Abstract
Purpose
The oncogenic microRNA miR-155 is upregulated in many human cancers and its expression is increased in more aggressive and therapy resistant tumors, but the molecular mechanisms through which miR-155 increases therapy resistance are not fully understood. The main objectives of this study were to determine the role of miR-155 in resistance to chemotherapy and to evaluate anti-miR-155 treatment to chemosensitize tumors.
Experimental Design
We performed in vitro studies on cell lines to investigate the role of miR-155 in therapy resistance. To assess the effects of miR-155 inhibition on chemoresistance, we used an in vivo orthotopic lung cancer model of athymic nude mice, which we treated with anti-miR-155 alone or in combination with chemotherapy. To analyze the association of miR-155 expression and the combination of miR-155 and TP53 expression with cancer survival, we studied 956 patients with lung cancer, chronic lymphocytic leukemia and acute lymphoblastic leukemia.
Results
We demonstrate that miR-155 induces resistance to multiple chemotherapeutic agents in vitro, and that downregulation of miR-155 successfully resensitizes tumors to chemotherapy in vivo. We show that miR-155 and TP53, the most frequently deregulated tumor suppressor, are linked in a negative feedback mechanism, and demonstrate that a combination of high expression of miR-155 and low expression of TP53 is significantly associated with shorter survival in lung cancer.
Conclusions
Our findings support the existence of a miR-155/TP53 feedback loop, which is involved in resistance to chemotherapy and which can be specifically targeted to overcome drug resistance, still the main cause of cancer-related deaths.
Keywords: microRNA, miR-155, TP53, lung cancer, leukemia
INTRODUCTION
Resistance to therapy is the leading cause of failure to respond to chemotherapeutic drugs that leads to the high mortality in cancer (1, 2). Despite decades of research, only modest advances have been made in developing strategies to overcome resistance (3). The addition of non-coding RNAs (ncRNAs) to the ever-expanding set of genes deregulated in cancer (4) offer the opportunity to deeper understand these mechanisms and the hope to eradicate chemoresistance. Non-small cell lung cancer (NSCLC) and chronic lymphocytic leukemia (CLL) are the most frequent adult solid and hematological malignancies in the Western world, respectively (2), and resistance to therapy is a very significant medical issue in these patients. Virtually all NSCLC patients will eventually develop resistance to the chemotherapeutic agents they are exposed to (5), and all CLL patients requiring treatment, including the standard of care chemotherapy-based fludarabine, cyclophosphamide and rituximab (FCR) treatment, are expected to relapse (6). The poorest prognosis CLL subgroup is characterized by deletions of chromosome 17p (del17p), the genomic locus of TP53, having an overall survival of less than 2 years (7). The tumor suppressor gene TP53 is frequently deleted or mutated in human cancers and is involved in the development of drug resistance by cancer cells (8).
MicroRNAs (miRNAs) are small ncRNAs that regulate the expression of protein coding genes (9). MiR-155 is a well-known oncogenic miRNA, which is upregulated in a wide variety of human cancers (10, 11), especially in more aggressive and therapy resistant tumors (12, 13). For example, we identified a signature of deregulated miRNAs in patients with CLL and 17p deletion, versus patients with normal genotype, having good prognosis (14). In the 17p deletion group, miR-155 was the most upregulated miRNA (14). Moreover, we and others have demonstrated that miR-155 has prognostic significance in multiple types of tumors, including leukemia (15, 16) and lung cancer (17, 18).
Overexpression of miR-155 has been associated with drug resistance in several human cancers, including breast cancer, B-cell lymphoma and colon cancer (12, 19, 20), but the molecular mechanisms through which miR-155 increases cancer cell resistance to treatment are not fully understood. Therefore, the main objectives of this study were to determine the molecular mechanism through which miR-155 induces resistance to chemotherapy and to evaluate anti-miR-155 treatment to chemosensitize tumors. We demonstrate that overexpression of miR-155 induces resistance to chemotherapy, which can be reversed upon miR-155 inhibition. We further identify a miR-155/TP53 negative regulatory feedback loop, which affects the development of cancer drug resistance. The inverse expression correlation between miR-155 and TP53 transcripts is additionally supported by survival data from four lung cancer cohorts, in which we show that high expression of miR-155 and low expression of TP53 are associated with shorter survival, further confirming the involvement of miR-155 in TP53-mediated resistance mechanisms.
MATERIALS AND METHODS
Patient samples
The origin of all patient datasets is presented in Table 1. The total number of patients included in the survival analyses was 956. Both analyzed CLL subgroups were previously described: CLL-NEJM (21) and CLL-Clin Cancer Res (22).
Table 1.
The analyzed patient datasets for survival analysis
| TP53 survival analysis | miR-155 survival analysis | Combined miR-155 and TP53 survival analysis | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||||
| Dataset | Reference | Total No. pts |
No. pts TP53 higha |
No. pts TP53 low |
Poor prognosis |
P- value |
No. pts miR- 155 high |
No. pts miR- 155 low |
Poor prognosis |
P- value |
No. pts miR-155 high/TP53 low |
No. pts miR-155 low/TP53 high |
Poor prognosis | P- value |
| CLL – NEJM | (21) | 94 | NA | NA | NA | NA | 39 | 55 | high miR-155 | 0.0337 | NA | NA | NA | NA |
| CLL – Clin Cancer Res | (22) | 193 | NA | NA | NA | NA | 87 | 106 | high miR-155 | 0.021 | NA | NA | NA | NA |
| ALL - MDACC | MDACC | 52 | NA | NA | NA | NA | 14 | 38 | high miR-155 | 0.0052 | NA | NA | NA | NA |
| NSCLC - Italy | IRST | 24 | 11 | 13 | / | 0.119 | 9 | 15 | / | 0.064 | 4 | 13 | high miR-155; low TP53 | 0.0161 |
| Lung adenocarcinoma - MDACC | MDACC | 58 | 22 | 36 | / | 0.06 | 34 | 24 | / | 0.22 | 26 | 14 | high miR-155; low TP53 | 0.0356 |
| Lung adenocarcinoma - TCGAa | TCGA | 343 | 216 | 127 | TP53 low | 0.019 | 236 | 107 | / | 0.19 | 90 | 70 | high miR-155; low TP53 | 0.0177 |
| Lung squamous cell carcinoma - TCGA | TCGA | 192 | 119 | 73 | / | 0.086 | 136 | 56 | / | 0.25 | 46 | 29 | high miR-155; low TP53 | 0.0243 |
|
| ||||||||||||||
| Total | 956 | 368 | 249 | 555 | 401 | 166 | 126 | |||||||
Abbreviations: CLL, chronic lymphocytic leukemia; NEJM, The New England Journal of Medicine; ALL, acute lymphoblastic leukemia; MDACC, The University of Texas MD Anderson Cancer Center; NSCLC, non-small cell lung cancer; TCGA, The Cancer Genome Atlas; pts, patients
The TCGA data were downloaded from the data portal at https://tcga-data.nci.nih.gov/tcga.
High and low expression of mir-155 and TP53 were determined with the log-rank test as indicated in the Statistical Analysis section of the Materials and Methods.
Twenty-four NSCLC samples were collected at the Istituto Scientifico Romagnolo per lo Studio e la Cura dei Tumori (IRST) IRCCS, Italy (NSCLC-Italy), 58 lung adenocarcinoma samples were collected at The University of Texas MD Anderson Cancer Center (lung adenocarcinoma-MDACC), and 52 ALL samples were collected at MDACC (ALL-MDACC). Clinical characteristics of both lung cancer datasets and the ALL dataset can be found in Supplementary Tables S1 and S2, respectively. All patients provided written informed consent prior to inclusion in the study, and collection of the samples was approved by the institutional review board at each institution (IRST Srl IRCCS, and MDACC). All the work described has been carried out in accordance with the Declaration of Helsinki. In addition, the TCGA datasets for lung adeno-carcinoma (n = 343) and lung squamous cell carcinoma (n = 192) were downloaded from the data portal at https://tcga-data.nci.nih.gov/tcga and survival analysis was performed (Table 1).
In vitro assays
A detailed description of the in vitro assays used, including cell lines, cell culture, transfection, drug treatment, quantitative real-time PCR, Western blotting, chromatin immunoprecipitation, luciferase assay, mutagenesis and drug resistance assays, can be found in the Supplementary Methods.
In vivo orthotopic mouse models
Liposomal control anti-miR and anti-miR-155 nanoparticle preparations, intrapulmonary injections, liposomal nanoparticle delivery, and cisplatin and doxorubicin treatment were carried out as previously described (23). The treatment schedules can be found in Figures 1A and Supplementary Figure S1A. MiR-155 expression in tissue sections was analyzed by in situ hybridization as previously described (24). Cell proliferation, angiogenesis and microvesicle density, and apoptosis were assessed by Ki-67 or CD31 immunostaining, or with the TUNEL assay as previously described (24). A more detailed description of the animal experiment can be found in the Supplementary Methods.
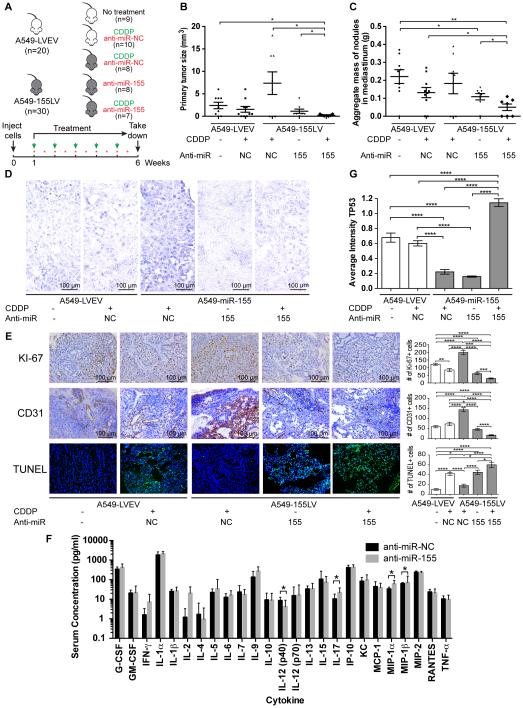
Figure 1. In vivo orthotopic lung cancer model for the role of miR-155 in chemoresistance.
(A) Injection and treatment schedule for CDDP (green arrows) and anti-miR negative control (NC) or anti-miR-155 liposomal nanoparticles (red stars) for five different treatment groups: mice that were injected with A549-LVEV cells and untreated (group 1), injected with A549-LVEV cells and treated with anti-miR-NC and CDDP (group 2), injected with A549-155LV cells and treated with anti-miR-NC and CDDP (group 3), injected with A549-155LV cells and treated with anti-miR-155 alone (group 4), and injected with A549-155LV cells and treated with anti-miR-155 and CDDP (group 5). (B-C) Graphs of the primary tumor size (B) and the aggregate mass of nodules in the mediastinum (C) for each of the five treatment groups. (D) In situ hybridization for miR-155 for each of the five treatment groups. (E) Immunohistochemical analyses for Ki-67 (proliferation) and CD31 (angiogenesis), as well as the TUNEL assay (apoptosis) for each of the five treatment groups. Quantifications are presented in the histograms at the right side of the pictures. (F) Cytokine assay detecting 25 pro-inflammatory cytokines on serum of mice (n=10/group) injected with either anti-miR-NC-DOPC or anti-miR-155-DOPC. (G) TP53 immunostaining for each of the five treatment groups. CDDP, cisplatin; LVEV, lentivirus empty vector; LV, lentivirus; NC, negative control. Error bars represent SEM (panels B, C, E and G) or SD (panel F). Scale bars in panels D and E represent 100 m. The number of mice in each group is indicated.
TCGA data analysis
Input data were downloaded from the publicly available data portal of The Cancer Genome Atlas Project (TCGA) at https://tcga-data.nci.nih.gov/tcga. Level 3 Illumina RNA-Seq and miRNA-Seq were used for the analysis of mRNA and miRNA expression, respectively. For miRNA-Seq data, we derived the “reads_per_million_miRNA_mapped” values for mature forms of each microRNA from the “isoform_quantification” files. Patient samples with survival data of 0 “days_to_last_follow_up” were excluded. Data for somatic mutations of TP53 in TCGA samples were downloaded from the cBio Portal at http://www.cbioportal.org/public-portal/.
Statistics
All patient-related analyses were carried out in the R statistical environment, version 3.0. (http:///www.r-project.org/). Survival analyses were performed as previously described (24) with some modifications. Briefly, for each cohort, a relationship between miR-155/TP53 expression and overall survival was assessed as follows. Patients were grouped into percentiles according to miR-155 and TP53 expression. The log-rank test was employed to determine the association between miRNA/mRNA expression and survival. The Kaplan-Meyer method was used to generate survival curves. The p-value and the cut-off to optimally separate the patients in high and low (min p-value) miR-155 and TP53 were recorded. We then considered whether combining inverse expression of miR-155 and TP53 would associate with survival. We used the following procedure. A fixed cut-off for miR-155 together with a fixed cut-off for TP53 splits the cohort in four groups corresponding to low or high miR-155 and low or high TP53 expression. For each pair of cut-offs we contrasted the two groups linked to a negative association: tumors with high levels of miR-155 and low levels of TP53 versus tumors with low levels of miR-155 and high levels of TP53. We recorded the best separation obtained (min p-value) for the pair and noticed that the difference in median survival time between the two groups contrasted is significantly larger than the difference between the groups classified into high/low based on the expression of miR-155 or TP53 alone. The relationship between survival and covariates (miR-155 and TP53 expression levels and available prognostic factors or other clinical parameters) was examined using a Cox proportional hazard model.
For lung adenocarcinoma cases with miR-155 expression, TP53 mutational status and survival information available, we checked for a relationship between miR-155 expression, TP53 expression and overall survival in patients with wild-type TP53 and mutated TP53 in a similar manner as described above. According to the TP53 mutational status, patients were divided into two groups: (i) those expressing wild-type TP53 (unmutated) or harboring TP53 mutations not affecting its protein function (according to the IARC TP53 database p53.iarc.fr), and (ii) those harboring TP53 mutations that affect TP53 protein function (according to the IARC TP53 database p53.iarc.fr). For each group, Kaplan-Meier overall survival curves were generated for high vs. low miR-155, and high miR-155 and low TP53 vs. low miR-155 and high TP53.
Statistical analysis of the in vitro and in vivo data was carried out with GraphPad Prism 6 software. To verify whether data followed a normal distribution, the Shapiro-Wilk normality test was performed, and an unpaired t-test (normal distribution) or non-parametric Mann-Whitney-Wilcoxon test (non-normal distribution) was applied to determine P-values. All tests were two-sided and P-values <0.05 were considered statistically significant. Statistical significances are presented as * according to the following scheme: *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
RESULTS
MiR-155 induces chemoresistance in vitro
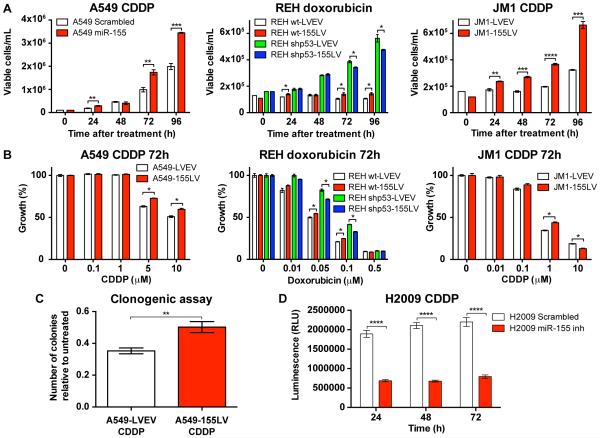
We treated three different lung cancer and leukemia cell lines with endogenous levels of miR-155 expression and after miR-155 overexpression (either by miR-155 precursor or miR-155 lentivirus) with chemotherapeutic agents commonly used to treat patients: the lung cancer cell line A549 with cisplatin (CDDP, cis-diamminodichloroplatinum) (5, 25), the acute lymphoblastic leukemia (ALL) cell line REH with doxorubicin (26) and the immunoblastic B-cell leukemia/lymphoma cell line JM1 with CDDP (27). As shown in Figure 2A-B, A549, REH and JM1 cells overexpresssing miR-155 grew significantly better and showed higher proliferation when undergoing treatment with CDDP or doxorubicin than cells expressing normal levels of miR-155. In addition, we performed a clonogenic assay for A549 cells stably overexpressing miR-155 (A549-155LV) and treated with CDDP vs. control cells (A549-LVEV) treated with CDDP. We observed a significant increase in the number of colonies when miR-155 was overexpressed, further demonstrating the chemoresistance induced by miR-155 (Figure 2C). Moreover, when we treated the H2009 lung cancer cell line with miR-155 inhibitor and CDDP, we found that these cells grew significantly less than H2009 cells treated with negative control inhibitor and CDDP (Figure 2D). Of note, when TP53 expression was abolished in REH cells by shRNA treatment, the protective effect to chemotherapeutic agents in cells overexpressing miR-155 disappeared (Figure 2A, middle panel and Figure 2B, middle panel). Finally, no difference in proliferation and cell growth was observed after fludarabine treatment and miR-155 overexpression in MEC1 and MEC2 cell lines, both of which carry a deletion of the TP53 locus (28) (Supplementary Figure S2). Altogether, these data suggest a role of miR-155 in drug resistance in various types of cancer, including lung cancer and leukemia, for multiple types of chemotherapy.
Figure 2. The effect of miR-155 modulation on drug resistance.
(A) Growth curves and (B) proliferation curves for A549 cells treated with CDDP (left graph), REH cells (wt and shp53) treated with doxorubicin (middle graph) and JM1 cells treated with CDDP (right graph). (C) Clonogenic assay of A549 cells treated with CDDP. (D) Viability assay for H2009 cells treated with CDDP. CDDP, cisplatin; wt, wild-type; shp53, short hairpin for TP53; LVEV, lentivirus empty vector; LV, lentivirus. Error bars represent SEM, and each assay was performed at least three times.
MiR-155-induced chemoresistance can be reversed in vivo by treatment with anti-miR-155-DOPC
To evaluate the in vivo involvement of miR-155 in therapy resistance, we established an orthotopic lung cancer mouse model by intrapulmonary injection of A549-LVEV (control) cells or with A549-155LV (miR-155 overexpressing) cells. Two independent experiments were carried out with four (Figure S1) and five (Figure 1) treatment groups, respectively, in which mice were treated with negative control anti-miR (anti-miR-NC) or with anti-miR-155 alone or in combination with CDDP, according to the schedule in Figure 1A and Supplementary Figure S1A. Mice injected with A549-LVEV cells and treated with CDDP and anti-miR-NC showed a decrease in number of tumors, reduced primary tumor size and a reduced aggregate mass of metastases when compared to untreated mice injected with A549-LVEV cells, although this decrease was not significant, indicating that these tumors are sensitive to CDDP, as was expected (Figure 1B-C and Supplementary Figure S1B-D). When miR-155 was overexpressed (through injection of A549-155LV cells), the tumors became resistant to CDDP treatment and the administration of anti-miR-155 alone significantly reduced number of tumors, tumor size and aggregate mass of metastases (Figure 1B-C). In addition, when anti-miR-155 was combined with CDDP treatment, the chemotherapy resistance was almost completely reversed (Figure 1B-C and Supplementary Figure S1B-D). In situ hybridization for miR-155 showed an increase of miR-155 expression in miR-155 overexpressing tumors treated with CDDP and anti-miR-NC, and miR-155 levels comparable to or lower than A549-LVEV tumors when miR-155 overexpressing tumors were treated with anti-miR-155 alone or in combination with CDDP (Figure 1D and Supplementary Figure S1E). Immunohistochemistry for Ki-67 (apoptosis), CD31 (angiogenesis) and the TUNEL (apoptosis) assay suggested that miR-155, even in the presence of CDDP, is able to induce cell proliferation and angiogenesis, and reduce apoptosis, effects that are completely abolished when miR-155 is inhibited (Figure 1E and Supplementary Figure S1F). Although treatment with anti-miR-155 alone resulted in a significant decrease in proliferation and angiogenesis, and increase in apoptosis, the effects are even more pronounced when anti-miR-155 is combined with CDDP therapy (Figure 1E). Therefore, the in vivo reversion of chemoresistance by anti-miR-155 administration is consistent and reproducible by independent sets of experiments.
We further investigated whether the anti-miR-155 molecule induces an immune response in vivo. We performed a cytokine assay detecting 25 pro-inflammatory cytokines on serum of mice injected with either anti-NC-DOPC or anti-miR-155-DOPC. With the exception of IL-12 (p40), IL-17, MIP-1α and MIP-1β, which showed marginally statistically significant differences, no activation of the immune system was observed (Figure 1F), suggesting that the therapeutic effects observed in our in vivo orthotopic mouse model are likely caused by targeting of miR-155, rather than immune induction.
Identification of a miR-155/TP53 negative feedback loop
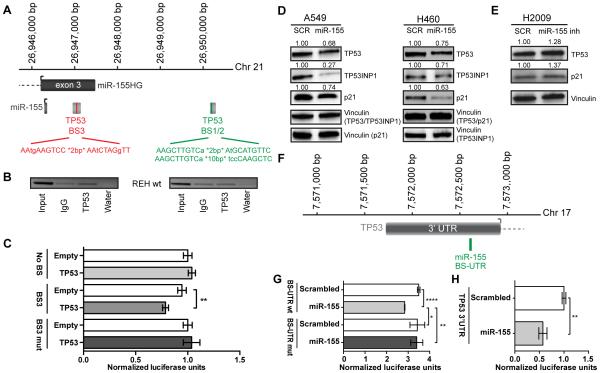
miR-155 is significantly overexpressed in patients with CLL and deletion of 17p, where the genomic TP53 locus resides (14), suggesting that TP53 might suppress the expression of miR-155. To assess this hypothesis, we performed chromatin immunoprecipitation (ChIP) for TP53 in the wild-type ALL cell line REH (REH wt) and showed that TP53 binds to one of three predicted binding sites (BS3) downstream of miR-155 (Figure 3A-B). A luciferase reporter assays for BS3 confirmed that TP53 inhibits the expression of miR-155 through direct binding in the region downstream of miR-155 (Figure 3C). The silencing effect was abrogated when BS3 was mutated, further confirming a direct binding of TP53 to BS3 (Figure 3C). To determine whether miR-155 is involved in a feedback loop, we checked whether overexpression of miR-155 affected TP53 expression. We transfected TP53 wild-type (wt) A549 and H460 cells (29) with miR-155 and observed reduced expression of TP53 protein, as well as of the known miR-155 target TP53INP1 (30, 31) and p21 (Figure 3D). When downregulating miR-155 in the H2009 lung cancer cell line harboring a mutation in TP53 that does not affect the miR-155 binding sites, we observed increased TP53 and p21 protein expression (Figure 3E). A luciferase reporter assay in the TP53 null cell line H1299 for two identified miR-155 binding sites in the 3’ untranslated region (3’ UTR) of TP53 mRNA (BS-UTR) (Figure 3F) and in the TP53 coding sequence (BS-CDS), respectively, showed a direct binding of miR-155 to BS-UTR (Figure 3G) but not to BS-CDS (data not shown). The silencing effect was abolished when BS-UTR was mutated (Figure 3G), indicating a direct binding of miR-155 to the 3’ UTR of TP53. Similar experiments in the TP53 wild-type cell line H460 showed a reduction in luciferase activity as well (Figure 3H). Finally, to assess the effects of miR-155 overexpression on TP53 expression in vivo, we performed TP53 immunostaining on the mouse tumors, and observed decrease in TP53 expression when miR-155 was overexpressed. Treatment with anti-miR-155 alone did not significantly affect TP53 expression, but a combination of anti-miR-155 with CDDP resulted in a significant increase of TP53 expression (Figure 1G and Supplementary Figure S1G). Altogether, these in vitro and in vivo data demonstrate a negative feedback loop between miR-155 and TP53, which is involved in resistance to chemotherapy.
Figure 3. In vitro validation of a miR-155/TP53 negative feedback loop.
(A) Schematic representation of three predicted TP53 binding sites in the downstream region of miR-155. (B) Chromatin immunoprecipitation for TP53 binding to BS1/2 and BS3 in REH cells with normal TP53 expression (REH wt). (C) Luciferase reporter assay and mutagenesis for the TP53 binding site BS3 downstream of miR-155 in A549 cells. (D) Western blot analysis of A549 and H460 cell lines with baseline miR-155 levels or overexpressing miR-155. (E) Western blot analysis of H2009 cells with relatively high basal miR-155 expression and after inhibiting miR-155. (F) Schematic representation of a predicted miR-155 binding site in the 3’ UTR of TP53 (BS-UTR). (G) Luciferase reporter assay and mutagenesis for BS-UTR in the TP53 null cell line H1299. (H) Luciferase reporter assay for the 3’ UTR of TP53 in the TP53 wild-type cell line H460. BS, binding site; UTR, untranslated region; SCR, scrambled. Error bars represent SD, and each assay was performed at least three times.
To understand the biological significance of the newly identified miR-155/TP53 feedback loop, and to determine how our findings fit in with other known functions and targets of miR-155, we performed integrated function and pathway analysis on 248 experimentally validated miR-155 target genes. Thirteen pathways (Supplementary Table S3) were significantly (p<0.01 and FDR<10%) enriched, the majority of which were related to cancer (pathways in cancer, colorectal cancer, pancreatic cancer), cell growth and death (cell cycle, apoptosis), as well as signal transduction pathways often deregulated in cancer and involved in drug resistance (Wnt signaling pathway, TGF-β signaling pathway, signaling by BMP, signaling by NGF). These pathways closely relate to the roles of miR-155 as an oncogene (32), TP53 as tumor suppressor and apoptosis inducer (8), and our novel findings of a miR-155/TP53 negative feedback loop involved in resistance to therapy.
High expression of miR-155 and low expression of TP53 are correlated with survival
MiR-155 was found to have prognostic impact in patients with various types of cancer (17) including lung cancer (18), leukemia (15, 16), breast cancer (33), renal cell carcinoma (34), glioma (35), colorectal cancer (36) and gallbladder carcinoma (37). We additionally assessed the correlation of miR-155 with survival in two independent and already published CLL cohorts (CLL-NEJM (21) and CLL-Clin Cancer Res (22)), in a new ALL cohort (ALL-MDACC), and in four lung cancer datasets (2 new cohorts, NSCLC-Italy and lung adenocarcinoma-MDACC, and the TCGA cohorts for lung adenocarcinoma and squamous cell carcinoma). To our surprise, we only found a correlation between high expression of miR-155 in the leukemia datasets (Supplementary Figure S3), but not in any of the lung cancer cohorts (Table 1). We previously showed that a combination of miR-520d-3p and its target EphA2 is a better prognostic factor for ovarian cancer than each gene by itself (24). To investigate whether this is also the case for our newly identified miR-155/TP53 negative feedback loop, we associated miR-155 and TP53 transcript expression with overall survival (OS) and time-to-progression (TTP) in four sets of lung cancer (Table 1). We used OS as a measure of resistance to therapy. In all cohorts, we found a significant decrease in survival when miR-155 expression was high and TP53 mRNA expression was low. Unfortunately, no TP53 expression data were available for any of the CLL and ALL datasets.
When TP53 mutation status was considered in the lung adenocarcinoma – TCGA subset, only in cases with unmutated (wild-type) TP53 or with TP53 mutations not affecting its function, high miR-155 expression was significantly associated with shorter OS (Figure 4A-B). In addition, only in cases with wild-type TP53 or TP53 mutations not affecting its function high miR-155 and low TP53 expression remained significantly associated with shorter OS (Figure 4C-D). Since all tumors in the NSCLC-Italy dataset were selected for having unmutated TP53, the same can be concluded for this dataset. Unfortunately, for the lung adenocarcinoma-MDACC and lung squamous cell carcinoma-TCGA datasets, too few patients were left to perform this analysis and get a reliable significance.
Figure 4. Clinical correlation of miR-155 and TP53 expression with survival in the lung adenocarcinoma – TCGA dataset when distinguishing between TP53 wild-type and TP53 mutated samples.
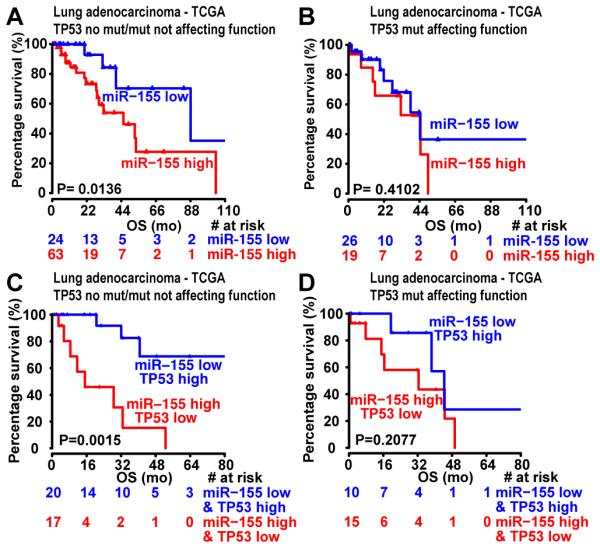
(A-B) Kaplan-Meier survival analysis for patients expressing high levels of miR-155 vs. low levels of miR-155 in samples that express wild-typeTP53 or harbor TP53 mutations that do not affect TP53 function (A), and in samples expressing mutated TP53 that affects TP53 function (B). (C-D) Kaplan-Meier survival analysis for patients expressing high levels of miR-155 and low levels of TP53 vs. low levels of miR-155 and high levels of TP53 in samples that express wild-type TP53 or harbor TP53 mutations that do not affect TP53 function (C), and in samples expressing mutated TP53 that affects TP53 function (D). The red and blue values below the curves represent patients at risk at the specified time points. OS, overall survival; mo, months; TCGA, The Cancer Genome Atlas; mut, mutation.
To assess whether expression of miR-155, TP53 and the combination of miR-155 and TP53 were independently associated with survival, uni- and multivariate analyses were carried out containing the miR-155 and TP53 expression data, as well as several known prognostic factors and available clinical parameters, as categorical variables (Table 2). In addition, the hazard ratio (HR) was calculated using the estimated parameters from the Cox models to compare survival between miR-155 high and TP53 low vs. miR-155 low and TP53 high groups (Supplementary Table S4). These analyses confirmed that high miR-155 and low TP53 mRNA expression or high miR-155 expression (when no TP53 expression data were available) were independently associated with survival in most datasets (Table 2 and Supplementary Table S4). This co-occurrence of high miR-155 expression with low TP53 mRNA expression appears to be important for predicting survival, as in all analyzed lung cancer datasets, miR-155 expression and TP53 mRNA expression by itself were not sufficient to be associated with survival. Interestingly, for the leukemia datasets (in which miR-155 expression alone was significantly associated with survival), when considering miR-155 as a continuous variable in the univariate analyses, the significance is lost for all cohorts, except CLL-Clin Cancer Res (Supplementary Table S51). This further supports our concept that a combination of both miR-155 and TP53 expression represents a better marker to predict survival.
Table 2.
Univariate and multivariate analyses of survival with patient characteristics and miR-155 and TP53 expression as categorical variables in different patient cohorts
| UNIVARIATE (CATEGORICAL) |
MULTIVARIATE (CATEGORICAL) |
||||
|---|---|---|---|---|---|
| Cohort | Variable | HR (95%CI) | p-value (log- rank) |
HR (95%CI) | p-value (wald) |
| CLL – Clin Cancer Res | IGHV (MUT vs UNM) | 3.8 (2.3-6.5) | <0.0001 | 2.4 (1.3-4.5) | 0.009 |
| CD38 (<30% vs > 30%) | 3.7 (2.2-6.2) | <0.0001 | 2.6 (1.4-4.8) | 0.002 | |
| FISH risk category (poor vs
intermediate/normal/favorable) |
4.6 (2.5-8.5) | <0.0001 | 1.9 (0.9-3.9) | 0.063 | |
| cMBL vs CLL | 2.5 (1.2-5.6) | 0.02 | 2.9 (1.3-6.5) | 0.01 | |
| miR-155 high vs miR-155 low | 2.1 (1.2-3.5) | 0.006 | 2.9 (1.3-3.6) | 0.01 | |
|
| |||||
| CLL – NEJM | IGHV (UNM vs MUT) | 4.21 (2.09-8.47) | <0.0001 | 3.97 (1.84-8.58) | 0.0005 |
| CD38 (>30% vs <30%) | 1.88 (0.92-3.84) | 0.0539 | 1.02 (0.50-2.10) | 0.9512 | |
| miR-155 high vs miR-155 low | 2.22 (1.14-4.34) | 0.0147 | 1.89 (0.97-3.68) | 0.0633 | |
|
| |||||
| ALL – MDACC | Age (>40 vs <40) | 2.16 (1.08-4.31) | 0.0258 | 1.86 (0.91-3.79) | 0.0881 |
| Gender (male vs female) | 1.11 (0.55-2.24) | 0.7698 | |||
| Molecular (BCR/ABL+ vs BCR/ABL-) | 0.61 (0.25-1.47) | 0.2651 | |||
| WBC (>50,000 vs <50,000) | 2.47 (1.14-5.33) | 0.0176 | 2.25 (1.03-4.93) | 0.0428 | |
| Cytogenetics (hyperdiploid (>50 chromosomes) vs rest) | 0.65 (0.25-1.71) | 0.3837 | |||
| miR-155 high vs miR-155 low | 3.42 (1.65-7.1) | 0.0005 | 2.61 (1.22-5.56) | 0.013 | |
|
| |||||
| NSCLC – Italy | Age (>64 vs <64) | 0.46 (0.05-4.52) | 0.4987 | ||
| Pathological stage (stage III-IV vs stage I-II) | 3.38 (0.54-21) | 0.1687 | |||
| TP53 low vs TP53 high | 2.71 (0.60- 12.33) |
0.1971 | |||
| miR-155 high vs miR-155 low | 5.25 (0.62- 44.36) |
0.1277 | |||
| miR-155 high and TP53 low vs miR-155 low and TP53
high |
6.87 (1.05- 63.08) |
0.0161 | |||
|
| |||||
| Lung adenocarcinoma – MDACC | Age (>60 vs <60) | 1.49 (0.52-4.3) | 0.4536 | ||
| Tobacco history (ever vs never) | 0.76 (0.167- 3.42) |
0.7165 | |||
| Pathological stage (stage III-IV vs stage I-II) | 2.53 (0.87-7.33) | 0.076 | |||
| TP53 low vs TP53 high | 2.32 (0.92-5.85) | 0.075 | |||
| miR-155 low vs miR-155 high | 1.67 (0.72-3.72) | 0.2354 | |||
| miR-155 high and TP53 low vs miR-155 low and TP53
high |
4.19 (1.03- 18.77) |
0.0356 | |||
|
| |||||
| Lung adenocarcinoma – TCGA | Age (>66 vs <66) | 1.03 (0.57-1.86) | 0.9203 | ||
| Tobacco history (ever vs never) | 0.77 (0.37-1.63) | 0.4995 | |||
| Pathological stage (stage III-IV vs stage I-II) | 1.7 (0.93-3.13) | 0.0823 | |||
| TP53 low vs TP53 high | 1.66 (1.09-2.54) | 0.019 | |||
| miR-155 high vs miR-155 low | 1.33 (0.85-2.09) | 0.2169 | |||
| miR-155 high and TP53 low vs miR-155 low and TP53
high |
2.03 (1.05-3.72) | 0.0177 | |||
|
| |||||
| Lung squamous cell carcinoma – TCGA |
Age (>70 vs <70) | 0.08 (0.37-1.8) | 0.6215 | ||
| Tobacco history (ever vs never) | 1.32 (0.49-3.58) | 0.585 | |||
| Pathological stage (stage III-IV vs stage I-II) | 1.21 (0.45-3.25) | 0.7078 | |||
| TP53 low vs TP53 high | 1.60 (0.96-2.68) | 0.0711 | |||
| miR-155 high vs miR-155 low | 1.41 (0.78-2.56) | 0.2537 | |||
| miR-155 high and TP53 low vs miR-155 low and TP53
high |
2.03 (1.05-3.72) | 0.0177 | |||
Abbreviations: HR, hazard ratio; CI, confidence interval; IGHV, immunoglobulin heavy chain variable region; MUT, mutated; UNM , unmutated; FISH, fluorescent in situ hybridization; cMBL, clinical monoclonal B-lymphocytosis; CLL, chronic lymphocytic leukemia; ALL, acute lymphoblastic leukemia; MDACC, The University of Texas MD Anderson Cancer Center; NSCLC, non-small cell lung cancer; WBC, white blood cells; TCGA, The Cancer Genome Atlas; MSI, microsatellite instable; MSS, microsatellite stable
DISCUSSION
Here, we showed for the first time that TP53 and miR-155 are linked in a new feedback mechanism. We further demonstrated that the miR-155/TP53 feedback loop is involved in resistance to multiple chemotherapeutic drugs used in treatment combinations in lung cancer (5) and leukemia (26, 38). Through miR-155 downregulation in vivo, we successfully resensitized the tumors to chemotherapy, and therefore, this miR-155/TP53 interactor loop could be exploited for miRNA-based therapeutic interventions in cancer patients (39). Others have shown that LNA-based and nanoparticle-based inhibition of miR-155 decreases tumor growth in mouse models of Waldenstrom macroglobulinemia and lymphoma, respectively (40-42). In addition, a recent publication showed that knockdown of miR-155 in the doxorubicin-resistant cell line A549/dox reversed doxorubicin resistance and restored doxorubicin-induced apoptosis and cell cycle arrest (43), further supporting that miR-155 might be a good target in chemosensitization of tumors.
When we took the TP53 mutational status into consideration for the survival analysis of the lung adenocarcinoma-TCGA cohort, we observed that miR-155 and the combination of miR-155 and TP53 are significantly associated with shorter OS, only in cases with unmutated TP53 or TP53 mutations not affecting its function. Similar conclusions could be drawn from the NSCLC-Italy cohort, since all patients were selected for unmutated TP53 status. In addition, we showed that overexpression of miR-155 in MEC1 and MEC2 cell lines (both carrying a deletion of the TP53 locus) does not induce chemoresistance to fludarabine treatment (Supplementary Figure S2), suggesting that there is a difference in response in the context of wild-type and mutant TP53 alleles. However, as the current data are very limited, further investigation is needed to assess the role of mutant TP53 vs. wild-type TP53 in the newly identified miR-155/TP53 feedback loop.
In contrast with some of the literature (17, 18, 33-37), we found that in most of the analyzed cancer datasets, miR-155 expression and TP53 mRNA expression by itself were not sufficient to be associated with OS (Table 1). In fact, significant correlations between miR-155 and survival could only be found in the leukemia cohorts. In addition, a recent meta-analysis evaluating miR-155 as a prognostic factor for survival in 1,557 NSCLC patients from 6 different studies suggested that high expression levels of miR-155 alone may not be significantly related to lung cancer prognosis, except for Asian and American patients (44). Our data further support the importance to consider miRNA (miR-155) and target mRNA (TP53) to predict survival. Actually, when combined, we found that high miR-155 and low TP53 expression significantly correlated with survival in 4 independent lung cancer datasets (Table 1), and that this combination remained independently associated with survival in the datasets analyzed in a multivariate analysis (Table 2 and Supplementary Table S4). We recently demonstrated that a combination of miR-520d-3p and its target EphA2 is a better prognostic factor for ovarian cancer than each gene by itself, and that simultaneous targeting of miRNA/mRNA (miR-520d-3p/EphA2) results in a remarkable therapeutic synergy as compared to either monotherapy (24).
In conclusion, our study is innovative due to multiple reasons. We show for the first time that the most frequently altered human tumor suppressor TP53 is directly targeted by one of the most oncogenic miRNAs, miR-155, and that TP53 directly regulates the expression of this miRNA as a feedback. Second, a combination of TP53 and miR-155 expression seems to be a much better classifier for overall survival of lung cancer and possibly also leukemia, than miR-155 alone. Third, miR-155 and TP53 and their downstream targets are involved in resistance to multiple types of chemotherapeutic regimens in various hystotypes. Finally, we propose to use anti-miR-155 as an additive to chemotherapy and not as a single agent, as was proposed by others (40-42). This means lower doses of drugs to be used and, consequently, less adverse reactions to occur in clinical trials. The identification of the miR-155/TP53 interaction will favor the advancement of new anti-miR-155 targeted therapies to overcome the development of drug resistance, still the main cause of cancer-related deaths.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Resistance to therapy is an important issue in cancer treatment and the main cause of cancer-related deaths. Despites decades of research into overcoming this resistance, only modest advances have been made and the resistance mechanisms remain poorly understood. This is the first report of a miR-155/TP53 negative feedback mechanism, in which there is a direct targeting of TP53 by miR-155, and which is involved in the resistance to multiple chemotherapeutic drugs used in the treatment of lung cancer and leukemias. The finding that treatment with anti-miR-155 can reverse chemoresistance in vivo supports a potential clinical use of anti-miR-155 therapy in human clinical trials of various cancer types as an addition to current chemotherapy regimens in order to overcome cancer-enacted resistance mechanisms.
ACKNOWLEDEGMENTS
The authors would like to thank Drs. Evan N Cohen and James M Ruben for their assistance with the cytokine assay. We further acknowledge the support of the RNAi and non-coding RNA Center of the UT MD Anderson Cancer Center.
Financial Support: This work was supported in part by a Developmental Research Award by Leukemia SPORE P50 CA100632. Dr Calin is The Alan M. Gewirtz Leukemia & Lymphoma Society Scholar. Work in Dr. Calin’s laboratory is supported in part by the National Institutes of Health/National Cancer Institute grants 1UH2TR00943-01 and 1 R01 CA182905-01, the UT MD Anderson Cancer Center SPORE in Melanoma grant from NCI (P50 CA093459), Aim at Melanoma Foundation and the Miriam and Jim Mulva research funds, the Brain SPORE (2P50CA127001), the Center for Radiation Oncology Research Project, the Center for Cancer Epigenetics Pilot project, a 2014 Knowledge GAP MDACC grant, a CLL Moonshot pilot project, the UT MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment, a SINF grant in colon cancer, the Laura and John Arnold Foundation, the RGK Foundation and the Estate of C. G. Johnson, Jr,. Dr. Fabbri is a St. Baldrick Foundation’s Scholar and is supported by the Concern Foundation, Hyundai Hope of Wheels, STOP Cancer, Alex’s Lemonade, the William Lawrence & Blanche Hughes Foundation, the Jean Perkins Foundation, the Nautica Malibu Triathlon Funds, the award number P30CA014089 from the National Cancer Institute at the National Institutes of Health, the Hugh and Audy Lou Colvin Foundation, and by a Shirley McKernan donation. Dr. Van Roosbroeck was a Henri Benedictus Fellow of the King Baudouin Foundation and the Belgian American Education Foundation (B.A.E.F.). Dr. Berindan-Neagoe was partially financed by a POSCCE grant (709/2010) entitled Clinical and Economical Impact of Proteome and Transcriptome Molecular Profiling in Neoadjuvant Therapy of Triple Negative Breast Cancer (BREASTIMPACT). Drs. Negrini, Neri and Morabito are partially funded by Associazione Italiana per la Ricerca sul Cancro (the Italian Association for Cancer Research (AIRC) 5xmille grant 9980). Part of this work was also supported by National Cancer Institute at the National Institutes of Health (grant number U54 CA151668) and by the Betty Anne Asche Murray Distinguished Professorship (Dr. Sood).
Footnotes
Conflict of Interest Disclosure: The authors declare no competing financial interests.
Authorship Contributions
Conceived and designed the experiments: KVR, FF, CVP, MJY, AF, RVD, IIW, AKS, GL, MF and GAC.
Performed the experiments: KVR, FF, TS, CI, CR, IV, RSR, LD, XZ, VG, RR, FM, AKS.
Analyzed and interpreted the data: KVR, FF, TS, CI, CR, IV, RSR, LD, XZ, MSN, SR, VG, RR, MF, FM, AN, DA, SC, LX, IB, MN, HMK, AKS, GL, MJK, MF and GAC.
Contributed reagents/materials/analysis tools: PPR, VVR, LA, RO and WP.
Wrote the first draft of the manuscript: KVR, FF, MF and GAC.
Contributed to the writing of the manuscript: KVR, FF, MF and GAC.
Statistical Analysis: KVR, CI, LX, MF, FM, AN, XW, MF and GAC.
Study Supervision: MF and GAC.
All authors critically reviewed the manuscript and approved the final version.
REFERENCES
- 1.Berindan-Neagoe I, Monroig Pdel C, Pasculli B, Calin GA. MicroRNAome genome: a treasure for cancer diagnosis and therapy. CA: a cancer journal for clinicians. 2014;64:311–36. doi: 10.3322/caac.21244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: a cancer journal for clinicians. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Doroshow JH. Overcoming resistance to targeted anticancer drugs. N Engl J Med. 2013;369:1852–3. doi: 10.1056/NEJMe1311325. [DOI] [PubMed] [Google Scholar]
- 4.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6:857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 5.Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer. 2011;71:3–10. doi: 10.1016/j.lungcan.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 6.Panovska A, Smolej L, Lysak D, Brychtova Y, Simkovic M, Motyckova M, et al. The outcome of chronic lymphocytic leukemia patients who relapsed after fludarabine, cyclophosphamide, and rituximab. Eur J Haematol. 2013;90:479–85. doi: 10.1111/ejh.12106. [DOI] [PubMed] [Google Scholar]
- 7.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 8.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nature reviews Cancer. 2009;9:701–13. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 9.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 10.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eastlack S, Alahari S. MicroRNA and Breast Cancer: Understanding Pathogenesis, Improving Management. Non-Coding RNA. 2015;1:17. doi: 10.3390/ncrna1010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong W, He L, Coppola M, Guo J, Esposito NN, Coppola D, et al. MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer. J Biol Chem. 2010;285:17869–79. doi: 10.1074/jbc.M110.101055. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Fabbri M, Bottoni A, Shimizu M, Spizzo R, Nicoloso MS, Rossi S, et al. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. Jama. 2011;305:59–67. doi: 10.1001/jama.2010.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rossi S, Shimizu M, Barbarotto E, Nicoloso MS, Dimitri F, Sampath D, et al. microRNA fingerprinting of CLL patients with chromosome 17p deletion identify a miR-21 score that stratifies early survival. Blood. 2010;116:945–52. doi: 10.1182/blood-2010-01-263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferrajoli A, Shanafelt TD, Ivan C, Shimizu M, Rabe KG, Nouraee N, et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood. 2013;122:1891–9. doi: 10.1182/blood-2013-01-478222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marcucci G, Maharry KS, Metzeler KH, Volinia S, Wu YZ, Mrozek K, et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J Clin Oncol. 2013;31:2086–93. doi: 10.1200/JCO.2012.45.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J, Zhang F, Wu Y, Zhang W, Zhu X, He X, et al. Prognostic role of microRNA-155 in various carcinomas: results from a meta-analysis. Dis Markers. 2013;34:379–86. doi: 10.3233/DMA-130984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu TP, Zhu CH, Zhang J, Xia R, Wu FL, Han L, et al. MicroRNA-155 Expression has Prognostic Value in Patients with Non-small Cell Lung Cancer and Digestive System Carcinomas. Asian Pac J Cancer Prev. 2013;14:7085–90. doi: 10.7314/apjcp.2013.14.12.7085. [DOI] [PubMed] [Google Scholar]
- 19.Kim JH, Kim WS, Park C. Epstein-Barr virus latent membrane protein-1 protects B-cell lymphoma from rituximab-induced apoptosis through miR-155-mediated Akt activation and up-regulation of Mcl-1. Leuk Lymphoma. 2012;53:1586–91. doi: 10.3109/10428194.2012.659736. [DOI] [PubMed] [Google Scholar]
- 20.Pu J, Bai D, Yang X, Lu X, Xu L, Lu J. Adrenaline promotes cell proliferation and increases chemoresistance in colon cancer HT29 cells through induction of miR-155. Biochem Biophys Res Commun. 2012;428:210–5. doi: 10.1016/j.bbrc.2012.09.126. [DOI] [PubMed] [Google Scholar]
- 21.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 22.Negrini M, Cutrona G, Bassi C, Fabris S, Zagatti B, Colombo M, et al. microRNAome expression in chronic lymphocytic leukemia: comparison with normal B-cell subsets and correlations with prognostic and clinical parameters. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:4141–53. doi: 10.1158/1078-0432.CCR-13-2497. [DOI] [PubMed] [Google Scholar]
- 23.Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, et al. Tumour angiogenesis regulation by the miR-200 family. Nature communications. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishimura M, Jung EJ, Shah MY, Lu C, Spizzo R, Shimizu M, et al. Therapeutic synergy between microRNA and siRNA in ovarian cancer treatment. Cancer Discov. 2013;3:1302–15. doi: 10.1158/2159-8290.CD-13-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nature reviews Cancer. 2007;7:573–84. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Manero G, Kantarjian HM. The hyper-CVAD regimen in adult acute lymphocytic leukemia. Hematol Oncol Clin North Am. 2000;14:1381–96. doi: 10.1016/s0889-8588(05)70192-1. x-xi. [DOI] [PubMed] [Google Scholar]
- 27.Hou Y, Wang HQ, Ba Y. Rituximab, gemcitabine, cisplatin, and dexamethasone in patients with refractory or relapsed aggressive B-cell lymphoma. Med Oncol. 2012;29:2409–16. doi: 10.1007/s12032-012-0211-2. [DOI] [PubMed] [Google Scholar]
- 28.Stacchini A, Aragno M, Vallario A, Alfarano A, Circosta P, Gottardi D, et al. MEC1 and MEC2: two new cell lines derived from B-chronic lymphocytic leukaemia in prolymphocytoid transformation. Leuk Res. 1999;23:127–36. doi: 10.1016/s0145-2126(98)00154-4. [DOI] [PubMed] [Google Scholar]
- 29.O'Connor PM, Jackman J, Bae I, Myers TG, Fan S, Mutoh M, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–300. [PubMed] [Google Scholar]
- 30.Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci U S A. 2007;104:16170–5. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang CM, Zhao J, Deng HY. MiR-155 promotes proliferation of human breast cancer MCF-7 cells through targeting tumor protein 53-induced nuclear protein 1. J Biomed Sci. 2013;20:79. doi: 10.1186/1423-0127-20-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faraoni I, Antonetti FR, Cardone J, Bonmassar E. miR-155 gene: a typical multifunctional microRNA. Biochim Biophys Acta. 2009;1792:497–505. doi: 10.1016/j.bbadis.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 33.Chen J, Wang BC, Tang JH. Clinical significance of microRNA-155 expression in human breast cancer. J Surg Oncol. 2012;106:260–6. doi: 10.1002/jso.22153. [DOI] [PubMed] [Google Scholar]
- 34.Shinmei S, Sakamoto N, Goto K, Sentani K, Anami K, Hayashi T, et al. MicroRNA-155 is a predictive marker for survival in patients with clear cell renal cell carcinoma. Int J Urol. 2013;20:468–77. doi: 10.1111/j.1442-2042.2012.03182.x. [DOI] [PubMed] [Google Scholar]
- 35.Sun J, Shi H, Lai N, Liao K, Zhang S, Lu X. Overexpression of microRNA-155 predicts poor prognosis in glioma patients. Med Oncol. 2014;31:911. doi: 10.1007/s12032-014-0911-x. [DOI] [PubMed] [Google Scholar]
- 36.Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79:313–20. doi: 10.1159/000323283. [DOI] [PubMed] [Google Scholar]
- 37.Zhang XL, Chen JH, Qin CK. MicroRNA-155 expression as a prognostic factor in patients with gallbladder carcinoma after surgical resection. International journal of clinical and experimental medicine. 2015;8:21241–6. [PMC free article] [PubMed] [Google Scholar]
- 38.Calin GA, Croce CM. Genomics of chronic lymphocytic leukemia microRNAs as new players with clinical significance. Seminars in oncology. 2006;33:167–73. doi: 10.1053/j.seminoncol.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12:847–65. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babar IA, Cheng CJ, Booth CJ, Liang X, Weidhaas JB, Saltzman WM, et al. Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma. Proc Natl Acad Sci U S A. 2012;109:E1695–704. doi: 10.1073/pnas.1201516109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Roccaro AM, Rombaoa C, Flores L, Obad S, Fernandes SM, et al. LNA-mediated anti-miR-155 silencing in low-grade B-cell lymphomas. Blood. 2012;120:1678–86. doi: 10.1182/blood-2012-02-410647. [DOI] [PubMed] [Google Scholar]
- 42.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–10. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lv L, An X, Li H, Ma L. Effect of miR-155 knockdown on the reversal of doxorubicin resistance in human lung cancer A549/dox cells. Oncology letters. 2016;11:1161–6. doi: 10.3892/ol.2015.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang F, Zhou J, Zhang Y, Wang Y, Cheng L, Bai Y, et al. The Value of MicroRNA-155 as a Prognostic Factor for Survival in Non-Small Cell Lung Cancer: A Meta-Analysis. PloS one. 2015;10:e0136889. doi: 10.1371/journal.pone.0136889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.