Abstract
We developed a systematic approach to map human genetic networks by combinatorial CRISPR-Cas9 perturbations coupled to robust analysis of growth kinetics. We targeted all pairs of 73 cancer genes with dual-guide RNAs in three cell lines, altogether comprising 141,912 tests of interaction. Numerous therapeutically relevant interactions were identified and these patterns replicated with combinatorial drugs at 75% precision. Based on these results we anticipate cellular context will be critical to synthetic-lethal therapies.
Simultaneous mutation of two genes can produce a phenotype that is unexpected in light of each mutation’s individual effect1. This phenomenon, known as genetic interaction, identifies an underlying functional relationship between the genes, such as contributions to the same protein complex or pathway2. Mapping these functional relationships in a systematic fashion has broad applicability for advancing our fundamental understanding of biological systems3–5. Genetic interactions also have implications for therapeutic development, for instance in cancer where negative or ‘synthetic-lethal’ interactions via simultaneous disruption of both genes causes cell killing6. The feasibility of this approach has been demonstrated with the recent approval of the drug olaparib, an inhibitor of PARP1/2, specifically for tumors with loss-of-function mutations in BRCA1 or BRCA2. However, further applications of synthetic-lethal cancer therapy have been limited by poor understanding of the important genetic interactions in a cancer cell, and how these vary from one cancer type to another or from patient to patient7,8.
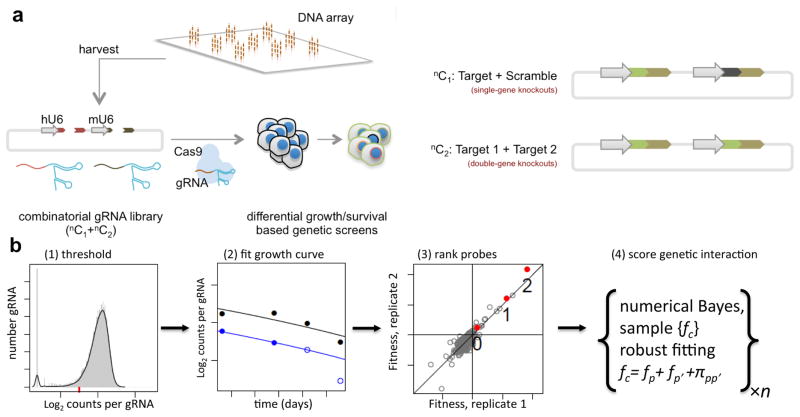
To enable systematic mapping of genetic interaction networks, we developed a CRISPR-Cas9 screening methodology for targeting single and pairs of genes in high-throughput. In the CRISPR-Cas9 system, a guide-RNA (gRNA), in complex with the Cas9 protein, targets genomic sequences homologous to the gRNA9,10. Targeting new genomic elements entails modifying the gRNA sequence, thus enabling many targeted genome editing and regulation capabilities9. Notably, Cas9 also enables facile multiplex targeting via delivery of multiple gRNAs per cell11. Here, we combined multiplex targeting with array-based oligonucleotide synthesis11–14 to create dual-gRNA libraries covering up to 105 defined gene pairs (Fig. 1A, Supplementary Figs. 1 and 2A–D). In these libraries, each construct bears two gRNAs, with each gRNA designed to target either a gene or a scrambled non-targeting sequence absent from the genome. Thus, all combinations of gene-gene (double gene perturbation) and gene-scramble (single gene perturbation) are exhaustively assayed for effects on cell growth. Notably, in our approach, both spacers for a dual-gRNA construct are directly specified during oligonucleotide synthesis thereby enabling the library constituents to be exactly defined to facilitate custom gRNA-pairing. By enabling determination and comparison of single-gene and dual-gene perturbation effects in the same assay, this approach allows for the systematic quantification of genetic interactions in humans.
Figure 1.
Experimental & analytical framework for identification of genetic interactions with combinatorial CRISPR knockout. (A) Schematic of overall experimental approach: Array-based oligonucleotide synthesis is used to create dual-gRNA libraries containing all gene-gene (double gene perturbation) and gene-scramble (single gene perturbation) combinations, which can then be assayed for effects on cell growth. (B) Schematic of computational analysis workflow: CRISPR screens are run as two independent replicate experiments, with cells harvested at four time points and gRNA frequencies determined by high throughput sequencing. All gRNAs below a threshold (red dash) are excluded from further analysis. Fitness is determined from a fit of log relative abundance over time; probes are subsequently ranked by absolute fitness and weighted, then a numerical Bayesian method is used to test for presence of a genetic interaction.
We conducted genetic interaction screens by transducing the dual-gRNA lentiviral library into a population of cells stably expressing Cas9, maintaining these cells in exponential growth over the course of four weeks, then sampling the relative changes in gRNAs at multiple time points: days 3, 14, 21 and 28 post-transduction (Methods). To robustly quantify gene fitness and genetic interactions, we developed a computational analysis framework that integrates all samples across the multiple days of the experiment. This method [1] detects and removes gRNA constructs with insufficient read coverage; [2] fits growth curves to the measured log2 abundances of each construct over time, the slopes of which reflect fitness; and [3] integrates data from the multiple gRNA constructs to derive a robust fitness value for disruption of each gene, fg, and gene pair, fg,g′. Finally [4], a genetic interaction score πgg′ is calculated as the difference of the observed from the expected fitness of the double gene knockout (Fig. 1B, Supplementary Fig. 3A–D). Significant departures from expected (π < 3σ or π > 3σ) are called as negative or positive genetic interactions, respectively. A negative interaction indicates slower-than-expected growth, suggesting synthetic sickness or lethality, while a positive interaction indicates faster-than-expected growth, suggesting epistasis.
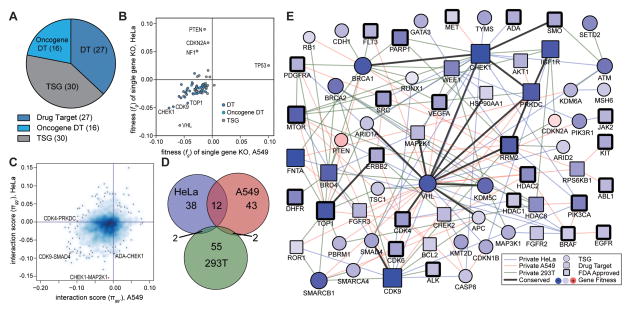
Using this method we evaluated all pairwise gene knockout combinations among a panel of 73 genes divided between tumor-suppressor genes (TSG) and cancer-relevant drug targets (DT), a subset of which were also verified oncogenes (Fig. 2A, Table S1)15. Experiments were performed in three cell lines: HeLa, a cervical cancer cell line driven by Human Papilloma Virus (HPV); A549, a lung cancer cell line driven by KRAS G12S mutation; and 293T, a human embryonic kidney cell line transformed by adenovirus that expresses the SV40 Large-T antigen. With nine gRNA pairs per combination, the library comprised 23,652 double gene knockout constructs and 657 single gene constructs; testing two replicates in each cell line yielded a total of 141,912 unique tests of interaction (Table S2A). Measurements of gene fitness (fg) were well correlated between biological replicates in the same cell line; (HeLa: Pearson r = 0.96, p = 4.2 × 10−40; A549: Pearson r = 0.94, p = 1.2 × 10−37; 293T: Pearson r = 0.97, p = 1.5 × 10−44) as were the π scores for significant genetic interactions (HeLa: r = 0.81 p = 4.7 × 10−18; A549: r = 0.65 p = 2.9 × 10−8; 293T: r = 0.79 p = 4.7 × 10−4; Supplementary Fig. 4A–F, Table S2B).
Figure 2. Genetic interactions in HeLa, A549 and 293T cancer cells.
(A) Genes selected for study included tumor suppressor genes (TSG) and cancer relevant drug targets (DT), which included many oncogenes. (B) Scatterplot of fitness of single gene knockout (KO) in HeLa vs. A549. (C) Scatterplot of interaction scores in HeLa vs. A549, using the smoothScatter R function with default settings. Density of gene pairs at each x.y location is represented by darkness of blue shading; single gene pairs in low density regions are marked by black dots. (D) Proportional Venn diagram summarizing the number of synthetic lethal interactions per cell line and the number conserved between each cell line pair. (E) Combined synthetic-lethal network for all three cell lines. Circles indicate TSG, squares DTs. Node colors indicate single gene knockout fitness effect, red: positive fitness effect, blue: negative fitness effect. Black edge around node indicates that the protein product of the gene is the target of an FDA approved drug. Color of edge indicates the cell line in which the interaction was identified, blue: HeLa, red: A549, green: 293T. Black edges were identified in multiple cell lines.
Moreover, we observed a significant correlation between the total number of genetic interactions identified for a gene and its single gene fitness (HeLa: r = 0.77, p = 3.4 × 10−10; A549: r = 0.45, p = 0.0018, 293T: r = 0.77, p = 9.0 × 10−10, Supplementary Fig. 4G), suggesting that network ‘hubs’ may have increased functional importance to cancer cells relative to genes with fewer interactions; such a relationship has been previously observed in model organisms but not reported before in humans5.
We next moved from comparison of replicates to comparison of the three cancer cell lines. First, we found lower but significant correlation of the single gene fitness scores across pairs of cell lines (Fig. 2B, Supplementary Fig. 4H,I, Supplementary Table 3A). Differences in these fitness scores recapitulated known biological differences, including the large positive growth effect of TP53 knockout in A549 but not HeLa or 293T, in which TP53 is already inactivated by viral proteins. Gene fitness scores did not significantly correlate with gene expression in any of the three cell lines (Supplementary Fig. 5A–C). Genes with very low or no expression had fitness scores very near the average for that cell line, consistent with a neutral growth effect.
Second, we found that the genetic interactions identified from these data were different between cell lines (Fig. 2C, Supplementary Fig. 4J,K). A total of 152 synthetic-lethal (negative) genetic interactions were identified in either HeLa, A549, or 293T cells (FDR ~ 0.3, Fig. 2D, Supplementary Fig. 6A–C, Supplementary Table S3B,C). Of these, 16 (10.5%) were identified in multiple cell lines and no interactions were seen in all three cell lines. The remaining 136 interactions were ‘private’ to a cell line (HeLa: 38 of 52, A549: 43 of 57, 293T: 55 of 59, Fig. 2E, Supplementary Fig. 6D–F). Additionally, there were eight positive genetic interactions (epistasis) identified in HeLa, two in 293T, and none in A549. Of all of these discoveries, we found that 28 interactions had been previously identified, including the therapeutically relevant interactions BRCA1-PARP16 and PTEN-MTOR16.
We next sought to validate these findings, and in particular the discrepancies across cell lines. We selected eight pairs of DT genes for which a synthetic-lethal genetic interaction had been identified in only HeLa or A549 cells. Rather than simply reproduce the dual CRISPR knockout experiment (gene-gene interaction), our goal was to examine the viability of cells exposed to drugs inhibiting the corresponding gene products (drug-drug interaction) – evaluating whether the interaction could be seen by an independent technology at the protein level, and whether it was also accessible therapeutically. In total, drug-drug assays validated six of eight interactions when tested in the cell line for which the interaction had been first observed by dual CRISPR (75% precision or positive predictive value). In contrast, for gene pairs tested in a cell line that had not been implicated by dual CRISPR knockout, only two of eight showed a negative genetic interaction by drug-drug assay (75% negative predictive value, Supplementary Figs. 7A–G and 8A–J, Table S4). Thus, the differences in genetic interaction across cell lines seen by systematic CRISPR could be largely reproduced as drug-drug interactions in small-scale assays.
Moving forward we envision that by allowing for genetic interaction mapping directly in eukaryotic cells our combinatorial CRISP-Cas9 technology will pave the way for systematic determination of cancer pathways, with two-fold application: improving our understanding of how networks of genes influence tumorigenesis, and aiding in the development of precision therapeutics via new druggable synthetic-lethal interactions. Recognizing that there may be great diversity in genetic interactions between different tumors, it will be important to perform these studies across a large number of samples, which is enabled by the high-throughput method we have presented. Consistent with prior studies, we have also noted the importance of gRNA efficacy and anticipate that improvements in gRNA design17–19 to increase editing rate and reduce false negatives, as well as using gRNA that specifically target functional protein domains20, will be critical to further scale these experiments and improve consistency. We note also that variability of Cas9 expression between individual cells, and from one cell line to another, could also affect perturbation efficiency. Moving forward, we believe integrating results from complementary perturbation strategies such as CRISPR inhibition and activation, as well proteomic and chemo-genetic studies, will enable generation of more comprehensive interaction maps7. Finally, this experimental and analytic framework is not unique to cancer cell cells, but can be readily applied to systematically map the genetic architecture of complex biological systems and diseases in any eukaryotic system amenable to lentiviral transduction and growth in culture.
ONLINE METHODS
Dual gRNA library cloning
Preparation of the dual-gRNA library involves a two-step cloning process whereby each synthesized oligonucleotide is assembled progressively with promoters and 3′ gRNA scaffolds (Supplementary Fig. 1)21. This multi-step protocol is critical since array-based oligonucleotides from commercial vendors have a maximum length of ~300 bp, while a dual-gRNA cassette is ~1000 bp in size; thus, additional steps of cloning are needed to reconstitute the full sequence. We optimized the library efficacy by eliminating large repeat sequences in the dual-gRNA vectors, as such repeats could potentially compromise both viral production and sequencing quality. Towards this goal we chose non-homologous polymerase III promoters (hU6, mU6) based on their comparable activity22. We also explored mutagenized gRNA scaffold sequences to further increase sequence diversity while maintaining the primary hairpin loops in the gRNA scaffold (via G-C vs. A-U interactions)23–25. Experiments showed that while engineered versions 2 and 3 were active, the wild-type scaffold and version 4 showed the most consistent activity (Supplementary Fig. 2A,B) and were thus used for all subsequent studies. We also confirmed that the two gRNA positions in the construct were equally functional (hU6-gRNA and mU6-gRNA), and thus the dual-gRNA libraries did not need to include both positions for each gRNA (Supplementary Fig. 2C). Additionally, we confirmed that the two gRNA were simultaneously active by targeting both EGFR and mCherry. There was a moderate reduction in activity when two guides are expressed in a dual-gRNA format, but each guide remains equally functional in both positions and with both gRNA scaffolds (Supplementary Fig. 2D).
To construct combinatorial libraries that exhaustively interrogate the network of genetic interactions among a panel of genes, our approach was to design three gRNAs against each gene. Additionally, three gRNAs were designed as non-targeting controls. Dual gRNA lentiviral constructs were then synthesized for all pairwise gRNA combinations between genes (double perturbations) and between genes and scrambles (single perturbations). This format resulted in nine pairwise gRNA constructs per gene-pair. The first step was to assemble the paired gRNAs into a backbone vector, and in the next step a fragment including both the first gRNA scaffold and a mouse U6 promoter inserted between the paired gRNAs.
Step I: paired gRNA cloning
The pooled oligonucleotide libraries were synthesized by CustomArray. Full-length oligonucleotides with dual gRNA spacers (i.e., 20-bp sequences used for targeting desired genes) were amplified by PCR using Kapa Hifi (Kapa Biosystems). PCR reactions were set up according to the manufacturer’s protocol, using 1 μL of synthesized oligonucleotide template (typically around 20 ng), an annealing temperature of 55 °C, and an extension time of 15 s. The numbers of cycles were tested to fall within the linear phase of amplification; 28 cycles were used in this experiment. Primer sequences were:
OLS_gRNA-SP_F: TATATATCTTGTGGAAAGGACGAAACACCG
OLS_gRNA-SP_R: CTTATTTTAACTTGCTATTTCTAGCTCT
To obtain high yield coverage of the PCR products, 10 repeats of 50 μL PCR reactions were performed for each library. The 144 bp amplicons were separated in 2% agarose gel electrophoresis and purified by QIAquick gel extraction kit (QIAGEN). Subsequently, the gRNA-LGP vector (Addgene 52963) was digested by BsmBI (NEB) via the following reaction at 55°C for 3 hours:
| gRNA-LGP vector | 4 μg |
| Buffer 3.1 | 5 μL |
| 10× BSA | 5 μL |
| BsmBI | 3 μL |
| H2O | Up to 50 μL |
After digestion, the vector was treated with 2 μL of Calf Intestinal Alkaline Phosphatase (NEB) at 37°C for 30 minutes, then purified by QIAquick PCR Purification Kit (Qiagen). To assemble the paired gRNAs into the vector, 10 Gibson assembly reactions were performed as follows:
| Linearized gRNA-LGP vector | 200 ng |
| Dual gRNA inserts | 36 ng (molar ratio 1:10) |
| 2× Gibson Assembly Master Mix (NEB) | 10 μL |
| H2O | Up to 20 μL |
After incubation at 50°C for 1 hour, the product was purified by QIAquick PCR Purification Kit (QIAGEN) and then transformed into One Shot Stbl3 Chemically Competent E. coli (Invitrogen). Twenty parallel transformations were performed to ensure adequate library representation. A small fraction (20–100 μL) of cultures was spread on carbenicillin (50 μg/ml) LB plates to calculate the library coverage, and the rest of the cultures were amplified overnight in 150 ml LB medium, ~100X library coverage was ensured. The plasmid DNA was then extracted using the HiSpeed Plasmid Maxi Kit (QIAGEN), and twenty independent clones were picked and Sanger-sequenced to estimate the overall quality of the library.
Step II: Insertion of the gRNA scaffold and the mouse U6 promoter
The step 1 library plasmids were digested by BsmBI (NEB) as per the following reaction at 55°C for 3 hours:
| Step 1 library | 4 μg |
| Buffer 3.1 | 5 μL |
| 10× BSA | 5 μL |
| BsmBI | 3 μL |
| H2O | Up to 50 μL |
After digestion, the linearized plasmids were treated with 2 μL of Calf Intestinal Alkaline Phosphatase (NEB) at 37°C for 30 minutes; cut plasmids were gel-purified via 0.6% agarose gel electrophoresis and QIAquick gel extraction (QIAGEN).
Concurrently, the step 2 inserts, synthesized commercially (Integrated DNA Technologies) and cloned into a TOPO vector, were digested by BsmBI (NEB) per the following reaction at 55 °C for 3 hours:
| Purified step 2 insert PCR product | 0.8 μg |
| Buffer 3.1 | 5 μL |
| 10× BSA | 5 μL |
| BsmBI | 3 μL |
| H2O | Up to 50 μL |
Sequence of the step 2 insert, with left-gRNA scaffold (underlined) and mU6 promoters (bold):
TATGAGGACGAATCTCCCGCTTATACGTCTCTGTTTCAGAGCTATGCTGGAAACTGCATAGCAAGTTGAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTTTTGTACTGAGTCGCCCAGTCTCAGATAGATCCGACGCCGCCATCTCTAGGCCCGCGCCGGCCCCCTCGCACAGACTTGTGGGAGAAGCTCGGCTACTCCCCTGCCCCGGTTAATTTGCATATAATATTTCCTAGTAACTATAGAGGCTTAATGTGCGATAAAAGACAGATAATCTGTTCTTTTTAATACTAGCTACATTTTACATGATAGGCTTGGATTTCTATAAGAGATACAAATACTAAATTATTATTTTAAAAAACAGCACAAAAGGAAACTCACCCTAACTGTAAAGTAATTGTGTGTTTTGAGACTATAAATATCCCTTGGAGAAAAGCCTTGTTTGAGAGACGGTACAAGCACACGTTTGTCAAGACC
Subsequently, the following ligation reaction was set up, involving overnight incubation at 16°C followed by heat inactivation at 65°C for 10 minutes:
| 10X T4 DNA Ligase Buffer | 2 μL |
| Step 1 library, digested | 100 ng |
| Step 2 insert, digested | 100 ng |
| T4 DNA Ligase (high concentration) | 1 μL |
| H2O | Up to 20 μL |
4 μL of the reaction was transformed into 100 μL of ElectroMAX Stbl4 Competent Cells (Invitrogen) according to the manufacturer’s protocol using an Eppendorf Electroporator. A small fraction (1–10 μL) of cultures were spread on carbenicillin (50 μg/ml) LB plates to calculate the library coverage, and the rest of the cultures were plated on ten 15 cm LB- carbenicillin plates and grown overnight at 37°C for amplification. Two transformations were required to get ~100X library coverage. The plasmid DNA was extracted using HiSpeed Plasmid Maxi Kit (Qiagen). Library diversity was determined by deep sequencing.
NGS library preparation
Harvested cell pellets were stored at −80°C until extraction of genomic DNA with the DNeasy Blood and Tissue Kit (Qiagen). The dual gRNA cassette was amplified and prepared for deep sequencing using two steps of PCR: The first step was performed as 10 separate 50 μL reactions with 2 μg input genomic DNA per reaction (total 20 μg for each sample) using Kapa Hifi. PCR primers were as follows:
NGS_dual-gRNA_SP_Lib_F: ACACTCTTTCCCTACACGACGCTCTTCCGATCT TATATATCTTGTGGAAAGGACGAAACACCG
NGS_dual-gRNA_SP_Lib_R: GACTGGAGTTCAGACGTGTGCTCTTCCGATCT CCTTATTTTAACTTGCTATTTCTAGCTCTA
Thermocycling parameters were: 95°C for 30 s, 21–26 cycles of (98°C for 15 s, 55°C for 15 s, 72°C for 45 s), and 72°C for 5 min. The numbers of cycles were tested to fall within the linear phase of amplification. Amplicons (600 bp) of 10 reactions for each sample were pooled, size-selected and purified by Agencourt AMPure XP beads at 0.8 ratio, followed by further purification using QIAquick PCR Purification (Qiagen). The second step of PCR was performed using 4 separate 50 μL reactions with 5 ng of first-step PCR product per reaction (total 20 ng for each sample) using Next® Multiplex Oligos for Illumina (New England Biosciences) to attach Illumina adaptors and indexes. The thermocycling parameters were: 95°C for 30 s, 7–8 cycles of (98°C for 15 s, 72°C for 45 s), and 72°C for 5 min. The amplicons from these 4 reactions for each sample were pooled, size-selected and purified twice using Agencourt AMPure XP beads at 0.8 ratio. The purified second step PCR library was quantified by real-time PCR using Illumina Library Quantification (Kapa Biosystems) and used for downstream sequencing using the Illumina HiSeq Rapid run platform.
Viral production and Cas9 cloning
HEK293T cells were maintained in DMEM medium supplemented with 10% fetal bovine serum. To produce lentivirus, HEK293T cells were seeded in 15 cm tissue culture dishes one day before transfection so that they were 70–80% confluent at the time of transfection. Prior to transfection, culture media was changed to pre-warmed DMEM medium supplemented with 10% fetal bovine serum. For each 15 cm dish, 36 μL of Lipofectamine 3000 (Life Technologies) was diluted in 1.2 mL OptiMEM (Life Technologies). Separately, 3 μg pMD2.G (Addgene #12259), 12 μg of pCMV delta R8.2 (Addgene #12263), 9 μg of lentiviral vector and 48 μL of P3000 Reagent were diluted in 1.2 mL OptiMEM. After incubation for 5 min, the Lipofectamine 3000 mixture and DNA mixture were combined and incubated at room temperature for 30 min. The mixture was then added dropwise to HEK293T cells. Viral particles were harvested 48 hours and 72 hours after transfection, further concentrated using Centricon Plus-20 centrifugal ultrafilters with a cutoff 100,000 NMWL (Millipore) to a final volume of 450 μL, and then aliquoted and frozen at −80°C.
For screening assays the CRISPR Cas9 nuclease was stably integrated into the human AAVS1 site in HeLa, 293T, and A549 cell lines. Cas9 cell-lines were obtained from GeneCopoeia, tested for mycoplasma contamination, and expanded and frozen in multiple aliquots so that subsequent experiments could be performed with low (<5) passage number. These were grown in DMEM supplemented with 10% fetal bovine serum and hygromycin (500 and 100 μg/ml, respectively). Nearly 100% killing was observed in cells without the Cas9 vector with these doses after 120 hours of exposure.
Design of gene constructs
A panel of 73 genes comprising 17 validated oncogenes, 30 validated tumor suppressor genes and 26 cancer-relevant druggable targets (DT) were selected for study (Figure 2A, Table S1). Priority was given to the genes most frequently mutated in human cancer26 and genes that are the target of an FDA approved drug. Three unique 20 bp gRNAs were designed for each target gene. A large number of gRNAs were designed to target the earliest exon of each gene and/or constitutive exons, as previously reported27. Poly-T sequences (i.e., more than two consecutive T’s) were avoided and, to avoid off-target editing, gRNAs were only used if they would require at least three substitutions in order to match any other sequence in the genome. After filtering all gRNA designs for the aforementioned criteria, three gRNAs were selected with one targeting the earliest exon and two targeting the earliest constitutive exons. Dual gRNA constructs were synthesized for all pairwise gRNA combinations between genes. In addition, twelve gRNAs were designed to be “non-targeters” that should not target any specific site in the genome. Three of these were randomly selected and paired with all targeting gRNAs to provide single knockout constructs. In addition, pairs of non-targeting gRNAs were included as negative controls. In total this resulted in 23,652 double gene knockout constructs, 657 single gene knockouts, and 600 negative controls.
Competitive growth experiments
The pooled library of double gRNA constructs was packaged into lentiviruses with each cell line infected at an MOI of 0.1–0.4 to ensure that each cell had 0–1 double gRNA constructs. Experiments were performed in three cell lines: HeLa, a cervical cancer cell line driven by Human Papilloma Virus (HPV); A549, a lung cancer cell line driven by KRAS G12S mutation; and 293T, a human embryonic kidney cell line transformed by adenovirus that expresses the SV40 Large-T antigen. To maintain adequate representation of all library elements (> 200 fold) the screen was started with 107 cells for HeLa and A549 cells. To accommodate this larger number of cells, 500 cm2 bioassay plates (Corning) were used. Puromycin selection was started 2 days after transduction and maintained throughout the course of the experiment to eliminate cells without gRNAs. The puromycin selection doses were 5 μg/ml for both cell lines. Following transduction, cells containing integrated gRNAs were maintained in exponential growth by harvesting and removing a fraction of the cells approximately every two to four days. A minimum of 5 × 106 cells were maintained in culture for all cell lines at each passage. DNA was extracted from cells harvested at 3, 14, 21 and 28-day time points following transduction using the Blood and Cell Culture DNA Mini Kit (Qiagen) according to manufacturer protocols. To assess the frequency of gRNAs before and after selection, integrated DNA encoding the gRNA sequence was PCR amplified and prepared for sequencing by HiSeq Rapid run, following manufacturer protocols. Standard Illumina sequencing primers were employed for library preparation and sequencing was conducted to generate 75 bp reads in a paired-end fashion. Following sequencing, data quality was assessed using FastQC.
Processing of paired-end reads
Analysis was performed using a software pipeline constructed from Python, R, and Jupyter Notebooks. FASTQ files were trimmed of scaffold sequence using cutadapt28, after which trimmed reads of unexpected lengths <19 or >21 bps were discarded. Remaining reads were truncated to 19 bases from the appropriate end, and reverse reads were reverse-complemented. Both reads in a pair were checked for sequence matches against gRNA sequences used in the library, allowing one mismatch anywhere in a read. Read pairs that matched to a known construct were aggregated to compute the total counts for that construct in the relevant sample, used for subsequent analysis.
Estimation of fitness of each construct
We assume that each subpopulation of cells expressing a particular construct c grows exponentially. In the continuous limit:
| (1) |
where Nc(t) is the number of cells in the population expressing construct c at time t; fc is the fitness of construct c measured in units of cell doublings per day (day−1); and f0 is the fitness of cells expressing a double null (control) construct. Pooled sequencing does not measure Nc directly, but estimates the relative abundance xc of each construct in the population:
| (2) |
Combining eqns. (1) and (2) yields:
| (3) |
which has linear (in time) and non-linear components. Here ac ≡ xc(0) is the initial condition. Note that while the same construct is expected to have the same fitness in replicate experiments, it may have different initial conditions. The non-linear term reflects effective interaction whereby relative frequency of one construct is modulated by growth of other constructs. Thus a particular xc may possibly decrease even when its fitness fc is positive. As we are working with relative frequencies, there is no need to “normalize” the raw counts in any way. By definition, log2 relative frequencies satisfy the constraint Σc2xc = 1 at all times.
Experimentally measured log2 relative frequencies Xc(t) deviate from the expected values xc(t). The parameters of the model are found by minimizing the sum of squares:
subject to the constraint Σc2ac = 1. Since E is invariant under the substitution fc → fc + δ where δ is an arbitrary constant, the single gene fitness is determined up to an overall additive constant, which can be fixed by setting the mean null probe fitness to zero. Formally, one has to find the minimum of the function Eλ ≡ E − λ(Σc2ac − 1), where λ is the Lagrange multiplier. In other words, we need to solve the system of non-linear equations: ∂Eλ/∂ac = ∂Eλ/∂fc = ∂Eλ/∂λ = 0. An analytical solution does not exist; however, and excellent approximation exists when the number of constructs is large, Σc 1 ≫ 1, in which case the solution is:
and
where the bar indicates mean over time points. The ac do not depend on the choice of δ.
To avoid fitting to spurious data, we use only data points above a certain threshold of raw sequencing reads. The threshold depends mainly on the size of the sample (number of cells) collected at a given time in relation to the size of the viral library and on the depth of sequencing. We note that the left-most peak in the histograms of (Supplementary Fig. 3A) contains severely under-sampled constructs with zero counts. Their x-coordinate corresponds to a pseudo-count of one introduced only for visualization purposes. It is arbitrary and therefore should not be used for fitting the model. Likewise, finite but very low counts are considered missing data. We set a threshold for every time point (red lines in Supplementary Fig. S3A). Note also that the right tail of these histograms moves to the right with time – this is due to the fastest-growing subpopulations becoming progressively larger fractions of the cells sampled. Also related is the fact that the peak of zero counts becomes taller as smaller subpopulations are outcompeted by faster growing ones and become under-sampled.
Estimation of gRNA fitness and gRNA-gRNA interactions
Once fc are known, we find the gRNA-level fitness and gRNA-level interactions as follows. As each construct contains two gRNA probes, p and p′, we write:
| (4) |
where πpp′ is the gRNA-level interaction. As there are n = 74 “genes” in the CV4 panel (73 genes and one null “gene”), each represented by three distinct probes, there are a total of 32n(n − 1)/2 = 24,309 constructs. Each gRNA is effectively replicated 3(n − 1) = 219 times as it appears in as many constructs. The gRNA-level π-scores are as unique as the construct fitnesses fc. The fp are found by robust fitting of Eq. (4). The gRNA-level π-scores are the residuals of the robust fit.
A negative interaction indicates slower-than-expected growth, suggesting synthetic sickness or lethality, while a positive interaction indicates faster-than-expected growth, suggesting epistasis29. Genes with very low or no expression had fitness scores very near the average for that cell line, consistent with a neutral growth effect. Consistent with prior competitive growth CRISPR knockout screens30, the average fitness effect of all genes was slightly negative in all three cell lines.
Supplementary Fig. 3C shows the replicate plot of gRNA-level fitness. Each gene is represented by three gRNA probes. We highlight three “genes” – the null gene as well as two genes with large positive and negative gRNA probe fitnesses. The origin is set to the center of mass of the null probes by a choice of δ. It is reassuring that the null probes cluster closely together. It is true of almost all genes that one of the three probes has almost zero fitness effect, and that the median probe splits the difference. In order to not dilute the signal with underperforming probes, we rank the probes as r(p) ∈ {0,1,2} in ascending order of |fp|. The ranks define weights for averaging as follows: the gene-level fitnesses are calculated as the weighted means of probe-level fitnesses with weights given by the squares of probe ranks, r2(p), and the gene-level interactions are calculated as the weighted means of gRNA-level interactions with weights given by products of gRNA probe ranks, r(p)r(p′) The means are over gRNA probes that represent the pair of interacting genes. The weights are designed so that probes with rank 0 do not contribute to the means, and the “best” gRNA probes have highest weights. This way, each gene-level fitness is determined by two gRNA probes, and each gene-level interaction is determined by four gRNA probe pairs. This heuristic may not be appropriate for other probe designs. For instance, if all three probes were performing well, it might be appropriate to choose equal weights.
Example fits are in Supplementary Fig. 3B. In the top panels, the fitted fc agrees well between replicates, but only after under sampled points had been dropped. In the bottom panels we show examples when fc does not agree well between replicates despite no obvious under sampling. These cases come in two flavors: those which the measured data have large variance (bottom left) and those in which the data have clear trends in both replicates but they disagree (bottom right). In the latter case we do not know whether there is a real biological difference between replicated experiments or whether this is just a random ordering of four data points with large variances into apparent trends; therefore, we take this variance at face value and incorporate it into a model that borrows power from both replicates. In this model we do not look for fc separately for each replicate. Rather, we find a single optimal fc from nc data points (nc = 2nt minus any number of points below the threshold). We are assuming fc does not change between experiments (the initial conditions ac may be different in each replicate). Each fc comes with a raw p-value calculated from the t-statistic
where
is the standard error of fc. Factor (nc − 2) is the number of degrees of freedom that can be between 1 and 2nt − 2 depending on the number of data points used for fitting. The raw p-values pc are transformed into posterior probabilities PPc using theory of Storey31,32 which connects p-values with Bayesian posterior probabilities in the context of the two-groups model. We find that roughly 2/3 of the posterior probabilities are zero, which means that about 2/3 of fc’s in Eq. (3) are likely truly zero. We avoid fitting to noise by designing a numerical Bayesian ensemble of experiments: In each member of the ensemble we assign a fitness value to construct c, which is either 0 with probability (1 − PPc), or a Gaussian-distributed random number with mean fc and standard deviation . The latter value of standard deviation includes both the experimental variance from sampling and counting, as well as possible biological variance. We typically create 102 samples, calculate gene-level quantities fg and πgg′ for each ensemble member, and report ensemble means and other statistics. We think the above sampling procedure is a reasonable data-driven solution to the bias-versus-variance problem. We calculate the z-scores by dividing raw values of πgg′ by the standard deviation of all interactions in a given experiment. We consider an interaction to be a candidate for further validation if it has a large absolute z-score, typically |z| > 3. We define the false discovery rate FDR(π) as the ratio of the observed number of interactions more extreme than π to the expected number of such interactions in the null model33, as adopted by other authors34. The null model is obtained from the Bayesian ensemble by mean-centering of the marginal distribution of every πgg′35. This null ensemble preserves correlations between gene pairs but is devoid of signal.
Replicate Correlation
To assess the variance between the two biological replicates for each cell line, single gene fitness (f) and genetic interaction (π) were separately calculated for each replicate. Standard Pearson correlation was used to compare single gene f from replicates 1 and 2 for both HeLa and A549 cells (Supplementary Fig. 4A–C). Given that genetic interaction is rare, the true value of most π scores is zero meaning that the measured values are driven entirely by noise. Therefore, correlation analysis was performed over the gene pairs with significant positive or negative interactions in at least one replicate (|z| > 3)) as has been proposed previously36. Additionally, the linear fit is constrained to pass through the origin (Supplementary Fig. 4D–F). For the calculation of genetic interaction scores for each cell line, the analysis pipeline combines the data from both biological replicates.
Gene Expression Analysis
RNA-Seq data for the HeLa, A549 and 293T cell lines were obtained from the ENCODE project (GSE30567)37. The reported Reads Per Kilobase of transcript per Million mapped reads (RPKM) values represent the average of two separate experiments.
Drug-drug interaction testing
We selected eight pairs of DT genes for which a synthetic-lethal genetic interaction had been identified in only HeLa or A549 cells. Rather than simply reproduce the dual CRISPR knockout experiment (gene-gene interaction), our goal was to examine the viability of cells exposed to drugs inhibiting the corresponding gene products (drug-drug interaction) – evaluating whether the interaction could be seen by an independent technology at the protein level, and whether it was also accessible therapeutically. Interactions were prioritized for validation testing by identifying the interactions with most negative Z-scores in either HeLa of A594 cells for which we could obtain specific chemical inhibitors of gene product. In certain cases, multiple drugs were tested for each gene to identify the drug that best phenocopied gene knockout. Either HeLa or A549 cells were seeded in clear 96 well plates and allowed to attach overnight. The next day drugs or solvent control (DMSO for all compounds except hydroxurea, which was dissolved in H2O) were added and the cells allowed to grow for 72 hours in the presence of drug, six replicates (individual wells, treated with drug by manual pipetting) were performed for each dose. After 72 hrs 20 uL of 10X Resazurin (450 μM) was added to each well and florescence was read on an Infinite F200 plate reader (Tecan) at excitation wavelength 565 nM and emission wavelength 590 nM. Each drug was initially run by itself to establish its single drug dose-response curve in each cell line. The drug hydroxyurea had a flat dose-response, such that doses well in excess of the reported IC50 for in vitro inhibition of RRM2 had minimal effect on cell viability; here a fixed dose expected to achieve near maximal inhibition of target as previously reported38 was chosen for combination experiments. For the other combinations where both drugs showed single agent toxicity, the second drug was tested at a fixed dose that inhibited growth by ~ 20% (IC20) as determined by its single agent dose-response (Supplementary Fig. 7). For two of the seven compounds the IC20 dose differed between HeLa and A549 (Table S4). To test for interaction between genes A and B, a dose-response curve was established for drug 1 (inhibitor of gene A) in presence and absence of drug 2 (inhibitor of gene B) at a fixed dose. Raw fluorescence values were normalized to either DMSO solvent wells (dose-response curve in absence of drug 2) or drug 2 alone (dose-response curve in presence of drug 2). Since the single agent activity of drug 2 was normalized to zero (i.e. defined as 100% normalized viability), dose-response curves with and without drug 2 will the same if there is only an additive effect. To assess for a synergistic effect, implying a synthetic lethal relationship between gene A and gene B, a four-parameter non-linear regression was used to fit a curve to each drug39. The IC50 of drug 1 alone was compared to the IC50 in presence of drug 2 using the sum-of-squares F test in the software package GraphPad Prism (GraphPad Software, Inc, Supplementary Fig. 8A–H).
Supplementary Material
Editorial summary.
A library of plasmids expressing two gRNAs allows the mapping of combinatorial genetic interactions with the CRISPR system. Results in cancer cells imply that cellular context is an important factor for the interaction network.
Acknowledgments
This work was generously supported by the following sources: UC San Diego School of Engineering and School of Medicine institutional funds (PM, TI, RS, AB), the Burroughs Wellcome Fund (1013926 to PM), March of Dimes Foundation (5-FY15-450 to PM), Sidney Kimmel Foundation (SKF-16-150 to PM), California Institute for Regenerative Medicine (TI, JPS), the National Cancer Institute (T32GM008806-16 to JL, R21 CA199292 to NEL, LRP to JPS), the National Institute for Environmental Health Sciences (NIEHS R01 ES014811 to TI), the National Institute for General Medical Sciences (NIGMS R01 GM084279 to TI and NK and P50 GM085764 to TI), the UC San Diego Clinical and Translational Research Institute Grant UL1TR001442 (RS, AB) and the Novo Nordisk Foundation Center for Biosustainability at DTU (NNF16CC0021858 to NEL).
Footnotes
Author contributions
J.P.S., D.Z., T.I. and P.M. conceived and supervised all experiments and wrote the paper. R.S. and J.L. performed computational analysis and wrote the paper. A.B., N.L., and A.C. performed computational analysis. J.P.S., D.Z., A.B.G, K.L., K.K., D.P., A.B., K.S., and P.M. performed experiments. D.D., A.R., N.K., and L.Q. provided technical advice. J.K. provided technical advice and wrote the paper.
Competing financial interests. All authors report no competing financial interests.
Data Availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files. Additionally networks and visualizations are freely available at www.ndexbio.org. Specific networks can be found here:
293T - http://www.ndexbio.org/#/newNetwork/199f9bb1-c3eb-11e6-8e29-06603eb7f303;
A549 - http://www.ndexbio.org/#/newNetwork/ec8bdae3-c3c9-11e6-8e29-06603eb7f303;
HeLa - http://www.ndexbio.org/#/newNetwork/e50ee3c2-c3d4-11e6-8e29-06603eb7f303.
Source code for the analysis pipeline is available at: http://ideker.ucsd.edu/papers/rsasik2017/
References
- 1.Mani R, St Onge RP, Hartman JLt, Giaever G, Roth FP. Defining genetic interaction. Proc Natl Acad Sci U S A. 2008;105:3461–6. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery L, Wasserman S. Ordering gene function: the interpretation of epistasis in regulatory hierarchies. Trends Genet. 1992;8:312–6. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins SR, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–10. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- 4.Bandyopadhyay S, et al. Rewiring of genetic networks in response to DNA damage. Science. 2010;330:1385–9. doi: 10.1126/science.1195618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Costanzo M, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353 doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lord CJ, Tutt AN, Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu Rev Med. 2015;66:455–70. doi: 10.1146/annurev-med-050913-022545. [DOI] [PubMed] [Google Scholar]
- 7.Krogan NJ, Lippman S, Agard DA, Ashworth A, Ideker T. The cancer cell map initiative: defining the hallmark networks of cancer. Mol Cell. 2015;58:690–8. doi: 10.1016/j.molcel.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srivas R, et al. A Network of Conserved Synthetic Lethal Interactions for Exploration of Precision Cancer Therapy. Molecular Cell. 2016;63:514–525. doi: 10.1016/j.molcel.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10:957–63. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014;159:647–61. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong ASL, et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:2544–2549. doi: 10.1073/pnas.1517883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bassik MC, et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell. 2013;152:909–22. doi: 10.1016/j.cell.2013.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horn T, et al. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nat Methods. 2011;8:341–6. doi: 10.1038/nmeth.1581. [DOI] [PubMed] [Google Scholar]
- 14.Laufer C, Fischer B, Billmann M, Huber W, Boutros M. Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nat Methods. 2013;10:427–31. doi: 10.1038/nmeth.2436. [DOI] [PubMed] [Google Scholar]
- 15.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neshat MS, et al. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci U S A. 2001;98:10314–9. doi: 10.1073/pnas.171076798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chari R, Mali P, Moosburner M, Church GM. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods. 2015;12:823–6. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–57. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi J, et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat Biotech. 2015;33:661–667. doi: 10.1038/nbt.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao D, Shen JP, Sasik R, Ideker T, Mali P. Combinatorial CRISPR-Cas9 Knockout Screen. Protocol Exchange. 2017 doi: 10.1038/protex.2017.063. [DOI] [Google Scholar]
- 22.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briner AE, et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–9. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 24.Dang Y, et al. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 2015;16:280. doi: 10.1186/s13059-015-0846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–8. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vogelstein B, et al. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–4. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. Journal. 2011:17. http://dx.doi.org/10.14806/ej.17.1.200.
- 29.Baryshnikova A, Costanzo M, Myers CL, Andrews B, Boone C. Genetic interaction networks: toward an understanding of heritability. Annu Rev Genomics Hum Genet. 2013;14:111–33. doi: 10.1146/annurev-genom-082509-141730. [DOI] [PubMed] [Google Scholar]
- 30.Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 31.Storey JD. The positive false discovery rate: a Bayesian interpretation and the q-value. The Annals of Statistics. 2003;31:2013–2035. [Google Scholar]
- 32.Storey JD, Bass AJ, Dabney A, Robinson D. qvalue: Q-value estimation for false discovery rate control. R package version 2.4.2. 2015 http://github.com/jdstorey/qvalue.
- 33.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 34.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pollard KS, van der Laan MJ. Choice of a null distribution in resampling-based multiple testing. Journal of Statistical Planning and Inference. 2004;125:85–100. [Google Scholar]
- 36.Baryshnikova A, et al. Quantitative analysis of fitness and genetic interactions in yeast on a genome scale. Nat Meth. 2010;7:1017–1024. doi: 10.1038/nmeth.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lori F, Lisziewicz J. Rationale for the Use of Hydroxyurea as an Anti-Human Immunodeficiency Virus Drug. Clinical Infectious Diseases. 2000;30:S193–S197. doi: 10.1086/313851. [DOI] [PubMed] [Google Scholar]
- 39.DeLean A, Munson P, Rodbard D. Simultaneous analysis of families of sigmoidal curves: application to bioassay, radioligand assay, and physiological dose-response curves. American Journal of Physiology - Gastrointestinal and Liver Physiology. 1978;235:G97–102. doi: 10.1152/ajpendo.1978.235.2.E97. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.