Abstract
Verticillium wilt, caused by the Verticillium dahliae phytopathogen, is a devastating disease affecting many economically important crops. A receptor-like protein (RLP) gene, Ve1, has been reported to confer resistance to V. dahliae in tomato plants, but few genes have been found to be involved in cotton Verticillium wilt resistance. Here, we cloned two RLP gene homologs, Gossypium barbadense resistance gene to Verticillium dahliae 1 (GbaVd1) and GbaVd2, from the Verticillium wilt-resistant cultivar G. barbadense cv. Hai7124. GbaVd1 and GbaVd2 display sequence divergence, but both encode typical RLPs. Virus-induced gene silencing of GbaVd1 or GbaVd2 compromised the resistance of cotton to V. dahliae, and both genes conferred Verticillium wilt resistance after interfamily transfer into Arabidopsis. Microarray analysis revealed that GbaVd1 and GbaVd2 participate in Verticillium wilt resistance in Arabidopsis through activation of defense responses, including the endocytosis process, signaling factors, transcription factors and reinforcement of the cell wall, as demonstrated by lignification in Arabidopsis transgenic plants. In addition, microarray analysis showed that GbaVd1 and GbaVd2 differentially mediate resistance signaling and activation of defense responses after overexpression in Arabidopsis. Thus, GbaVd1 and GbaVd2 encode RLPs and act as disease resistance genes that mediate the defense response against V. dahliae in cotton.
Keywords: cotton, Verticillium wilt, receptor-like proteins, virus-induced gene silencing, transgenic, microarray analysis
Introduction
Verticillium dahliae Kleb. is a soil-borne vascular wilt fungus characterized by a broad host range of over 200 dicotyledonous species (Fradin and Thomma, 2006; Klosterman et al., 2009). Improving genetic resistance is a preferred method to manage Verticillium wilt in several crop plants (Schaible et al., 1951; Putt, 1964; Huang, 2003; Simko et al., 2004; Bolek et al., 2005). However, only the tomato Verticillium (Ve) locus has been efficiently used in molecular breeding (Kawchuk et al., 2001; Fradin et al., 2009). The tomato Ve locus contains two closely linked, inversely oriented genes: Ve1 and Ve2. These genes encode cell surface receptors that belong to a group of extracellular receptor-like proteins (RLPs), which typically include leucine-rich repeat (LRR) domains, a single-pass transmembrane domain, and a short cytoplasmic tail that lacks obvious motifs for directly activating intracellular signal transduction (Kawchuk et al., 2001). The RLP genes involved in plant disease resistance have been widely identified and include tomato leaf mold resistance Cf genes (Dixon et al., 1996, 1998; Parniske et al., 1997; Rivas and Thomas, 2005), the apple scab resistance gene HcrVf2 (Vinatzer et al., 2001), and the tomato LeEix1 and LeEix2 genes encoding the fungal elicitor ethylene-inducing xylanase of Trichoderma viride (Ron and Avni, 2004).
The Ve gene mediates disease resistance against V. dahliae race 1 strains, but is not functional against race 2 stains (Schaible et al., 1951; Fradin et al., 2009). More recent research has found that only Ve1, but not Ve2, has functional Verticillium wilt resistance activity in tomato plants (Fradin and Thomma, 2006; Fradin et al., 2009). Additionally, the interfamily transfer of Ve1 to Arabidopsis thaliana provides fully functional Verticillium wilt resistance (Fradin et al., 2011). Comparative genomic analysis of race 1 and race 2 strains has identified an effector named Ave1, a small secreted protein with four cysteines that contributes to full virulence in tomato plants lacking Ve1 but acts as an avirulence factor recognized by Ve1, and activates Ve1-mediated resistance (de Jonge et al., 2012).
The resistance mechanism of RLPs has been well described in the Cf genes, which mediate tomato resistance to Cladosporium fulvum. RLPs use LRR domains to recognize effectors that are secreted by pathogens and activate the defense response through the cooperation of the cytoplasmic tail with diverse signal factors (Joosten and de Wit, 1999). Several studies have shown that a signal factor binding to the cytoplasmic tail is required for RLP function, such as the interactions of vesicle-associated protein 27 (VAP27) and Cf-9 interacting thioredoxin (CITRX) with the Cf-9 cytoplasmic tail (Laurent et al., 2000; Rivas et al., 2004). Moreover, many signal factors have also been shown to participate in defense responses mediated by RLPs, such as the protein kinase Avr9/Cf-9 induced kinase 1 (ACIK1) (Rowland et al., 2005), the NB-LRR required for HR-associated cell death-1 (NRC1) (Gabriëls et al., 2006, 2007), the U-box protein of Cys, Met, Pro, and Gly protein 1 (CMPG1) (González-Lamothe et al., 2006), the Lycopersicon esculentum mitogen-activated protein kinase 1 (LeMPK1), LeMPK2, and LeMPK3 (Stulemeijer et al., 2007), and the F-box protein Avr9/Cf-9 rapidly elicited protein 189 (ACRE189) (van den Burg et al., 2008). The endocytosis signal motif in the cytoplasmic tail also plays an important role in the signal transduction mediated by RLPs. Vesicle-associated protein 27 (VAP27) possesses an endocytosis signal with a KKX motif that is involved in membrane trafficking (Theriot, 1996; Laurent et al., 2000; Bonifacino and Traub, 2003). LeEix2 contains a YXXø (ø is an amino acid with a hydrophobic side chain) motif that stimulates RLP-mediated endocytosis, and its ability to transport the elicitor into the cytoplasm via endocytosis is lost after deletion of this motif (Ron and Avni, 2004). Genetic dissection has shown that several signal factors are also required for the Verticillium wilt resistance mediated by tomato Ve1, including enhanced disease susceptibility 1 (EDS1), non-race-specific disease resistance 1 (NDR1), BRI1-associated kinase 1 (BAK1), and mitogen-activated protein kinase kinase 2 (MKK2) (Fradin et al., 2009).
In cotton plants, Verticillium wilt is difficult to control because of the lack of resistance germplasm resources. Recently, several Ve1 gene homologs have been isolated from island cotton Gossypium barbadense, including GbVe, GbVe1, and Gbvdr3, and have been shown to confer Verticillium wilt resistance by virus-induced gene silencing (VIGS) in cotton or overexpression in Arabidopsis (Zhang et al., 2011, 2012; Chen T. et al., 2015; Yang et al., 2015). Here, two RLP gene homologs, GbaVd1 and GbaVd2, were cloned from the Verticillium wilt-resistant cultivar G. barbadense cv. Hai7124. GbaVd1 and GbaVd2 encode typical RLPs, conditionaly enhanced Verticillium wilt resistance susceptibility against V. dahliae through VIGS in cotton plants, and enhanced Verticillium wilt resistance after interfamily transfer into Arabidopsis. Finally, the GbaVd1- and GbaVd2-mediated resistance signal transduction mechanisms and activation of defense responses were investigated by microarray analysis in transgenic Arabidopsis plants.
Materials and methods
Plant material, fungal culture, and inoculation
Seedlings of the resistant cultivar G. barbadense cv. Hai7124 and six cotton species (Gossypium anomalum, Gossypium trilobum, Gossypium aridum, Gossypium davidsonii, Gossypium thurberi, and Gossypium hirsutum) were planted in a 12-cm pot with sterilized soil at 28°C with a 14/10 light-dark photoperiod for 2 weeks, as previously described (Chen and Dai, 2010). The highly virulent V. dahliae strain Vd991 was cultured on potato dextrose agar (PDA) medium at 25°C for 10 days, the conidia were then washed with sterile water and the concentration was adjusted to 2 × 107 conidia/mL. Six 2-week-old seedlings were each inoculated with 5 mL of conidial suspension by a soil drenching method. To prepare the samples for detection of GbaVd1 and GbaVd2 expression, the root tissues were collected 2, 6, 12, 24, and 48 h after inoculation.
Gene cloning
RNA samples from inoculated plants were used for gene cloning. The RNA was extracted by using the cetyltrimethylammonium bromide (CTAB) method (Bekesiova et al., 1999). After tracing DNA in the RNA samples was removed by treatment with DNase I, cDNA was synthesized using a RevertAid™ First Strand cDNA Synthesis Kit from MBI (Fermentas, Glen Burnie, Maryland, USA). The primers specific to the cotton RLP gene homologs, GbaVd1 and GbaVd2 (cloned in our lab by homology-based cloning, the accession numbers is GU299533 and GU299534, respectively), were designed and used to amplify the cDNA samples. Primers used for gene cloning are listed in Table S1.
Bioinformatics analysis of Gbavd1 and Gbavd2
The open reading frames (ORFs) of GbaVd1 and GbaVd2 were determined by using ORF Finder (NCBI), and the protein sequences were deduced on the basis of gene sequences. The identities between GbaVd1 and GbaVd2 were determined by using the Blast program (http://www.ncbi.nlm.nih.gov/Blast.cgi) and Vector NTI software (Lu and Moriyama, 2004). Signal peptides and membrane-spanning structures were predicted by using SignalP 4.0 (Petersen et al., 2011) and TMHMM 2.0 (Sonnhammer et al., 1998), respectively. The primary protein structure was determined by protein sequence alignment with known RLPs, including Cf-2.2 (U42445), Cf-5 (AF053993), Cf-4 (AJ002235), Cf-9 (AJ002236), HcrVf1 (AJ297739), HcrVf2 (AJ297740), HcrVf3 (AJ297741), LeEix1 (AY359965), LeEix2 (AY359966), Ve1 (AF365929), and Ve2 (AF272366). ClustalX 1.83 software was used for multiple sequence alignment (Thompson et al., 1997), and data were exported with Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html, written by K. Hofmann and M. Baron). Phylogenetic trees were constructed in Mega 6.0 with the Maximum Parsimony method, using the Jones-Taylor-Thornton (JTT) model and performing 1000 bootstrap replicates (Tamura et al., 2013).
VIGS in cotton and detection of Verticillium wilt resistance
For the VIGS assays, GbaVd1 and GbaVd2 were integrated into a vector and introduced into A. tumefaciens GV3101. Agrobacterium the strains harboring the pTRV2-GbaVd1/GbaVd2 plasmid combined with strains harboring the pTRV1 vector were mixed in a 1:1 ratio and co-infiltrated into the cotyledon leaves of 2-week-old cotton plants. The effectiveness of the VIGS assay was evaluated by using the control cotton CLA1 gene as previously described (Gao and Shan, 2013). Approximately 14 days after the VIGS procedure, a visible phenotype of white-colored leaves were observed in plants in which the CLA1 gene had been targeted by VIGS, and all of the plantlets were subjected to V. dahliae inoculation with 5 mL of conidial suspension (2 × 107 conidia/mL). The Verticillium wilt symptoms were investigated 3 weeks after inoculation. Fungal biomass in cotton were determined using a method described previously (Santhanam et al., 2013). qPCR was performed using a qPCR SYBR premix Ex Taq II kit (TaKaRa, Japan) with primers specific to the cotton 18S gene and V. dahliae elongation factor 1-α (EF-1α) (Table S1).
Arabidopsis transformation and evaluation of transgenic disease resistance
According to the sequences of GbaVd1 and GbaVd2, the ORF fragments were amplified with primers containing BamH I and BstE II enzyme sites. The fragments of GbaVd1 and GbaVd2 were integrated into the binary vector pCAMBIA1303 under control of the cauliflower mosaic virus (CaMV) 35S promoter. The recombinant vectors were transferred into A. tumefaciens strain LBA4404, and this was followed by genetic transformation of A. thaliana ecotype Col-0 via the floral dip transformation method (Clough and Bent, 1998). Homozygous transgenic Arabidopsis (T3) plants were screened and used for this study. Transgenic plants were selected on MS medium containing 50 mg/L hygromycin and were identified by RT-PCR. The RT-PCR conditions consisted of an initial 94°C denaturation step for 10 min, which was followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and ubiquitin 4 (Ubi4, NM_122069.3) was used as a control. Two random transgenic plant lines carrying GbaVd1 and GbaVd2 were selected for the Verticillium wilt resistance assay. The inoculation method was performed as in cotton. In brief, 4-week-old seedlings were inoculated with 5 mL of conidial suspension (5 × 106 conidia/ml). The inflorescence height was detected 4 weeks after inoculation. Likewise, the development of fungal biomass in transgenic plants was detected by qPCR, with primers to RNA-binding family proteins in Arabidopsis (Table S1).
Microarray analysis
Microarray analysis was performed using Arabidopsis Gene Expression Microarray Version 4.0 (43,803 probes) (Agilent Technologies, USA). Three independent transgenic lines of GbaVd1 and GbaVd2 overexpression were used for microarray analysis. Two-week-old seedling transgenic plants without inoculation were collected for RNA extraction, and the wild type (Col-0) was used as a control. Total RNA was extracted from the root and labeled with Cy-3 using a Low RNA Input Linear Amplification/Labeling Kit, One-color (Agilent Technologies), hybridized, and washed according to the manufacturer's instructions. Hybridized microarrays were then scanned with an Agilent Microarray Scanner (Agilent Technologies). Statistical data extraction processes were performed on three biological replicates for each treatment according to the instructions (Agilent, GeneSpring GX9 software package). Fold changes in expression levels in response to each transgenic plant were compared with those of the respective wild-type, P-values were adjusted for multiple comparisons using the Bonferroni correction. Sequence homologies of the deduced amino acids were determined using the Arabidopsis Information Resource (TAIR) database (http://www.Arabidopsis.org/). Functional annotation of GO and metabolic pathways were performed using the GeneSpring GX9 package (Agilent Technologies, USA), and statistically for overrepresented GO terms. The significance of GO catalog for differentially expressed genes were identified using a Fisher's Exact Test (filtered with FDR ≤ 0.05) as previously described (Chen et al., 2016).
Expression of Gbavd1 and Gbavd2
The cDNA samples were used for qRT-PCR analysis with SYBR Green (Invitrogen, Carlsbad, California, USA), and the PCR was performed using a thermocycler (ABI PRISM 7500, Applied Biosystems) with the following program: 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s for 40 cycles. The cotton 18S gene was used as an endogenous control. Relative transcript levels of GbaVd1 and GbaVd2 were determined using the 2−ΔΔCT method (Livak and Schmittgen, 2001), with three independent determinations.
Lignin histochemical staining
Four-week-old seedlings of Arabidopsis transgenic lines were inoculated with 5 mL of conidial suspension (5 × 106 conidia/ml). The internode of shoot tissue between the second and third nodes was used for cell wall reinforcement detection at 4 weeks after inoculation. The samples were stained with phloroglucinol solution (3% w/v phloroglucinol in 95% ethanol) for 2 min after slice treatment and were then immersed in concentrated HCl for a few seconds and covered with glycerol (Nakano and Meshitsuka, 1992). The tissue slices were observed with a Leitz Diaplan light microscope and photographed immediately.
Results
Isolation of Gbavd1 and Gbavd2 from cotton
The full-length of GbaVd1 and GbaVd2 were cloned from the cDNA sample of G. barbadense (cv. Hai7124) inoculated with V. dahliae by PCR (Figure S1A). Sequences analysis showed that GbaVd1 and GbaVd2 are 3,262 and 3,466 bp in length, respectively, without any introns, and each has a single open reading frame (ORF) of 1,020 or 1,082 amino acids (Figure S1B), respectively. BlastP analysis revealed that both contain an LRR ribonuclease inhibitor domain, a potential signal peptide and a transmembrane region (Figure S2), and both were predicted to be located in the plasma membrane (the testk used for kNN was 14, and the Nearest Neighbors numbers of GbaVd1 and GbaVd2 related to the plasma membrane were 9 and 8, respectively). Sequence alignment indicated that the protein-coding sequences of GbaVd1 and GbaVd2 are similar to those of known plant RLP genes, with GbaVd2 showing the highest identities to Ve genes, but with GbaVd1 displaying the highest identities to HcrVf genes (Figure 1A). Likewise, the alignment of encoded protein sequences also confirmed that GbaVd1 and GbaVd2 are most homologous to Ve and HcrVf genes (Figure 1A), respectively. Sequence alignment showed that GbaVd2 is a homolog to Ve-like proteins from G. barbadense, and is nearly identical to the previously cloned and published Gbvdr5 (Yang et al., 2015) with only one conservative amino acid change (Figure S3). GbaVd1 protein sequence showed high identity to a RLP from cotton (Figure S4). In addition, six allelic variants of the GbaVd1 and GbaVd2 were isolated from six different cotton species, and two of them are susceptible to V. dahliae. A DNA sequence alignment showed a total of 125 and 264 single nucleotide polymorphisms (SNPs) among the allelic variants compared with the reference sequence of GbaVd1 and GbaVd2 (Figures S5A,C), resulting in 74 and 177 amino acid changes (Figures S5B,D), respectively. However, the association of the amino acid substitutions with the Verticillium wilt resistance of cotton species was not found among these allelic variants (Figures S5B,D). Phylogenetic analysis showed a close relationship between GbaVd1 and HcrVfs, which clustered into the same branch, and between GbaVd2 and Ve genes (Figure 1B). These results suggested that both GbaVd1 and GbaVd2 display homology to the known RLPs but also show sequence divergence when compared with each other.
Figure 1.
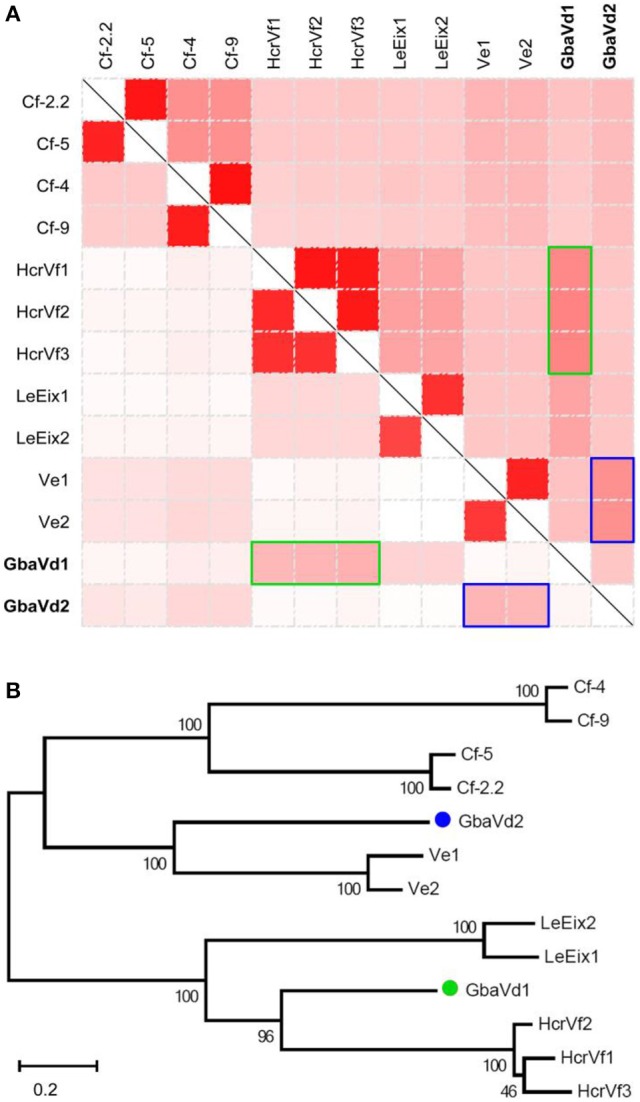
Analysis of GbaVd1 and GbaVd2 sequence characteristics. (A) Homology analysis of GbaVd1 and GbaVd2 with known RLPs. The upper-right and bottom-left represent identical nucleotide and encoding protein sequences among RLPs, respectively. The colors, from white to red, represent the percentage identities, from 0 to 100%. The green and blue rectangles represent the best hits of RLPs with GbaVd1 and GbaVd2, respectively. (B) Phylogenetic tree of GbaVd1 and GbaVd2 with homologs from other plant species. The analysis was performed using the package MEGA6 with the Maximum Parsimony method.
Gbavd1 and Gbavd2 encode typical RLPs
Although the sequence identities between GbaVd1 or GbaVd2 and known RLPs were relatively low (Figure 1A), the alignment results showed that the amino acid residues involved in the LRR structure are conserved (Figure S6). Sequence alignments between GbaVd1 or GbaVd2 and known RLPs indicated that both contain six conserved domains, Domains A through F (Figure 2). Domain A consists of the signal peptide and its cleavage site at the N-terminus of the protein. Domain B belongs to the N-terminus of the mature protein. Domain C contains LRR structures, with 32 and 33 LRRs in GbaVd1 and GbaVd2, respectively. Domain C is usually divided into two sections, domains C1 and C2, which are linked by a small peptide, and the number of LRRs (four) in domain C2 is identical between GbaVd1 and GbaVd2. Domains B and C also contain 22 and 26 putative N-glycosylation sites Nx(S/T) in GbaVd1 and GbaVd2, respectively. Domain D is a negatively charged extracytoplasmic domain predicted to play a role in orienting and anchoring the protein to the cell membrane and links domain C with the transmembrane structure of domain E. Domain F is a small cytoplasmic peptide with positively charged residues and is linked to domain E. In addition, the cytoplasmic domain of GbaVd1 possesses a YXXø signal sequence (YFRI), and GbaVd2 contains a KKX motif (KKH), both of which probably participate in the endocytosis process (Figure 2). A sequence similarity search using GbaVd1 showed high identity to a protein annotated as a LRR receptor-like serine/threonine-protein kinase FLS2 from cotton (Figure S4). However, there is no kinase domain in the small cytoplasmic region, suggesting improper annotation of the FLS2 homolog and that GbaVd1 encodes a RLP (not an RLK). Therefore, similar to the known RLPs, GbaVd1 and GbaVd2 encode extracytoplasmic glycoproteins in which the majority of the extracytoplasmic domain consists of multiple LRRs.
Figure 2.
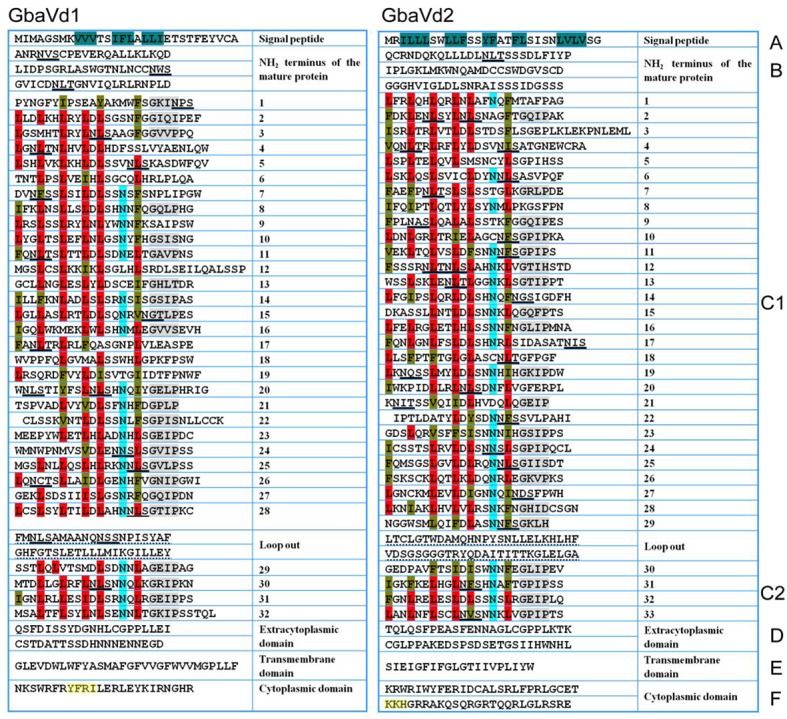
GbaVd1 and GbaVd2 encoded typical RLPs. The primary structures were divided into domains (A–F). Domain (A) is the putative signal peptide, residues in dark green are hydrophobic amino acids; domain (B) is the putative N-terminus of the mature protein; domain (C) (divided into C1,C2) is the LRR domain, in which conserved leucine residues (L, red) are often replaced by isoleucine (I), phenylalanine (F), or valine (V) (brown), conserved N-termini are highlighted in light blue, and the conserved GxIP motif is depicted in gray; domain (D) is a negatively charged extracytoplasmic peptide with neutral and acidic amino acids; domain (E) is the transmembrane-associated domain; domain (F) is the small cytoplasmic peptide with neutral and basic amino acids, with the endocytosis signal motifs highlighted in yellow. Putative N-glycosylation sites Nx(S/T) present in domains (B,C) are underlined with black lines.
Silencing Gbavd1 and Gbavd2 compromise resistance to V. dahliae in cotton
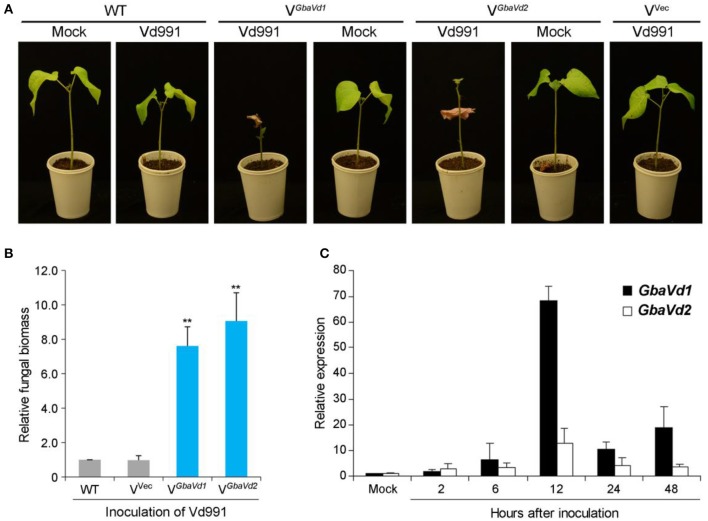
To identify the roles of GbaVd1 and GbaVd2 in Verticillium wilt resistance, tobacco rattle virus (TRV)-based virus-induced gene silencing was used in the island cotton cv. Hai7124. The cotton gene cloroplastos alterados 1 (CLA1), which is essential for chloroplast development, was used as a positive control to monitor VIGS efficiency. Plants in which the GbaVd1 or GbaVd2 expression was silenced by TRV-based VIGS were challenged with V. dahliae after all of the CLA1-silenced cotton plants presented an albino phenotype on their newly emerged leaves. At 21 days post-inoculation with the V. dahliae strain V991, plants treated with TRV-based VIGS of GbaVd1 or GbaVd2 but not the wild-type plants presented clear Verticillium wilt symptoms of leaf chlorosis, including withering and dwarfing (Figure 3A), thus indicating that the resistant cotton cv. Hai7124 displayed compromised resistance to V. dahliae after silencing of GbaVd1 or GbaVd2 by VIGS. Real-time quantitative polymerase chain reaction (qPCR) quantification of gene expression related to fungal biomass development demonstrated significantly increased V. dahliae strain Vd991 propagation in GbaVd1- or GbaVd2-silenced plants compared with the wild-type and empty vector-treated plants (Figure 3B). Moreover, GbaVd1 and GbaVd2 transcript accumulation was quickly and strongly induced 2–48 h after inoculation with V. dahliae and reached the highest expression levels (>10-fold change) 12 h after inoculation (Figure 3C). Thus, it can be concluded that the functions of GbaVd1 and GbaVd2 are closely associated with Verticillium wilt resistance in cotton.
Figure 3.
The GbaVd1 and GbaVd2 genes play essential roles in Verticillium wilt resistance in cotton. (A) Determination of Verticillium wilt resistance in GbaVd1- or GbaVd2-silenced cotton plants. Approximately 14 days after the VIGS procedure in the resistant cotton G. barbadense cv. Hai7124, the GbaVd1- or GbaVd2-silenced plants were inoculated with 5 mL of conidial suspension (2 × 107 conidia/mL) from V. dahliae strain Vd991. The Verticillium wilt phenotypes were determined and photographed 3 weeks after inoculation. WT, the wild type of G. barbadense cv. Hai7124; Mock, inoculation with sterile water; VGbaVd1/VGbaVd2, the GbaVd1- or GbaVd2-silenced plants, respectively; Vvec, VIGS positive control of infiltration with the empty vector pTRV2. (B) Real-time PCR quantification of fungal biomass in GbaVd1- or GbaVd2-silenced plants. Error bars represent standard errors, **significant differences (P ≤ 0.01), according to unpaired Student's t-test. (C) Expression patterns of GbaVd1 or GbaVd2 in cotton after inoculation with V. dahliae. Relative gene quantifications over time were calculated using the comparative threshold (2−ΔΔCT) method, and three independent biological replicates were analyzed. Error bars represent standard errors.
Heterologous overexpression of Gbavd1 and Gbavd2 confer Verticillium wilt resistance in Arabidopsis
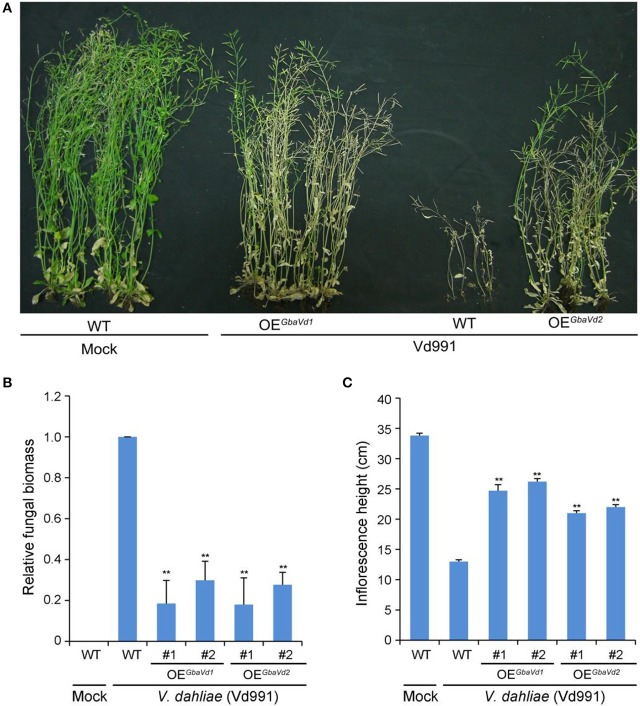
To identify the Verticillium wilt resistance functions of GbaVd1 and GbaVd2, both were interfamily-transferred into the Arabidopsis genome by using the Agrobacterium tumefaciens-mediated transformation method. Ten GbaVd1 and nine GbaVd2 independent Arabidopsis transgenic lines were obtained by hygromycin-resistance selection and verified by reverse transcription PCR (RT-PCR), and the integrated genes were successfully expressed, except in one GbaVd2 Arabidopsis transgenic line (Figure S7). Two random transgenic plant lines of GbaVd1 and GbaVd2 were selected for Verticillium wilt resistance identification with the root dip method and were assessed for Verticillium wilt resistance by evaluation of the extent of leaf chlorosis and inflorescence heights. Compared with the wild-type Col-0 ecotype, overexpressed GbaVd1 or GbaVd2 in Arabidopsis improved the resistance of the transgenic plants to V. dahliae strain Vd991, as indicated by significant decreases in leaf chlorosis, withering and dwarfing (Figure 4A). More importantly, unlike the wild-type Arabidopsis, which lost the capacity to set seed, the transgenic plants presented with a normal seed set 4 weeks after inoculation with V. dahliae (Figure 4A), a result strongly suggesting that overexpression of GbaVd1 or GbaVd2 conferred Verticillium wilt resistance to Arabidopsis. Further examination by qPCR confirmed that the V. dahliae significantly developed less fungal biomass in GbaVd1 or GbaVd2 transgenic plants than in the wild-type plants (Figure 4B). In addition, plant dwarfing, a typical Verticillium wilt symptom, was also significantly alleviated in transgenic lines after inoculation with V. dahliae (Figure 4C). Together, these results strongly suggested that GbaVd1 and GbaVd2 function as resistance genes and that interfamily transfer of these genes confers Verticillium wilt resistance in Arabidopsis.
Figure 4.
GbaVd1 and GbaVd2 conferred Verticillium wilt resistance in transgenic Arabidopsis. (A) Identification of Verticillium wilt resistance after interfamily transfer of GbaVd1 and GbaVd2 in Arabidopsis. Four-week-old seedlings of homozygote transgenic Arabidopsis (T3) were inoculated with 5 mL of conidial suspension (5 × 106 conidia/mL). The Verticillium wilt phenotypes were determined and photographed 4weeks after inoculation. WT, the wild-type Col-0; Mock, inoculation with sterile water; OEGbaVd1/OEGbaVd2, transgenic plants overexpressing GbaVd1 or GbaVd2, respectively. (B) Real-time PCR quantification of fungal biomass in GbaVd1 or GbaVd2 transgenic plants. Error bars represent standard errors, **significant differences (P ≤ 0.01), according to unpaired Student's t-test. (C) Investigation of the inflorescence height after inoculation with V. dahliae. Error bars represent standard errors, **significant differences (P ≤ 0.01), according to unpaired Student's t-test.
Microarray analysis of gene expression profiles in the Gbavd1 and Gbavd2 transgenic lines
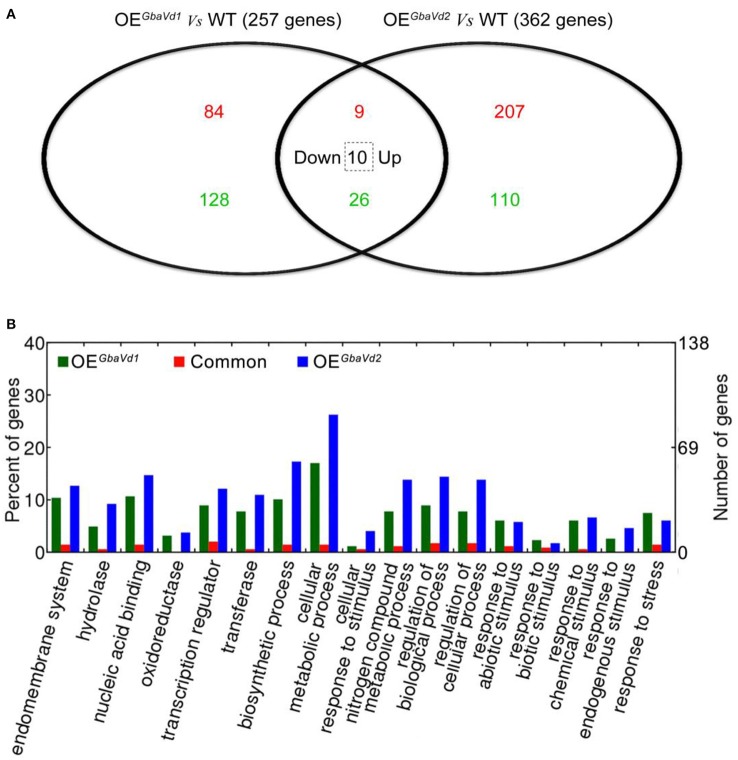
To identify the defense responses mediated by GbaVd1 and GbaVd2, the transcriptome of transgenic lines were assayed by using an Agilent 44 K custom oligo microarray system with three independent biological replicates. In total, 4,153 and 4,484 probes were detected in GbaVd1 and GbaVd2 transgenic lines (Supplementary Data S1), respectively. Of these detected genes, 257 (93 genes up-regulated and 164 genes down-regulated) showed differential expression in the GbaVd1 transgenic line (fold-change ≥1.5, p < 0.01), and 362 (226 genes up-regulated and 136 genes down-regulated) were differentially expressed in the GbaVd2 transgenic line (fold-change ≥1.5, p < 0.01) (Figure 5A; Table S2). A further comparison of the two differentially expressed gene sets revealed 45 common genes between the GbaVd1 and GbaVd2 transgenic lines, of which 35 displayed the same regulation patterns (nine genes up-regulated and 26 genes down-regulated), and the other 10 of which showed down-regulation in the GbaVd1 transgenic line but up-regulation in the GbaVd2 transgenic line (Figure 5A; Table S2). These results indicated that the regulation patterns mediated by GbaVd1 and GbaVd2 display some of the same aspects, but that each has unique characteristics. Gene ontology (GO) analysis indicated that subsets of the differentially expressed genes activated by GbaVd1 and GbaVd2 were involved in the categories cellular metabolic process (statistically in overrepresented, filtered with FDR ≤ 0.05), regulation of cellular process, response to stress, and transcription regulation (Figure 5B). These results suggested that GbaVd1 and GbaVd2 have similar functions in mediating the defense response after overexpression in Arabidopsis.
Figure 5.
Microarray analysis of differentially expressed genes in GbaVd1 and GbaVd2 transgenic lines. (A) The Venn diagram represents the differentially expressed genes between the transgenic and wild-type (WT) lines under normal conditions. The red numbers represent the up-regulated genes, the green numbers represent the down-regulated genes, and the number in the dotted box represents genes down-regulated in the GbaVd1 transgenic line but up-regulated in the GbaVd2 transgenic line. (B) GO analysis of differentially expressed genes in GbaVd1 and GbaVd2 transgenic lines.
Overexpression of Gbavd1 and Gbavd2 altere the expression of defense-related genes in Arabidopsis
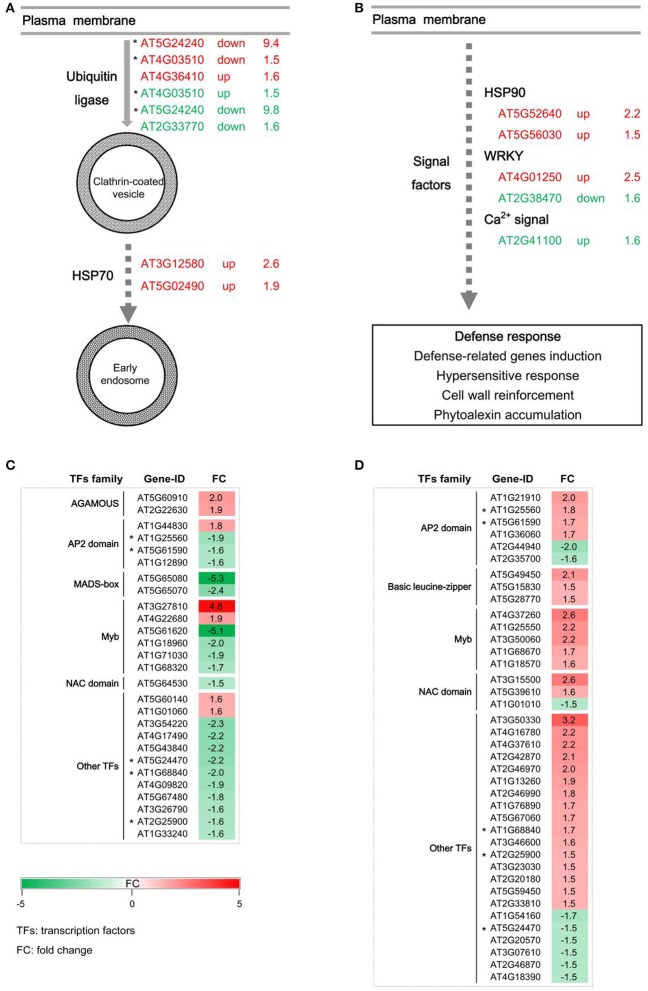
Generally, the endocytosis process and signal transduction are necessary for the disease resistance mediated by RLPs. GbaVd1 and GbaVd2 contain a conserved motif that is probably involved in the endocytosis process (Figure 2). As expected, several genes associated with endocytosis were differentially expressed after GbaVd1 and GbaVd2 overexpression in Arabidopsis, including genes encoding ubiquitin ligase family proteins and heat shock protein 70 (HSP70) (Figure 6A; Table S3). Specifically, there are three genes encoding ubiquitin ligase family proteins, two of which were commonly activated in the GbaVd1 and GbaVd2 transgenic lines. These proteins facilitate formation of clathrin-coated vesicles during endocytosis (Figure 6A; Table S3). In addition, two HSP70 genes (AT3G12580 and AT5G02490) involved in the formation of early endosomes were also differentially expressed in the GbaVd1 transgenic line (Figure 6A; Table S3). These results suggested that the endocytosis process associated with plant-pathogen interactions was affected after GbaVd1 and GbaVd2 overexpression in Arabidopsis. In addition, several genes encoding signal factors involved in plant-pathogen interactions were also differentially expressed in the GbaVd1 and GbaVd2 transgenic lines (Figure 6B; Table S3). In the GbaVd1 transgenic line, the genes encoding two heat shock protein 90 (HSP90) isoforms and the transcription factor WRKY33 were up-regulated, but two other genes encoding calmodulin-like (CaM/CML) protein 12 and the transcription factor WRKY22 were activated in the GbaVd2 transgenic line (Figure 6B). Moreover, 27 and 39 genes encoding transcription factors were significantly differentially expressed after overexpression of GbaVd1 and GbaVd2, respectively; these included members of the AP2 domain-containing protein family and the myeloblastosis (Myb) family (Figures 6C,D; Table S3). However, most transcription factor genes were specifically differentially expressed in either the GbaVd1 or GbaVd2 transgenic lines, except for five common genes (Figures 6C,D; Table S3). Together, these results indicated that the interfamily transfer of cotton GbaVd1 and GbaVd2 provided the capacity to differentially expressed defense response-related genes and mediate Verticillium wilt resistance in Arabidopsis.
Figure 6.
Pathway enrichment analysis of differentially expressed genes in GbaVd1 and GbaVd2 transgenic plants. (A) Differentially expressed genes involved in the endocytosis process. The differentially expressed genes associated with the GbaVd1 and GbaVd2 transgenic plants are shown in red and green, respectively. (B) Differentially expressed genes involved in signal transduction. (C,D) Activation of genes encoding transcription factors after GbaVd1 (C) or GbaVd2 (D) overexpression in Arabidopsis. The red and green blocks represent the up- and down-regulated patterns, respectively, and genes with asterisks represent the common activation genes between the GbaVd1 and GbaVd2 transgenic lines.
Overexpression of Gbavd1 and Gbavd2 result in cell wall reinforcement in Arabidopsis
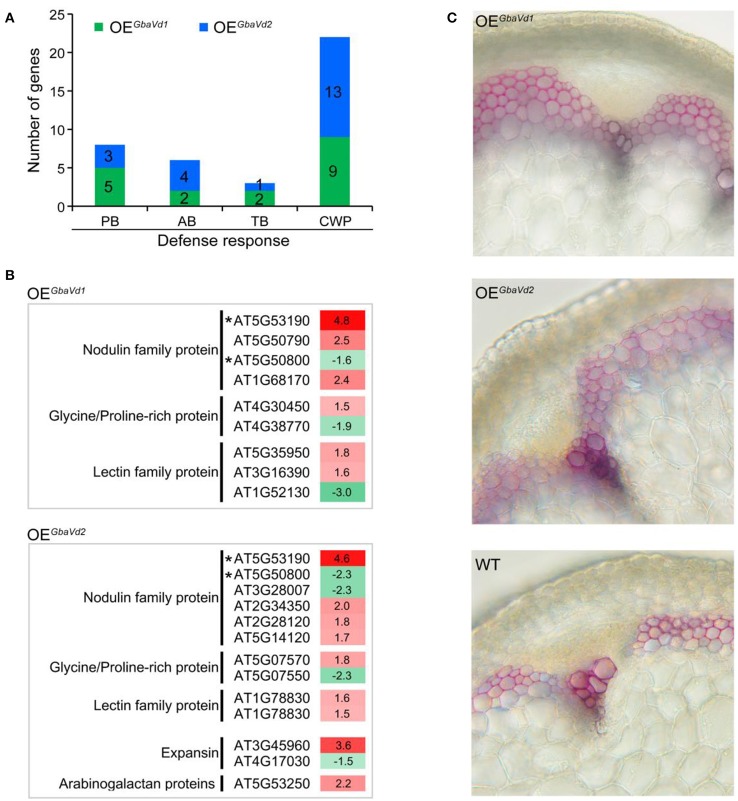
Plants usually enhance disease resistance by activation of defense responses that are mediated by the plant-pathogen interaction pathway (PATHWAY: map04626, KEGG database). In this study, the plant-pathogen interaction pathway was significantly activated after GbaVd1 or GbaVd2 overexpression in Arabidopsis, thus resulting in the induction of defense responses, including the hypersensitive response, phytoalexin accumulation, and cell wall reinforcement (Figures 6A,B). Microarray analysis confirmed that several differentially expressed genes that were significantly enriched were involved in the phytoalexin (phenylpropanoids, alkaloids, terpenoids, etc.) biosynthesis pathway or encode cell wall-related proteins (Figure 7A; Table S4).
Figure 7.
Analysis of the activation of defense responses in GbaVd1 and GbaVd2 transgenic lines. (A) Enrichment analysis pathway of differentially expressed genes participating in the defense response. PB, phenylpropanoid biosynthesis; AB, alkaloid biosynthesis; TB, terpenoid biosynthesis; CWP, cell wall-related protein. (B) List of cell wall-related proteins activated after GbaVd1 or GbaVd2 overexpression in Arabidopsis. Genes with asterisks represent the common activation genes between GbaVd1 and GbaVd2 transgenic lines. (C) Detection of the lignification of GbaVd1 and GbaVd2 transgenic lines. The transgenic plant shoot tissues between the second and third nodes were collected 4 weeks after inoculation with V. dahliae strain Vd991. The lignification phenotypes were determined and photographed after staining via the phloroglucinol-HCl method.
Functional analysis of differentially expressed genes revealed that 9 and 13 genes encoding cell wall-related proteins were differentially expressed by overexpression of GbaVd1 or GbaVd2, respectively, in Arabidopsis (Figure 7A). Of these genes, the most encoded were nodulin family proteins, glycine/proline-rich proteins, and lectin family proteins, and were mainly up-regulated in transgenic lines (Figure 7B). Specifically, for the nodulin family, four and six genes were significantly activated in the GbaVd1 and GbaVd2 transgenic lines, respectively (Figure 7B). Moreover, the identification of cell wall structures confirmed that the degree of lignification was significantly enhanced by the transcription of genes encoding cell wall-related proteins (Figure 7C), thus indicating that the cell wall is reinforced after GbaVd1 or GbaVd2 overexpression in Arabidopsis. In addition, of these cell wall-related genes, only two genes were commonly regulated between the GbaVd1 and GbaVd2 transgenic lines (Figure 7B), thus suggesting that the cell wall-related proteins were differentially activated in the two transgenic lines.
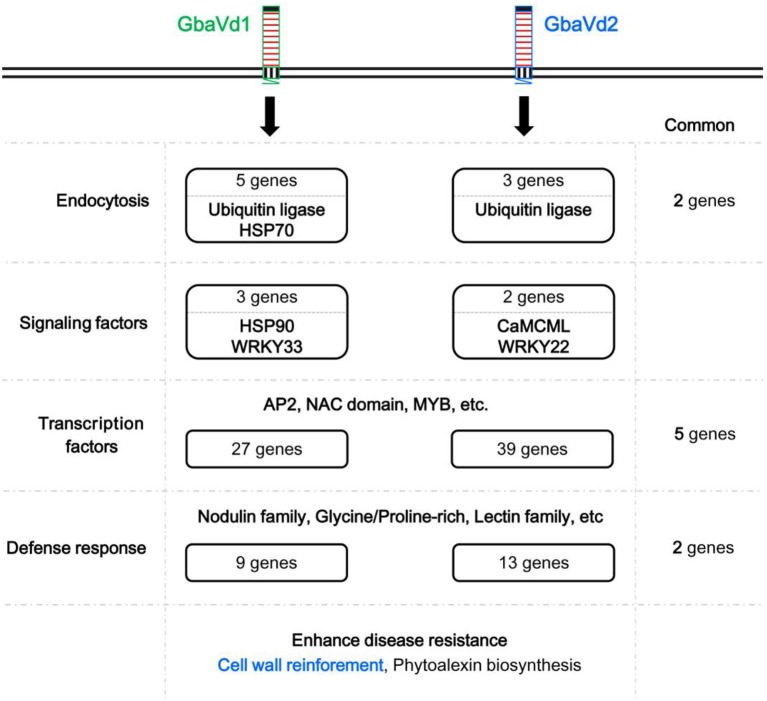
Gbavd1 and Gbavd2 differentially regulate defense responses, enhancing Verticillium wilt resistance
Although both GbaVd1 and GbaVd2 encode RLPs, the sequences and primary structures display divergence (Figures 1, 2), thus suggesting that the functions of GbaVd1 and GbaVd2 might diverge during plant-pathogen interactions. Microarray analysis revealed that at least four layers of defense responses were differentially activated after GbaVd1 or GbaVd2 overexpression in Arabidopsis (Figures 6, 7). Further analysis of the differentially expressed genes revealed that the defense responses mediated by GbaVd1 and GbaVd2 displayed significant differences that only five genes encoding transcription factors and two genes encoding cell wall-related proteins were common between the two transgenic lines (Figure 8). The activation of genes involved in endocytosis appeared to be similar between the GbaVd1 and GbaVd2 transgenic lines, with two common genes out of six differentially expressed genes, but more genes were activated in the GbaVd1 transgenic plants (Figure 8). Together, these results suggested that GbaVd1 and GbaVd2 mediate different resistance signaling and activation of defense responses to confer Verticillium wilt resistance after interfamily overexpression in Arabidopsis.
Figure 8.
Schematic overview of Verticillium wilt resistance mediated by GbaVd1 and GbaVd2.
Discussion
Verticillium wilt resistance genes have been well studied in tomato plants, in which the Ve locus and the functional gene Ve1 mediate the disease resistance to V. dahliae (Schaible et al., 1951; Fradin and Thomma, 2006; Fradin et al., 2009). Recently, several Ve1 gene homologs, belonging to the RLP family, have also been isolated from cotton plants and demonstrated to be functional in Verticillium wilt resistance (Zhang et al., 2011, 2012; Yang et al., 2015; Chen et al., 2016). The Verticillium wilt resistance of cotton may be controlled by multi-dominant resistance genes. Previous studies have shown that the cotton genome encodes an extremely large RLP family protein that contains at least 144 RLPs in diploid cotton (Chen J. Y. et al., 2015), thus indicating that other RLP genes probably participate in Verticillium wilt resistance in cotton. In this study, two RLP genes, GbaVd1 and GbaVd2, were cloned from the Verticillium wilt-resistant cultivar G. barbadense cv. Hai7124, which proved to be involved in Verticillium wilt resistance via the activation of diverse defense responses.
RLPs referred to as LRR-TMs, usually function as disease resistance genes in plants (Kruijt et al., 2005a). Like the primary structures of known RLPs (Dixon et al., 1996, 1998; Parniske et al., 1997; Vinatzer et al., 2001; Ron and Avni, 2004; Kruijt et al., 2005a; van der Hoorn et al., 2005), GbaVd1 and GbaVd2 contain signal peptides, LRR domains, transmembrane domains and small cytoplasmic tails (Figure 2; Figures S2, S3). GbaVd1 lack of any kinase characteristic in the small cytoplasmic domain. An orthologous gene of GbaVd1, Gorai.003G112100.1 (Accession number: KJB19646.1), was designated as a disease resistance family protein_LRR due to the lack of kinase motif (Paterson et al., 2012), and we annotated it as RLP in cotton (Chen J. Y. et al., 2015). LRRs are typical domains of RLPs with the consensus sequence LxxLxxLxxLxLxxNxLxGxIPxx, and the number of LRRs may dictate the specific recognition of pathogen effectors (Jones et al., 1994; Jones and Jones, 1997; van der Hoorn et al., 2005). Moreover, the LRR domains of RLPs usually contain many potential N-glycosylation sites important to gene function; for example, Cf-9 contains 22 PGSs, and most are glycosylated (van der Hoorn et al., 2005). GbaVd1 and GbaVd2 also contain different numbers of LRRs and potential N-glycosylation sites (Figure 2), thus possibly indicating that there are two kinds of RLPs in cotton. Like other RLPs, GbaVd1 and GbaVd2 contain a predicted small cytoplasmic tail that is necessary for signal transduction, as demonstrated by the interaction of the cytoplasmic peptide of Cf-9 with VAP27 and CITRX (Laurent et al., 2000; Nekrasov et al., 2006). Moreover, similar to the YXXø motif of LeEix2 and the KKX motif of Cf-4/9, which play important roles in the endocytotic process (Thomas et al., 1997; Ron and Avni, 2004; Sharfman et al., 2011), the cytoplasmic domain of GbaVd1 possesses a YXXø (YFRI) motif, and GbaVd2 contains a KKX motif (KKH) (Figure 2), which stimulates receptor-mediated endocytosis and mammalian cell-surface receptor degradation (Letourneur and Klausner, 1992; Benghezal et al., 2000; Bonifacino and Traub, 2003). Previous studies showed that the RLPs generally displayed significant divergence, which correspond to the fast evolution of pathogen (Kruijt et al., 2005a). Similarity, the sequence of allelic variants of GbaVd1 and GbaVd2 among the cotton species were also divergent, as the Cf-4 and Cf-9 in tomato (Kruijt et al., 2005b), but the polymorphisms do not obviously associate with the Verticillium wilt resistance (Figure S4). Thus, GbaVd1 and GbaVd2 are likely to encode typical RLPs with similar primary structures that contain extracytoplasmic LRR motifs and cytoplasmic tails.
RLPs play significant roles in plant defense against pathogens and plant development (Kruijt et al., 2005a). The interfamily transfer of tomato Ve1, a classical RLP, confers Verticillium wilt resistance in Arabidopsis (Fradin et al., 2011). Likewise, Cf proteins in tomato plants prevent the foliar leaf mold pathogen Cladosporium fulvum (Dixon et al., 1996, 1998; Parniske et al., 1997; Belfanti et al., 2004), and HcrVf proteins from Malus domestica enhance resistance to the scab fungus Venturia inaequalis (Vinatzer et al., 2001; Malnoy et al., 2008). Recently, several Ve1 gene homologs identified in island cotton have been reported to confer resistance to V. dahliae, including GbVe (Zhang et al., 2011), Gbve1 (Zhang et al., 2012), Gbvdr5 (Yang et al., 2015), and Gbvdr3 (Chen et al., 2016). In this study, GbaVd1 and GbaVd2 also conferred resistance to V. dahliae Vd991 stains (race 2) that lacks Ave1 (Yang et al., 2015), as demonstrated by VIGS in cotton and the interfamily transfer of the genes in Arabidopsis (Figures 3, 4). The ectopic overexpression of GbaVd1 and GbaVd2 was driven by the constitutive CaMV 35S promoter, which resulted in enhancing Verticillium wilt resistance in Arabidopsis. Previous studies showed that the ectopic expression of Ve1 driven by the tomato native promoter is less effective than the CaMV 35S promoter in conferring Verticillium wilt resistance in Arabidopsis (Fradin et al., 2011). In addition, many other studies also found that overexpression of RLP genes from cotton by CaMV 35S promoter can significantly enhance the Verticillium wilt resistance in Arabidopsis (Zhang et al., 2011; Chen T. et al., 2015; Yang et al., 2015). Similarity, several resistance genes conferring resistance after overexpression in transgenic lines have been driven by constitutive promoter (Liu et al., 2005; Schoonbeek et al., 2015; Yeh et al., 2015). These results also support by previous findings that the Ve-like gene Gbvdr5 conferred Verticillium wilt resistance in cotton and Arabidopsis transgenic plants (Yang et al., 2015), which only contains a residue divergence in GbaVd2 (Figure S3). In addition, several genes encoding RLP family proteins confer Verticillium wilt resistance, thus indicating that the inheritance of cotton resistance to V. dahliae may be controlled by multi-dominant resistance genes. Subsequently, except for the race 2 strain as the Vd991 used in this study, we need to further test the performance of GbaVd1 or transgenic lines after inoculation with different V. dahliae genotypes, which facilitate us to understand the Verticillium wilt resistance functional GbaVd1 and GbaVd2.
The genetics of RLP-mediated disease resistance is significantly associated with the endocytosis process, which plays a key role in plant-pathogen interactions (Gabriëls et al., 2006; Postma et al., 2016). Several RLPs contain an endocytosis motif (such as the YXXΦ endocytosis signature, where Φ represents an amino acid with a hydrophobic side chain, and X represents any amino acid) that can stimulate receptor endocytosis and initiate plant immunity (Jones et al., 1994; Thomas et al., 1997; Bonifacino and Traub, 2003; Ron and Avni, 2004; Sharfman et al., 2011). In addition, many signaling factors are recruited to participate in the resistance mediated by RLPs, which has been most extensively studied by exploiting the tomato Cf genes and the Ve1 gene (Rowland et al., 2005; Gabriëls et al., 2006, 2007; González-Lamothe et al., 2006; Stulemeijer et al., 2007; van den Burg et al., 2008; Fradin et al., 2009). Similarly, GbaVd1 and GbaVd2 contain endocytosis motifs (Figure 2), and we found that the endocytosis process and signaling factors involved in the plant-pathogen interaction pathway were significantly activated after interfamily transfer into Arabidopsis (Figure 6), thereby resulting in an enhancement to Verticillium wilt resistance by initiating the defense response, as expected (Figure 7). Therefore, GbaVd1 and GbaVd2, similarly to the known RLP genes, likely to provide resistance against V. dahliae by activation of the endocytosis process and recruitment of signaling factors.
Previous studies showed that EDS1, NRC1, ACIF, and MEK2 are required for the defense response mediated by Ve1 and Cfs. However, Serk3/Bak1 has not been tested for their requirement in Cfs signaling, while Sgt1 has not been tested for its requirement in Ve1 signaling, thus indicating that the RLP, Ve1 and Cf proteins, differentially require downstream signaling components (Fradin et al., 2009). Similarity, GbaVd1 and GbaVd2 mediated different regulation patterns and only a few genes were commonly regulated between the two transgenic plants (Figure 8). This means that the sequence and primary structures divergence of GbaVd1 and GbaVd2 may require different signaling factor (Figures 1, 2). It suggested that GbaVd2 (Gbvdr5) recognizes a new effector to activate the defense response due to the lack of Ave1 (race 2) in test strain Vd991 (de Jonge et al., 2012; Yang et al., 2015). We expected the defense mediated by GbaVd1 to display differences, due to the low sequence identity with GbaVd2 (Figures S3, S4). Plants generally employ several resistance genes to activate defense responses during pathogen infection (Wiesner-Hanks and Nelson, 2016), and several RLP genes have been identified in cotton that confer Verticillium wilt resistance (Zhang et al., 2011, 2012; Yang et al., 2015; Chen et al., 2016). Therefore, our study suggests that both GbaVd1 and GbaVd2 function as disease resistance proteins, but require different downstream signaling components to protect against V. dahliae.
RLP-mediated disease resistance activates a multilayered defense response, such as the production of reactive oxygen species (Piedras et al., 1998) and induction of the hypersensitive response (Ron and Avni, 2004), and drives phenylalanine-ammonia lyase gene expression (Gayoso et al., 2010). In this study, defense-related genes were also found to be activated in transgenic plants, including genes involved in enhancing the cell wall or phytoalexin biosynthesis (Figures 7A,B). Specifically, the genes encoding cell wall proteins were significantly affected in transgenic plants, thus resulting in reinforcement of plant cell walls in Arabidopsis (Figure 7C). The cell wall is an important defensive structure for avoiding pathogen infection (Hückelhoven, 2007; Hématy et al., 2009). In the defense response mediated by Cfs in tomato plants, the plant cell wall-related genes are activated, thus leading to the reinforcement of the cell wall (Stergiopoulos and de Wit, 2009). In cotton, the resistant cultivar exhibited higher and earlier-induced levels of enzyme activity and lignin-like polymers compared with the susceptible cultivar (Smit and Dubery, 1997). Therefore, GbaVd1 and GbaVd2 probably possess the capacity to regulate the genes encoding cell wall proteins, resulting in enhancing Verticillium wilt resistance in Arabidopsis.
It is known that the Ve-like gene Gbvdr5 confers resistance to Verticillium dahliae in transgenic Arabidopsis and upland cotton, which can be induced by hormones and activated defense response, including callose deposition, regulated defense-related genes expression, and hypersensitivity reaction (HR)-mimic cell death (Yang et al., 2015). Coincidently, the Ve-like genes cloned in our study, GbaVd2 only displays a residue divergence at position 598 (Valine to leucine) compares to Gbvdr5 (Figure S3). Our study confirmed that the polymorphism occurs between GbaVd2 and Gbvdr5, and suggested that that GbaVd2 and Gbvdr5 are the same genes in G. barbadense, due to a random mutation between different starting materials. However, we found some novel defense response mediated by GbaVd2 (Gbvdr5) in this study, including differentially expressed the endocytosis process and signaling factor, and enhanced cell wall reinforcement (Figure 8), which facilitate us to further understand the mechanism of GbaVd2 (Gbvdr5) confers Verticillium wilt resistance in cotton.
In conclusion, two genes encoding RLPs, GbaVd1 and GbaVd2, were cloned from the resistant cultivar of G. barbadense in this study. We revealed that GbaVd1 and GbaVd2 play essential roles in Verticillium wilt resistance, a result also demonstrated by VIGS in cotton and interfamily transfer into Arabidopsis. Similar to the Verticillium wilt resistance mediated by typical RLPs, genes associated with the endocytosis process, signaling factors, transcription factors and defense response were significantly activated after GbaVd1 or GbaVd2 overexpression, thereby resulting in cell wall reinforcement in Arabidopsis. In addition, microarray analysis revealed that GbaVd1 and GbaVd2 differentially mediated resistance signaling and activation of defense responses, corresponding to the divergence of GbaVd1 and GbaVd2 in terms of sequence, primary protein structure, and phylogeny.
Author contributions
XD designed the experiments and revised the paper. JC performed the microarray analysis and gene cloning. NL, XM, TL, and DZ performed the experiment of gene function identification. JC, VG, and DZ wrote and critically reviewed this article.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by CAAS (an Agricultural Science and Technology Innovation Program grant to XD), the Special Public Welfare Industry Research on Agriculture (201503109), the National Natural Science Foundation of China (31471759) and the China Major Projects for Transgenic Breeding (2011ZX08005).
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00844/full#supplementary-material
Sequence data from this article have been deposited with the GenBank data libraries under accession numbers GU299533 (GbaVd1) and GU299534 (GbaVd2). The microarray data are available via online.
References
- Bekesiova I., Nap J. P., Mlynarova L. (1999). Isolation of high quality DNA and RNA from leaves of the Carnivorous plant Drosera rotundifolia. Plant Mol. Biol. Rep. 17, 269–277. 10.1023/A:1007627509824 [DOI] [Google Scholar]
- Belfanti E., Silfverberg-Dilworth E., Tartarini S., Patocchi A., Barbieri M., Zhu J., et al. (2004). The HcrVf2 gene from a wild apple confers scab resistance to a transgenic cultivate dvariety. Proc. Natl. Acad. Sci. U.S.A. 101, 886–890. 10.1073/pnas.0304808101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benghezal M., Wasteneys G. O., Jones D. A. (2000). The C-terminal dilysine motif confers endoplasmic reticulum localization to type I membrane proteins in plants. Plant Cell 12, 1179–1201. 10.1105/tpc.12.7.1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolek Y., El-Zik K. M., Pepper A. E., Bell A. A., Magill C. W., Thaxton P. M., et al. (2005). Mapping of Verticillium wilt resistance genes in cotton. Plant Sci. 168, 1581–1590. 10.1016/j.plantsci.2005.02.008 [DOI] [Google Scholar]
- Bonifacino J. S., Traub L. M. (2003). Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447. 10.1146/annurev.biochem.72.121801.161800 [DOI] [PubMed] [Google Scholar]
- Chen J. Y., Dai X. F. (2010). Cloning and characterization of the Gossypium hirsutum major latex protein gene and functional analysis in Arabidopsis thaliana. Planta 231, 861–873. 10.1007/s00425-009-1092-2 [DOI] [PubMed] [Google Scholar]
- Chen J. Y., Huang J. Q., Li N. Y., Ma X. F., Wang J. L., Liu C., et al. (2015). Genome-wide analysis of the gene families of resistance gene analogues in cotton and their response to Verticillium wilt. BMC Plant Biol. 15:148. 10.1186/s12870-015-0508-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J. Y., Xiao H. L., Gui Y. J., Zhang D. D., Li L., Bao Y. M., et al. (2016). Characterization of the Verticillium dahliae exoproteome involves in pathogenicity from cotton-containing medium. Front. Microbiol. 28:1709 10.3389/fmicb.2016.01709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Kan J., Yang Y., Ling X., Chang Y., Zhang B. (2015). A Ve homologous gene from Gossypium barbadense, Gbvdr3, enhances the defense response against Verticillium dahliae. Plant Physiol. Biochem. 98, 101–111. 10.1016/j.plaphy.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- de Jonge R., van Esse H. P., Maruthachalam K., Bolton M. D., Santhanam P., Saber M. K., et al. (2012). Tomato immune receptor Ve1 recognizes effector of multiple fungal pathogens uncovered by genome and RNA sequencing. Proc. Natl. Acad. Sci. U.S.A. 109, 5110–5115. 10.1073/pnas.1119623109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M. S., Hatzixanthis K., Jones D. A., Harrison K., Jones J. D. (1998). The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell. 10, 1915–1925. 10.1105/tpc.10.11.1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon M. S., Jones D. A., Keddie J. S., Thomas C. M., Harrison K., Jones J. D. (1996). The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84, 451–459. 10.1016/S0092-8674(00)81290-8 [DOI] [PubMed] [Google Scholar]
- Fradin E. F., Abd-El-Haliem A., Masini L., van den Berg G. C., Joosten M. H., Thomma B. P. (2011). Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265. 10.1104/pp.111.180067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fradin E. F., Thomma B. P. H. J. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum. Mol. Plant Pathol. 7, 71–86. 10.1111/j.1364-3703.2006.00323.x [DOI] [PubMed] [Google Scholar]
- Fradin E. F., Zhang Z., Ayala J. C. J., Castroverde C. D., Nazar R. N., Robb J., et al. (2009). Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332. 10.1104/pp.109.136762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriëls S. H., Takken F. L., Vossen J. H., de Jong C. F., Liu Q., Turk S. C., et al. (2006). cDNA-AFLP combined with functional analysis reveals novel genes involved in the hypersensitive response. Mol. Plant Microbe Interact. 19, 567–576. 10.1094/MPMI-19-0567 [DOI] [PubMed] [Google Scholar]
- Gabriëls S. H., Vossen J. H., Ekengren S. K., Ooijen G. V., Abd-El-Haliem A. M., Berg G., et al. (2007). An NB-LRR protein required for HR signaling mediated by both extra- and intracellular resistance proteins. Plant J. 50, 14–28. 10.1111/j.1365-313X.2007.03027.x [DOI] [PubMed] [Google Scholar]
- Gao X., Shan L. (2013). Functional genomic analysis of cotton genes with agrobacterium-mediated virus-induced gene silencing. Methods Mol. Biol. 975, 157–165. 10.1007/978-1-62703-278-0_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayoso C., Pomar F., Novo-Uzal E., Merino F., de Ilárduya O. M. (2010). The Ve-mediated resistance response of the tomato to Verticillium dahliae involves H2O2, peroxidase and lignins and drives PAL gene expression. BMC Plant Biol. 10:232. 10.1186/1471-2229-10-232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Lamothe R., Tsitsigiannis D. I., Ludwig A. A., Panicot M., Shirasu K., Jones J. D. (2006). The U-box protein CMPG1 is required for efficient activation of defense mechanisms triggered by multiple resistance genes in tobacco and tomato. Plant Cell 18, 1067–1083. 10.1105/tpc.106.040998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K., Cherk C., Somerville S. (2009). Host-pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 12, 406–413. 10.1016/j.pbi.2009.06.007 [DOI] [PubMed] [Google Scholar]
- Huang H. C. (2003). Verticillium wilt of alfalfa: epidemiology and control strategies. Can. J. Plant Pathol. 25, 328–338. 10.1080/07060660309507088 [DOI] [Google Scholar]
- Hückelhoven R. (2007). Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45, 101–127. 10.1146/annurev.phyto.45.062806.094325 [DOI] [PubMed] [Google Scholar]
- Jones D. A., Jones J. D. G. (1997). The role of leucine rich repeat proteins in plant defenses. Adv. Bot. Res. 24, 89–167. 10.1016/S0065-2296(08)60072-5 [DOI] [Google Scholar]
- Jones D. A., Thomas C. M., Hammond-Kosack K. E., Balint-Kurti P. J., Jones J. D. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. 10.1126/science.7973631 [DOI] [PubMed] [Google Scholar]
- Joosten M., de Wit P. (1999). The tomato-Cladosporium fulvum interaction: a versatile experimental system to study plant-pathogen interactions. Annu. Rev. Phytopathol. 37, 335–367. 10.1146/annurev.phyto.37.1.335 [DOI] [PubMed] [Google Scholar]
- Kawchuk L. M., Hachey J., Lynch D. R., Kulcsar F., van Rooijen G., Waterer D. R., et al. (2001). Tomato Ve disease resistance genes encode cell surface-like receptors. Proc. Natl. Acad. Sci. U.S.A. 98, 6511–6515. 10.1073/pnas.091114198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosterman S. J., Atallah Z. K., Vallad G. E., Subbarao K. V. (2009). Diversity, pathogenicity, and management of Verticillium species. Annu. Rev. Phytopathol. 47, 39–62. 10.1146/annurev-phyto-080508-081748 [DOI] [PubMed] [Google Scholar]
- Kruijt M., de Kock M. J., de Wit P. J. (2005a). Receptor-like proteins involved in plant disease resistance. Mol. Plant Pathol. 6, 85–97. 10.1111/j.1364-3703.2004.00264.x [DOI] [PubMed] [Google Scholar]
- Kruijt M., Kip D. J., Joosten M. H., Brandwagt B. F., de Wit P. J. (2005b). The Cf-4 and Cf-9 resistance genes against Cladosporium fulvum are conserved in wild tomato species. Mol. Plant Microbe Interact. 18, 1011–1021. 10.1094/MPMI-18-1011 [DOI] [PubMed] [Google Scholar]
- Laurent F., Labesse G., de Wit P. (2000). Molecular cloning and partial characterization of a plant VAP33 homologue with a major sperm protein domain. Biochem. Biophys. Res. Commun. 270, 286–292. 10.1006/bbrc.2000.2387 [DOI] [PubMed] [Google Scholar]
- Letourneur F., Klausner R. D. (1992). A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell 69, 1143–1157. 10.1016/0092-8674(92)90636-Q [DOI] [PubMed] [Google Scholar]
- Liu G., Holub E. B., Alonso J. M., Ecker J. R., Fobert P. R. (2005). An Arabidopsis NPR1-like gene, NPR4, is required for disease resistance. Plant J. 41, 304–318. 10.1111/j.1365-313x.2004.02296.x [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lu G., Moriyama E. N. (2004). Vector, N. T. I., a balanced all-in-one sequence analysis suite. Brief. Bioinform. 5, 378–388. 10.1093/bib/5.4.378 [DOI] [PubMed] [Google Scholar]
- Malnoy M., Xu M., Borejsza-Wysocka E., Korban S. S., Aldwinckle H. S. (2008). Two receptor-like genes, Vfa1 and Vfa2, confer resistance to the fungal pathogen Venturia inaequalis inciting apple scab disease. Mol. Plant Microbe. Interact. 21, 448–458. 10.1094/MPMI-21-4-0448 [DOI] [PubMed] [Google Scholar]
- Nakano J. M., Meshitsuka G. (1992). The detection of lignin, in Methods in Lignin Chemistry, eds Lin S. Y., Dence C. W. (Berlin: Springer; ), 23–32. [Google Scholar]
- Nekrasov V., Ludwig A. A., Jones J. D. (2006). CITRX thioredoxin is a putative adaptor protein connecting Cf-9 and the ACIK1 protein kinase during the Cf-9/Avr9- induced defence response. FEBS Lett. 580, 4236–4241. 10.1016/j.febslet.2006.06.077 [DOI] [PubMed] [Google Scholar]
- Parniske M., Hammond-Kosack K. E., Golstein C., Thomas C. M., Jones D. A., Harrison K., et al. (1997). Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91, 821–832. 10.1016/S0092-8674(00)80470-5 [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Wendel J. F., Gundlach H., Guo H., Jenkins J., Jin D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. 10.1038/nature11798 [DOI] [PubMed] [Google Scholar]
- Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. 10.1038/nmeth.1701 [DOI] [PubMed] [Google Scholar]
- Piedras P., Hammond-Kosack K. E., Harrison K., Jones J. D. G. (1998). Rapid, Cf-9- and Avr9-dependent production of active oxygen species in tobacco suspension cultures. Mol. Plant Microbe Interact. 11, 1155–1166. 10.1094/MPMI.1998.11.12.1155 [DOI] [Google Scholar]
- Postma J., Liebrand T. W., Bi G., Evrard A., Bye R. R., Mbengue M., et al. (2016). Avr4 promotes Cf-4 receptor-like protein association with the BAK1/SERK3 receptor-like kinase to initiate receptor endocytosis and plant immunity. New Phytol. 210, 627–642. 10.1111/nph.13802 [DOI] [PubMed] [Google Scholar]
- Putt E. D. (1964). Breeding behavior of resistance to leaf mottle or Verticillium in sunflower. Crop Sci. 4, 177–179. 10.2135/cropsci1964.0011183X000400020016x [DOI] [Google Scholar]
- Rivas S., Rougon-Cardoso A., Smoker M., Schauser L., Yoshioka H., Jones J. D. (2004). CITRX thioredoxin interacts with the tomato Cf-9 resistance protein and negatively regulates defence. EMBO J. 23, 2156–2165. 10.1038/sj.emboj.7600224 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rivas S., Thomas C. M. (2005). Molecular interactions between tomato and the leaf mold pathogen Cladosporium fulvum. Annu. Rev. Phytopathol. 43, 395–436. 10.1146/annurev.phyto.43.040204.140224 [DOI] [PubMed] [Google Scholar]
- Ron M., Avni A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell. 16, 1604–1615. 10.1105/pc.022475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland O., Ludwig A. A., Merrick C. J., Baillieul F., Tracy F. E., Durrant W. E., et al. (2005). Functional analysis of Avr9/Cf9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf9-dependent disease resistance in tomato. Plant Cell. 17, 295–310. 10.1105/tpc.104.026013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santhanam P., van Esse H. P., Albert I., Faino L., Nürnberger T., Thomma B. P. (2013). Evidence for functional diversification within a fungal NEP1-Like protein family. Mol. Plant Microbe Interact. 26, 278–286. 10.1094/MPMI-09-12-0222-R [DOI] [PubMed] [Google Scholar]
- Schaible L., Cannon O. S., Waddoups V. (1951). Inheritance of resistance to Verticillium wilt in a tomato cross. Phytopathology 41, 986–990. [Google Scholar]
- Schoonbeek H. J., Wang H. H., Stefanato F. L., Craze M., Bowden S., Wallington E., et al. (2015). Arabidopsis EF-Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 206, 606–613. 10.1111/nph.13356 [DOI] [PubMed] [Google Scholar]
- Sharfman M., Bar M., Ehrlich M., Schuster S., Melech-Bonfil S., Ezer R., et al. (2011). Endosomal signaling of the tomato leucine-rich repeat receptor-like protein LeEix2. Plant J. 68, 413–423. 10.1111/j.1365-313X.2011.04696.x [DOI] [PubMed] [Google Scholar]
- Simko I., Costanzo S., Haynes K. G., Christ B. J., Jones R. W. (2004). Linkage disequilibrium mapping of a Verticillium dahliae resistance quantitative trait locus in tetraploid potato (Solanum tuberosum) through a candidate gene approach. Theor. Appl. Genet. 108, 217–224. 10.1007/s00122-003-1431-9 [DOI] [PubMed] [Google Scholar]
- Smit F., Dubery L. A. (1997). Cell wall reinforcement in cotton hypocotyls in response to a Verticillium dahliae elicitor. Phytochemistry 44, 811–815. 10.1016/S0031-9422(96)00595-X [DOI] [Google Scholar]
- Sonnhammer E. L., von Heijne G., Krogh A. (1998). A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182. [PubMed] [Google Scholar]
- Stergiopoulos I., de Wit P. J. G. M. (2009). Fungal effector proteins. Annu. Rev. Phytopathol. 47, 233–263. 10.1146/annurev.phyto.112408.132637 [DOI] [PubMed] [Google Scholar]
- Stulemeijer I. J., Stratmann J. W., Joosten M. H. (2007). Tomato mitogen-activated protein kinases LeMPK1, LeMPK2, and LeMPK3 are activated during the Cf4/Avr4-induced hypersensitive response and have distinct phosphorylation specificities. Plant Physiol. 144, 1481–1494. 10.1104/pp.107.101063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot J. A. (1996). Worm sperm and advances in cell locomotion. Cell 84, 1–4. 10.1016/S0092-8674(00)80068-9 [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Jones D. A., Parniske M., Harrison K., Balint-Kurti P. J., Hatzixanthis K., et al. (1997). Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell. 9, 2209–2224. 10.1105/tpc.9.12.2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl. Acids Res. 25, 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Burg H. A., Tsitsigiannis D. I., Rowland O., Lo J., Rallapalli G., MacLean D., et al. (2008). The F-box protein ACRE189/ACFI1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell. 20, 697–719. 10.1105/tpc.107.056978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Hoorn R. A., Wulff B. B., Rivas S., Durrant M. C., van der Ploeg A., de Wit P. J., et al. (2005). Structure-function analysis of Cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell. 17, 1000–1015. 10.1105/tpc.104.028118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinatzer B. A., Patocchi A., Gianfranceschi L., Tartarini S., Zhang H. B., Gessler C., et al. (2001). Apple contains receptor-like genes homologous to the Cladosporium fulvum resistance gene family of tomato with a cluster of genes cosegregating with Vf apple scab resistance. Mol. Plant Microbe Interact. 14, 508–515. 10.1094/MPMI.2001.14.4.508 [DOI] [PubMed] [Google Scholar]
- Wiesner-Hanks T., Nelson R. (2016). Multiple disease resistance in plants. Annu. Rev. Phytopathol. 54, 229–252. 10.1146/annurev-phyto-080615-100037 [DOI] [PubMed] [Google Scholar]
- Yang Y., Ling X., Chen T., Cai L., Liu T., Wang J., et al. (2015). A cotton Gbvdr5 gene encoding a leucine-rich-repeat receptor-like protein confers resistance to Verticillium dahliae in transgenic Arabidopsis and upland cotton. Plant Mol. Biol. Rep. 33, 987–1001. 10.1007/s11105-014-0810-5 [DOI] [Google Scholar]
- Yeh Y. H., Chang Y. H., Huang P. Y., Huang J. B., Zimmerli L. (2015). Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front. Plant Sci. 6:322. 10.3389/fpls.2015.00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Yang Y., Chen T., Yu W., Liu T., Li H., et al. (2012). Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS ONE 7:e51091. 10.1371/journal.pone.0051091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang X., Yang S., Chi J., Zhang G., Ma Z. (2011). Cloning and characterization of a Verticillium wilt resistance gene from Gossypium barbadense and functional analysis in Arabidopsis thaliana. Plant Cell Rep. 30, 2085–2096. 10.1007/s00299-011-1115-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.