A high-content quantitative imaging screen identifies regulators of the post-Golgi transport of the phospholipid flippase Neo1. Neo1 is a new Snx3 cargo protein that is required for sorting of other Snx3 cargo. Thus flippase activity may generate curvature to promote the formation of Snx3-recycling tubules.
Abstract
P4-ATPases are a family of putative phospholipid flippases that regulate lipid membrane asymmetry, which is important for vesicle formation. Two yeast flippases, Drs2 and Neo1, have nonredundant functions in the recycling of the synaptobrevin-like v-SNARE Snc1 from early endosomes. Drs2 activity is needed to form vesicles and regulate its own trafficking, suggesting that flippase activity and localization are linked. However, the role of Neo1 in endosomal recycling is not well characterized. To identify novel regulators of Neo1 trafficking and activity at endosomes, we first identified mutants with impaired recycling of a Snc1-based reporter and subsequently used high-content microscopy to classify these mutants based on the localization of Neo1 or its binding partners, Mon2 and Dop1. This analysis identified a role for Arl1 in stabilizing the Mon2/Dop1 complex and uncovered a new function for Vps13 in early endosome recycling and Neo1 localization. We further showed that the cargo-selective sorting nexin Snx3 is required for Neo1 trafficking and identified an Snx3 sorting motif in the Neo1 N-terminus. Of importance, the Snx3-dependent sorting of Neo1 was required for the correct sorting of another Snx3 cargo protein, suggesting that the incorporation of Neo1 into recycling tubules may influence their formation.
INTRODUCTION
Vesicle trafficking is a highly conserved process that requires the intricate coordination of cargo recruitment, coat formation, and lipid dynamics. Phospholipid flippases are members of a family of P4-type ATPases that translocate phospholipids from the luminal to the cytosolic leaflet of the membrane bilayer and are important for trafficking at the Golgi and endosomes (Muthusamy et al., 2009). By selectively increasing the number of phospholipids in one leaflet, flippases induce membrane bending, which promotes vesicle budding. The flipping of the negatively charged phospholipid phosphatidylserine (PS) also changes membrane electrostatics, which is needed to recruit specific components of the transport machinery (Graham, 2004; van Meer et al., 2008; Muthusamy et al., 2009; Sebastian et al., 2012; Xu et al., 2013; Takeda et al., 2014).
The yeast Saccharomyces cerevisiae has five putative flippases—Dnf1, Dnf2, Dnf3, Drs2, and Neo1 (Catty et al., 1997; Halleck et al., 1998; Hua et al., 2002)—which have partially overlapping steady-state localizations. Dnf1 and Dnf2 are found largely on the plasma membrane and are important for maintaining cell polarity (Liu et al., 2007; Sartorel et al., 2014). Conversely, Neo1, Drs2, and Dnf3 are present in internal structures that colocalize with Golgi and/or endosomal markers and may regulate vesicle trafficking (Hua et al., 2002; Wicky et al., 2004; Furuta et al., 2007; Liu et al., 2008; Hanamatsu et al., 2014; Wu et al., 2016). Despite their partially overlapping distributions, yeast flippases are believed to be functionally distinct (Baldridge and Graham, 2011, 2012; Baldridge et al., 2013; Takar et al., 2016).
Drs2 is the only yeast flippase shown to translocate lipids in a cell-free system (Natarajan et al., 2004). However, other yeast flippases maintain lipid asymmetry in vivo and exhibit different specificities (Pomorski et al., 2003; Zhou and Graham, 2009; Baldridge et al., 2013). Neo1 primarily regulates phosphatidylethanolamine (PE) asymmetry, and may also act on PS (Takar et al., 2016), whereas Drs2 preferentially flips PS but has some activity toward PE (Natarajan et al., 2004, 2009; Zhou and Graham, 2009). Dnf1 and Dnf2 prefer phosphatidylcholine (PC), PE, and their lyso-PE and lyso-PC derivatives (Pomorski et al., 2003; Riekhof and Voelker, 2006; Baldridge and Graham, 2011). A single point mutation that changes the lipid preference of Dnf1 from PE to PS partially compensates for loss of Drs2 (Baldridge and Graham, 2011). This suggests that flippases have distinct roles in the cell that are partially determined by their lipid specificity.
The activity and trafficking of Drs2 appear to be closely linked. Drs2 has an autoinhibitory C-terminal tail that interacts with regulators at the Golgi and endosomes. At the Golgi, binding of PI4P and the ArfGEF Gea2 to adjacent regions of this C-terminal tail synergistically stimulates Drs2 flippase activity (Natarajan et al., 2009; Zhou et al., 2013). Drs2 activity is needed for the formation of the AP-1 vesicles that carry Drs2 from Golgi to endosomes (Chantalat et al., 2004; Liu et al., 2008; Muthusamy et al., 2009). At endosomes, the recycling protein Rcy1 binds at a similar site within the Drs2 C-terminus. Rcy1 binding and Drs2 activity are needed for Drs2 trafficking out of endosomes and for the recycling of other endosomal proteins (Furuta et al., 2007; Hanamatsu et al., 2014). Thus flippases appear to regulate both vesicle formation and their own transport.
Several flippases contribute to the endosomal recycling of the vesicle-soluble N-ethylmaleimide–sensitive factor attachment protein receptor (v-SNARE) Snc1. Triple deletion of Dnf1/2/3 can cause Snc1-recycling defects; however, mutations in either Drs2 or Neo1 cause internal accumulation of Snc1 on their own (Hua et al., 2002; Takeda et al., 2014; Yamagami et al., 2015), suggesting that they have a more significant role in Snc1 trafficking. Although the role of Drs2 in post-Golgi transport is relatively well studied, the endosomal requirement for Neo1 is less well understood and provides an interesting avenue for further study.
Few Neo1 regulators have been identified. Neo1 forms a complex with Dop1 and Mon2, which are also needed for Snc1 recycling, suggesting that these proteins regulate Neo1 activity or trafficking (Wicky et al., 2004; Efe et al., 2005; Gillingham et al., 2006; Barbosa et al., 2010; Yamagami et al., 2015). We predicted that other proteins that control the post-Golgi trafficking or activity of Neo1 would also cause Snc1 trafficking defects. To systematically identify Neo1 regulators at endosomes, we used a genome-wide screen to find mutants that accumulate intracellular Snc1 and assessed the localization of Neo1 in these mutants using a high-content microscopy screen. This approach identified several regulators of Neo1 transport, including the sorting nexin Snx3. Disrupting the Snx3-mediated sorting of Neo1 caused defects in the trafficking of another Snx3-dependent protein, supporting a model in which flippase activity at the site of vesicle formation is needed to stimulate vesicle transport.
RESULTS
A quantitative Snc1-based assay identifies potential regulators of Neo1
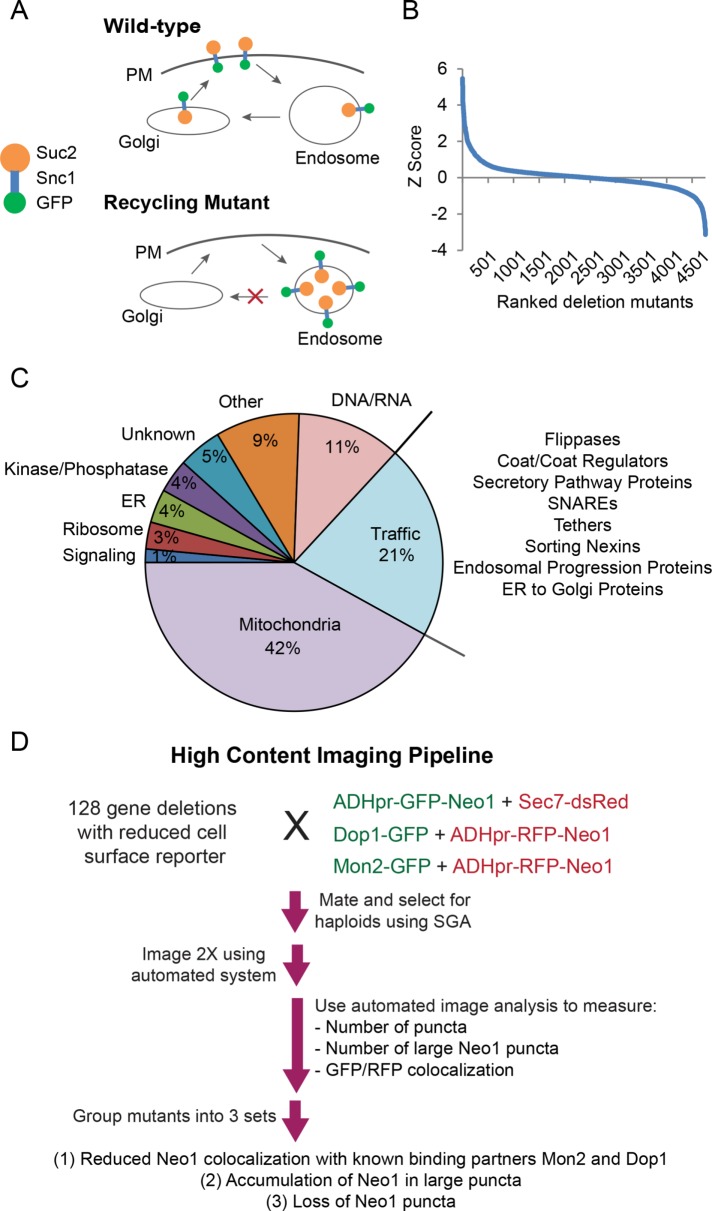
Mutating or depleting Neo1 causes defects in COPI retrograde transport from the Golgi to the endoplasmic reticulum (ER) and impairs the Golgi/endosome trafficking of Snc1 (Hua and Graham, 2003; Wicky et al., 2004; Mioka et al., 2014; Takeda et al., 2014; Yamagami et al., 2015). Neo1’s role at early endosomes, as opposed to its ER/Golgi function, is not expected to be essential for viability (Takeda et al., 2014). To find genes that regulate the post-Golgi trafficking of Neo1, we first used a genome-wide screen to identify the set of viable mutants that cause internal accumulation of an Snc1-based reporter (Figure 1A). Levels of the green fluorescent protein (GFP)–Snc1-Suc2 (GSS) chimera can be quantified by measuring the activity of invertase (Suc2) exposed at the cell surface using an agar plate–based assay (Burston et al., 2008; Dalton et al., 2015). The set of mutants with higher cell surface reporter levels were previously used to uncover endocytosis regulators (Burston et al., 2009). Here we focused on the subset of mutants with reduced cell surface levels of the Snc1 chimeric reporter (Figure 1B and Supplemental Table S1).
FIGURE 1:
A genome-wide Snc1 recycling screen and high-content microscopy analysis identifies potential Neo1 regulators. (A) The GFP-Snc1-Suc2 (GSS) chimera is trafficked to the plasma membrane, internalized, and recycled to the Golgi. A block in recycling from early endosomes reduces surface levels of the chimera, which is detected by an invertase (Suc2) activity assay. (B) Cell surface levels of the GSS reporter were quantified in yeast deletion strains and reported as Z-scores. A positive Z-score indicates mutants have reduced surface levels of the chimera. (C) Functional classification of top-scoring mutants. The proportional representation of each functional category was graphed, and subgroups in the trafficking category are listed at right. (D) High-content microscopy workflow and follow-up analysis.
We identified 275 top-scoring mutants using Cutoff Linked to Interaction Knowledge (CLIK) analysis, which sets a threshold based on the enrichment of genetic and physical interactions (Supplemental Table S2; Dittmar et al., 2013). Several of these mutants corresponded to known Snc1 regulators, including proteins involved in vesicle formation (sorting nexins Snx4 and Atg20, epsins Ent3 and Ent5), vesicle tethering (Golgi-associated retrograde protein complex [GARP] subunits), and the Drs2 noncatalytic subunit Cdc50 (Hua et al., 2002; Conibear et al., 2003; Hettema et al., 2003; Zimmermann et al., 2010). We also identified the flippase Dnf3, its noncatalytic subunit Crf1 (Saito et al., 2004), and a variety of other proteins, including kinases, phosphatases, and vesicle biogenesis proteins not previously associated with Snc1 transport (Figure 1C). Other hits, which corresponded to known regulators of DNA/RNA synthesis or mitochondrial function, may have indirect effects on Snc1 or Neo1 trafficking and were not pursued further, leaving 128 candidate mutants (Supplemental Table S2).
We next used high-throughput microscopy and quantitative image analysis to identify which of these 128 mutants altered Neo1 trafficking or the localization of the Neo1-interacting proteins Dop1 and Mon2 (Figure 1D). Neo1 was tagged with GFP at its N-terminus, which was previously determined not to alter its function (Wicky et al., 2004), and expressed from the ADH promoter at the chromosomal locus, which provided a bright signal without noticeably affecting localization (Wu et al., 2016). Synthetic genetic array (SGA; Tong and Boone, 2006) techniques were used to generate a set of haploid mutants coexpressing ADHpr-GFP-Neo1 and the Golgi marker Sec7-dsRed (McDonold and Fromme, 2014). Parallel mutant arrays were constructed to compare the localization of ADHpr-RFP-Neo1 with Dop1-GFP or Mon2-GFP. High-throughput imaging of haploid arrays containing each reporter combination was performed twice to reduce anomalies and ensure full coverage of the hit list.
Mutations that disrupt Neo1 transport at endosomes may result in its depletion from Golgi compartments and accumulation in the vacuole or endosomal structures. Other mutations may change the extent of Neo1 colocalization with Dop1 and Mon2. To detect these changes, we created in-house automated image quantitation pipelines that assessed the degree of Neo1 colocalization with cellular markers in an unbiased manner and determined the number of puncta per cell labeled by each marker. For all measurements, a normalized score (t-statistic) representing the deviation from the average value was calculated and is reported in Supplemental Table S3. Negative scores indicate reduced colocalization or reduced number of puncta, whereas positive scores indicate increases in these values. These scores were used to define three distinct sets of mutants (Figure 1D) that were investigated in detail.
Arl1 promotes stability of Dop1 complexes
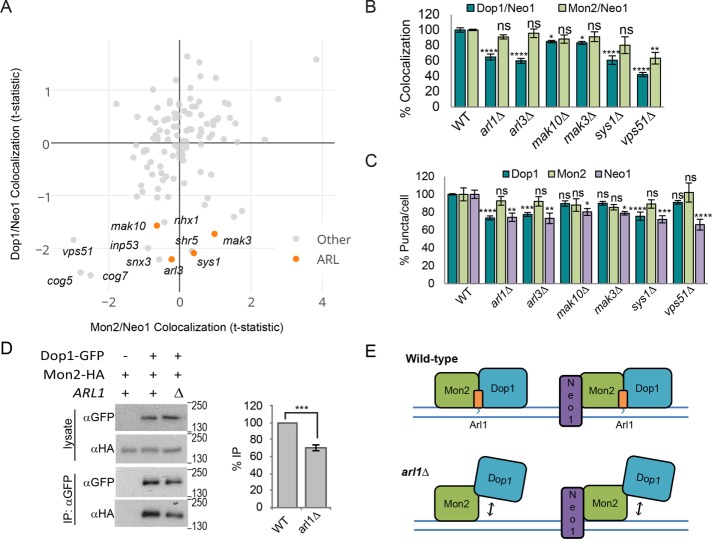
The first set of mutants was selected based on altered association of Mon2 and/or Dop1 with Neo1 (Figure 2A). Mutation of GARP and conserved oligomeric Golgi complex (COG) subunits caused a strong reduction in the colocalization of Neo1 with both Dop1 and Mon2. Of interest, a subset of mutants showed a specific decrease in the localization of Dop1 at Neo1-containing structures that was not seen with Mon2 (Figure 2A). This subset was enriched for components of the Arl pathway, including the small ARF-like GTPase Arl3, the Arl3 recruitment factor Sys1, and two components of the NatC acetyltransferase that modify Arl3 to enhance its binding to Sys1 (Mak10 and Mak3; Behnia et al., 2004). The specific effect of Arl pathway mutations on the localization of Dop1 at Neo1 puncta was retested in the same strains and found to be significant, with p values ranging from 0.0128 (mak10∆) to <0.0001 (arl1Δ; Figure 2B).
FIGURE 2:
Arl1 promotes stability of the Dop1–Mon2 complex. (A) Plot of Mon2 vs. Dop1 localization at Neo1-labeled puncta from the high-content imaging analysis. Labels indicate Arl pathway mutants and mutants with a Dop1/Neo1 colocalization t-statistic <1.5. (B) Secondary validation by small-scale fluorescence microscopy shows that in comparison with wild type (WT), Arl pathway mutants exhibit significantly reduced localization of Dop1-GFP, but not Mon2-GFP, at ADHpr-RFP-Neo1 puncta. Error bars report SEM; >300 cells/trial, n = 3. (C) Analysis of ADHpr-GFP-Neo1, Dop1-GFP, and Mon2-GFP puncta per cell from small-scale imaging shows that the number of Dop1 puncta is reduced in Arl pathway mutants but not vps51∆ strains. Error bars report SEM; >300 cells/trial, n = 3. One-way ANOVA with a Dunnett-corrected post hoc test: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (D) Absence of Arl1 decreases the amount of Mon2-HA that coimmunoprecipitates with Dop1-GFP. Loading of immunoprecipitate relative to lysate was 1.3× (left). Recovery of Mon2-HA relative to Dop1-GFP was quantified by densitometry, and values were normalized to WT (right). Error bars report SEM; n = 3. Two-tailed unpaired t test, ***p < 0.001. (E) Model illustrating the contribution of Arl1 to the stability of the Dop1/Mon2/Neo1 and/or Dop1/Mon2 complex.
To determine whether the reduced overlap was due to changes in Neo1 or Dop1 localization to membranes, we analyzed the average number of Dop1, Mon2, and Neo1 puncta/cell in these mutants. Loss of the GARP subunit Vps51 reduced Neo1 puncta without significantly affecting the number of Dop1 or Mon2 puncta, which is consistent with an observed missorting of Neo1 to fragmented vacuole structures in vps51Δ mutants, whereas Mon2 and Dop1 recruitment to membranes was unaffected (unpublished data). This suggests that GARP regulates Neo1 recycling at endosomes, as described for other flippases (Takagi et al., 2012), and that Dop1 and Mon2 may be recruited to membranes independently of Neo1. Alternatively, GARP mutants may retain a small pool of Neo1 at Golgi/endosomes that is sufficient for Dop1/Mon2 recruitment.
The number of Dop1 puncta/cell was significantly reduced in arl1Δ, arl3Δ, and sys1Δ mutants when RFP-Neo1 was moderately overexpressed from the ADH promoter (Figure 2C) but not in arl mutants expressing untagged Neo1 (unpublished data), suggesting that Dop1 membrane recruitment is sensitive to alteration of both Neo1 and Arl1. The Arl pathway mutants also exhibited a reduced number of Neo1 puncta/cell (Figure 2C). Whereas some Neo1 appeared to be missorted to the vacuole in Arl pathway mutants, a substantial pool of Neo1 was still present in puncta that colocalized with Mon2 (Figure 2, A and B), consistent with previous reports that Mon2 localization is not affected by loss of Arl1 (Gillingham et al., 2006).
The observation that defects in the Arl pathway reduce Dop1 recruitment was unexpected. Mon2 is needed to recruit Dop1 to the Golgi (Gillingham et al., 2006; Barbosa et al., 2010). Because loss of Arl1 selectively affected Dop1, we hypothesized that Arl1 contributes to the stable association of Dop1 and Mon2. We found that the amount of Dop1 that coprecipitated with Mon2 was reduced by ∼30% in an arl1Δ mutant (Figure 2D), which supports the idea that loss of Arl1 impairs Mon2–Dop1 binding. These results show that our systematic imaging analysis can detect subtle changes in localization that reflect altered binding of regulatory proteins. Based on these observations, we hypothesize that the Arl1–Mon2 complex is needed to stabilize the localization of Dop1 at the membrane (Figure 2E).
Vps13 and PI3K regulate Neo1 transit through endosomes
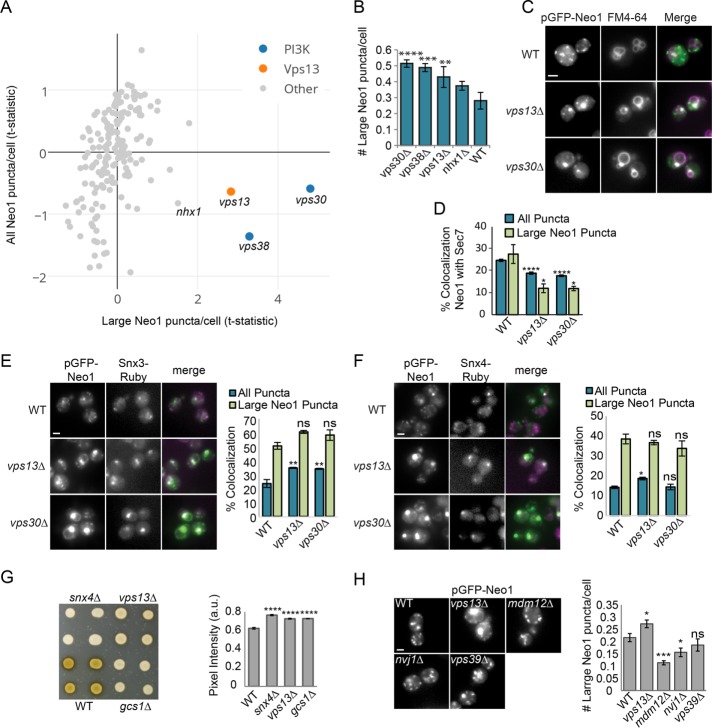
Defects in trafficking at endosomes can cause proteins to accumulate in endosomal compartments or result in vacuolar missorting. In wild-type cells, Neo1 is largely present in small puncta that colocalize with the Golgi marker Sec7 and is less frequently found in large puncta that may represent endosomes. To identify the set of mutants that cause Neo1 to accumulate in endosomes, we defined an initial group of 10 mutants with a reduced number of total Neo1 puncta/cell (t-statistic < −0.33) and an increased number of large Neo1 puncta (t-statistic ≥ 0.25; Figure 3A). Reimaging these 10 mutants identified three that exhibit a statistically significant increase in the number of large Neo1 puncta/cell relative to a wild-type strain: vps13Δ, vps30Δ, and vps38Δ (Figure 3B). Large puncta were similarly observed when a plasmid expressing GFP-Neo1 from its native promoter was introduced into vps13Δ and vps30Δ mutants, indicating that the phenotype was not due to Neo1 overexpression (Figure 3C).
FIGURE 3:
Vps13 and the PI3K subunits Vps30/38 are required for Neo1 sorting at endosomes. (A) The number of large Neo1 puncta/cell was compared with all Neo1 puncta/cell to identify mutants with intracellular accumulation of Neo1. The t-statistic values represent the number of puncta/cell relative to the mean. The indicated mutants were subject to further analysis. (B) Small-scale reimaging shows significant accumulation of large Neo1 puncta in vps13Δ, vps30Δ, and vps38Δ mutants compared with WT. Error bars report SEM; >700 cells/trial, n = 3. One-way ANOVA with Dunnett-corrected test: **p < 0.01, ***p < 0.001, ****p < 0.0001. (C) Live-cell fluorescence microscopy of WT and mutant strains expressing GFP-Neo1 from its native promoter (pLD27) shows that Neo1 accumulates in endosomes that label with FM4-64. Scale bar, 2 μm. (D) Colocalization of integrated ADHpr-GFP-Neo1 with the Golgi marker Sec7-dsRed was quantified in WT and mutant strains. Error bars report SEM; >700 cells/trial, n = 3. One-way ANOVA with a Dunnett-corrected post hoc test: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (E, F) Representative images (left) and quantification (right) of (E) Snx3-Ruby and (F) Snx4-Ruby localization to pADHpr-GFP-Neo1 (pMD124) puncta. Error bars report SEM; >700 cells/trial, n = 3. One-way ANOVA with a Dunnett-corrected post hoc comparisons to WT: ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Scale bar, 2 μm. (G) Invertase activity overlay assay shows reduced GSS cell surface levels in vps13Δ, snx4Δ, and gcs1Δ colonies (left), which were quantified by densitometry (right). Error bars report SEM; a.u., arbitrary units; n = 3. One-way ANOVA with Dunnett-corrected post hoc test: ****p < 0.0001. (H) Neo1 does not accumulate in large puncta in contact-site mutants. Left, live-cell fluorescence microscopy of WT and mutant strains expressing pADHpr-GFP-Neo1 (pMD124). Scale bar, 2 μm. Right, number of large Neo1 puncta/cell. Error bars report SEM; >300 cells/trial, n > 6. One-way ANOVA with a Dunnett-corrected post hoc test: ns, not significant; *p < 0.05, **p < 0.01, ***p < 0.001.
The large punctate structures often colabeled with FM4-64 (Figure 3C). To determine whether these large puncta are aberrant endosomes, we compared the overlap of Neo1 puncta with Sec7 and the endosome markers Snx4 and Snx3 (Strochlic et al., 2007). In vps30Δ and vps13Δ mutants, Neo1 showed a significantly reduced colocalization with Sec7 (p < 0.0001), which was more pronounced for large puncta (p < 0.015), indicating that these structures are not enlarged Golgi compartments (Figure 3D). Instead, we observed a striking colocalization of the large Neo1 puncta with Snx3 and Snx4 (Figure 3E). The total area of all Neo1 puncta also showed an increased overlap with Snx3 in vps13Δ and vps30Δ mutants (p < 0.008; Figure 3E) and with Snx4 in a vps13Δ mutant (p = 0.029; Figure 3F). Together these data suggest that Vps13 and Vps30/38 are important regulators of Neo1 trafficking at endosomes.
Vps13 is required for sorting at early and late endosomes
Vps30/38 are two subunits of the type I Vps34 phosphoinositide 3-kinase (PI3K) complex important for maintaining phosphatidylinositol 3-phosphate (PI3P) at endosomes (Kihara et al., 2001), but the function of Vps13 is largely unknown. Vps13 is required for the recycling of Vps10 at late endosomes (Robinson et al., 1988; Rothman et al., 1989; Redding et al., 1996). We confirmed that a vps13Δ mutant had significantly reduced levels of the GSS reporter at the cell surface (p < 0.0001), similar to mutants lacking Gcs1 and Snx4, which are known regulators of Snc1 trafficking (Hettema et al., 2003; Xu et al., 2013; Figure 3G). This suggests that vps13Δ mutants have altered sorting at early endosomes.
Recent work suggests that Vps13 has a role in the function or maintenance of membrane contact sites (Lang et al., 2015; Park et al., 2016). However, we did not find that loss of known contact-site proteins caused reduced cell surface levels of the Snc1 reporter in our genome-wide screen (Supplemental Table S1). We also determined that mutation of genes important for the formation of different membrane contact sites, including nvj1Δ (NVJ), mdm12Δ (ERMES), and vps39Δ (v-CLAMP), did not cause the accumulation of Neo1 in large puncta (Figure 3H). Finally, we found that missense mutations in Vps13 that blocked Vps13 localization to the NVJ and rescued ERMES defects, possibly by enhancing v-CLAMP function (Lang et al., 2015), were equally effective in rescuing the defective trafficking of the Snc1 reporter or GFP-Neo1 in the vps13∆ strain (p < 0.0001; Supplemental Figure S1). Together these results support a new role for Vps13 in the endosomal recycling of Neo1 and the Snc1-based reporter GSS that appears to be independent of its role at membrane contact sites.
Identification of factors that regulate Neo1 recycling from endosomes
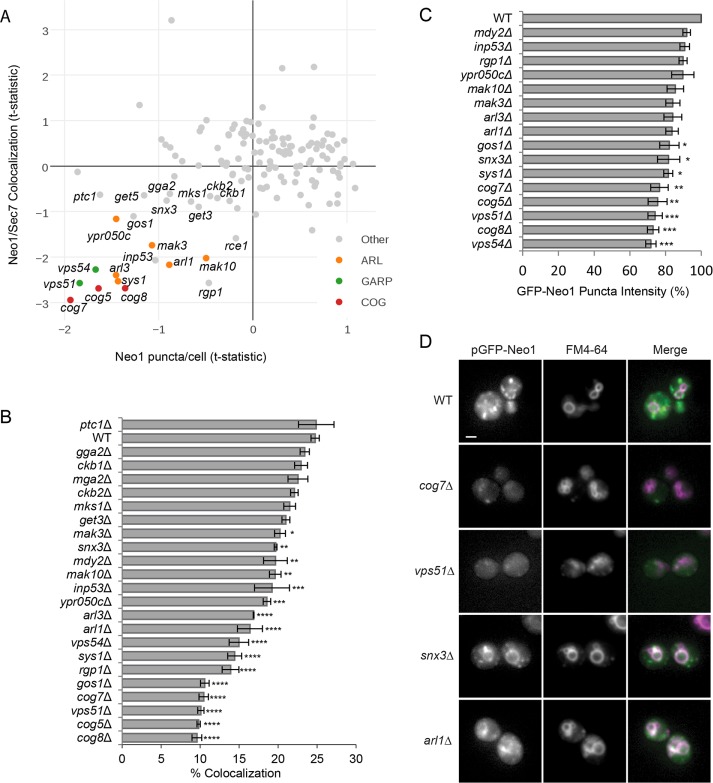
Our final analysis identified mutants that altered the Golgi localization of Neo1. A few mutants, including the Rab GTPase Ypt31 and Trs33, a subunit of the TRAPP-II complex that regulates GTP exchange on Ypt31 (Tokarev et al., 2009), caused Neo1 to accumulate at the Golgi (Supplemental Figure S2), suggesting defects in the anterograde trafficking of Neo1. To find the set of mutants that missort Neo1 to the vacuole, we first selected deletion strains with reduced localization of Neo1 at the Golgi, as defined by a decrease in the total number of Neo1 puncta/cell (t-statistic < −0.33) and reduced colocalization with Sec7 (t-statistic < −0.67; Figure 4A). This list was further refined by eliminating mutants that accumulate Neo1 in endosomes (large puncta; t-statistic < 1).
FIGURE 4:
Identification of mutants that cause Neo1 missorting to the vacuole. (A) Plot of Sec7 localization to Neo1 puncta compared with the average number of Neo1 puncta/cell. Indicated mutants were subject to further analysis. (B, C) Quantitation after small-scale reimaging identifies a subset of mutants with (B) reduced Sec7 colocalization with Neo1-labeled puncta and (C) reduced Neo1 puncta intensity compared with WT. Error bars report SEM; >300 cells/trial, n = 3. One-way ANOVA with a Dunnett-corrected post hoc test: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (D) Live-cell fluorescence microscopy of WT and mutant strains expressing pGFP-Neo1 under its native promoter shows that Neo1 mislocalizes to vacuoles that are labeled with FM4-64. Scale bar, 2 μm.
This set of 22 mutants was reimaged and reanalyzed, identifying 16 with significantly reduced colocalization between Neo1 and Sec7 relative to a wild-type strain (Figure 4B). A subset of the 16 mutants also showed a significant reduction in the intensity of Neo1 puncta (Figure 4C), which suggests that they have more-severe missorting defects. Some of these mutations disrupt components of endosome-to-Golgi trafficking pathways, such as the COG and GARP tethering complexes (Conibear and Stevens, 2000; Suvorova et al., 2002; Fotso et al., 2005), the synaptojanin-like protein Inp53 (Ha et al., 2003), the v-SNARE Gos1 (McNew et al., 1998), and the GET complex, which is needed for membrane insertion of SNAREs and other tail-anchored proteins (Mateja et al., 2015). We confirmed that Neo1 expressed from its native promoter is mislocalized to FM4-64–labeled vacuolar membranes in a subset of these mutants (Figure 4D). For example, in mutants lacking the sorting nexin Snx3, Neo1 is found in vacuolar rings, as well as puncta, suggesting a partial defect in retrograde trafficking of Neo1 (Figure 4D).
Taken together, these mutants describe a recycling pathway involving SNAREs, SNARE regulators, and tethering complexes that aid in vesicle docking and fusion with the Golgi. Snx3 was the only cargo-selective protein identified by this analysis, suggesting that it may be responsible for recruiting Neo1 into retrograde carriers at endosomes.
Snx3-dependent sorting of Neo1 requires a short amino acid motif in the N-terminal tail
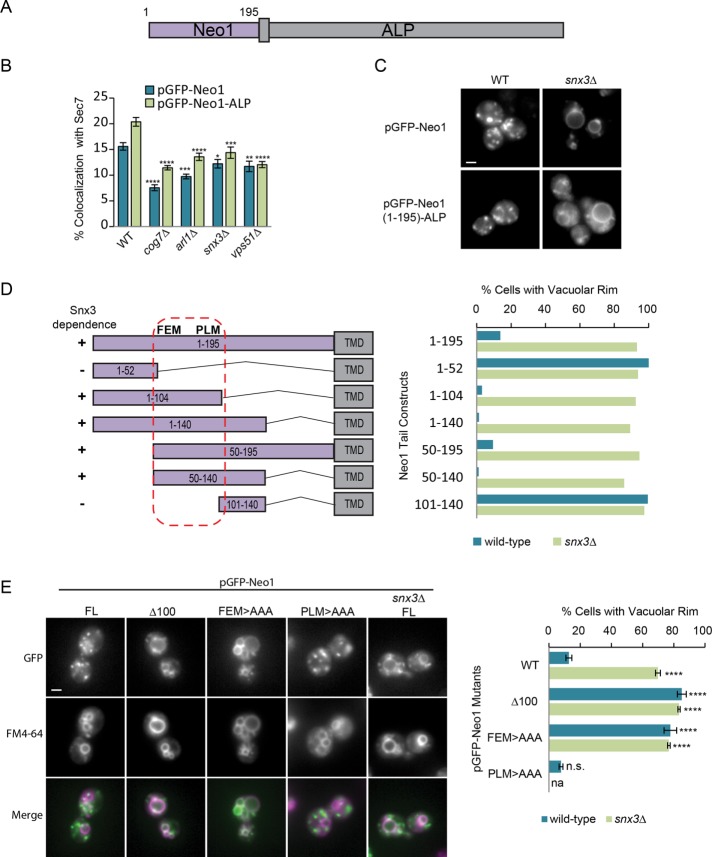
Relatively few proteins (Ste13, Kex2, Pep12, and Ftr1) have been shown to require the cargo-selective sorting nexin Snx3 for recycling from endosomes to the Golgi (Voos and Stevens, 1998; Hettema et al., 2003; Strochlic et al., 2007). If Snx3 is responsible for sorting Neo1 into retrograde tubules, Neo1 may contain a cytosolic signal that is recognized by Snx3. Members of the P4-ATPase family have a conserved structure, containing nucleotide-binding, actuator, phosphorylation, and translocation domains and regulatory N- and C-terminal cytosolic tails (Catty et al., 1997; Andersen et al., 2016). For example, the related flippase Drs2 interacts with trafficking effectors through its C-terminal tail (Chantalat et al., 2004; Hanamatsu et al., 2014), and the N-terminus is needed for endocytosis of Dnf1 and Drs2 (Liu et al., 2007). Neo1 has a 195–amino acid (aa) N-terminal cytosolic tail with several conserved motifs and a much shorter C-terminal tail (21 aa).
To determine whether the N-terminal tail of Neo1 contains signals that confer Snx3-dependent sorting, we fused amino acids 1–195 of Neo1 to the transmembrane and luminal portions of alkaline phosphatase (ALP), creating a chimeric GFP-Neo1-ALP protein expressed under the ADH promoter (Figure 5A). We found that GFP-Neo1-ALP exhibited significantly reduced colocalization with Sec7 in snx3∆, arl1∆, vps51∆ (GARP), and cog7∆ mutants, similar to that of GFP-Neo1, suggesting that this N-terminal tail contains information that directs endosome–Golgi sorting (Figure 5B). GFP-Neo1 and GFP-Neo1-ALP were also both missorted to the vacuolar rim in snx3∆ mutants (Figure 5C). This suggests that the Neo1 N-terminal tail contains an Snx3 sorting signal that specifies its Golgi/endosome recycling.
FIGURE 5:
A signal in the Neo1 N-terminal tail is needed for Snx3-dependent sorting. (A) Schematic of the Neo1-ALP chimera. The first 195 amino acids of Neo1 are fused to the transmembrane and luminal portions of ALP. (B) Quantitation of GFP-Neo1 (pMD124) or GFP-Neo1-ALP (pMD192) colocalization with the Golgi marker Sec7-dsRed in WT and selected mutants. Error bars report SEM; >200 cells/trial, n = 3. One-way ANOVA with a post hoc test, comparisons to WT using Dunnett correction: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. (C) Live-cell fluorescence microscopy of GFP-Neo1 and GFP-Neo1-ALP in WT and snx3∆ strains indicates that the Neo1 N-terminus confers Snx3-dependent Golgi localization. Scale bar, 2 μm. (D) Truncation analysis suggests that the sorting signal lies between amino acids 50 and 140. Left, schematic representation of truncation mutants. Right, percentage of cells showing GFP-Neo1-ALP at the vacuolar rim; >100 cells, n = 1. (E) Live-cell fluorescence microscopy of cells expressing full-length (FL) and mutant derivatives of pADHpr-GFP-Neo1 shows that mutation of the FEM motif causes missorting to FM4-64-labeled vacuoles. Left, representative images. Right, percentage of cells showing GFP-Neo1-ALP at the vacuolar rim. Error bars report SEM; >100 cells/trial, n = 3. One-way ANOVA with a Dunnett-corrected post hoc test: *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
To find the Snx3 signal, we fused fragments of the Neo1 N-terminal tail to the ALP transmembrane and luminal domains and compared the number of cells exhibiting vacuolar rim localization in wild-type and snx3Δ cells (Figure 5D and Supplemental Figure S3, A–C). Deletion of 54 or 80 membrane-proximal residues had no effect on the Snx3-dependent sorting of GFP-Neo1-ALP, nor did the removal of the N-terminal 50 residues. However, further deletions caused localization to the vacuole rim in wild-type cells, suggesting that the Snx3 sorting signal lies between amino acid residues 50 and 104 (Figure 5D and Supplemental Figure S3A). This region contains two short stretches of conservation surrounded by poorly conserved sequences (Supplemental Figure S4). We confirmed that removal of the first 100 amino acids of GFP-Neo1 caused mislocalization to the vacuole (Figure 5E). We then mutated conserved residues in each candidate motif to alanines in full-length GFP-Neo1. We observed a significant relocalization of Neo1-F65EM > AAA to the vacuolar rim, whereas mutation of Neo1-P94LM > AAA had no effect (Figure 5E). Thus the conserved FEM residues may be part of an Snx3 sorting signal.
The Neo1-F65EM > AAA mutation caused significant mislocalization of the 1–140 chimera to the vacuolar rim. Surprisingly, when this F65EM motif was mutated in the 1–195 chimera, there was little increase in vacuolar rim localization (Supplemental Figure S3B). We also found that the GFP-Neo1-ALP chimera lacking the first 100 amino acids was competent for sorting (Supplemental Figure S3C), but a similar deletion blocked the Snx3-dependent transport of GFP-Neo1 (Figure 5E). These results suggest that a membrane-proximal portion of the N-terminal tail contains a second Snx3 sorting signal, but this signal is masked or nonfunctional in the context of the native Neo1 protein. Taken together, these results show that the conserved FEM motif is required for sorting of Neo1 by Snx3.
Neo1 sorting by Snx3 is functionally important
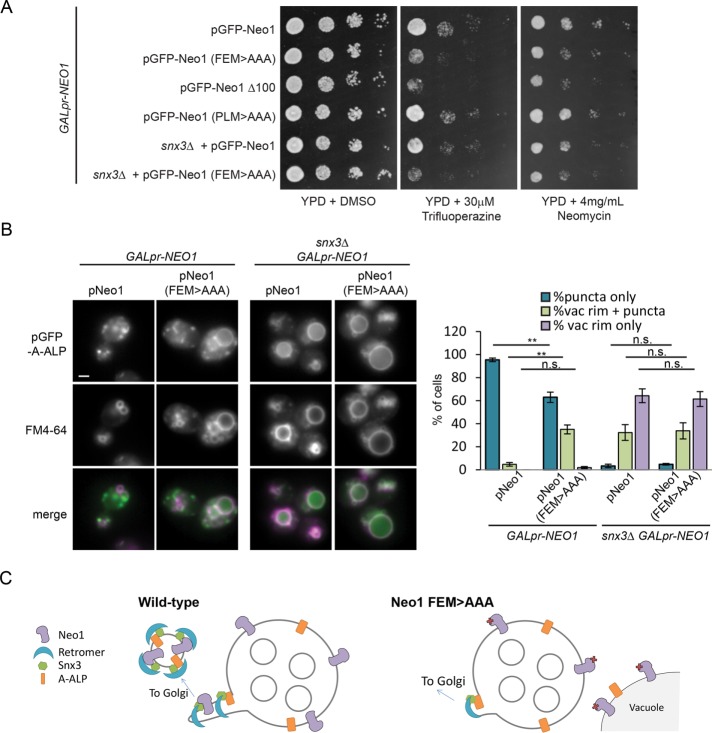
The loss of Snx3 causes partial mislocalization of Neo1 to the vacuole rim. To determine whether mutating the Snx3 sorting motif affects viability, we introduced Neo1-F65EM > AAA and Neo1-P94LM > AAA plasmids into a strain expressing NEO1 from the GAL1 promoter. When grown on dextrose to repress GALpr-NEO1 expression, the wild-type and mutant plasmids could support cell growth, but strains with the empty plasmid were inviable (Supplemental Figure S5). Thus these sequences are not required for the essential function of Neo1. However, strains expressing Neo1-F65EM > AAA, but not wild-type Neo1 or the Neo1-P94LM > AAA mutant, exhibited sensitivity to neomycin and trifluoperazine, drugs that cause reduced growth of a heterozygous Neo1 mutant (Figure 6A; Hoepfner et al., 2014).
FIGURE 6:
The sorting of Neo1 by Snx3 is needed for sorting of other Snx3 cargo. (A) Neo1 mutants lacking the FEM sorting motif are sensitive to neomycin and trifluoperazine. We grew 10× serial dilutions of strains expressing the indicated WT and mutant forms of pGFP-Neo1 under the native promoter on glucose-containing medium (YPD ± inhibitors) to repress expression of GALpr-NEO1. (B) The model Snx3 cargo protein pA-ALP (pMD130) shows increased missorting to the vacuolar rim in strains expressing pNeo1 (FEM>AAA). Left, representative images. Scale bar, 2 μm. Right, percentage of cells showing GFP-A-ALP at the vacuolar rim. Error bars report SEM; >100 cells/trial, n = 3. Unpaired t test: ns, not significant; **p < 0.01. (C) Model of Neo1 function in Snx3-dependent recycling. Sorting of Neo1 into Snx3 tubules may induce curvature needed for tubule formation and efficient sorting of other Snx3 cargoes.
The Neo1-F65EM > AAA and snx3Δ mutants showed a similar pattern of drug sensitivity, and this sensitivity was not further enhanced in a Neo1-F65EM > AAA snx3Δ double mutant, suggesting that the snx3 mutant phenotypes result from the perturbation of Neo1 localization and function. Removing the putative scramblase Any1 is believed to compensate for reduced Neo1 flippase activity by preserving lipid asymmetry originating from flippases at other cellular locations (van Leeuwen et al., 2016). Because the trifluoperazine sensitivity of snx3Δ and Neo1-F65EM > AAA mutants was suppressed by loss of ANY1 (Supplemental Figure S6), this drug-sensitivity phenotype likely reflects loss of lipid asymmetry generated by Neo1 flippase activity.
Drs2 sorting into vesicles at the Golgi and endosomes is believed to contribute to the formation of these vesicles (Furuta et al., 2007; Liu et al., 2007; Hanamatsu et al., 2014). To test whether Neo1 inclusion in Snx3 vesicles is important for the trafficking of other Snx3 cargoes, we examined the sorting of the well-characterized model Snx3 cargo protein A-ALP (Nothwehr et al., 1993). GFP-A-ALP was missorted to the vacuolar rim in ∼90% of snx3∆ cells (p < 0.0001). A partial but significant defect in A-ALP sorting was also seen for cells expressing Neo1-F65EM > AAA (∼30% of cells, p < 0.0001; Figure 6B). No further increase in A-ALP missorting was observed in a snx3Δ Neo1-F65EM > AAA double mutant, suggesting that they work in the same pathway. The A-ALP sorting defect in cells expressing the Neo1-F65EM > AAA mutant was not due to a general disruption of endosome function, as these cells did not exhibit CPY missorting (Supplemental Figure S7) or vacuole fragmentation (Figure 6B). This suggests that the sorting of Neo1 into Snx3 retrograde carriers aids in the trafficking of other proteins by that pathway. From these results, we propose a model in which Snx3 concentrates Neo1 in recycling tubules, where its lipid flipping activity contributes to the formation and/or function of these tubules (Figure 6C).
DISCUSSION
Identification of novel endosome-trafficking regulators
A genome-wide screen uncovered new and known regulators of Snc1 recycling at endosomes. We hypothesized that some of these proteins may influence Snc1 trafficking indirectly by sorting one or more flippases responsible for lipid asymmetry. Drs2 and its regulators have well-documented roles in regulating Snc1 recycling from early endosomes (Saito et al., 2004; Furuta et al., 2007; Hanamatsu et al., 2014). In contrast, the role of the essential flippase Neo1 in this process (Yamagami et al., 2015) has not been studied in detail.
Here we used high-content imaging to identify regulators of Neo1 trafficking with a high degree of sensitivity. We found that many known regulators of Snc1 sorting at endosomes, including the coat proteins Ent3/5 and Snx4/Atg20, did not influence Neo1 localization. Loss of the Drs2-Cdc50 flippase complex also altered the trafficking of Snc1 but not Neo1, consistent with previous studies (Barbosa et al., 2010). These results suggest that Snc1 and Neo1 are differentially sorted into recycling carriers at endosomes.
Previous studies demonstrated that Drs2’s flippase activity is required for vesicle formation, and this activity is also needed for its own sorting (Chantalat et al., 2004; Liu et al., 2008; Hanamatsu et al., 2014). However, not all proteins that regulate Neo1 activity at a single trafficking step are expected to alter its steady-state distribution. Overall localization may be unaffected if an alternative or redundant trafficking pathway exists. For example, Drs2 uses a plasma membrane bypass pathway to maintain its Golgi localization when AP-1 vesicle formation is blocked (Liu et al., 2008). Furthermore, the kinases Fpk1 and Fpk2 regulate the activity but not the localization of Dnf1 and Dnf2 (Nakano et al., 2008). Neo1 is not phosphorylated by these kinases (Nakano et al., 2008). It will be interesting to determine whether Neo1 is regulated by other kinases and phosphatases identified in our screen and whether other factors were missed due to bypass trafficking pathways.
The Neo1 binding partners Mon2 and Dop1 also bind Arl1 (Efe et al., 2005; Gillingham et al., 2006; Barbosa et al., 2010). We found that Mon2 recruits Dop1 to the Golgi (unpublished data), as noted by others (Gillingham et al., 2006). Our analysis further indicated that Arl1 regulates the localization of Dop1 by stabilizing the Dop1–Mon2 interaction. Mon2 binds Arl1 directly (Jochum et al., 2002), and genetic suppression experiments suggest Dop1 can bind Arl1 in the absence of Mon2 (Barbosa et al., 2010), consistent with the formation of a trimeric Mon2-Arl1-Dop1 complex. Our results could explain why Mon2 was identified as a negative regulator of constitutively active Arl1 (Manlandro et al., 2012): it is possible that, by enhancing levels of the Mon2/Dop1/Arl1 complex, overactive Arl1 sequesters the essential Dop1 protein, an effect that would be reversed by deletion of MON2.
We identified several genes that regulate the trafficking of both Snc1 and Neo1. Mutations in subunits of the COG or GARP tethering complexes or the Ypt6 exchange factor Rgp1, which direct fusion with the Golgi, caused missorting of Neo1 to the vacuole. We also uncovered mutations that cause Neo1 to accumulate in endosomal structures. Loss of Vps30 and Vps38, two subunits of the PI3K complex, reduces levels of PI3P needed for endosomal function (Odorizzi et al., 2000; Burda et al., 2002). However, the role of Vps13 is unclear. Recent work indicates that Vps13 is present at membrane contact sites (Lang et al., 2015; Park et al., 2016), and yet mutation of other contact-site components did not block the recycling of Neo1 or Snc1 at endosomes. Vps13 could work redundantly with other contact-site proteins or else have a function in early and late endosome recycling that is distinct from its role at contact sites.
Neo1 is a new Snx3 cargo protein
We discovered that Neo1 is a new Snx3 cargo protein, which is consistent with a recent report that Neo1 is missorted in retromer mutants (Wu et al., 2016). Relatively few Snx3 cargo proteins have been identified. In yeast, these include the iron transporter Ftr1, the SNARE protein Pep12, and two membrane-bound proteases, Ste13 and Kex2 (Voos and Stevens, 1998; Hettema et al., 2003; Strochlic et al., 2007), whereas the Wnt receptor Wntless and the transferrin receptor are sorted by Snx3 in metazoans (Harterink et al., 2011; Chen et al., 2013). The short conserved sorting motif that we identified in the cytoplasmic N-terminal tail of Neo1 contains a core FEM tripeptide that has some similarity to signals in Ste13 and Kex2 (Wilcox et al., 1992; Nothwehr et al., 1993; Voos and Stevens, 1998) and fits a recently defined Y/F-x-Φ consensus (where Φ is a bulky hydrophobic residue) implicated in retromer and Snx3-based sorting in a large-scale study (Bean et al., 2017). This tripeptide is frequently associated with conserved acidic and hydrophobic residues. Although a downstream aspartate residue contributes to Ste13 sorting, mutating other nearby residues had little effect (Nothwehr et al., 1993), suggesting that acidic and/or hydrophobic residues flanking the Y/F-x-Φ motif are not essential but may contribute to optimal signal recognition for some cargoes.
Neo1 activity may promote recycling from endosomes
Of importance, we found that recruitment of Neo1 into Snx3 carriers contributes to the sorting of another Snx3 cargo protein, suggesting that Neo1 lipid flippase activity might help drive the formation of Snx3-dependent recycling tubules. Two models, which are not mutually exclusive, might explain how flippase activity promotes vesicle (or tubule) formation (Graham, 2004). First, lipid flipping could cause curvature needed for vesicle biogenesis due to accumulation of lipids on one side of the leaflet (Sheetz and Singer, 1974; Natarajan et al., 2004). Consistent with this model, loss of Drs2 activity does not prevent recruitment of the AP-1 coat but instead appears to block vesicle budding (Chantalat et al., 2004; Liu et al., 2008; Muthusamy et al., 2009). By extension, Neo1 activity may generate an initial curvature that is stabilized by SNX-BAR proteins, which are believed to be important drivers of curvature at endosomes (Mim and Unger, 2012; van Weering et al., 2012). In metazoans, in which Snx3 is believed to function independently of retromer-associated SNX-BAR proteins (Cullen and Korswagen, 2012), flippases may be even more important for generating curvature formation.
Second, lipid flipping by Neo1 could alter the charge, packing, and/or curvature of the endosomal membrane to recruit effectors important for the formation of Snx3 carriers (Graham, 2004). Drs2-mediated translocation of PS recruits the ARFGAP Gcs1 to promote recycling at early endosomes in yeast (Xu et al., 2013), and in mammals, the Drs2-related flippase ATP8A1 recruits EHD1 to recycling endosomes, which is important for the scission of retromer tubules (Lee et al., 2015). By extension, Neo1 could play a similar role at the yeast late endosome. Although Neo1 activity has not yet been reconstituted in a cell-free system, recent work suggests that it primarily flips PE (Takar et al., 2016), consistent with studies showing that the Neo1 homologue Tat-5 is required for PE asymmetry in worms (Wehman et al., 2011). Because PE is uncharged and conical, increasing the concentration of PE on the cytosolic face is predicted to cause lipid-packing defects without altering membrane electrostatics. This could enhance recruitment of a different set of effectors relative to those recruited by Drs2, which could explain why high levels of Drs2 do not compensate for loss of Neo1 function (Takar et al., 2016).
Conservation in higher cells
Recent work suggests that Neo1 family flippases are also important for endosomal sorting in metazoans. Mammals have two Neo1 homologues: ATP9A, which is found at early/recycling endosomes and the trans-Golgi network (TGN), and ATP9B, which is localized to the TGN (Takatsu et al., 2011). Although the role of ATP9B is unclear, ATP9A is needed for exit of the transferrin receptor and other cargo from Vps35-containing endosomes, whereas vacuolar fusion is not affected (Tanaka et al., 2016). The transferrin receptor requires Snx3 for its sorting (Chen et al., 2013). Of interest, ATP9A is required for sorting of Wntless by Snx3 (P. Cullen, personal communication), consistent with the idea that ATP9A, like Neo1, participates in a Snx3-dependent sorting pathway in mammals.
The activity of many P-type ATPases is regulated by their cytosolic N or C-terminal domains (Zhou et al., 2013; Andersen et al., 2016). For example, binding of phospholipids and coat proteins to the autoinhibitory C-terminal domain of Drs2 could ensure that its activity is coupled to vesicle formation (Zhou et al., 2013; Hanamatsu et al., 2014). Neo1 has a very short C-terminus and an extended N-terminus that contains the Snx3 sorting motif. The N-terminal tails of ATP9A and 9B also specify their differential localization, suggesting this region of the protein binds regulatory proteins (Takatsu et al., 2011). Although the yeast Snx3 sorting motif is not conserved in ATP9A or ATP9B, other sequences found in Neo1 are present within a region of ATP9B that is necessary and sufficient for Golgi targeting (Takatsu et al., 2011). Thus future studies may identify new conserved regulators that associate with the N-terminal tail of Neo1 family flippases.
MATERIALS AND METHODS
Yeast strains and plasmids
All yeast strains listed in Supplemental Table S4 were made by homologous recombination (Longtine et al., 1998; Janke et al., 2004; Sheff and Thorn, 2004; Lee et al., 2013; Slubowski et al., 2015). Strains not listed were made by SGA using parental strains listed in Supplemental Table S4; Tong and Boone, 2006). Strains were propagated in rich medium (YPD: 1% yeast extract, 2% peptone, 2% dextrose) or synthetic medium (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% synthetic complete mix, 2% dextrose) supplemented with the appropriate amino acids.
All plasmids (Supplemental Table S5) were made by homologous recombination in yeast, rescued in Escherichia coli, and confirmed by sequencing. pMD124 was made by PCR-amplifying NAT::ADHpr-GFP-NEO1 from genomic DNA and cotransforming it with linearized pRS413. pMD195 was made by PCR-amplifying NAT::ADHpr-GFP-NEO1(1-195) and PHO8(34-567) using primers that introduce overlapping homology, and cotransformation with linearized pMD124. To make pMD151, the ADH promoter from pMD124 was replaced with the 800–base pair 5′ untranslated region (UTR) of Neo1. pMD192 was made by switching the HIS3 marker with the URA3 gene from pRS416. pLD39 was made by removing sequences encoding GFP from pMD192. To make pLG1, VPS13 as well as the 5′ and 3′ UTR, was PCR-amplified and introduced into the multiple cloning site of pRS416 by homologous recombination. Subsequent mutations were made by homologous recombination using PCR products encoding these mutations and digested with pLG1.
Genome-wide invertase screen
Raw images from the genome-wide invertase-based GSS reporter screen (Burston et al., 2009) were reanalyzed using an in-house script, and values were subjected to background subtraction, normalized to the plate median, and filtered to eliminate absent or very slow-growing strains. Raw values were converted to a z-score and the mean MATa and MATα value was used for the final reported ranking. CLIK analysis (www.rothsteinlab.com/tools/clik) based on BIOGRID version 3.4.128 on 1 October 2015 was used to select a top hit list of 275 unique genes, which were subsequently filtered to remove genes with roles in mitochondrial, ribosomal, DNA, or RNA biology.
High-content imaging screen
Fluorescent reporters were introduced into 128 MATa deletion mutants using SGA technology (Tong and Boone, 2006) and a pinning robot (S&P Robotics, Toronto, Canada). Haploid strains were selected on synthetic complete –LEU –LYS –ARG –HIS/URA (strain dependent) medium containing l-canavanine (Sigma-Aldrich, St. Louis, MO), S-(2-aminoethyl)-l-cysteine (Sigma-Aldrich), and nourseothricin-dihydrogen sulfate (Werner Bioagents, Jena, Germany). Haploid arrays were grown overnight, with shaking, in 96-well plates containing 150 µl of synthetic complete medium and a 5 mm glass bead (Fisher Scientific) and then diluted 1:10 and grown for 3 h before transfer to 384-well glass bottom Matriplates (Brooks, Chelmsford, MA) treated with 0.1 mg/ml type V concanavalin A (inner surface) and SigmaCote (bottom surface; both from Sigma-Aldrich). Images were acquired at room temperature using a Leica TCS SP8 3X microscope with Adaptive Focus Control (Leica Microsystems, Wetzlar, Germany) with an HC PL APO 63×/1.30 Glyc CORR CS objective (Leica) and an ORCA-Flash4.0 digital camera (Hamamatsu Photonics, Hamamatsu City, Japan). Imaging was automated using MetaMorph 7.7 software (MDS Analytical Technologies, Sunnyvale, CA).
Small-scale imaging was carried out as described using concanavalin A–treated 96-well plates. Labeling with 4 µM FM4-64 (Life Technologies, Carlsbad, CA) was carried out in synthetic medium for 20 min at room temperature and chased for >20 min. Images were adjusted using MetaMorph 7.7 and Photoshop (Adobe, San Jose, CA).
Quantitative image analysis
Three parameters were quantified for each image: number of live cells, number of puncta per cell, and colocalization of puncta identified in the green and red channels. Image analysis was carried out using custom MetaMorph 7.7 journals. Live cells were identified and counted using the Count Nuclei application in one channel, using two different thresholds: 1) a low threshold to identify the number and location of all cells, and 2) a high threshold to identify dead cells. All puncta were identified using the Granularity application. Small or large Neo1 puncta were identified with 0.6–1.5 μm or 1.7–2.5 μm size thresholds. To find puncta in live cells, the Granularity application was applied to images in which the dead cells were masked using an inverted image from the high-threshold Count Nuclei step. The number of puncta was then divided by the number of live cells to obtain the number of puncta per cell.
Using the Colocalization application, the overlap of green and red puncta identified in previous steps was measured and divided by the total puncta area (either green or red channel).
All nonsupplemental figures report the t-statistic based on the percentage area of overlapping puncta divided by the total Neo1 puncta area. Because the brightness of the fluorescently tagged proteins and dyes varied significantly from day to day, all comparisons were made between samples acquired on a given day, and an analysis of variance (ANOVA) was used to assess the statistical significance of differences between mutants and wild-type. The t-statistic is reported for all measurements. This is a normalized score in which the raw value is subtracted from the mean of the sample set and divided by the SD of the sample set. A value of zero indicates that the selected mutant behaves similarly to the average, and nonzero numbers indicate the SD from the mean.
Small-scale invertase overlay assay
Strains containing a chromosomally integrated GFP-Snc1-Suc2 reporter were grown overnight in synthetic medium with 2% fructose (SF). Overnight cultures were adjusted to 0.1 OD600/ml, spotted on SF plates, and incubated at 30°C for 24 h. Images were taken before and after incubation with overlay reagents (125 mM sucrose, 100 mM NaOAC, pH 5.5, 0.4 mM N-ethylmaleimide, 0.01 mg/ml horseradish peroxidase, 8 U/ml glucose oxidase, 0.6 mg/ml O-dianisidine, and 0.7% agarose) using a Canon Rebel T3I camera (Canon, Tokyo, Japan). Densitometry of each colony was assessed using the Cell Profiler MeasureObjectIntensity module, and the intensity of each set of four was averaged for each replicate.
Sequence alignments
Closely related fungal homologues of Neo1 were identified using NCBI BlastP and aligned using T-Coffee (Waterhouse et al., 2009; Di Tommaso et al., 2011). An image of the resulting alignment was created using Jalview version 2 (Waterhouse et al., 2009).
Coimmunoprecipitation
Coimmunoprecipitation was conducted essentially as described in Whitfield et al. (2016) with the following changes: frozen spheroplasts were lysed in lysis buffer (50 mM Tris, pH 7.5, 100 mM NaCl, 1% CHAPSO, 1 mM phenylmethylsulfonyl fluoride, and 1× protease inhibitors [ThermoFisher]). Lysates were incubated 60 min with rabbit polyclonal anti-GFP (Luc Berthiaume, University of Alberta, Edmonton, Canada) and another 60 min with Protein A–Sepharose beads (GE Healthcare, Mississauga, Canada). Beads were suspended in Thorner buffer (8 M urea, 5% SDS, 50 mM Tris-Cl, pH 6.8, 0.1 M EDTA, 0.4 mg/ml bromophenol blue, and 1% β-mercaptoethanol) and subjected to SDS–PAGE.
Supplementary Material
Acknowledgments
We acknowledge Luc Berthiaume for his generous gift of rabbit anti-GFP serum. We thank Tony Cooke from Leica Microsystems for helping us develop our automated imaging system. This work was supported by Canadian Institutes for Health Research Grants 247169 and 365914 and the Canada Foundation for Innovation (Leading Edge Fund #30636).
Abbreviations used:
- COG
conserved oligomeric Golgi complex
- ER
endoplasmic reticulum
- GARP
Golgi-associated retrograde protein complex
- GFP
green fluorescent protein
- GSS
GFP-Snc1-Suc2 chimera
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PS
phosphatidylserine
- v-SNARE
vesicle-soluble N-ethylmaleimide–sensitive factor attachment protein receptor.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-11-0772) on April 12, 2017.
REFERENCES
- Andersen JP, Vestergaard AL, Mikkelsen SA, Mogensen LS, Chalat M, Molday RS. P4-ATPases as phospholipid flippases structure, function, and enigmas. Front Physiol. 2016;7:275. doi: 10.3389/fphys.2016.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge RD, Graham TR. Identification of residues defining phospholipid flippase substrate specificity of type IV P-type ATPases. Proc Natl Acad Sci USA. 2011;109:E290–E298. doi: 10.1073/pnas.1115725109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge RD, Graham TR. Two-gate mechanism for phospholipid selection and transport by type IV P-type ATPases. Proc Natl Acad Sci USA. 2012;110:E358–E367. doi: 10.1073/pnas.1216948110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge RD, Xu P, Graham TR. Type IV p-type ATPases distinguish mono-versus diacyl phosphatidylserine using a cytofacial exit gate in the membrane domain. J Biol Chem. 2013;288:19516–19527. doi: 10.1074/jbc.M113.476911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa S, Pratte D, Schwarz H, Pipkorn R, Singer-Krüger B. Oligomeric Dop1p is part of the endosomal Neo1p-Ysl2p-Arl1p membrane remodeling complex. Traffic. 2010;11:1092–1106. doi: 10.1111/j.1600-0854.2010.01079.x. [DOI] [PubMed] [Google Scholar]
- Bean BDM, Davey M, Conibear E. Cargo selectivity of yeast sorting nexins. Traffic. 2017;18:110–122. doi: 10.1111/tra.12459. [DOI] [PubMed] [Google Scholar]
- Behnia R, Panic B, Whyte JRC, Munro S. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat Cell Biol. 2004;6:405–413. doi: 10.1038/ncb1120. [DOI] [PubMed] [Google Scholar]
- Burda P, Padilla SM, Sarkar S, Emr SD. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J Cell Sci. 2002;115:3889–3900. doi: 10.1242/jcs.00090. [DOI] [PubMed] [Google Scholar]
- Burston HE, Davey M, Conibear E. Genome-wide analysis of membrane transport using yeast knockout arrays. In: Vancura A., editor. Membrane Trafficking. Totowa, NJ: Humana Press; 2008. pp. 29–39. [DOI] [PubMed] [Google Scholar]
- Burston HE, Maldonado-Báez L, Davey M, Montpetit B, Schluter C, Wendland B, Conibear E. Regulators of yeast endocytosis identified by systematic quantitative analysis. J Cell Biol. 2009;185:1097–1110. doi: 10.1083/jcb.200811116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catty P, De Kerchove D’Exaerde A, Goffeau A. The complete inventory of the yeast Saccharomyces cerevisiae P-type transport ATPases. FEBS Lett. 1997;409:325–332. doi: 10.1016/s0014-5793(97)00446-8. [DOI] [PubMed] [Google Scholar]
- Chantalat S, Park S.-K, Hua Z, Liu K, Gobin R, Peyroche A, Rambourg A, Graham TR, Jackson CL. The Arf activator Gea2p and the P-type ATPase Drs2p interact at the Golgi in Saccharomyces cerevisiae. J Cell Sci. 2004;117:711–722. doi: 10.1242/jcs.00896. [DOI] [PubMed] [Google Scholar]
- Chen C, Garcia-Santos D, Ishikawa Y, Seguin A, Li L, Fegan KH, Hildick-Smith GJ, Shah DI, Cooney JD, Chen W, et al. Snx3 regulates recycling of the transferrin receptor and iron assimilation. Cell Metab. 2013;17:343–352. doi: 10.1016/j.cmet.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Cleck JN, Stevens TH. Vps51p Mediates the Association of the GARP (Vps52/ 53/54) Complex with the Late Golgi t-SNARE Tlg1p. Mol Biol Cell. 2003;14:1610–1623. doi: 10.1091/mbc.E02-10-0654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. Vps52p , Vps53p, and Vps54p Form a novel multisubunit complex required for protein sorting at the yeast late golgi. Mol Biol Cell. 2000;11:305–323. doi: 10.1091/mbc.11.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Korswagen HC. Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol. 2012;14:29–37. doi: 10.1038/ncb2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton L, Davey M, Conibear E. Large-scale analysis of membrane transport in yeast using invertase reporters. In: Tang BL, editor. Membrane Trafficking, 2nd ed. New York: Springer; 2015. pp. 395–409. [DOI] [PubMed] [Google Scholar]
- Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:13–17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmar JC, Pierce S, Rothstein R, Reid RJD. Physical and genetic-interaction density reveals functional organization and informs significance cutoffs in genome-wide screens. Proc Natl Acad Sci USA. 2013;110:7389–7394. doi: 10.1073/pnas.1219582110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotso P, Koryakina Y, Pavliv O, Tsiomenko AB, Lupashin VV. Cog1p plays a central role in the organization of the yeast conserved oligomeric Golgi complex. J Biol Chem. 2005;280:27613–27623. doi: 10.1074/jbc.M504597200. [DOI] [PubMed] [Google Scholar]
- Furuta N, Fujimura-Kamada K, Saito K, Yamamoto T, Tanaka K. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p – Rcy1p pathway. Mol Biol Cell. 2007;18:295–312. doi: 10.1091/mbc.E06-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Whyte JRC, Panic B, Munro S. Mon2, a relative of large Arf exchange factors, recruits Dop1 to the Golgi apparatus. J Biol Chem. 2006;281:2273–2280. doi: 10.1074/jbc.M510176200. [DOI] [PubMed] [Google Scholar]
- Graham TR. Flippases and vesicle-mediated protein transport. Trends Cell Biol. 2004;14:670–677. doi: 10.1016/j.tcb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Ha S, Torabinejad J, DeWald DB, Wenk MR, Lucast L, De Camilli P, Newitt RA, Aebersold R, Nothwehr SF. Synaptojanin-like protein Inp53/Sjl3 functions with clathrin in a yeast TGN-to-endosome pathway distinct from the GGA protein-dependent pathway. Mol Biol Cell. 2003;14:1319–1333. doi: 10.1091/mbc.E02-10-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halleck MS, Pradhan D, Blackman C, Berkes C, Williamson P, Schlegel RA. Multiple members of a third subfamily of P-type ATPases identified by genomic sequences and ESTs. Genome Res. 1998;8:354–361. doi: 10.1101/gr.8.4.354. [DOI] [PubMed] [Google Scholar]
- Hanamatsu H, Fujimura-Kamada K, Yamamoto T, Furuta N, Tanaka K. Interaction of the phospholipid flippase Drs2p with the F-box protein Rcy1p plays an important role in early endosome to trans-Golgi network vesicle transport in yeast. J Biochem. 2014;155:51–62. doi: 10.1093/jb/mvt094. [DOI] [PubMed] [Google Scholar]
- Harterink M, Port F, Lorenowicz MJ, Mcgough IJ, Silhankova M, Betist MC, van Weering JRT, van Heesbeen RGHP, Middelkoop TC, Basler K, et al. A SNX3-dependent retromer pathway mediates retrograde transport of the Wnt sorting receptor Wntless and is required for Wnt secretion. Nat Cell Biol. 2011;13:914–923. doi: 10.1038/ncb2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hettema EH, Lewis MJ, Black MW, Pelham HRB. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003;22:548–557. doi: 10.1093/emboj/cdg062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D, Helliwell SB, Sadlish H, Schuierer S, Filipuzzi I, Brachat S, Bhullar B, Plikat U, Abraham Y, Altorfer M, et al. High-resolution chemical dissection of a model eukaryote reveals targets, pathways and gene functions. Microbiol Res. 2014;169:107–120. doi: 10.1016/j.micres.2013.11.004. [DOI] [PubMed] [Google Scholar]
- Hua Z, Fatheddin P, Graham TR. An essential subfamily of Drs2p-related P-Type ATPases is required for protein trafficking between golgi complex and endosomal/vacuolar system. Mol Biol Cell. 2002;13:3162–3177. doi: 10.1091/mbc.E02-03-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z, Graham TR. Requirement for Neo1p in retrograde transport from the Golgi complex to the endoplasmic reticulum. Mol Biol Cell. 2003;14:4971–4983. doi: 10.1091/mbc.E03-07-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe JA, Plattner F, Hulo N, Kressler D, Emr SD, Deloche O. Yeast Mon2p is a highly conserved protein that functions in the cytoplasm-to-vacuole transport pathway and is required for Golgi homeostasis. J Cell Sci. 2005;118:4751–4764. doi: 10.1242/jcs.02599. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Jochum A, Jackson D, Schwarz H, Pipkorn R, Singer-Krüger B. Yeast Ysl2p, homologous to Sec 7 domain guanine nucleotide exchange factors, functions in endocytosis and maintenance of vacuole integrity and interacts with the Arf-Like small GTPase Arl1p. Mol Cell Biol. 2002;22:4914–4928. doi: 10.1128/MCB.22.13.4914-4928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;153:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AB, Peter ATJ, Walter P, Kornmann B. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J Cell Biol. 2015;210:883–890. doi: 10.1083/jcb.201502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lim WA, Thorn KS. Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae. PLoS One. 2013;8:4–11. doi: 10.1371/journal.pone.0067902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Uchida Y, Wang J, Matsudaira T, Nakagawa T, Kishimoto T, Mukai K, Inaba T, Kobayashi T, Molday RS, et al. Transport through recycling endosomes requires EHD1 recruitment by a phosphatidylserine translocase. EMBO J. 2015;34:669–688. doi: 10.15252/embj.201489703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Hua Z, Nepute JA, Graham TR. Yeast P4-Atpases Drs2p and Dnf1p are essential cargos of the NPFXD/Sal1p endocytic pathway. Mol Biol Cell. 2007;18:2991–3001. doi: 10.1091/mbc.E06-07-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Surendhran K, Nothwehr SF, Graham TR. P4-ATPase requirement for AP-1/clathrin function in protein transport from the trans-Golgi network and early endosomes. Mol Biol Cell. 2008;19:3526–3535. doi: 10.1091/mbc.E08-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Manlandro CMA, Palanivel VR, Schorr EB, Mihatov N, Antony AA, Rosenwald AG. Mon2 is a negative regulator of the monomeric G protein, Arl1. FEMS Yeast Res. 2012;12:637–650. doi: 10.1111/j.1567-1364.2012.00814.x. [DOI] [PubMed] [Google Scholar]
- Mateja A, Paduch M, Chang H.-Y, Szydlowska A, Kossiakoff AA, Hegde RS, Keenan RJ. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science. 2015;347:1152–1155. doi: 10.1126/science.1261671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonold CM, Fromme JC. Four GTPases differentially regulate the Sec 7 Arf-GEF to direct traffic at the trans-golgi network. Dev Cell. 2014;30:759–767. doi: 10.1016/j.devcel.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew JA, Coe JG, Sogaard M, Zemelman BV, Wimmer C, Hong W, Sollner TH. Gos1p, a Saccharomyces cerevisiae SNARE protein involved in Golgi transport. FEBS Lett. 1998;435:89–95. doi: 10.1016/s0014-5793(98)01044-8. [DOI] [PubMed] [Google Scholar]
- Mim C, Unger VM. Membrane curvature and its generation by BAR domains. Trends Biochem Sci. 2012;37:526–533. doi: 10.1016/j.tibs.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mioka T, Fujimura-Kamada K, Tanaka K. Asymmetric distribution of phosphatidylserine is generated in the absence of phospholipid flippases in Saccharomyces cerevisiae. Microbiologyopen. 2014;3:803–821. doi: 10.1002/mbo3.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy BP, Natarajan P, Zhou X, Graham TR. Linking phospholipid flippases to vesicle-mediated protein transport. Biochim Biophys Acta. 2009;1791:612–619. doi: 10.1016/j.bbalip.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Yamamoto T, Kishimoto T, Noji T, Tanaka K. Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol Biol Cell. 2008;19:1783–1797. doi: 10.1091/mbc.E07-07-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P, Liu K, Patil DV, Sciorra VA, Jackson CL, Graham TR. Regulation of a Golgi flippase by phosphoinositides and an ArfGEF. Nat Cell Biol. 2009;11:1421–1426. doi: 10.1038/ncb1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P, Wang J, Hua Z, Graham TR. Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc Natl Acad Sci USA. 2004;101:10614–10619. doi: 10.1073/pnas.0404146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothwehr SF, Roberts CJ, Stevens TH. Membrane protein retention in the yeast Golgi apparatus: dipeptidyl aminopeptidase A is retained by a cytoplasmic signal containing aromatic residues. J Cell Biol. 1993;121:1197–1209. doi: 10.1083/jcb.121.6.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci. 2000;25:229–235. doi: 10.1016/s0968-0004(00)01543-7. [DOI] [PubMed] [Google Scholar]
- Park JS, Thorsness MK, Policastro R, McGoldrick LL, Hollingsworth NM, Thorsness PE, Neiman AM. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol Biol Cell. 2016;27:2435–2449. doi: 10.1091/mbc.E16-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomorski T, Lombardi R, Riezman H, Devaux PF, van Meer G, Holthuis JCM. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol Biol Cell. 2003;14:1240–1254. doi: 10.1091/mbc.E02-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redding K, Brickner JH, Marschall LG, Nichols JW, Fuller RS. Allele-specific suppression of a defective trans-Golgi network (TGN) localization signal in Kex2p identifies three genes involved in localization of TGN transmembrane proteins. Mol Cell Biol. 1996;16:6208–6217. doi: 10.1128/mcb.16.11.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riekhof WR, Voelker DR. Uptake and utilization of lyso-phosphatidylethanolamine by Saccharomyces cerevisiae. J Biol Chem. 2006;281:36588–36596. doi: 10.1074/jbc.M608851200. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JH, Howald I, Stevens TH. Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO J. 1989;8:2057–2065. doi: 10.1002/j.1460-2075.1989.tb03614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Fujimura-Kamada K, Nobumichi F, Kato U, Umeda M, Tanaka K. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid Translocation in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorel E, Barrey E, Lau RK, Thorner J. Plasma membrane aminoglycerolipid flippase function is required for signaling competence in the yeast mating pheromone response pathway. Mol Biol Cell. 2014;26:134–150. doi: 10.1091/mbc.E14-07-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian TT, Baldridge RD, Xu P, Graham TR. Phospholipid flippases: building asymmetric membranes and transport vesicles. Biochim Biophys Acta. 2012;1821:1068–1077. doi: 10.1016/j.bbalip.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz MP, Singer SJ. Biological membranes as bilayer couples. A molecular mechanism of drug-erythrocyte interactions. Proc Natl Acad Sci USA. 1974;71:4457–4461. doi: 10.1073/pnas.71.11.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- Slubowski CJ, Funk AD, Roesner JM, Paulissen SM, Huang LS. Plasmids for C-terminal tagging in Saccharomyces cerevisiae that contain improved GFP proteins, Envy and Ivy. Yeast. 2015;32:379–387. doi: 10.1002/yea.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strochlic TI, Setty TG, Sitaram A, Burd CG. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J Cell Biol. 2007;177:115–125. doi: 10.1083/jcb.200609161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorova ES, Duden R, Lupashin VV. The Sec 34/Sec 35p complex, a Ypt1p effector required for retrograde intra-Golgi trafficking, interacts with Golgi SNAREs and COPI vesicle coat proteins. J Cell Biol. 2002;157:631–643. doi: 10.1083/jcb.200111081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi K, Iwamoto K, Kobayashi S, Horiuchi H, Fukuda R, Ohta A. Involvement of Golgi-associated retrograde protein complex in the recycling of the putative Dnf aminophospholipid flippases in yeast. Biochem Biophys Res Commun. 2012;417:490–494. doi: 10.1016/j.bbrc.2011.11.147. [DOI] [PubMed] [Google Scholar]
- Takar M, Wu Y, Graham TR. The essential Neo1 from budding yeast plays a role in establishing aminophospholipid asymmetry of the plasma membrane. J Biol Chem. 2016;291:15727–15739. doi: 10.1074/jbc.M115.686253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsu H, Baba K, Shima T, Umino H, Kato U, Umeda M, Nakayama K, Shin HW. ATP9B, a P4-ATPase (a putative aminophospholipid translocase), localizes to the trans-Golgi network in a CDC50 protein-independent manner. J Biol Chem. 2011;286:38159–38167. doi: 10.1074/jbc.M111.281006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda M, Yamagami K, Tanaka K. Role of phosphatidylserine in phospholipid flippase-mediated vesicle transport in Saccharomyces cerevisiae. Eukaryot. Cell. 2014;13:363–375. doi: 10.1128/EC.00279-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Ono N, Shima T, Tanaka G, Katoh Y, Nakayama K, Takatsu H, Shin HW. The phospholipid flippase ATP9A is required for recycling pathway from endosomes to the plasma membrane. Mol Biol Cell. 2016;27:3883–3893. doi: 10.1091/mbc.E16-08-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokarev AA, Taussig D, Sundaram G, Lipatova Z, Liang Y, Mulholland JW, Segev N. TRAPPII complex assembly requires Trs33 or Trs65. 2009;10:1831–1844. doi: 10.1111/j.1600-0854.2009.00988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AHY, Boone C. Synthetic genetic array analysis in Saccharomyces cerevisiae. In: Xiao W, editor. Yeast Protocols. Clifton, NJ: Humana Press; 2006. pp. 171–192. [DOI] [PubMed] [Google Scholar]
- van Leeuwen J, Pons C, Mellor JC, Yamaguchi TN, Friesen H, Koschwanez J, Ušaj MM, Pechlaner M, Takar M, Ušaj M, et al. Exploring genetic suppression interactions on a global scale. Science. 2016;354:aag0839. doi: 10.1126/science.aag0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Weering JRT, Sessions RB, Traer CJ, Kloer DP, Bhatia VK, Stamou D, Carlsson SR, Hurley JH, Cullen PJ. Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J. 2012;31:4466–4480. doi: 10.1038/emboj.2012.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voos W, Stevens TH. Retrieval of resident late-Golgi membrane proteins from the prevacuolar compartment of Saccharomyces cerevisiae is dependent on the function of Grd19p. J Cell Biol. 1998;140:577–590. doi: 10.1083/jcb.140.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2-A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehman AM, Poggioli C, Schweinsberg P, Grant BD, Nance J. The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr Biol. 2011;21:1951–1959. doi: 10.1016/j.cub.2011.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ST, Burston HE, Bean BDM, Raghuram N, Maldonado-Baez L, Davey M, Wendland B, Conibear E. The alternate AP-1 adaptor subunit Apm2 interacts with the Mil1 regulatory protein and confers differential cargo sorting. Mol Biol Cell. 2016;27:588–598. doi: 10.1091/mbc.E15-09-0621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky S, Schwarz H, Singer-Krüger B. Molecular interactions of yeast Neo1p, an essential member of the Drs2 family of aminophospholipid translocases, and its role in membrane trafficking within the endomembrane system. Mol Cell Biol. 2004;24:7402–7418. doi: 10.1128/MCB.24.17.7402-7418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox CA, Redding K, Wright R, Fuller RS. Mutation of a tyrosine localization signal in the cytosolic tail of yeast Kex2 protease disrupts Golgi retention and results in default transport to the vacuole. Mol Biol Cell. 1992;3:1353–1371. doi: 10.1091/mbc.3.12.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Takar M, Cuentas-Condori AA, Graham TR. Neo1 and phosphatidylethanolamine contribute to vacuole membrane fusion in Saccharomyces cerevisiae. Cell Logist. 2016;6:e1228791. doi: 10.1080/21592799.2016.1228791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Baldridge RD, Chi RJ, Burd CG, Graham TR. Phosphatidylserine flipping enhances membrane curvature and negative charge required for vesicular transport. J Cell Biol. 2013;202:875–886. doi: 10.1083/jcb.201305094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagami K, Yamamoto T, Sakai S, Mioka T, Sano T, Igarashi Y, Tanaka K. Inositol depletion restores vesicle transport in yeast phospholipid flippase mutants. PLoS One. 2015;10:e0120108. doi: 10.1371/journal.pone.0120108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Graham TR. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc Natl Acad Sci USA. 2009;106:16586–16591. doi: 10.1073/pnas.0904293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Sebastian TT, Graham TR. Auto-inhibition of Drs2p, a yeast phospholipid flippase, by its carboxyl-terminal tail. J Biol Chem. 2013;288:31807–31815. doi: 10.1074/jbc.M113.481986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann J, Chidambaram S, Fischer von Mollard G. Dissecting Ent3p: the ENTH domain binds different SNAREs via distinct amino acid residues while the C-terminus is sufficient for retrograde transport from endosomes. Biochem J. 2010;431:123–134. doi: 10.1042/BJ20100693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.