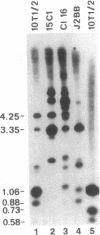
Abstract
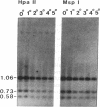
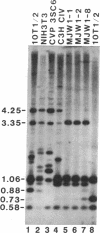
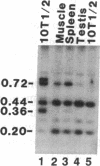
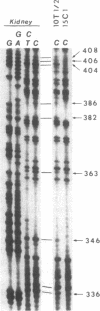
CpG dinucleotides are unevenly distributed in the vertebrate genome. Bulk DNA is depleted of CpGs and most of the cytosines in the dinucleotide in this fraction are methylated. On the other hand, CpG islands, which are often associated with genes, are unmethylated at testable sites in all normal tissues with the exception of genes on the inactive X chromosome. We used Hpa II/Msp I analysis and ligation-mediated polymerase chain reaction to examine the methylation of the MyoD1 CpG island in adult mouse tissues, early cultures of mouse embryo cells, and immortal fibroblastic cell lines. The island was almost devoid of methylation at CCGG sites in adult mouse tissues and in low-passage mouse embryo fibroblasts. In marked contrast, the island was methylated in 10T 1/2 cells and in six other immortal cell lines showing that methylation of this CpG island had occurred during escape from senescence. The island became even more methylated in chemically transformed derivatives of 10T 1/2 cells. Thus, CpG islands not methylated in normal tissues may become modified to an abnormally high degree during immortalization and transformation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antequera F., Macleod D., Bird A. P. Specific protection of methylated CpGs in mammalian nuclei. Cell. 1989 Aug 11;58(3):509–517. doi: 10.1016/0092-8674(89)90431-5. [DOI] [PubMed] [Google Scholar]
- Bestor T. H., Ingram V. M. Two DNA methyltransferases from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor T., Laudano A., Mattaliano R., Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. The carboxyl-terminal domain of the mammalian enzymes is related to bacterial restriction methyltransferases. J Mol Biol. 1988 Oct 20;203(4):971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- Bird A. P. CpG-rich islands and the function of DNA methylation. Nature. 1986 May 15;321(6067):209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Borrello M. G., Pierotti M. A., Donghi R., Bongarzone I., Cattadori M. R., Traversari C., Mondellini P., Della Porta G. Modulation of the human Harvey-ras oncogene expression by DNA methylation. Oncogene Res. 1988;2(2):197–203. [PubMed] [Google Scholar]
- Buschhausen G., Wittig B., Graessmann M., Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex virus thymidine kinase gene. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotti D., Palitti F., Lavia P., Strom R. In vitro methylation of CpG-rich islands. Nucleic Acids Res. 1989 Nov 25;17(22):9219–9229. doi: 10.1093/nar/17.22.9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler L. A., Ghazi H., Jones P. A., Boukamp P., Fusenig N. E. Allele-specific methylation of the human c-Ha-ras-1 gene. Cell. 1987 Aug 28;50(5):711–717. doi: 10.1016/0092-8674(87)90329-1. [DOI] [PubMed] [Google Scholar]
- Chen J., Jones P. Determination genes. Curr Opin Cell Biol. 1989 Dec;1(6):1075–1080. doi: 10.1016/s0955-0674(89)80053-5. [DOI] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compere S. J., Palmiter R. D. DNA methylation controls the inducibility of the mouse metallothionein-I gene lymphoid cells. Cell. 1981 Jul;25(1):233–240. doi: 10.1016/0092-8674(81)90248-8. [DOI] [PubMed] [Google Scholar]
- Davis R. L., Weintraub H., Lassar A. B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987 Dec 24;51(6):987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987 Jul 20;196(2):261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gebara M. M., Drevon C., Harcourt S. A., Steingrimsdottir H., James M. R., Burke J. F., Arlett C. F., Lehmann A. R. Inactivation of a transfected gene in human fibroblasts can occur by deletion, amplification, phenotypic switching, or methylation. Mol Cell Biol. 1987 Apr;7(4):1459–1464. doi: 10.1128/mcb.7.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P. A., Laug W. E., Gardner A., Nye C. A., Fink L. M., Benedict W. F. In vitro correlates of transformation in C3H/10T1/2 clone 8 mouse cells. Cancer Res. 1976 Aug;36(8):2863–2867. [PubMed] [Google Scholar]
- Jones P. A., Wolkowicz M. J., Harrington M. A., Gonzales F. Methylation and expression of the Myo D1 determination gene. Philos Trans R Soc Lond B Biol Sci. 1990 Jan 30;326(1235):277–284. doi: 10.1098/rstb.1990.0011. [DOI] [PubMed] [Google Scholar]
- Keith D. H., Singer-Sam J., Riggs A. D. Active X chromosome DNA is unmethylated at eight CCGG sites clustered in a guanine-plus-cytosine-rich island at the 5' end of the gene for phosphoglycerate kinase. Mol Cell Biol. 1986 Nov;6(11):4122–4125. doi: 10.1128/mcb.6.11.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolsto A. B., Kollias G., Giguere V., Isobe K. I., Prydz H., Grosveld F. The maintenance of methylation-free islands in transgenic mice. Nucleic Acids Res. 1986 Dec 22;14(24):9667–9678. [PMC free article] [PubMed] [Google Scholar]
- Lock L. F., Takagi N., Martin G. R. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987 Jan 16;48(1):39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- Monk M., Boubelik M., Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987 Mar;99(3):371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Mueller P. R., Wold B. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science. 1989 Nov 10;246(4931):780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Nesnow S., Heidelberger C. The effect of modifiers of microsomal enzymes on chemical oncogenesis in cultures of C3H mouse cell lines. Cancer Res. 1976 May;36(5):1801–1808. [PubMed] [Google Scholar]
- Pfeifer G. P., Grünwald S., Boehm T. L., Drahovsky D. Isolation and characterization of DNA cytosine 5-methyltransferase from human placenta. Biochim Biophys Acta. 1983 Aug 2;740(3):323–330. doi: 10.1016/0167-4781(83)90141-0. [DOI] [PubMed] [Google Scholar]
- Pfeifer G. P., Steigerwald S. D., Mueller P. R., Wold B., Riggs A. D. Genomic sequencing and methylation analysis by ligation mediated PCR. Science. 1989 Nov 10;246(4931):810–813. doi: 10.1126/science.2814502. [DOI] [PubMed] [Google Scholar]
- Razin A., Szyf M. DNA methylation patterns. Formation and function. Biochim Biophys Acta. 1984 Sep 10;782(4):331–342. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- Reznikoff C. A., Bertram J. S., Brankow D. W., Heidelberger C. Quantitative and qualitative studies of chemical transformation of cloned C3H mouse embryo cells sensitive to postconfluence inhibition of cell division. Cancer Res. 1973 Dec;33(12):3239–3249. [PubMed] [Google Scholar]
- Reznikoff C. A., Brankow D. W., Heidelberger C. Establishment and characterization of a cloned line of C3H mouse embryo cells sensitive to postconfluence inhibition of division. Cancer Res. 1973 Dec;33(12):3231–3238. [PubMed] [Google Scholar]
- Selker E. U. DNA methylation and chromatin structure: a view from below. Trends Biochem Sci. 1990 Mar;15(3):103–107. doi: 10.1016/0968-0004(90)90193-f. [DOI] [PubMed] [Google Scholar]
- Stewart C. L., Stuhlmann H., Jähner D., Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TODARO G. J., GREEN H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963 May;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor S. M., Jones P. A. Changes in phenotypic expression in embryonic and adult cells treated with 5-azacytidine. J Cell Physiol. 1982 May;111(2):187–194. doi: 10.1002/jcp.1041110210. [DOI] [PubMed] [Google Scholar]
- Taylor S. M., Jones P. A. Multiple new phenotypes induced in 10T1/2 and 3T3 cells treated with 5-azacytidine. Cell. 1979 Aug;17(4):771–779. doi: 10.1016/0092-8674(79)90317-9. [DOI] [PubMed] [Google Scholar]
- Thayer M. J., Tapscott S. J., Davis R. L., Wright W. E., Lassar A. B., Weintraub H. Positive autoregulation of the myogenic determination gene MyoD1. Cell. 1989 Jul 28;58(2):241–248. doi: 10.1016/0092-8674(89)90838-6. [DOI] [PubMed] [Google Scholar]
- Toniolo D., Martini G., Migeon B. R., Dono R. Expression of the G6PD locus on the human X chromosome is associated with demethylation of three CpG islands within 100 kb of DNA. EMBO J. 1988 Feb;7(2):401–406. doi: 10.1002/j.1460-2075.1988.tb02827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth M., Lichtenberg U., Doerfler W. Genomic sequencing reveals a 5-methylcytosine-free domain in active promoters and the spreading of preimposed methylation patterns. Proc Natl Acad Sci U S A. 1989 May;86(10):3728–3732. doi: 10.1073/pnas.86.10.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venolia L., Gartler S. M., Wassman E. R., Yen P., Mohandas T., Shapiro L. J. Transformation with DNA from 5-azacytidine-reactivated X chromosomes. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2352–2354. doi: 10.1073/pnas.79.7.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Tapscott S. J., Davis R. L., Thayer M. J., Adam M. A., Lassar A. B., Miller A. D. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V. L., Jones P. A. DNA methylation decreases in aging but not in immortal cells. Science. 1983 Jun 3;220(4601):1055–1057. doi: 10.1126/science.6844925. [DOI] [PubMed] [Google Scholar]
- Wise T. L., Harris M. Deletion and hypermethylation of thymidine kinase gene in V79 Chinese hamster cells resistant to bromodeoxyuridine. Somat Cell Mol Genet. 1988 Nov;14(6):567–581. doi: 10.1007/BF01535311. [DOI] [PubMed] [Google Scholar]
- Wolf S. F., Migeon B. R. Clusters of CpG dinucleotides implicated by nuclease hypersensitivity as control elements of housekeeping genes. Nature. 1985 Apr 4;314(6010):467–469. doi: 10.1038/314467a0. [DOI] [PubMed] [Google Scholar]
- de Bustros A., Nelkin B. D., Silverman A., Ehrlich G., Poiesz B., Baylin S. B. The short arm of chromosome 11 is a "hot spot" for hypermethylation in human neoplasia. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5693–5697. doi: 10.1073/pnas.85.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]