Abstract
Clonal mosaicism arises when a post-zygotic mutational event is detectable in subpopulations of cells as an alternative genotype while not present in the germline genome. Although described in a subset of pediatric disorders, new genomic technologies have detected higher than anticipated frequencies of clonal mosaicism in adult population studies, stimulating investigation as to how clonal mosaicism could contribute to chronic human diseases, such as cancer, diabetes and neurodegenerative disorders. It has also been postulated to be an important mechanism for functional cellular diversity, including the brain. Early studies have characterized the spectrum of detectable mosaic alterations and have begun to investigate whether detectable mosaicism could be important as an overall biomarker for risk or in the case of hematologic cancers, identification of preleukemic clones.
Introduction and Background
Classically, genetic mosaicism is the presence of one or more distinct populations of cells within an individual with acquired genomic alteration(s) that differ from the inherited, germline genome (Figure 1)[1,2]. Mosaicism is distinct from de novo mutations, namely those detected in the offspring, but not the parents; in some pediatric diseases, de novo mutations account for an increasing, measurable fraction of the identified mutational events, mainly due to next generation sequencing[3,4]. Genetic mosaicism can be differentiated from chimerism because the former arises post-zygotically (e.g., somatic) whereas the later refers to the presence of cells from another individual, as in fetal cells detectable in the circulating blood of post-partum women[5]. Genetic mosaicism is not considered to be constitutional, but instead occurs in tissue compartments that undergo somatic mutation post-fertilization and either disrupt normal cellular function or as we are learning in larger population studies, can be tolerated with no immediate health consequences[6–9]. It is important to point out that acquired somatic mutations are important hallmarks of the cancer genome, driving abnormal proliferation as well as clinically dangerous disease[10]; in rare cases, somatic alterations can drive noncancerous diseases, particularly neurologic disorders[11]. It is notable that not all somatic alterations detected result in abnormal proliferation or cell survival. In fact, it has been suggested that mosaic events could contribute to normal development of the brain[12–14].
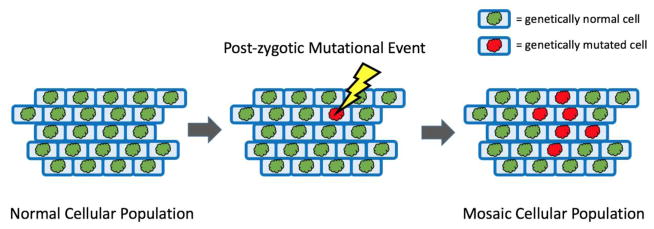
Figure 1.
Depiction of a genetically normal cellular population that acquires a somatic mutation and clonally expands to daughter cells to form a mosaic cellular population.
The new tools of genomic sciences, namely the massively parallel platforms that enable genotyping of single nucleotide polymorphisms (SNP) using microarrays and next generation sequencing, have led to the detection of a spectrum of genetic mosaic events. The majority of large survey studies examining detectable genetic mosaicism have characterized events in leukocytes DNA isolated from blood or buccal swabs. Examples of genetic mosaicism include mosaic single nucleotide substitutions[8,9], mosaic structural deletions larger than 1 megabase (Mb)[15–17] or mosaic gains of an entire chromosome[18]. Genetic mosaicism has been described in all chromosomes, including the sex chromosomes at a higher frequency[19–21] than the autosomes[6,7,17,18]; so far, large surveys have not yet provided an accurate estimate of the frequency of mosaic mitochondrial DNA[22,23]. Current technologies permit detection of cellular fractions with alternative genotypes in between 5 and 95% of circulating leukocytes and have shown these fractions can increase by approximately 1% per year in adult cohorts[18].
Instructive rare case reports and syndromes have provided sufficient evidence that acquired somatic events early in development can result in pediatric disorders. Mosaic trisomy 21 accounts for 2–4% of Down’s syndrome cases, which are distinct from complete trisomy in that the former can have less severe intellectual and developmental manifestations[24]. Mosaic Turner’s syndrome, also known as mosaic X loss, also manifests in a less severe phenotype than classical Turner’s syndrome[25]. McCune-Albright syndrome is an example of a potentially embryonic lethal genetic mutation in the GNAS1 gene that can be present in the mosaic state[26]. Likewise, Proteus syndrome is believed to be caused by early mosaic mutations in the AKT1 oncogene[27], an oncogene implicated in several solid tumors[28,29]. Additionally, embryonic mosaic mutations in IDH1 and IDH2 have been linked to Ollier disease and Maffucci syndrome[30] and early mosaic mutations in HRAS, KRAS, and NRAS have been associated with nevus sebaceous[31], Schimmelpenning syndrome[31], and keratinocytic epidermal nevus syndrome[32].
The genetics of mosaicism
How and why mosaic events arise remains a challenging question, but the evidence so far points towards three issues, none of which can be adequately reviewed here: (1) errors in DNA replication in different tissues have distinct errors rates and mechanisms[33,34]; (2) timing of the mutational event- does it occur early or later in life- as part of senescence; and (3) the capacity to be tolerated and actually increase with age. The initiating event is likely related to errors in background DNA repair, transcription coupled repair or nondisjunction in cell division. In rare and informative cases, ‘endogenous’ events can be the result of inherited mutations in genomic maintenance (eg. CEP57, BUB1B and NF1)[35–37], breakdowns in DNA repair and stability pathways or age-related telomere attrition[38]. For example, familial truncating mutations in CEP57 result in variegated aneuploidy and predisposition to cancer[35].
Genomic aberrations can occur early in development when cells are rapidly undergoing cell division to expand from a zygote into differentiated organs and tissues[39]. These rapidly dividing cells are most susceptible to acquiring errors in transcription as well as undergoing damage by environmental agents. An initiating event can also be driven by exogenous factors, such as tobacco smoke[20,40], radiation[41] or chemotherapy[42] exposure that, in turn, lead to DNA damage. So far, large prospective birth cohorts have not progressed to the age at which genetic mosaicism is more readily detected; consequently, we must speculate as to whether some or all events occur early in life, remain below detection and at a later time, steadily increase as a fraction of the overall cell population (Figure 2a). Alternatively, it is plausible that mutational events can accumulate and increase with age, as senescence sets in and the host capacity to remove cells with alternative genotypes is hampered (Figure 2b). We must also consider that mosaic subpopulations can populate more efficiently because the mutation can confer a selective advantage such as when a cell produces daughter cells at an elevated rate compared to other normal, non-mutated cells of the same tissue type, or subpopulations could arise due to the depletion of the pool of tissue progenitor cells such that the population of cells in the tissue are comprised of a detectible portion of mutated daughter cells. Understanding the temporality of when mosaic events arise and what aspects govern their clonal expansion is an important aspect to evaluate when considering the use of genetic mosaicism as a marker for disease risk.
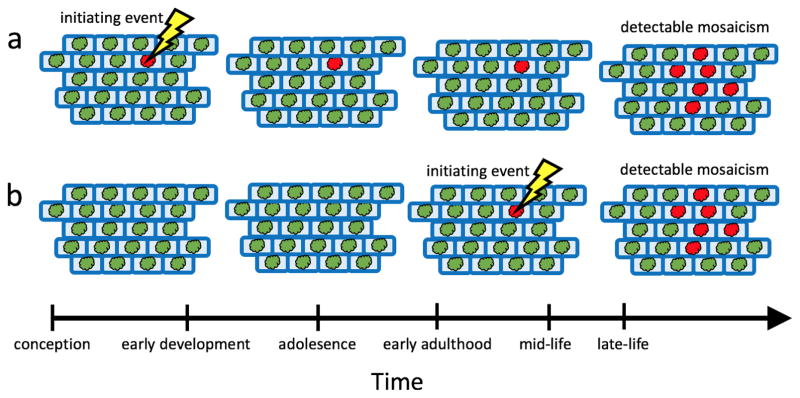
Figure 2.
Exploring two biologically plausible models for the acquisition of mosaicism. In the first model (a), a somatic event is acquired early in development during periods of rapid cellular growth and division, but the aberrant cells remain at low cellular proportions until later in life when changes in the cellular environment confer a selective advantage of aberrant clones over normal cells allowing them to expand to a detectable proportion of the cellular population. In the second model (b), somatic events are acquired later in life and soon after clonally expand to become a detectable proportion of the cellular population.
Based on the surveys of genetic mosaicism in large-population-based studies, there are four attributes that could contribute to its possible impact on health[2,43]. First, the genomic location of the event could result in alterations affecting important housekeeping or cell cycle genes that have greater potential to impact health. Examples already have been described in pediatric diseases for years, such as mosaic trisomy 18 and mosaic trisomy 21 (e.g., Down’s syndrome). Similarly, there is evidence that it could contribute to neurodevelopment[11]; rare, informative cases point towards mosaic events as an explanation for neurodevelopmental problems, such as hemimegalencephaly due to mosaicism of the AKT3 gene as well as others in the mTOR pathway[44]. Second, the proportion of cells with an alternative genotype may need to reach a critical fraction at which point deleterious consequences arise, suggesting that low mosaic proportions could have sub-clinical or be tolerated with no apparent health effects. Third, the consequences of the altered genotypes could vary by specific tissue types, as seen in Proteus Syndrome[27]. For example, phenotypic manifestations of a mutated melanocyte could be quite benign, but the same mutation in a vital tissue or organ could have life altering or potentially fatal outcomes; examples of this has been described for TP53 and KRAS mutations in the skin[32,45]. Fourth, the timing of the mutation has important developmental consequences, as observed in pediatric disorders cited above.
Detectible clonal mosaicism in the population
Recently, new bioinformatic algorithms have been applied to SNP microarrays analyzed in large adult population-based studies, providing estimates of the frequencies of different types of events[46,47]. Examination of B allele frequency and log R ratio intensity signals yields high-resolution karyotypes capable of detecting large (>2 Mb) structural mosaicism in samples of tens of thousands of individuals[46,48–50]. Unexpectedly, the initial studies of mosaic events discovered large structural genetic changes in the autosomes of approximately 1% of adult participants, nearly half of whom were healthy and asymptomatic, suggesting these events are tolerated[6,7,17]. Evidence suggests a striking relationship between increasing age and increasing frequency of mosaicism with frequencies in excess of 2% in individuals over 80 years of age[6,7,16]. A combined analysis of DNA from leukocytes from nearly 130,000 individuals provided additional evidence for the association of mosaicism with increasing age as well as suggested elevated frequencies of mosaicism in males[18].
Hot spot regions of recurrent mosaicism have also been identified in regions commonly altered in hematologic malignancies such as mosaic 13q14 deletions and 20q deletions[15,18]. Studies have established a strong link between clonal mosaicism detected in the blood and risk of future hematologic malignancies[6,7,51], with mosaic hematologic cancer associated mutations present in blood derived DNA as many as 14 years prior to diagnosis. Another study also suggests large structural clonal mosaicism may be associated with type 2 diabetes, and in particular, associated with vascular complications[52].
SNP array studies have identified elevated frequencies of mosaicism of the sex chromosomes. Mosaicism on the X chromosome is 4 times more common than autosomal mosaicism and the female inactivated X chromosome is preferentially affected, interestingly similar to the higher rate of somatic alterations observed in the inactivated X chromosome in many cancers[19,53]. Mosaic chromosome Y loss is more common than autosomal and X mosaicism, reaching frequencies in excess of 15% in men over 80 years old[20,21]. Smoking is an important risk factor for mosaic Y loss and its effect attenuates after smoking cessation[20,40]. It is not clear that mosaic Y loss represents a risk factor for cancer overall[20,21]. New evidence suggests a connection between mosaic Y loss and Alzheimer’s risk[54], although further replication is needed. Interestingly, a germline variant near TCL1A has been found to be associated with susceptibility to mosaic Y loss[20], suggesting a heritable predisposition to at least mosaicism of the Y chromosome.
A combined analysis of large scale exome (e.g., all of the coding exons in the genome) sequencing projects has yielded new insights into the frequency and class of genes frequently displaying mosaicism of single nucleotide variants (SNVs) in leukocytes. Age-related frequencies of mosaic SNVs were elevated in individuals over 60 years old (approximately 10% frequency) as compared to individuals younger than 50 years of age (approximately 1% frequency)[8,9]. Mosaic SNVs were associated with hematologic cancer risk as well as coronary heart disease and ischemic stroke[8,9]. As with large-scale mosaic events >2 Mb in size, mosaicism of single nucleotides preferentially clusters in certain genomic regions. Genes in which mosaic SNVs are commonly detected include DNMT3A, TET2, and ASXL1[8,9,55–57]. These are key genes commonly mutated in hematologic cancers and therefore cells harboring such mutations, while still phenotypically normal, may have greater potential to progress to precancerous or cancerous states. Similarly, an analysis of DNA derived from normal skin biopsies also detected high burdens of mosaic point mutations in a panel 74 cancer genes[45]. Very high sequencing coverage (average of 500X) detected an average of 140 mosaic mutations per square centimeter of skin, suggesting aged, sun-exposed skin is a patchwork of mosaic clones that carry mutations in cancer driver genes, but yet still retains normal physiological characteristics.
Detection of genetic mosaicism as a biomarker
The use of SNP microarrays and next generation sequencing platforms has yielded new opportunities to investigate whether detectable genetic mosaicism in leukocytes as well as in buccal swabs or skin could serve as an effective biomarker for chronic diseases associated with aging (e.g., cancer, diabetes or neurodegenerative disorders). Examples of such biomarkers may be to search for mosaic DNMT3A mutations in leukocytes or mosaic KRAS mutations in epidermal cells as a means to detect precancerous lesions. However, it is still too early to determine whether the presence of specific somatic alterations directly leads to disease or perhaps is a barometer of overall genomic integrity. In this regard, a higher burden of events could stochastically increase the likelihood of one or more events that can drive a cancer or lead to complications of diabetes or neurodegenerative disorders[10]. In preliminary studies, it has not been possible to identify large structural mosaic alterations in the tumors that exactly correspond to what was observed in peripheral leukocytes[17]. Large structural mosaic deletions of 13q14 or 20q in leukocyte-derived DNA have been associated with increased risk for hematological neoplasms[6,7,15] and could be used as an example of how detection of events that are characteristic of a hematologic neoplasm could be used as an early biomarker. For instance, in a prospective cohort, it was possible to detect a mosaic 13q14 deletion fourteen years before the diagnosis of chronic lymphocytic leukemia (CLL)[7]. However, not all individuals with 13q14 deletion mosaicism develop CLL, which suggests that some can tolerate this event, or at least until other drivers arise in the same subpopulation that eventually lead to CLL. Further prospective studies with sampling at multiple time points are needed to follow individuals with mosaic mutations in genes or regions commonly associated with cancer over time so precise measures of risk can be determined and used to inform clinical management.
Cataloging mosaic mutation profiles could lead to accurate and informative predictors for early detection and thus reduce severity of disease. Past studies have suggested associations of mosaicism with a host of rare disorders as well as adult onset diseases such as hematologic cancers[6,7], type 2 diabetes[52], Alzheimer’s disease[54], coronary heart disease[8] and ischemic stroke[8]. For example, sequencing data from a large-scale case-control study suggests mosaic protein truncating mutations in PPM1D may be a risk factor for breast and ovarian cancer[58], although further prospective studies are needed to verify this relationship. Additionally, a recent investigation of uninvolved margin tissue from breast cancer cases suggests mosaic copy number aberrations are present in over a third of cases and often include mosaic gains of ERBB2 and growth factor receptor genes[59]. Better understanding which events are important for future disease risk and the penetrance of such events will be important as the field moves forward in assessing clonal mosaicism as markers of risk for common diseases.
Key opportunities for future investigation
Genetic mosaicism has great potential to be used as an informative genetic intermediate between normal and disease states, nevertheless much remains to be understood before genetic mosaicism can be effectively used as a biomarker for early detection, as part of ‘precision prevention’[60]. Further methodological work is needed to improve the accuracy of detection before testing in larger, prospective cohorts to assess the possible impact on risk for a spectrum of diseases. Methods exist to detect mosaic SNVs and very large structural events, but current methods fail to provide good estimates on the frequency and distribution of intermediate-sized mosaic events, which is further confounded by difficulty in filtering inherited copy number variants. To date, most studies have focused on DNA from accessible tissues such as blood or buccal cells, but to fully investigate the role of genetic mosaicism in disease multiple tissue types, particularly tissues affected by solid tumors or other common diseases, need to be interrogated. Serial sampling from large, prospectively collected cohort studies with thorough characterization of exposures and phenotypes should yield new insights and serve as the foundation to explore the role of genetic mosaicism in human health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strachan T, Read AP, Strachan T. Human molecular genetics. 4. New York: Garland Science; 2011. [Google Scholar]
- 2.Machiela MJ, Chanock SJ. Detectable clonal mosaicism in the human genome. Semin Hematol. 2013;50:348–359. doi: 10.1053/j.seminhematol.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Veltman JA, Brunner HG. De novo mutations in human genetic disease. Nat Rev Genet. 2012;13:565–575. doi: 10.1038/nrg3241. [DOI] [PubMed] [Google Scholar]
- 4.Hoischen A, Krumm N, Eichler EE. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat Neurosci. 2014;17:764–772. doi: 10.1038/nn.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianchi DW, Zickwolf GK, Weil GJ, Sylvester S, DeMaria MA. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc Natl Acad Sci U S A. 1996;93:705–708. doi: 10.1073/pnas.93.2.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laurie CC, Laurie CA, Rice K, Doheny KF, Zelnick LR, McHugh CP, Ling H, Hetrick KN, Pugh EW, Amos C, et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet. 2012;44:642–650. doi: 10.1038/ng.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jacobs KB, Yeager M, Zhou W, Wacholder S, Wang Z, Rodriguez-Santiago B, Hutchinson A, Deng X, Liu C, Horner MJ, et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet. 2012;44:651–658. doi: 10.1038/ng.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. A scan of exome sequencing data for mosaic point mutations in genes commonly mutated in hematologic malignancies. Findings suggest mosaic SNVs increase with age and may be associated with hematologic cancer and all-cause mortality. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. Identified mosaic SNVs in exome sequencing data based on unusual allelic fractions. Found evidence mosaic SNVs are age-related and associated with hematologic cancers and death. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441:1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- 14.Baillie JK, Barnett MW, Upton KR, Gerhardt DJ, Richmond TA, De Sapio F, Brennan PM, Rizzu P, Smith S, Fell M, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machiela MJ, Zhou W, Caporaso N, Dean M, Gapstur SM, Goldin L, Stevens VL, Yeager M, Chanock SJ. Mosaic 13q14 deletions in peripheral leukocytes of non-hematologic cancer cases and healthy controls. J Hum Genet. 2016;61:411–418. doi: 10.1038/jhg.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forsberg LA, Rasi C, Razzaghian HR, Pakalapati G, Waite L, Thilbeault KS, Ronowicz A, Wineinger NE, Tiwari HK, Boomsma D, et al. Age-related somatic structural changes in the nuclear genome of human blood cells. Am J Hum Genet. 2012;90:217–228. doi: 10.1016/j.ajhg.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Santiago B, Malats N, Rothman N, Armengol L, Garcia-Closas M, Kogevinas M, Villa O, Hutchinson A, Earl J, Marenne G, et al. Mosaic uniparental disomies and aneuploidies as large structural variants of the human genome. Am J Hum Genet. 2010;87:129–138. doi: 10.1016/j.ajhg.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18*.Machiela MJ, Zhou W, Sampson JN, Dean MC, Jacobs KB, Black A, Brinton LA, Chang IS, Chen C, Chen C, et al. Characterization of large structural genetic mosaicism in human autosomes. Am J Hum Genet. 2015;96:487–497. doi: 10.1016/j.ajhg.2015.01.011. This study of over 120,000 participants is the largest to date to characterize the frequency and distribution of autosomal mosaicism. Associations with age are replicated, evidence suggests males have elevated rates and an inverse association is observed between event size and population frequency. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Machiela MJ, Zhou W, Karlins E, Sampson JN, Freedman ND, Yang Q, Hicks B, Dagnall C, Hautman C, Jacobs KB, et al. Female chromosome X mosaicism is age-related and preferentially affects the inactivated X chromosome. Nat Commun. 2016;7:11843. doi: 10.1038/ncomms11843. The first study to characterize mosaicism on female X chromosomes. Estimated frequencies are four fold higher than on autosomes and a preference is observed for mosaicism to occur on the inactivated X chromosome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.Zhou W, Machiela MJ, Freedman ND, Rothman N, Malats N, Dagnall C, Caporaso N, Teras LT, Gaudet MM, Gapstur SM, et al. Mosaic loss of chromosome Y is associated with common variation near TCL1A. Nat Genet. 2016;48:563–568. doi: 10.1038/ng.3545. Mosaic Y loss is investigated in prospective cohort studies. While no strong associations are observed with cancer risk or mortality, a significant association is observed between a variant near TCL1A and risk of mosaic Y loss suggesting genetic susceptibility to mosaicism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Forsberg LA, Rasi C, Malmqvist N, Davies H, Pasupulati S, Pakalapati G, Sandgren J, Diaz de Stahl T, Zaghlool A, Giedraitis V, et al. Mosaic loss of chromosome Y in peripheral blood is associated with shorter survival and higher risk of cancer. Nat Genet. 2014;46:624–628. doi: 10.1038/ng.2966. First study to estimate the frequency of mosaic Y loss to be approximately 8%. Found evidenct to suggest mosaic Y loss is associated with all-cause and non-hematologic cancer mortality. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 24.Papavassiliou P, York TP, Gursoy N, Hill G, Nicely LV, Sundaram U, McClain A, Aggen SH, Eaves L, Riley B, et al. The phenotype of persons having mosaicism for trisomy 21/Down syndrome reflects the percentage of trisomic cells present in different tissues. Am J Med Genet A. 2009;149A:573–583. doi: 10.1002/ajmg.a.32729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sybert VP, McCauley E. Turner’s syndrome. N Engl J Med. 2004;351:1227–1238. doi: 10.1056/NEJMra030360. [DOI] [PubMed] [Google Scholar]
- 26.Weinstein LS, Shenker A, Gejman PV, Merino MJ, Friedman E, Spiegel AM. Activating mutations of the stimulatory G protein in the McCune-Albright syndrome. N Engl J Med. 1991;325:1688–1695. doi: 10.1056/NEJM199112123252403. [DOI] [PubMed] [Google Scholar]
- 27.Lindhurst MJ, Sapp JC, Teer JK, Johnston JJ, Finn EM, Peters K, Turner J, Cannons JL, Bick D, Blakemore L, et al. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med. 2011;365:611–619. doi: 10.1056/NEJMoa1104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 29.Testa JR, Tsichlis PN. AKT signaling in normal and malignant cells. Oncogene. 2005;24:7391–7393. doi: 10.1038/sj.onc.1209100. [DOI] [PubMed] [Google Scholar]
- 30.Amary MF, Damato S, Halai D, Eskandarpour M, Berisha F, Bonar F, McCarthy S, Fantin VR, Straley KS, Lobo S, et al. Ollier disease and Maffucci syndrome are caused by somatic mosaic mutations of IDH1 and IDH2. Nat Genet. 2011;43:1262–1265. doi: 10.1038/ng.994. [DOI] [PubMed] [Google Scholar]
- 31.Groesser L, Herschberger E, Ruetten A, Ruivenkamp C, Lopriore E, Zutt M, Langmann T, Singer S, Klingseisen L, Schneider-Brachert W, et al. Postzygotic HRAS and KRAS mutations cause nevus sebaceous and Schimmelpenning syndrome. Nat Genet. 2012;44:783–787. doi: 10.1038/ng.2316. [DOI] [PubMed] [Google Scholar]
- 32.Hafner C, Groesser L. Mosaic RASopathies. Cell Cycle. 2013;12:43–50. doi: 10.4161/cc.23108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gensler HL, Bernstein H. DNA damage as the primary cause of aging. Q Rev Biol. 1981;56:279–303. doi: 10.1086/412317. [DOI] [PubMed] [Google Scholar]
- 34.De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–185. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- 35.Snape K, Hanks S, Ruark E, Barros-Nunez P, Elliott A, Murray A, Lane AH, Shannon N, Callier P, Chitayat D, et al. Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat Genet. 2011;43:527–529. doi: 10.1038/ng.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schliekelman M, Cowley DO, O’Quinn R, Oliver TG, Lu L, Salmon ED, Van Dyke T. Impaired Bub1 function in vivo compromises tension-dependent checkpoint function leading to aneuploidy and tumorigenesis. Cancer Res. 2009;69:45–54. doi: 10.1158/0008-5472.CAN-07-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kehrer-Sawatzki H, Kluwe L, Sandig C, Kohn M, Wimmer K, Krammer U, Peyrl A, Jenne DE, Hansmann I, Mautner VF. High frequency of mosaicism among patients with neurofibromatosis type 1 (NF1) with microdeletions caused by somatic recombination of the JJAZ1 gene. American Journal of Human Genetics. 2004;75:410–423. doi: 10.1086/423624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–528. doi: 10.1038/nature08982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez LC, Torres M, Real FX. Somatic mosaicism: on the road to cancer. Nat Rev Cancer. 2016;16:43–55. doi: 10.1038/nrc.2015.1. [DOI] [PubMed] [Google Scholar]
- 40*.Dumanski JP, Rasi C, Lonn M, Davies H, Ingelsson M, Giedraitis V, Lannfelt L, Magnusson PK, Lindgren CM, Morris AP, et al. Mutagenesis. Smoking is associated with mosaic loss of chromosome Y. Science. 2015;347:81–83. doi: 10.1126/science.1262092. This study details an association between smoking and mosaic Y loss providing evidence that modifiable environmental exposures are risk factors for mosaic Y loss. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vilenchik MM, Knudson AG., Jr Inverse radiation dose-rate effects on somatic and germ-line mutations and DNA damage rates. Proc Natl Acad Sci U S A. 2000;97:5381–5386. doi: 10.1073/pnas.090099497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pharoah PD, Song H, Dicks E, Intermaggio MP, Harrington P, Baynes C, Alsop K, Bogdanova N, Cicek MS, et al. Australian Ovarian Cancer Study G. PPM1D Mosaic Truncating Variants in Ovarian Cancer Cases May Be Treatment-Related Somatic Mutations. J Natl Cancer Inst. 2016:108. doi: 10.1093/jnci/djv347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Youssoufian H, Pyeritz RE. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet. 2002;3:748–758. doi: 10.1038/nrg906. [DOI] [PubMed] [Google Scholar]
- 44.Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, Lehtinen MK, Hills LB, Heinzen EL, Hill A, Hill RS, et al. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron. 2012;74:41–48. doi: 10.1016/j.neuron.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martincorena I, Roshan A, Gerstung M, Ellis P, Van Loo P, McLaren S, Wedge DC, Fullam A, Alexandrov LB, Tubio JM, et al. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez JR, Rodriguez-Santiago B, Caceres A, Pique-Regi R, Rothman N, Chanock SJ, Armengol L, Perez-Jurado LA. A fast and accurate method to detect allelic genomic imbalances underlying mosaic rearrangements using SNP array data. BMC Bioinformatics. 2011;12:166. doi: 10.1186/1471-2105-12-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47**.Vattathil S, Scheet P. Extensive Hidden Genomic Mosaicism Revealed in Normal Tissue. Am J Hum Genet. 2016;98:571–578. doi: 10.1016/j.ajhg.2016.02.003. With a novel detection method that integrates haplotype data with current approaches, the authors demonstrate there is a substantial portion of low-frequency mosaicism that is missed when using standard detection approaches. Rates of large structural autosomal mosaicism may be nearly 3 times higher than previously estimated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pique-Regi R, Caceres A, Gonzalez JR. R-Gada: a fast and flexible pipeline for copy number analysis in association studies. BMC Bioinformatics. 2010;11:380. doi: 10.1186/1471-2105-11-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang K, Bucan M. Copy Number Variation Detection via High-Density SNP Genotyping. CSH Protoc. 2008;2008 doi: 10.1101/pdb.top46. pdb top46. [DOI] [PubMed] [Google Scholar]
- 50.Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SF, Hakonarson H, Bucan M. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schick UM, McDavid A, Crane PK, Weston N, Ehrlich K, Newton KM, Wallace R, Bookman E, Harrison T, Aragaki A, et al. Confirmation of the reported association of clonal chromosomal mosaicism with an increased risk of incident hematologic cancer. PLoS One. 2013;8:e59823. doi: 10.1371/journal.pone.0059823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonnefond A, Skrobek B, Lobbens S, Eury E, Thuillier D, Cauchi S, Lantieri O, Balkau B, Riboli E, Marre M, et al. Association between large detectable clonal mosaicism and type 2 diabetes with vascular complications. Nat Genet. 2013;45:1040–1043. doi: 10.1038/ng.2700. [DOI] [PubMed] [Google Scholar]
- 53.Jager N, Schlesner M, Jones DT, Raffel S, Mallm JP, Junge KM, Weichenhan D, Bauer T, Ishaque N, Kool M, et al. Hypermutation of the inactive X chromosome is a frequent event in cancer. Cell. 2013;155:567–581. doi: 10.1016/j.cell.2013.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dumanski JP, Lambert JC, Rasi C, Giedraitis V, Davies H, Grenier-Boley B, Lindgren CM, Campion D, Dufouil C, et al. European Alzheimer’s Disease Initiative I. Mosaic Loss of Chromosome Y in Blood Is Associated with Alzheimer Disease. Am J Hum Genet. 2016;98:1208–1219. doi: 10.1016/j.ajhg.2016.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Busque L, Patel JP, Figueroa ME, Vasanthakumar A, Provost S, Hamilou Z, Mollica L, Li J, Viale A, Heguy A, et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet. 2012;44:1179–1181. doi: 10.1038/ng.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McKerrell T, Park N, Moreno T, Grove CS, Ponstingl H, Stephens J, Crawley C, Craig J, Scott MA, et al. Understanding Society Scientific G. Leukemia-associated somatic mutations drive distinct patterns of age-related clonal hemopoiesis. Cell Rep. 2015;10:1239–1245. doi: 10.1016/j.celrep.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med. 2014;20:1472–1478. doi: 10.1038/nm.3733. Analyzed TCGA exome sequencing data from blood-derived DNA and found mosaic mutations in leukemia or lymphoma associated genes in 2% of individuals. Higher frequencies of mutations were observed in individuals over 70 years. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruark E, Snape K, Humburg P, Loveday C, Bajrami I, Brough R, Rodrigues DN, Renwick A, Seal S, Ramsay E, et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature. 2013;493:406–410. doi: 10.1038/nature11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forsberg LA, Rasi C, Pekar G, Davies H, Piotrowski A, Absher D, Razzaghian HR, Ambicka A, Halaszka K, Przewoznik M, et al. Signatures of post-zygotic structural genetic aberrations in the cells of histologically normal breast tissue that can predispose to sporadic breast cancer. Genome Res. 2015;25:1521–1535. doi: 10.1101/gr.187823.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rebbeck TR. Precision prevention of cancer. Cancer Epidemiol Biomarkers Prev. 2014;23:2713–2715. doi: 10.1158/1055-9965.EPI-14-1058. [DOI] [PubMed] [Google Scholar]