H15 HA. The mean signal and standard error were calculated from six independent replicates on the array. α2-3-Linked sialosides are shown as yellow bars (glycans 11 to 79 on the x axis), and α2-6-linked sialosides are shown as green bars (glycans 80 to 135). Glycans 1 to 10 are nonsialylated control glycans and shown in gray. Glycans imprinted on the array are listed in Table S1.
DISCUSSION
More than 100 different combinations of HA and NA subtypes of IAV have been isolated from wild aquatic birds, the natural reservoir of avian IAV. Yet, influenza A viruses isolated from humans have been reported for only a very few combinations of these subtypes. Several criteria are required for avian influenza virus to cross the host specificity barrier and efficiently infect and transmit in the human population. In the last 2 decades, an apparent increase has been reported for cases of human infection by new avian influenza A subtypes (e.g., H5N1, H5N6, H9N2, H7N7, H7N2, H6N1, and H10N8) that are antigenically distinct from the circulating seasonal strains (21, 24, 45–50). In the majority of these cases, only sporadic infections have occurred with no documented human-to-human transmission. Although these human-infecting avian IAV have so far retained their host specificity, they are still a major threat to public health as demonstrated by H5N1 and H7N9 viruses. The first case of human infection by the avian influenza H5N1 A virus was reported in 1997 in Hong Kong (46, 50). Since then 856 cases have been documented in Asia, Africa, Europe, and North America with a mortality rate of ∼53% for those reported cases (as of 14 February 2017, http://www.who.int/influenza/human_animal_interface/H5N1_cumulative_table_archives/en/). For H7N9, 1,223 laboratory-confirmed cases have been documented since the first case in March 2013 in China with a mortality rate of ∼31% (as of 14 February 2017, http://www.who.int/influenza/human_animal_interface/HAI_Risk_Assessment/en).
Although the isolation of H15 IAV is quite rare and geographically limited to Australia and only recently to western Siberia, we do not yet know what the outcome will be of zoonotic infections by H15 IAV. The H15 HAs are members of the HA subgroup that also include the H7N9 and H10N8 subtypes, which infected humans in 2013 (Fig. 1A). A recent study that compared the virulence of avian IAV HA from different subtypes to that of the H1N1 1918 virus revealed that the avian H7, H10, and H15 viruses (together with H1 and H6) contain mammalian virulence factors that help cause severe disease in mice (26). On the basis of this study, we can assume that the avian viruses from the H7-H10-H15 subgroup share common features that seem to increase their virulence in humans compared to some other avian HAs. One of the characteristics of the H7-H10-H15 subgroup is an extension of the 150-loop in the vicinity of the HA RBS that can contribute to antigenic variation of these HAs and perturb the binding of some broadly neutralizing antibodies that bind to the HA head domain (Fig. 4).
Over the years, different techniques and methods have been developed to elucidate the receptor specificity of the HA (for a review, see reference 42). The glycan array results for shWA79 H15 HA reveal, as expected, a clear preference for binding of avian-type receptors with no detectable binding of human-type receptors. Within avian-type receptors, H15 HA shows enhanced avidity for short, sulfated, linear glycans and also long, branched, O-linked and N-linked glycans (Fig. 1C) that is reminiscent of the binding profile of H7N9 HAs (38; R. P. de Vries, W. Peng, O. C. Grant, X. Zhu, K. M. Bouwman, I. N. Ambepitiya Wickramasinghe, C. A. M. de Haan, W. Yu, R. McBride, R. J. Woods, M. H. Verheije, I. A. Wilson, and J. C. Paulson, submitted for publication). For H7 HAs, it was previously shown that they bound sulfated receptor analogs, and this was recognized as a characteristic feature of the receptor specificity of H7 poultry IAV (51, 52). The glycan array results indicate a similar favored binding of H15 HAs to sulfated receptors, although there are no reports of H15 virus isolation in poultry or terrestrial birds.
In an attempt to explain this phenotype, it was suggested that binding of sulfated analogs is correlated with a basic side chain at position 193 (Lys) (51, 52). However, the crystal structure of the H7N9 HA from a human isolate (38) revealed interaction of the sulfate group with the main chain of Ser227 and carboxyl side chain of Glu190, but not with Lys193. For H15 HAs, a Lys at position 193 is conserved in all viruses isolated in Australia (Fig. 2B; see Fig. S1A in the supplemental material); notwithstanding, mutagenesis of H15 RBS Gln226 and Gly228 resulted in loss of binding to sulfated analogs in the glycan array (Fig. 6).
Mutating the same RBS residues in H15 HA that were required to switch receptor specificity of the H1N1, H2N2, and H3N2 pandemic strains was not sufficient to change receptor specificity for the H15 HA. These results together with comparable studies on H7N9 and H10N8 receptor specificity (53; de Vries et al., submitted) indicate that additional substitutions are required for these avian IAV isolated to attain human HA specificity and concomitantly lose avian HA binding. Indeed, studies on the H5N1 that included transmission experiments between ferrets (54–56) show that additional substitutions are required to acquire the ability to transmit in the human population. Although adaptive substitutions that result in changing receptor specificity of IAV viruses are rare events, they nevertheless can occur as reported for A/Shanghai/1/2013 H7N9. The first H7N9 virus isolated from a human contains the avian-specific Gln at position 226, whereas more-recent viruses contain Ile or Leu (38, 57), signifying a need for continued surveillance of these avian IAV. Taking all the data together, although avian H15 viruses currently appear to be of low risk to the human population, it is necessary to continue to monitor such viruses, especially since they seem to be increasing their geographical range in the last few years.
MATERIALS AND METHODS
Expression and purification of the H15 HAs for crystallization.
Codon-optimized H15 HA-encoding cDNAs (Genscript, USA) of A/shearwater/Western Australia/2576/1979 (shWA79) (H15N9) [GISAID accession number EPI129343], was cloned into a pFastBac vector. The H15 HA was expressed in Hi5 insect cells with an N-terminal gp67 signal peptide, a C-terminal thrombin cleavage site, a foldon trimerization sequence, and a His6 tag, as described previously (42). The expressed HA0s were purified through a His tag affinity purification step, dialyzed against 20 mM Tris-HCl–100 mM NaCl (pH 8.0) and then cleaved by thrombin to remove the foldon and His6 tag. The digested protein was purified further by gel filtration chromatography using a Superdex-200 column (Pharmacia). The protein eluted as an HA0 trimer, even after trypsin treatment, and was concentrated to 5 mg/ml.
Expression and purification of HAs for the glycan microarray.
The H15 HA expression was similar to that used for the crystallization experiments. The expressed HA0 was purified via His tag affinity chromatography, dialyzed against phosphate-buffered saline (PBS), and concentrated to 1 mg/ml prior to binding assays.
Crystallization and structural determination of the H15 HA.
Crystals of the H15 HA were obtained using the vapor diffusion sitting drop method at 20°C against a reservoir solution containing 20% (wt/vol) polyethylene glycol 3500 (PEG 3500) and 0.2 M potassium nitrate (pH 6.9). The HA receptor complex was obtained by soaking HA crystals in the reservoir solution that contained receptor analog 3′-SLN at a final concentration of 5 mM. Prior to data collection, the crystals were flash cooled in liquid nitrogen. Diffraction data were collected at the Advanced Photon Source (APS) (Table 1). Data were integrated and scaled using HKL2000 (58). The H15 apo structure was solved by molecular replacement method using Phaser (59) with the H7N9 HA structure (PDB ID 4N5J) as a search model. The refined apo structure was used as the starting model for determination of the H15 HA-glycan complex structure. Structure refinement was carried out in Phenix (60), and model building was performed with COOT (61). Final refinement statistics are summarized in Table 1.
Glycan microarray analysis of HAs expressed in insect cells.
Purified, soluble trimeric HA was precomplexed with horseradish peroxidase (HRP)-linked anti-His tag mouse antibody and with Alexa Fluor 488-linked anti-mouse IgG (4:2:1 molar ratio) for 15 min on ice in 100 μl of PBS with Tween 20 (PBS-T), and incubated on the array surface in a humidified chamber for 90 min. The slides were subsequently washed by successive rinses with PBS-T, PBS, and deionized H2O. Washed arrays were dried by centrifugation and immediately scanned for fluorescein isothiocyanate (FITC) signal on a Perkin-Elmer ProScanArray Express confocal microarray scanner. The fluorescent-signal intensity was measured using Imagene (Biodiscovery), and the mean intensity minus mean background was calculated and graphed using Microsoft Excel. For each glycan, the mean signal intensity was calculated from six replicate spots. The highest and lowest signals of the six replicates were removed, and the remaining four replicates were used to calculate the mean signal, standard deviation (SD), and standard error of the mean (SEM). Bar graphs show the averaged mean signal minus background for each glycan sample, and error bars are the SEM value. A list of glycans on the microarray is included in Table S1 in the supplemental material.
Accession number(s).
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession codes 5TG8 for H15 HA in the apo form and 5TG9 in complex with 3′-SLN.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by National Institutes of Health grants R56 AI117675 (to I.A.W.) and R01 AI114730 (to J.C.P.) and a grant from the Kuang Hua Educational Foundation (to J.C.P.). We thank Robyn Stanfield, X. Dai, and M. Elsliger for crystallographic and computational support, Henry Tien of the Robotics Core at the Joint Center for Structural Genomics and Wilson lab for automated crystal screening (supported by NIH grant U54 GM094586), the staff at the Advanced Photon Source beamline 23ID-B (GM/CA CAT). GM/CA CAT is funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and NIGMS (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy (DOE), Basic Energy Sciences, Office of Science, under contract DE-AC02-06CH11357.
This project was designed by N.T., J.C.P., and I.A.W. X-ray structure determination and analysis were performed by N.T. Glycan array studies were performed by N.T., C.M.N., W.P., and R.M. The manuscript was written by N.T., J.C.P., and I.A.W.
Footnotes
This article is manuscript 29334 from The Scripps Research Institute.
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00046-17.
REFERENCES
- 1.Reed KD, Meece JK, Henkel JS, Shukla SK. 2003. Birds, migration and emerging zoonoses: West Nile virus, Lyme disease, influenza A and enteropathogens. Clin Med Res 1:5–12. doi: 10.3121/cmr.1.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Munster VJ, Baas C, Lexmond P, Waldenstrom J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WE, Schutten M, Olsen B, Osterhaus AD, Fouchier RA. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog 3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B, Osterhaus AD. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79:2814–2822. doi: 10.1128/JVI.79.5.2814-2822.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamblin SJ, Skehel JJ. 2010. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem 285:28403–28409. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skehel JJ, Wiley DC. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 69:531–569. doi: 10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 6.Air GM. 1981. Sequence relationships among the hemagglutinin genes of 12 subtypes of influenza A virus. Proc Natl Acad Sci U S A 78:7639–7643. doi: 10.1073/pnas.78.12.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nobusawa E, Aoyama T, Kato H, Suzuki Y, Tateno Y, Nakajima K. 1991. Comparison of complete amino acid sequences and receptor-binding properties among 13 serotypes of hemagglutinins of influenza A viruses. Virology 182:475–485. doi: 10.1016/0042-6822(91)90588-3. [DOI] [PubMed] [Google Scholar]
- 8.Li Q, Sun X, Li Z, Liu Y, Vavricka CJ, Qi J, Gao GF. 2012. Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus. Proc Natl Acad Sci U S A 109:18897–18902. doi: 10.1073/pnas.1211037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO. 2013. New World bats harbor diverse influenza A viruses. PLoS Pathog 9:e1003657. doi: 10.1371/journal.ppat.1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, Wu Y, Tefsen B, Shi Y, Gao GF. 2014. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol 22:183–191. doi: 10.1016/j.tim.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu X, Yang H, Guo Z, Yu W, Carney PJ, Li Y, Chen LM, Paulson JC, Donis RO, Tong S, Stevens J, Wilson IA. 2012. Crystal structures of two subtype N10 neuraminidase-like proteins from bat influenza A viruses reveal a diverged putative active site. Proc Natl Acad Sci U S A 109:18903–18908. doi: 10.1073/pnas.1212579109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu X, Yu W, McBride R, Li Y, Chen LM, Donis RO, Tong S, Paulson JC, Wilson IA. 2013. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc Natl Acad Sci U S A 110:1458–1463. doi: 10.1073/pnas.1218509110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matrosovich M, Stech J, Klenk HD. 2009. Influenza receptors, polymerase and host range. Rev Sci Tech 28:203–217. [DOI] [PubMed] [Google Scholar]
- 14.Rogers GN, Pritchett TJ, Lane JL, Paulson JC. 1983. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology 131:394–408. doi: 10.1016/0042-6822(83)90507-X. [DOI] [PubMed] [Google Scholar]
- 15.Hansbro PM, Warner S, Tracey JP, Arzey KE, Selleck P, O'Riley K, Beckett EL, Bunn C, Kirkland PD, Vijaykrishna D, Olsen B, Hurt AC. 2010. Surveillance and analysis of avian influenza viruses, Australia. Emerg Infect Dis 16:1896–1904. doi: 10.3201/eid1612.100776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hurt AC, Hansbro PM, Selleck P, Olsen B, Minton C, Hampson AW, Barr IG. 2006. Isolation of avian influenza viruses from two different transhemispheric migratory shorebird species in Australia. Arch Virol 151:2301–2309. doi: 10.1007/s00705-006-0784-1. [DOI] [PubMed] [Google Scholar]
- 17.Muzyka D, Pantin-Jackwood M, Spackman E, Stegniy B, Rula O, Shutchenko P. 2012. Avian influenza virus wild bird surveillance in the Azov and Black Sea regions of Ukraine (2010-2011). Avian Dis 56:1010–1016. doi: 10.1637/10157-040912-ResNote.1. [DOI] [PubMed] [Google Scholar]
- 18.Sivay MV, Baranovich T, Marchenko VY, Sharshov KA, Govorkova EA, Shestopalov AM, Webby RJ. 2013. Influenza A (H15N4) virus isolation in Western Siberia, Russia. J Virol 87:3578–3582. doi: 10.1128/JVI.02521-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention. 2013. Emergence of avian influenza A(H7N9) virus causing severe human illness - China, February-April 2013. MMWR Morb Mortal Wkly Rep 62:366–371. [PMC free article] [PubMed] [Google Scholar]
- 20.Centers for Disease Control, ROC (Taiwan). 21 June 2013. Laboratory-confirmed case of human infection with avian influenza A(H6N1) virus in Taiwan recovered; Taiwan CDC urges public to take precautions to stay healthy. Centers for Disease Control, ROC (Taiwan), Taipei City, Taiwan (Republic of China). [Google Scholar]
- 21.Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J, Zou S, Yang L, Chen T, Dong L, Bo H, Zhao X, Zhang Y, Lan Y, Bai T, Dong J, Li Q, Wang S, Zhang Y, Li H, Gong T, Shi Y, Ni X, Li J, Zhou J, Fan J, Wu J, Zhou X, Hu M, Wan J, Yang W, Li D, Wu G, Feng Z, Gao GF, Wang Y, Jin Q, Liu M, Shu Y. 2014. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet 383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- 22.Garcia-Sastre A, Schmolke M. 2014. Avian influenza A H10N8 - a virus on the verge? Lancet 383:676–677. doi: 10.1016/S0140-6736(14)60163-X. [DOI] [PubMed] [Google Scholar]
- 23.Shi W, Shi Y, Wu Y, Liu D, Gao GF. 2013. Origin and molecular characterization of the human-infecting H6N1 influenza virus in Taiwan. Protein Cell 4:846–853. doi: 10.1007/s13238-013-3083-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei SH, Yang JR, Wu HS, Chang MC, Lin JS, Lin CY, Liu YL, Lo YC, Yang CH, Chuang JH, Lin MC, Chung WC, Liao CH, Lee MS, Huang WT, Chen PJ, Liu MT, Chang FY. 2013. Human infection with avian influenza A H6N1 virus: an epidemiological analysis. Lancet Respir Med 1:771–778. doi: 10.1016/S2213-2600(13)70221-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan J, Zhang L, Kan X, Jiang L, Yang J, Guo Z, Ren Q. 2013. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin Infect Dis 57:1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- 26.Qi L, Pujanauski LM, Davis AS, Schwartzman LM, Chertow DS, Baxter D, Scherler K, Hartshorn KL, Slemons RD, Walters KA, Kash JC, Taubenberger JK. 2014. Contemporary avian influenza A virus subtype H1, H6, H7, H10, and H15 hemagglutinin genes encode a mammalian virulence factor similar to the 1918 pandemic virus H1 hemagglutinin. mBio 5:e02116. doi: 10.1128/mBio.02116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng W, de Vries RP, Grant OC, Thompson AJ, McBride R, Tsogtbaatar B, Lee PS, Razi N, Wilson IA, Woods RJ, Paulson JC. 2017. Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe 21:23–34. doi: 10.1016/j.chom.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohm C, Zhou N, Suss J, Mackenzie J, Webster RG. 1996. Characterization of a novel influenza hemagglutinin, H15: criteria for determination of influenza A subtypes. Virology 217:508–516. doi: 10.1006/viro.1996.0145. [DOI] [PubMed] [Google Scholar]
- 29.Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O'Neil RE, Faynboym AM, Horowitz M, Horowitz L, Ward AB, Palese P, Webby R, Lerner RA, Bhatt RR, Wilson IA. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489:526–532. doi: 10.1038/nature11414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA. 2014. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, Vogels R, Brakenhoff JP, Kompier R, Koldijk MH, Cornelissen LA, Poon LL, Peiris M, Koudstaal W, Wilson IA, Goudsmit J. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, van der Vlugt R, Lamrani M, Korse HJ, Geelen E, Sahin O, Sieuwerts M, Brakenhoff JP, Vogels R, Li OT, Poon LL, Peiris M, Koudstaal W, Ward AB, Wilson IA, Goudsmit J, Friesen RH. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337:1343–1348. doi: 10.1126/science.1222908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu R, Ekiert DC, Krause JC, Hai R, Crowe JE Jr, Wilson IA. 2010. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science 328:357–360. doi: 10.1126/science.1186430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu X, Guo YH, Jiang T, Wang YD, Chan KH, Li XF, Yu W, McBride R, Paulson JC, Yuen KY, Qin CF, Che XY, Wilson IA. 2013. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol 87:12619–12635. doi: 10.1128/JVI.01577-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin T, Wang G, Li A, Zhang Q, Wu C, Zhang R, Cai Q, Song W, Yuen KY. 2009. The hemagglutinin structure of an avian H1N1 influenza A virus. Virology 392:73–81. doi: 10.1016/j.virol.2009.06.028. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Stevens DJ, Haire LF, Walker PA, Coombs PJ, Russell RJ, Gamblin SJ, Skehel JJ. 2009. Structures of receptor complexes formed by hemagglutinins from the Asian influenza pandemic of 1957. Proc Natl Acad Sci U S A 106:17175–17180. doi: 10.1073/pnas.0906849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiong X, Coombs PJ, Martin SR, Liu J, Xiao H, McCauley JW, Locher K, Walker PA, Collins PJ, Kawaoka Y, Skehel JJ, Gamblin SJ. 2013. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 497:392–396. doi: 10.1038/nature12144. [DOI] [PubMed] [Google Scholar]
- 38.Xu R, de Vries RP, Zhu X, Nycholat CM, McBride R, Yu W, Paulson JC, Wilson IA. 2013. Preferential recognition of avian-like receptors in human influenza A H7N9 viruses. Science 342:1230–1235. doi: 10.1126/science.1243761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, McCauley JW, Collins PJ, Walker PA, Skehel JJ, Gamblin SJ. 2013. Receptor binding by an H7N9 influenza virus from humans. Nature 499:496–499. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, de Vries RP, Tzarum N, Zhu X, Yu W, McBride R, Paulson JC, Wilson IA. 2015. A human-infecting H10N8 influenza virus retains a strong preference for avian-type receptors. Cell Host Microbe 17:377–384. doi: 10.1016/j.chom.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. 2000. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol 74:8502–8512. doi: 10.1128/JVI.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevens J, Blixt O, Paulson JC, Wilson IA. 2006. Glycan microarray technologies: tools to survey host specificity of influenza viruses. Nat Rev Microbiol 4:857–864. doi: 10.1038/nrmicro1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, Garcia-Sastre A. 2007. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315:655–659. doi: 10.1126/science.1136212. [DOI] [PubMed] [Google Scholar]
- 44.Connor RJ, Kawaoka Y, Webster RG, Paulson JC. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 45.Centers for Disease Control and Prevention. 2012. Notes from the field: highly pathogenic avian influenza A (H7N3) virus infection in two poultry workers – Jalisco, Mexico, July 2012. MMWR Morb Mortal Wkly Rep 61:726–727. [PubMed] [Google Scholar]
- 46.Claas EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, Krauss S, Shortridge KF, Webster RG. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472–477. doi: 10.1016/S0140-6736(97)11212-0. [DOI] [PubMed] [Google Scholar]
- 47.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 48.Guo Y, Li J, Cheng X. 1999. Discovery of men infected by avian influenza A (H9N2) virus. Chinese J Exp Clin Virol 13:105–108. (In Chinese.) doi: 10.1016/S1386-6532(99)00027-X. [DOI] [PubMed] [Google Scholar]
- 49.Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, Bosman A. 2004. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 363:587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- 50.Yuen KY, Chan PK, Peiris M, Tsang DN, Que TL, Shortridge KF, Cheung PT, To WK, Ho ET, Sung R, Cheng AF. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467–471. doi: 10.1016/S0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 51.Gambaryan AS, Matrosovich TY, Philipp J, Munster VJ, Fouchier RA, Cattoli G, Capua I, Krauss SL, Webster RG, Banks J, Bovin NV, Klenk HD, Matrosovich MN. 2012. Receptor-binding profiles of H7 subtype influenza viruses in different host species. J Virol 86:4370–4379. doi: 10.1128/JVI.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gambaryan AS, Tuzikov AB, Pazynina GV, Desheva JA, Bovin NV, Matrosovich MN, Klimov AI. 2008. 6-Sulfo sialyl Lewis X is the common receptor determinant recognized by H5, H6, H7 and H9 influenza viruses of terrestrial poultry. Virol J 5:85. doi: 10.1186/1743-422X-5-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tzarum N, de Vries RP, Peng W, Thompson AJ, Bouwman KM, McBride R, Yu W, Zhu X, Verheije MH, Paulson JC, Wilson IA. 2017. The 150-loop restricts the host specificity of human H10N8 influenza virus. Cell Rep 19:235–245. doi: 10.1016/j.celrep.2017.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen LM, Blixt O, Stevens J, Lipatov AS, Davis CT, Collins BE, Cox NJ, Paulson JC, Donis RO. 2012. In vitro evolution of H5N1 avian influenza virus toward human-type receptor specificity. Virology 422:105–113. doi: 10.1016/j.virol.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, Li C, Kawakami E, Yamada S, Kiso M, Suzuki Y, Maher EA, Neumann G, Kawaoka Y. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486:420–428. doi: 10.1038/nature10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi Y, Zhang W, Wang F, Qi J, Wu Y, Song H, Gao F, Bi Y, Zhang Y, Fan Z, Qin C, Sun H, Liu J, Haywood J, Liu W, Gong W, Wang D, Shu Y, Wang Y, Yan J, Gao GF. 2013. Structures and receptor binding of hemagglutinins from human-infecting H7N9 influenza viruses. Science 342:243–247. doi: 10.1126/science.1242917. [DOI] [PubMed] [Google Scholar]
- 58.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 59.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr 61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 60.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 62.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.