Abstract
Purpose
Hormone receptor-positive (HR+) breast cancer is clinically and biologically heterogeneous and subgroups with different prognostic and treatment sensitivities need to be identified.
Experimental design
Research-based PAM50 subtyping and expression of additional genes was performed on 63 patients with HR+/HER2- disease randomized to neoadjuvant multi-agent chemotherapy versus endocrine therapy in a phase II trial. The biology associated with treatment response was used to derive a PAM50-based Chemo-Endocrine Score (CES). CES’s predictive ability was evaluated in 4 independent neoadjuvant datasets (n=675) and 4 adjuvant datasets (n=1,505). The association of CES, intrinsic biology and PAM50 risk of relapse (ROR) was explored across 6,007 tumors.
Results
Most genes associated with endocrine sensitivity were also found associated with chemotherapy resistance. In the chemotherapy test/validation datasets, CES was independently associated with pathological complete response (pCR), even after adjusting for intrinsic subtype. pCR rates of the CES endocrine sensitive (CES-E), uncertain (CES-U) and chemotherapy sensitive (CES-C) groups in both datasets combined were 25%, 11% and 2%, respectively. In the endocrine test/validation datasets, CES was independently associated with response. Compared to ROR, >90% of ROR-low and ROR-high tumors were identified as CES-E and CES-C, respectively; however, each CES-group represented >25% of ROR-intermediate disease. In terms of survival outcome, CES-C was associated with poor relapse-free survival in patients with ROR-intermediate disease treated with either adjuvant endocrine therapy-only or no adjuvant systemic therapy, but not in patients treated with (neo)adjuvant chemotherapy.
Conclusions
CES is a genomic signature capable of estimating chemo-endocrine sensitivity in HR+ breast cancer beyond intrinsic subtype and risk of relapse.
Keywords: breast cancer, PAM50, intrinsic subtype, chemotherapy, predictor
Introduction
Approximately 70% of invasive breast cancers at diagnosis are hormone receptor-positive and HER2-negative (HR+/HER2-)1,2. However, HR+/HER2- disease is clinically and biologically heterogeneous and further subclassifications are needed to better tailor current and future treatments3–5.
Over the last decade, molecular characterization studies have identified and extensively investigated the two main molecular subtypes within HR+/HER2- disease (i.e. Luminal A and B)1,2,6. Luminal A tumors have an improved prognosis at 5- and 10-year follow-up compared with Luminal B tumors irrespective of classical clinical-pathological variables (e.g. tumor size and nodal status) and (neo)adjuvant treatment (i.e. endocrine and chemotherapy)1,2,7,8. In terms of treatment sensitivity, Luminal A tumors achieve significant lower rates of pathological complete response (pCR) than Luminal B tumors following neoadjuvant multi-agent chemotherapy9–12. However, less clear is the difference in endocrine sensitivity between the two luminal subtypes13,14.
Today, adjuvant endocrine therapy for 5–10 years is recommended for all patients with HR+/HER2- early breast cancer, whereas chemotherapy is recommended for patients with intermediate and high risk tumors15. However, the relationship between therapy and risk warrants further study considering that risk is associated with both factors related to tumor biology and clinical-pathological features such as tumor size and nodal status, whereas therapy responsiveness is generally considered to be independent of clinical-pathological factors.
Methods and Materials
GEICAM/2006-03 clinical trial
Pre-treatment core biopsy samples from patients recruited in the luminal cohort of the GEICAM/2006-03 phase II neoadjuvant clinical trial were evaluated16. In this study, 95 patients with estrogen receptor (ER)-positive (Allred 3–8), progesterone receptor (PR)-positive (Allred 3–8), HER2- (according to the ASCO/CAP guidelines17), and cytokeratin 8/18-positive breast cancer were randomly assigned to receive 24 weeks of neoadjuvant chemotherapy or endocrine therapy. Chemotherapy consisted of epirubicin combined with cyclophosphamide for 4 cycles followed by docetaxel for 4 cycles. Endocrine therapy consisted of oral exemestane. Premenopausal patients received goserelin for 6 doses.
GEICAM 2006-03 pathological response end-point
The 5-point scale Miller and Payne histological grading system18 was used to measure tumor response. In this study, the Miller and Payne scale was reduced to a 3-point scale in order to have a fair number of cases in each category and arm: no response (grade 1 and 2), intermediate response (Grade 3) and high response (Grade 4 and 5).
GEICAM 2006-03 gene expression analysis
Sixty-three of 95 pre-treatment tumor samples were available for gene expression analyses. Total RNA was purified to measure the expression of 543 breast cancer-related genes, 5 house-keeping genes and 14 negative and positive controls using the nCounter platform (Nanostring Technologies, Seattle, WA, US)19. Raw gene expression can be found in Supplemental Material.
Independent/testing datasets
Gene expression and response data were evaluated from 4 independent neoadjuvant datasets (Supplemental Material)14,20–23,24. Gene expression and survival data were evaluated from 4 independent datasets of patients with early breast cancer (Supplemental Material)2,20,25,26.
Intrinsic subtype assignment
All tumors were assigned to an intrinsic molecular subtype of breast cancer (Luminal A, Luminal B, HER2-enriched, Basal-like) and the normal-like group using the research-based PAM50 subtype predictor27,28, except for the Malaga cohort where the PAM50 standardized and commercial nCounter-based assay was used. Before subtyping, each individual dataset was normalized accordingly as previously reported29, except for the Malaga cohort that was normalized by Nanostring according to their algorithm. Of note, the Edinburgh microarray-based dataset is composed of ER+ samples-only and proper centering for intrinsic subtyping calling was not possible30. In this dataset, CES vas evaluated as a continuous variable since it is not affected by centering.
Combined cohort of primary breast cancer
To evaluate the relationship between PAM50 subtype calls, prognosis (ROR-P) and CES, we combined PAM50 data from 7 independent and previously reported cohorts1,2,11,20,31–33 representing a total of 6,007 primary tumor samples. CES was evaluated in each individual cohort, and a combined matrix was created (Supplemental Material).
Statistical analysis
Biologic analysis of gene lists was performed with DAVID 6.7 annotation tool34 using the 543-gene list as background. Association between the expression of each gene and Miller-Payne response (3 categories) was assessed by a quantitative Significance Analysis of Microarrays (SAM)35. In both testing datasets, association between each variable and pCR or clinical/radiological response was assessed by univariate and multivariable logistic regression analyses. The predictive performance of CES was evaluated using receiver operating characteristic (ROC) curve analysis. Estimates of survival were from the Kaplan-Meier curves and tests of differences by the log-rank test. Univariate and multivariable Cox-models were used to test the independent prognostic significance of each variable. Reported P values are two-sided.
Results
GEICAM 2006-03 dataset
Sixty-three pre- and post-menopausal patients were evaluated in this study (Table 1). Most patients presented ductal carcinomas (83%), tumor sizes of 2–5 cm (76%), histological grade 3 tumors (59%), clinical node-negative disease (54%) and luminal disease by PAM50 (84%).
Table 1.
Clinical-pathological characteristics and subtype distribution in the GEICAM 2006-03 study*.
| CT | % | ET | % | P-value | ||
|---|---|---|---|---|---|---|
| Num. | 32 | - | 31 | - | - | |
| Age (mean) | 53.7 | - | 52.3 | - | 0.596 | |
| Menopausal status | ||||||
| Pre-menopausal | 14 | 44% | 14 | 45% | 1.000 | |
| Post-menopausal | 18 | 56% | 17 | 55% | ||
| Tumor stage | ||||||
| T1 | 1 | 3% | 2 | 6% | 0.420 | |
| T2 | 23 | 72% | 25 | 81% | ||
| T3 | 8 | 25% | 4 | 13% | ||
| Mean tumor size (cm) | 4.2 | 3.8 | 0.278 | |||
| Node | ||||||
| N0 | 15 | 47% | 19 | 61% | 0.501 | |
| N1 | 16 | 50% | 11 | 35% | ||
| N2 | 1 | 3% | 1 | 3% | ||
| Grade | ||||||
| G1 | 0 | 0% | 0 | 0% | ||
| G2 | 8 | 25% | 6 | 19% | 0.862 | |
| G3 | 18 | 56% | 19 | 61% | ||
| G4 | 6 | 19% | 6 | 19% | ||
| Histological Type | ||||||
| Ductal | 26 | 81% | 26 | 84% | 1.000 | |
| Lobular | 2 | 6% | 2 | 6% | ||
| Others | 4 | 13% | 3 | 10% | ||
| Ki-67 IHC (mean) | 31.1 | 33.5 | 0.720 | |||
| Miller-Payne Response (mean) | 2.6 | 2.2 | 0.124 | |||
| PAM50 | ||||||
| Luminal A | 16 | 50% | 13 | 42% | 0.564 | |
| Luminal B | 11 | 34% | 13 | 42% | ||
| HER2-E | 0 | 0% | 1 | 3% | ||
| Basal-like | 2 | 6% | 0 | 0% | ||
| Normal-like | 3 | 9% | 4 | 13% | ||
CT, chemotherapy arm; ET, endocrine therapy arm.
Following chemotherapy, Luminal B tumors showed higher Miller-Payne response than Luminal A disease (mean 2.0 vs. 1.4, P=0.048). However, no difference in response between the two luminal subtypes was observed following endocrine therapy (P=0.407). In addition, no statistical significant interaction (P=0.429) between subtype and treatment (endocrine vs. chemotherapy) for tumor response was observed. Interestingly, the only patient that achieved a pCR (i.e. Miller-Payne Grade 5) had a Basal-like tumor and was contained within the chemotherapy arm.
Gene expression association with treatment sensitivity
To understand the biology associated with either chemotherapy or endocrine sensitivity within HR+/HER2- disease, we explored the association between the expression of 543 breast cancer-related genes and Miller-Payne response in each treatment arm. High expression of 70 (12.9%) and 17 (3.1%) genes was found significantly associated (P<0.05 uncorrected for multiple comparisons) with response after endocrine therapy and chemotherapy, respectively. The gene list associated with endocrine therapy response was enriched for the following biological processes (Supplemental Material): vasculature development (e.g. AKT1 and catenin beta 1), tube development (e.g. FOXA1 and gremlin 1) and cell growth (e.g. androgen receptor and fibroblast growth factor receptor 1). On the other hand, the gene list associated with chemotherapy response was enriched for cell cycle (e.g. EXO1 and MKI67) and extracellular matrix (e.g. netrin 4 and thrombospondin 1).
We then evaluated the interaction between the expressions of each individual gene with response to therapy (endocrine vs. chemotherapy). Interestingly, 41 of the 70 genes associated with response to endocrine therapy, and 8 of 17 genes associated with chemotherapy response, showed a significant interaction with treatment (P<0.05 uncorrected for multiple comparisons). Thus, the biological factors associated with endocrine sensitivity seemed to be associated, at the same time, with chemotherapy resistance, and vice versa. Indeed, an overall inverse pattern was observed between expression of most genes and response to treatment (Fig. 1A).
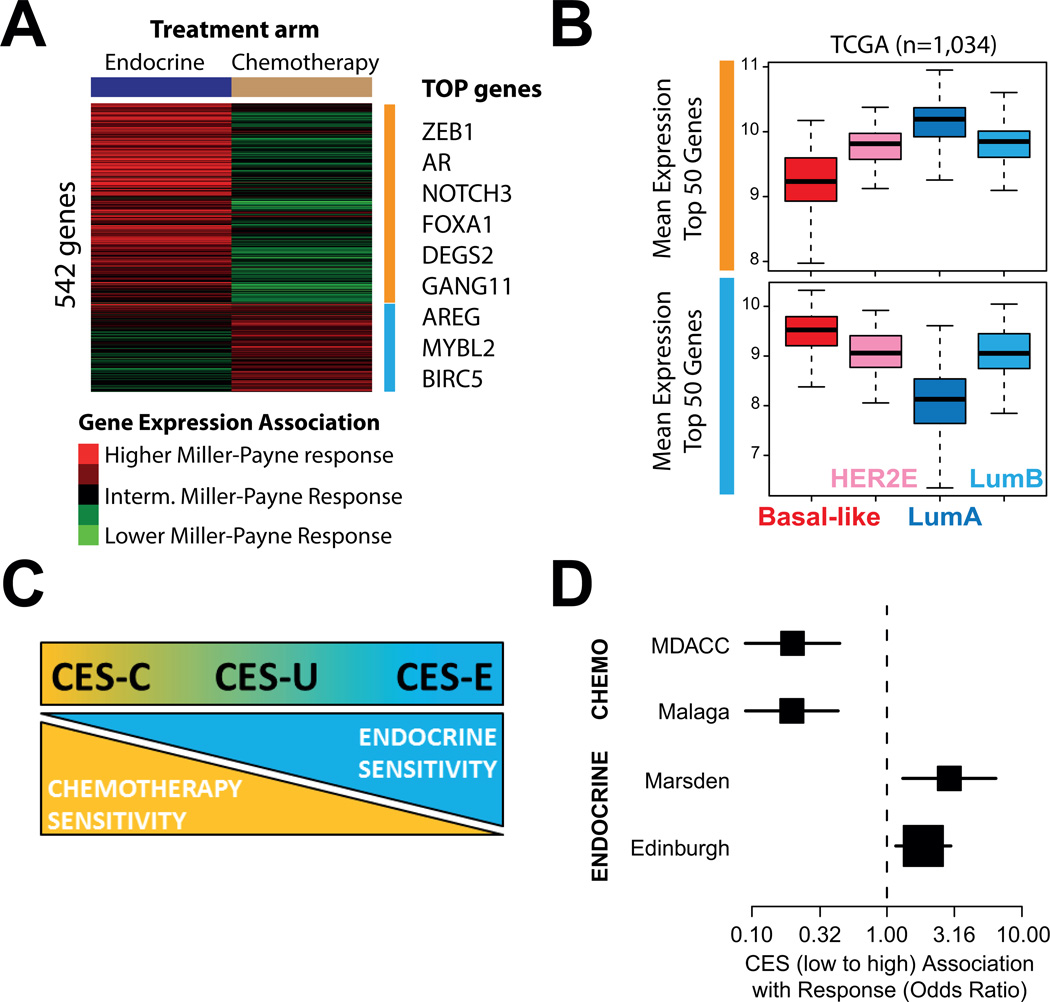
Fig 1.
Gene expression association with either chemotherapy or endocrine therapy sensitivity. (A) Association between the expression of each individual gene (n=542) and Miller-Payne response in each arm of the GEICAM 2006-03 trial. Selected top genes who’s expression is found significantly associated with response are shown on the right. (B) Mean expression of the Top 50 genes associated with endocrine sensitivity (upper panel) and chemotherapy sensitivity (bottom panel) in the GEICAM 2006-03 trial across the intrinsic subtypes of breast cancer. The RNAseq-based gene expression data has been obtained from the The Cancer Genome Atlas breast cancer project data portal (https://tcga-data.nci.nih.gov/tcga/). (C) Significance and scoring of the CES.
To further understand the biological factors associated with treatment response, we evaluated the mean expression of genes associated with high endocrine but low chemotherapy sensitivity, or low endocrine but high chemotherapy sensitivity, across 1,034 primary tumors representing all intrinsic molecular subtypes of breast cancer (Fig. 1B). The results revealed that the biology associated with chemo-endocrine sensitivity is mostly driven by the Luminal A (i.e. high endocrine but low chemotherapy sensitive) vs. Basal-like biology (i.e. low endocrine but high chemotherapy sensitive).
Development of a PAM50-based CES
Our previous results suggested that capturing the relative differences in the Luminal A vs. Basal-like biology within HR+/HER2- could help better predict endocrine and chemotherapy sensitivity. To capture this biological state in each tumor, we obtained, from the PAM50 classification algorithm, the correlation coefficients (CC) of each sample to the PAM50 Luminal A and Basal-like subtype centroids, and then subtracted the 2 values to create the Chemo-Endocrine Score (CES=CC to Luminal A – CC to Basal-like). Thus, samples with a positive score were identified as being more endocrine sensitive than chemotherapy sensitive, whereas samples with a negative score were identified as being more chemotherapy sensitive (CES-C) than endocrine sensitive (CES-E) (Fig. 1C). From GEICAM 2006-03 samples, cutoffs based on tertiles groups were determined (CES-E vs. CES uncertain [CES-U] group, cutoff = 0.70; CES-U vs. CES-C group, cutoff=0.30). The interaction of the CES score (as a continuous variable) with treatment in GEICAM 2006-03 trial provides some evidence of association (P=0.059).
MDACC-based dataset
We evaluated a combined dataset of 272 patients with HR+/HER2- disease treated with anthracycline/taxane-based neoadjuvant chemotherapy across several neoadjuvant trials (Table 2). In this dataset, 51.5%, 25.8% and 22.7% of the samples were identified as CES-E, -U and -C, respectively. The rates of pCR across the CES-E, -U and -C groups were 2.4%, 9.0% and 23.7%, respectively (P<0.0001), and were found to be similar even if non-luminal tumors were removed (2.2%, 8.8% and 25.0%). The neoadjuvant chemotherapy predictive ability of CES was independent of clinical-pathological variables and intrinsic subtype (Table 3 and Table S1). Similar results were obtained when residual cancer burden was used as the endpoint (Tables S2–3).
Table 2.
Clinical-pathological characteristics and subtype distribution of the 4 testing sets*.
| MDACC | Malaga | Marsden | Edinburgh | |||||
|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | |
| Treatment | CT | CT | ET | ET | ||||
| N | 272 | 180 | 103 | 120 | ||||
| HER2 status¥ | ||||||||
| HER2-negative | 272 | 100% | 180 | 100% | 89 | 86% | 31 | 69% |
| HER2-positive | 0 | 0% | 0 | 0% | 14 | 14% | 14 | 31% |
| Age (mean) | 50.1 | 50.0 | 53.7 | 76.1 | ||||
| Menopausal status | ||||||||
| Pre-menopausal | NA | 108 | 60% | 0 | 0% | 0 | 0% | |
| Post-menopausal | NA | 72 | 40% | 103 | 100% | 120 | 100% | |
| Tumor stage | ||||||||
| T0-T1 | 19 | 7% | 18 | 10% | 60 | 58% | 10 | 9% |
| T2 | 142 | 52% | 115 | 67% | 42 | 36% | ||
| T3-T4 | 111 | 41% | 39 | 23% | 43 | 42% | 63 | 55% |
| Node | ||||||||
| N0 | 96 | 35% | 67 | 37% | 61 | 59% | 86 | 72% |
| N1 | 133 | 49% | 61 | 34% | 39 | 38% | 34 | 28% |
| N2-N3 | 43 | 16% | 52 | 29% | 3 | 3% | ||
| Grade | ||||||||
| G1 | 28 | 11% | 27 | 16% | 15 | 15% | 13 | 11% |
| G2 | 136 | 53% | 96 | 57% | 63 | 62% | 82 | 68% |
| G3 | 91 | 36% | 46 | 27% | 24 | 23% | 25 | 21% |
| ET response rate§ | NA | NA | 53% | 72% | ||||
| CT response rate | ||||||||
| pCR breast/axilla | 8.8% | 6.7% | NA | NA | ||||
| PAM50 | ||||||||
| Luminal A | 141 | 52% | 54 | 30% | 37 | 36% | - | - |
| Luminal B | 102 | 38% | 105 | 58% | 20 | 19% | - | - |
| HER2-E | 6 | 2% | 7 | 4% | 12 | 12% | - | - |
| Basal-like | 7 | 2% | 14 | 8% | 4 | 4% | - | - |
| Normal-like | 16 | 6% | - | - | 30 | 29% | - | - |
ET, endocrine therapy; CT, chemotherapy.
Edinburgh dataset has 75 patients without clinical HER2 status.
The definition of ET response is different in the Marsden and Edinburgh datasets. Clinical tumor response (complete and partial response versus stable and progressive disease) was used as the endpoint in the Marsden dataset. Response was evaluated by imaging ultrasound in the Edinburgh dataset. Clinical tumor response was defined as tumor volume shrinkage of at least 70% by 90 days of treatment.
Table 3.
CES association with chemotherapy sensitivity in the MDACC-based dataset.
| Univariate Analysis |
Multivariable Analysis |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Signatures | N | pCR rate |
OR | Lower 95% |
Upper 95% |
p- value |
OR | Lower 95% |
Upper 95% |
p- value |
| Age (cont. variable) | - | - | 1.0 | 0.93 | 1.02 | 0.251 | 1.0 | 0.92 | 1.02 | 0.205 |
| Tumor size | ||||||||||
| T0-T2 | 153 | 8% | 1.0 | - | - | - | 1.0 | - | - | - |
| T3-T4 | 107 | 9% | 1.1 | 0.47 | 2.63 | 0.813 | 0.6 | 0.22 | 1.70 | 0.341 |
| Nodal status | ||||||||||
| N0 | 96 | 7% | 1.0 | - | - | - | 1.0 | - | - | - |
| N1 | 125 | 9% | 1.2 | 0.46 | 3.29 | 0.685 | 0.9 | 0.30 | 2.78 | 0.882 |
| N2-3 | 39 | 13% | 1.9 | 0.56 | 6.29 | 0.312 | 1.0 | 0.24 | 4.51 | 0.965 |
| Grade | ||||||||||
| 1 | 26 | 4% | 1.0 | - | - | - | 1.0 | - | - | - |
| 2 | 130 | 4% | 1.0 | 0.11 | 8.93 | 1.000 | 0.7 | 0.07 | 6.88 | 0.753 |
| 3 | 89 | 17% | 5.1 | 0.64 | 40.34 | 0.125 | 1.8 | 0.18 | 18.42 | 0.608 |
| PAM50 | ||||||||||
| Luminal A | 134 | 3% | 1.0 | - | - | - | 1.0 | - | - | - |
| Luminal B | 99 | 15% | 5.8 | 1.86 | 18.08 | 0.002 | 1.2 | 0.25 | 6.28 | 0.792 |
| HER2-E | 6 | 0% | 0.0 | - | - | 0.989 | 0.0 | - | - | 0.991 |
| Basal-like | 7 | 29% | 13.0 | 1.91 | 88.50 | 0.009 | 0.4 | 0.02 | 9.97 | 0.586 |
| Normal-like | 14 | 14% | 5.4 | 0.90 | 32.69 | 0.065 | 1.7 | 0.23 | 12.75 | 0.602 |
| CES | ||||||||||
| CES-E | 134 | 2% | 1.0 | - | - | - | - | - | - | - |
| CES-U | 67 | 9% | 4.3 | 1.04 | 17.75 | 0.044 | - | - | - | - |
| CES-C | 59 | 24% | 13.6 | 3.73 | 49.46 | <0.001 | - | - | - | - |
| CES (cont. variable) | - | - | 0.2 | 0.08 | 0.40 | <0.001 | 0.2 | 0.03 | 0.77 | 0.022 |
Six gene expression-based signatures (i.e. PAM50 proliferation score, ROR-P, genomic grade index, SET index, chemopredictor, DLDA30 and residual cancer burden [RCB] predictor) have been previously reported in this dataset20. In addition, we applied a microarray-based version of OncotypeDX Recurrence Score36,37. Here, we evaluated the performance of CES to predict pCR within HR+/HER2- disease compared with these 7 gene signatures. Interestingly, CES provided the highest aROC (Table S4–12) either as a continuous variable (aROC=0.770) or as group categories (aROC=0.765). The second most predictive signature was the RCB predictor (aROC=0.740). Of note, RCB predictor was trained using 165 of 272 (60.7%) HR+/HER2- samples from this data set (i.e. the training dataset). When these training samples were removed, CES showed a higher performance either as a continuous variable (aROC=0.805) or as group categories (aROC=0.786) than RCB predictor (aROC=0.640).
Malaga-based dataset
We evaluated a dataset of 180 patients with HR+/HER2- disease treated with anthracycline/taxane-based neoadjuvant chemotherapy (Table 2). In this dataset, 46.1%, 16.1% and 37.8% of the samples were identified as CES-E, -U and -C, respectively. The pCR and RCB 0/1 rates across the CES-E, -U and -C groups were 2.4%/9.6%, 3.4%/17.2% and 13.2%/30.9%, respectively (P=0.022 and 0.004).
To test the ability of CES to predict chemotherapy response independently of known clinical-pathological variables and intrinsic subtype, we performed a multivariable logistic regression analysis using RCB (0/1 vs. 2/3) as the endpoint since only 12 samples achieved a RCB 0 (i.e. pCR) in this dataset. The results revealed that CES provided independent predictive information beyond intrinsic subtype (Table 4), Ki-67 by IHC (Table S13) and PAM50 ROR score (Table S14). The aROC of CES for predicting RCB 0/1 was 0.746. Finally, we observed a significant association between CES and Miller-Payne response data (Fig. S1).
Table 4.
CES association with chemotherapy sensitivity in the Malaga dataset.
| Univariate Analysis |
Multivariable Analysis |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Signatures | N | RCB0/1 rate |
OR | Lower 95% |
Upper 95% |
p- value |
OR | Lower 95% |
Upper 95% |
p-value |
| Age (cont. variable) | - | - | 1.0 | 0.95 | 1.02 | 0.331 | 1.0 | 0.96 | 1.07 | 0.599 |
| Tumor size | ||||||||||
| T0-T2 | 133 | 22% | 1.0 | - | - | - | 1.0 | - | - | - |
| T3-T4 | 39 | 10% | 0.4 | 0.13 | 1.25 | 0.116 | 0.4 | 0.09 | 1.90 | 0.260 |
| Grade | ||||||||||
| 1 | 27 | 7% | 1.0 | - | - | - | 1.0 | - | - | - |
| 2 | 96 | 16% | 2.3 | 0.50 | 10.82 | 0.286 | 1.6 | 0.26 | 9.31 | 0.625 |
| 3 | 46 | 35% | 6.7 | 1.40 | 31.82 | 0.017 | 3.0 | 0.40 | 23.34 | 0.283 |
| PAM50 | ||||||||||
| Luminal A | 54 | 9% | 1.0 | - | - | - | 1.0 | - | - | - |
| Luminal B | 105 | 20% | 2.4 | 0.87 | 6.91 | 0.090 | 0.9 | 0.19 | 4.34 | 0.905 |
| HER2-E | 7 | 14% | 1.6 | 0.16 | 16.43 | 0.677 | 0.1 | 0.00 | 3.19 | 0.188 |
| Basal-like | 14 | 50% | 9.8 | 2.43 | 39.51 | 0.001 | 0.1 | 0.00 | 3.40 | 0.214 |
| CES | ||||||||||
| CES-E | 83 | 10% | 1.0 | - | - | - | - | - | - | - |
| CES-U | 29 | 17% | 2.0 | 0.58 | 6.54 | 0.277 | - | - | - | - |
| CES-C | 68 | 31% | 4.2 | 1.72 | 10.22 | 0.002 | - | - | - | - |
| CES (cont. variable) | - | - | 0.2 | 0.09 | 0.44 | <0.001 | 0.2 | 0.07 | 0.76 | 0.016 |
Marsden-based dataset: CES and endocrine sensitivity
We evaluated a dataset of 103 post-menopausal patients with HR+ disease treated with anastrozole for 16 weeks in the neoadjuvant setting (Table 2). In this dataset, 23.5%, 34.3% and 42.2% of samples were identified as CES-E, -U and -C, respectively. Clinical tumor response (complete and partial response versus stable and progressive disease) was used as the endpoint. No pCR was observed in this dataset. The rates of clinical tumor response across the CES-E, -U and -C groups were 75.0%, 48.6% and 44.2%, respectively (P=0.043). CES was found to be the only variable significantly associated with response (Table S15), independently of HER2 status (Tables S15–16).
Edinburgh-based dataset: CES and endocrine sensitivity
We evaluated a dataset of 120 post-menopausal patients with HR+ disease treated with letrozole for at least 12 weeks in the neoadjuvant setting (Fig. S2A). Two patients of 120 achieved a complete response. Similar to previous results, CES as a continuous variable was found to be the only variable significantly associated with a ≥70% reduction in tumor volume by 90 days (Fig. S2B), even within HER2-negative disease (Fig. S2C).
Prognosis, intrinsic subtype and chemo-endocrine sensitivity
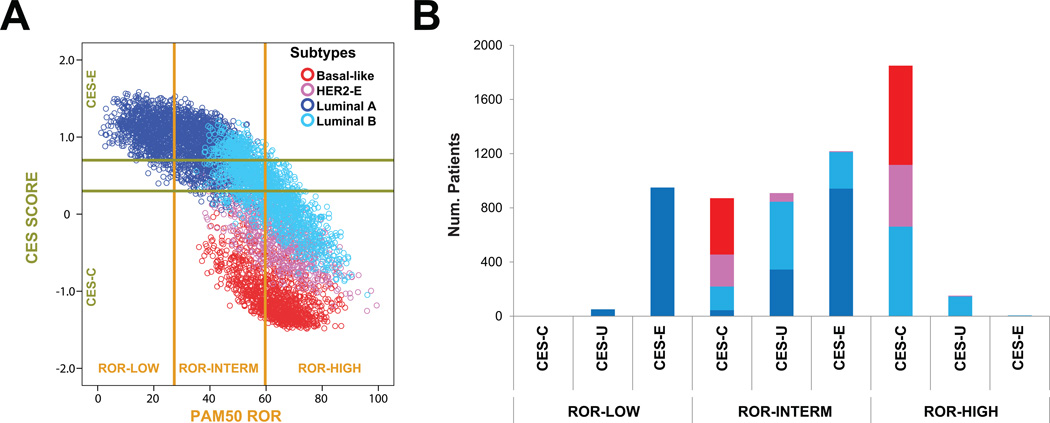
To better understand the relationship between prognosis, intrinsic biology and chemo-endocrine sensitivity, we pulled together PAM50 data from many different datasets for a total of 6,007 primary breast cancers representing all subtypes (Fig. 2). The results revealed that in the ROR-low group, 94.9% of cases were identified as CES-E and 100% were of the Luminal A subtype. In the ROR-high, 92.1% of the samples were identified as CES-C; non-luminal and Luminal B subtypes represented 64.3% and 35.7% of the ROR-high/CES-C cases, respectively.
Fig 2.
Prognosis (PAM50 ROR), intrinsic subtype and CES in 6,007 primary breast cancers. (A) A scatter plot of CES score and ROR score, colored by subtype, is shown. The two horizontal lines indicate the cutoffs of each CES group. The two vertical lines indicate the cutoffs of each PAM50 ROR group. (B) Number of patients in each CES group based on ROR. Each bar is colored by subtype.
In the ROR-intermediate group, high heterogeneity was observed. In terms of intrinsic biology, Luminal A, Luminal B and non-Luminal subtypes represented 44.4%, 31.5% and 24.1%, respectively. In terms of chemo/endocrine-sensitivity, CES-E, CES-U and CES-C represented 40.6%, 30.3% and 29.1%, respectively. As expected, the vast majority of ROR-intermediate/CES-E samples (77.3%) were of the Luminal A subtype.
Survival outcome of CES within HR+/ROR-intermediate disease
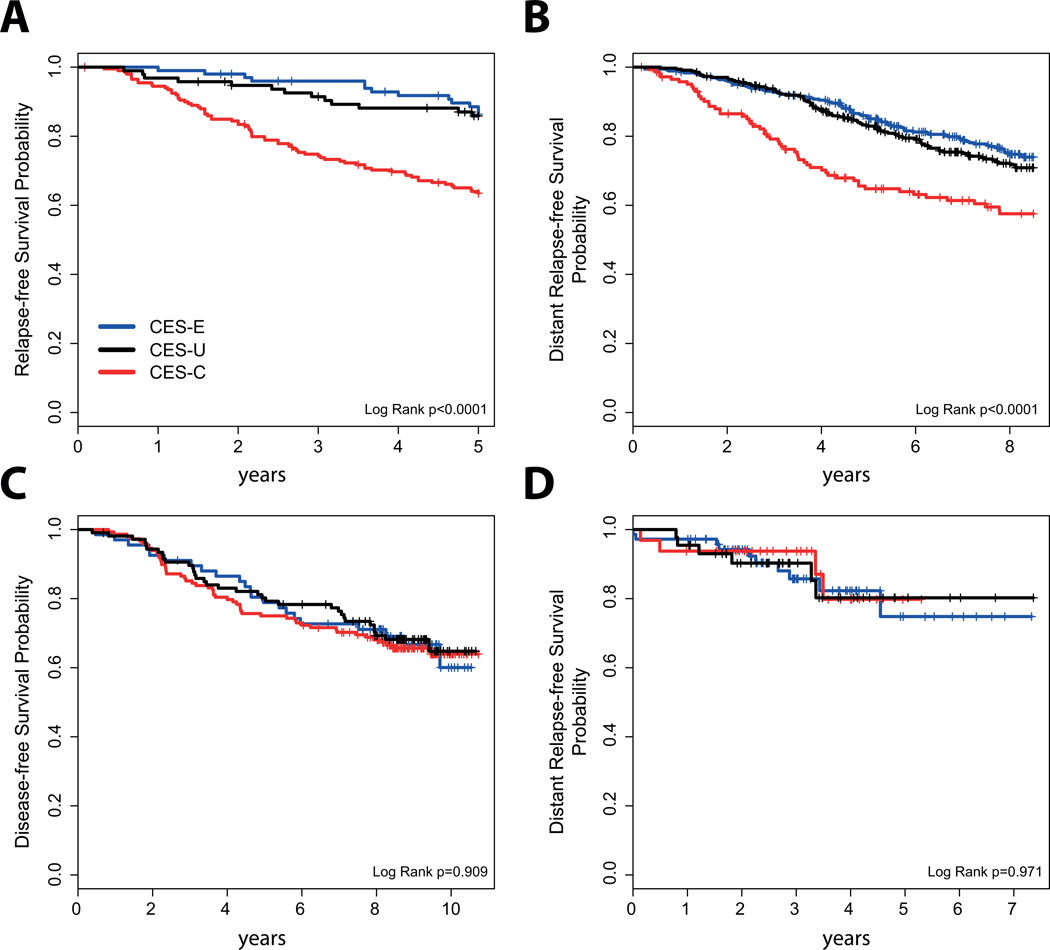
To continue exploring the value of CES within HR+/ROR-intermediate disease, we evaluated the association of CES with survival outcome in HR+/ROR-intermediate early breast cancer in 4 independent datasets of patients treated with no adjuvant systemic therapy (n=189), adjuvant tamoxifen-only (n=846) or adjuvant chemotherapy and endocrine therapy (n=322 and n=148).
In patients with node-negative disease treated without adjuvant systemic therapy, CES (as a continuous variable or as group categories) was found significantly associated with distant relapse-free survival (Fig. 3A). The hazard ratio between the CES-C group vs the CES-E group was 2.68 (0.163–0.858 95% confidence interval). Similar results were obtained in the dataset where patients were treated with adjuvant tamoxifen-only (Fig. 3B). However, CES (as a continuous variable or as group categories) was not found significantly associated with survival outcome in 2 independent cohorts of patients treated with (neo)adjuvant chemotherapy and endocrine therapy (Fig. 3C and D).
Fig 3.
Survival outcomes in HR+ early breast cancer with ROR-intermediate disease. (A) Node-negative disease treated without adjuvant systemic therapy. (B) Node-negative and node-positive disease treated with adjuvant tamoxifen-only. (C) Node-positive disease treated with adjuvant chemotherapy and endocrine therapy in the GEICAM/9906 clinical trial. (D) Node-negative and node-positive disease treated with neoadjuvant chemotherapy and adjuvant endocrine therapy.
Discussion
Our results are the first to confirm, in a randomized setting, an inverse relationship of endocrine and chemotherapy sensitivity in ER+ breast cancer. Previous evidence has suggested an inverse relationship of proliferation- and ER-related biological processes regarding endocrine and chemotherapy sensitivity of ER+ breast cancer. For example, two independent studies showed an inverse correlation between a 200-gene ER reporter score, or between TAU expression, an ER-related gene, and endocrine sensitivity and chemosensitivity38,39. In addition, high recurrence score measured by Oncotype DX (Genomic Health, Inc., Redwood, CA) predicted little or no benefit from adjuvant tamoxifen therapy in the NSABP-B14 trial, but at the same time also predicted substantial benefit from adjuvant CMF chemotherapy in the NSABP-B20 trial40,41. These results fit with our results showing that virtually all patients with ROR-high disease are identified as CES-C; however, our data also highlights that within ROR-high/CES-C disease not all ER+/HER2- samples are luminal (i.e. Luminal A or B) since non-luminal disease (i.e. Basal-like and HER2-enriched) can also be identified. According to our results (Fig. 2), the chemotherapy benefit of ROR-high/non-luminal tumors within HR+/HER2- disease is likely even greater than ROR-high/Luminal B tumors.
Our results also suggest that a main driver of endocrine therapy sensitivity and chemotherapy sensitivity within ER+/HER2- disease is the Basal-like versus Luminal A intrinsic biology. To capture both biological states in each individual sample, we calculated the correlation coefficients of each sample to both PAM50-centroids (i.e. Luminal A and Basal-like) and then subtracted both coefficients. Thus, instead of choosing a gene signature (e.g. a proliferation-based signature) of the many signatures that can discriminate between both subtypes in one way or another, we decided to incorporate into a score the Basal-like vs. Luminal A intrinsic state of each tumor as identified by the PAM50 subtype predictor. Of note, the PAM50 genes were originally selected for their ability to capture the intrinsic biology displayed by 1,900 genes (i.e. the so-called intrinsic gene list). In fact, in the TCGA, intrinsic subtype defined by PAM50 captured the vast majority of the biological diversity displayed by most molecular data-types analyzed1.
From a clinical perspective, our results support current breast cancer guidelines for the systemic treatment of early HR+/HER2- breast cancer. On one hand, patients with a low-ROR score and a low tumor burden (i.e. <10% risk of distant relapse at 10 years) are recommended to be treated with endocrine therapy-only42. Indeed, our results suggest that these patients have tumors that are highly endocrine sensitive and have low chemotherapy sensitivity. On the other hand, patients with high-risk HR+/HER2- disease are recommended to be treated with endocrine therapy and chemotherapy. According to our analysis, this group is the one with high chemotherapy benefit and low endocrine benefit. Regarding endocrine therapy in this group, the main issue is that we do not have survival data suggesting that CES-C tumors do not benefit at all from endocrine therapy. Therefore, withdrawal of a potentially efficacious treatment strategy such as endocrine therapy in a patient with an ER+ tumor (as defined by the ASCO/CAP guidelines) that is identified as CES-C or ROR-high should not be recommended today, although in patients whose tumors contain low levels of ER (1% to 10%), ASCO/CAP recommend to discuss the pros and cons of endocrine therapy. A large randomized adjuvant trial involving thousands of patients to answer this particular question is unlikely to happen.
Although the clinical implications of CES in low and high risk HR+/HER2- disease are minimal, the observation that intermediate risk HR+/HER2- disease, which represents ~30% of newly diagnosed breast cancer, is biologically heterogeneous with a range of chemotherapy sensitivities might have implications for the interpretation of two ongoing prospective clinical trials. In the TailorX phase III trial, 4,500 patients with HR+/HER2- node-negative early breast cancer with intermediate RS have been randomized to adjuvant chemotherapy or no chemotherapy. According to our analysis, this intermediate group might be composed of at least 3 groups with different chemotherapy sensitivities. Of note, the CES-U group seems to be a genuine grey area where decisions regarding the need of chemotherapy might be difficult. A similar situation might occur in the RxPONDER phase III clinical trial where patients with HR+/HER2- early breast cancer, and 1–3-positive lymph nodes, with low/intermediate risk are being randomized to adjuvant chemotherapy or not. A potential explanation is that OncotypeDX RS, as well as other prognostic gene expression-based tests, such as PAM50 ROR or MammaPrint43, have been specifically designed or trained to predict outcome and not intrinsic tumor biology or treatment sensitivity. Although a strong negative correlation is observed between ROR (risk) and CES (drug sensitivity), there are substantial differences between them at the individual level (~40% discordance).
There are several caveats to our study. First, this is a retrospective study involving heterogeneous patient populations and the results need to be confirmed in a prospective clinical trial(s). Second, although the data presented here validates CES from a clinical perspective, further analytical validation will be needed since in most datasets, except the Malaga set, the research-based version of PAM50 was used. However, the fact that CES (as a continuous variable and the 2 cut-points) predicted pCR in the Malaga set suggests that analytical validation of this biomarker is feasible. Third, we did not evaluate the association of CES with survival data from a randomized clinical trial of adjuvant chemotherapy vs no adjuvant chemotherapy, or adjuvant endocrine therapy versus no adjuvant endocrine therapy. Thus, the predictive value of these signatures was only evaluated in the neoadjuvant setting where different tumor response endpoints were evaluated, most of which have been associated with patient survival18,44. Fourth, some of the signatures evaluated in the MDACC-based dataset, such as OncotypeDX recurrence score or genomic grade index, were derived from microarray-based data and thus are not the commercially available versions. Fifth, we were not able to demonstrate a consistent association of CES with endocrine response in HR+ disease after excluding the HER2-positive cases. In the Edinburgh dataset, HER2 status was not available for all patients. Although we derived an ERBB2 expression-based surrogate definition of HER2 status and showed that CES is independently associated with response, this was not prespecified and does not meet REMARK guidelines. In addition, the association of CES with endocrine response did not reach statistical significance (p=0.09) in patients with HR+/HER2-negative disease in the Marsden dataset. Finally, patients from each of the datasets received different anthracycline/taxane-based chemotherapy regimens, schedules and doses, and thus the ability of the signatures to predict response to particular chemotherapeutics or treatment regimens could not be tested.
Another important consideration of our study is that we did not attempt to identify an optimal cutoff(s) for CES but rather focused on the association of the continuous expression of CES with each endpoint. The main reason is that different gene expression-based platforms and protocols were used in each cohort and thus, standardization of a biomarker cut-point would have been difficult to achieve and most likely unreliable. In any case, the fact that all four testing sets gave very similar associations, and were found independently of the platform/protocol used, argues in favor of a robust finding.
To conclude, CES is a single genomic signature capable of measuring chemo-endocrine sensitivity in HR+/HER2- breast cancer beyond intrinsic subtype, other genomic signatures, and the standard pathology variables. CES could be of particular clinical value in patients with HR+/HER2- intermediate risk disease where the benefit of adjuvant multi-agent chemotherapy is unclear.
Supplementary Material
Statement of translational relevance.
Hormone receptor-positive (HR+) breast cancer is clinically and biologically heterogeneous and subgroups with different prognostic and treatment sensitivities need to be identified. Here, we present the development and clinical validation across multiple studies of a gene expression-based predictor, based on the well-known PAM50 assay, is associated with chemotherapy and endocrine therapy response in early breast cancer beyond PAM50 Risk of Relapse (ROR) and intrinsic subtypes. The potential clinical utility of this PAM50-based chemo-endocrine score (CES) predictor might be in the PAM50 ROR-intermediate, where the proportion of each CES group (endocrine-sensitive, intermediate and chemo-sensitive) is more than 25%. This is important as we await the results of large adjuvant clinical trials such as TailorX or RxPonder that have randomized patients with intermediate risk to adjuvant chemotherapy or not.
Acknowledgments
This work was supported by funds from the NCI Breast SPORE program (P50-CA58223-09A1), by RO1- CA148761 (C.M.P.), by Instituto de Salud Carlos III - PI13/01718 (A.P.), by a Career Catalyst Grant from the Susan Komen Foundation (A.P.), by Breast Cancer Now (formerly Breakthrough Breast Cancer), by the Royal Marsden NIHR Biomedical Research Centre, by Banco Bilbao Vizcaya Argentaria (BBVA) Foundation (A.P.) and by the Breast Cancer Research Foundation. This work was also supported by funds from FEDER (RETICC): RD12/0036/0076 (JA), RD12/0036/0051 (JA), RD12/0036/0070 (AL) and RD12/0036/0076 (MM). The funders did not have any role in the study design, conduct, or decision to submit the manuscript for publication.
Footnotes
Additional files: One supplemental file.
Competing interests. C.M.P is an equity stock holder of BioClassifier LLC and University Genomics. C.M.P. has filed a patent on the PAM50 assay. Uncompensated advisory role of A.P. and M.M. for Nanostring Technologies. The other authors declare that they have no competing interests.
Authors contributions. Study conception and design: AP, EA and CMP. Acquisition of data: all authors. Analysis and interpretation of data: all authors. All authors were involved in drafting the article or revising it critically for important intellectual content, and approved the final version of the manuscript.
References
- 1.TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prat A, Cheang MCU, Martín M, Parker JS, Carrasco E, Caballero R, et al. Prognostic Significance of Progesterone Receptor-Positive Tumor Cells Within Immunohistochemically Defined Luminal A Breast Cancer. Journal of Clinical Oncology. 2013;31:203–209. doi: 10.1200/JCO.2012.43.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ades F, Zardavas D, Bozovic-Spasojevic I, Pugliano L, Fumagalli D, de Azambuja E, et al. Luminal B Breast Cancer: Molecular Characterization, Clinical Management, and Future Perspectives. Journal of Clinical Oncology. 2014;32:2794–2803. doi: 10.1200/JCO.2013.54.1870. [DOI] [PubMed] [Google Scholar]
- 4.Prat A, Perou CM. Deconstructing the molecular portraits of breast cancer. Mol Oncol. 2011;5:5–23. doi: 10.1016/j.molonc.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prat A, Ellis MJ, Perou CM. Practical implications of gene-expression-based assays for breast oncologists. Nat Rev Clin Oncol. 2012;9:48–57. doi: 10.1038/nrclinonc.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 7.Martín M, Prat A, Rodríguez-Lescure Á, Caballero R, Ebbert MW, Munárriz B, et al. PAM50 proliferation score as a predictor of weekly paclitaxel benefit in breast cancer. Breast Cancer Research and Treatment. 2013;138:457–466. doi: 10.1007/s10549-013-2416-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prat A, Carey LA, Adamo B, Vidal M, Tabernero J, Cortés J, et al. Molecular Features and Survival Outcomes of the Intrinsic Subtypes Within HER2-Positive Breast Cancer. Journal of the National Cancer Institute. 2014;106 doi: 10.1093/jnci/dju152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Usary J, Zhao W, Darr D, Roberts PJ, Liu M, Balletta L, et al. Predicting Drug Responsiveness in Human Cancers Using Genetically Engineered Mice. Clinical Cancer Research. 2013;19:4889–4899. doi: 10.1158/1078-0432.CCR-13-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Minckwitz G, Untch M, Blohmer J-U, Costa SD, Eidtmann H, Fasching PA, et al. Definition and Impact of Pathologic Complete Response on Prognosis After Neoadjuvant Chemotherapy in Various Intrinsic Breast Cancer Subtypes. Journal of Clinical Oncology. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 11.Prat A, Parker JS, Fan C, Perou CM. PAM50 assay and the three-gene model for identifying the major and clinically relevant molecular subtypes of breast cancer. Breast Cancer Research and Treatment. 2012;135:301–306. doi: 10.1007/s10549-012-2143-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prat A, Fan C, Fernández A, Hoadley KA, Martinello R, Vidal M, et al. Response and survival of breast cancer intrinsic subtypes following multi-agent neoadjuvant chemotherapy. BMC Medicine. 2015;13:1–11. doi: 10.1186/s12916-015-0540-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellis MJ, Suman VJ, Hoog J, Lin L, Snider J, Prat A, et al. Randomized Phase II Neoadjuvant Comparison Between Letrozole, Anastrozole, and Exemestane for Postmenopausal Women With Estrogen Receptor-Rich Stage 2 to 3 Breast Cancer: Clinical and Biomarker Outcomes and Predictive Value of the Baseline PAM50-Based Intrinsic Subtype—ACOSOG Z1031. Journal of Clinical Oncology. 2011;29:2342–2349. doi: 10.1200/JCO.2010.31.6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunbier AK, Anderson H, Ghazoui Z, Salter J, Parker JS, Perou CM, et al. Association between breast cancer subtypes and response to neoadjuvant anastrozole. Steroids. 2011;76:736–740. doi: 10.1016/j.steroids.2011.02.025. [DOI] [PubMed] [Google Scholar]
- 15.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thürlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Annals of Oncology. 2013 doi: 10.1093/annonc/mdt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alba E, Calvo L, Albanell J, De la Haba JR, Arcusa Lanza A, Chacon JI, et al. Chemotherapy (CT) and hormonotherapy (HT) as neoadjuvant treatment in luminal breast cancer patients: results from the GEICAM/2006-03, a multicenter, randomized, phase-II study. Annals of Oncology. 2012 doi: 10.1093/annonc/mds132. [DOI] [PubMed] [Google Scholar]
- 17.Wolff AC, Hammond MEH, Schwartz JN, Hagerty KL, Allred DC, Cote RJ, et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. Journal of Clinical Oncology. 2006;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 18.Ogston KN, Miller ID, Payne S, Hutcheon AW, Sarkar TK, Smith I, et al. A new histological grading system to assess response of breast cancers to primary chemotherapy: prognostic significance and survival. The Breast. 2003;12:320–327. doi: 10.1016/s0960-9776(03)00106-1. [DOI] [PubMed] [Google Scholar]
- 19.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotech. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 20.Hatzis C, Pusztai L, Valero V, et al. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. Jama. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prat A, Galván P, Jimenez B, Buckingham W, Jeiranian HA, Schaper C, et al. Prediction of Response to Neoadjuvant Chemotherapy Using Core Needle Biopsy Samples with the Prosigna Assay. Clinical Cancer Research. 2015 doi: 10.1158/1078-0432.CCR-15-0630. [DOI] [PubMed] [Google Scholar]
- 22.Dunbier AK, Anderson H, Ghazoui Z, Folkerd EJ, A’Hern R, Crowder RJ, et al. Relationship Between Plasma Estradiol Levels and Estrogen-Responsive Gene Expression in Estrogen Receptor-Positive Breast Cancer in Postmenopausal Women. Journal of Clinical Oncology. 2010;28:1161–1167. doi: 10.1200/JCO.2009.23.9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith IE, Walsh G, Skene A, Llombart A, Mayordomo JI, Detre S, et al. A Phase II Placebo-Controlled Trial of Neoadjuvant Anastrozole Alone or With Gefitinib in Early Breast Cancer. Journal of Clinical Oncology. 2007;25:3816–3822. doi: 10.1200/JCO.2006.09.6578. [DOI] [PubMed] [Google Scholar]
- 24.Turnbull AK, Arthur LM, Renshaw L, Larionov AA, Kay C, Dunbier AK, et al. Accurate Prediction and Validation of Response to Endocrine Therapy in Breast Cancer. Journal of Clinical Oncology. 2015 doi: 10.1200/JCO.2014.57.8963. [DOI] [PubMed] [Google Scholar]
- 25.Fan C, Prat A, Parker JS, Liu Y, Carey LA, Troester MA, et al. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Medical Genomics. 2011;4:1–15. doi: 10.1186/1755-8794-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prat A, Parker JS, Fan C, Cheang MCU, Miller LD, Bergh J, et al. Concordance among gene expression-based predictors for ER-positive breast cancer treated with adjuvant tamoxifen. Annals of Oncology. 2012;23:2866–2873. doi: 10.1093/annonc/mds080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen TO, Parker JS, Leung S, Voduc D, Ebbert M, Vickery T, et al. A Comparison of PAM50 Intrinsic Subtyping with Immunohistochemistry and Clinical Prognostic Factors in Tamoxifen-Treated Estrogen Receptor-Positive Breast Cancer. Clin Cancer Res. 2010;16:5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prat A, Lluch A, Albanell J, Barry WT, Fan C, Chacon JI, et al. Predicting response and survival in chemotherapy-treated triple-negative breast cancer. Br J Cancer. 2014;111:1532–1541. doi: 10.1038/bjc.2014.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prat A, Ellis MJ, Perou CM. Practical implications of gene-expression-based assays for breast oncologists. Nat Rev Clin Oncol. 2012;9 doi: 10.1038/nrclinonc.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Horak CE, Pusztai L, Xing G, Trifan OC, Saura C, Tseng L-M, et al. Biomarker Analysis of Neoadjuvant Doxorubicin/Cyclophosphamide Followed by Ixabepilone or Paclitaxel in Early-Stage Breast Cancer. Clinical Cancer Research. 2013;19:1587–1595. doi: 10.1158/1078-0432.CCR-12-1359. [DOI] [PubMed] [Google Scholar]
- 33.Fan C, Prat A, Parker J, Liu Y, Carey L, Troester M, et al. Building prognostic models for breast cancer patients using clinical variables and hundreds of gene expression signatures. BMC Medical Genomics. 2011;4:3. doi: 10.1186/1755-8794-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: database for annotation, visualization, and Integrated discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- 35.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fan C, Oh DS, Wessels L, Weigelt B, Nuyten DSA, Nobel AB, et al. Concordance among Gene-Expression-Based Predictors for Breast Cancer. New England Journal of Medicine. 2006;355:560–569. doi: 10.1056/NEJMoa052933. [DOI] [PubMed] [Google Scholar]
- 37.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A Multigene Assay to Predict Recurrence of Tamoxifen-Treated, Node-Negative Breast Cancer. New England Journal of Medicine. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 38.Symmans WF, Hatzis C, Sotiriou C, Andre F, Peintinger F, Regitnig P, et al. Genomic Index of Sensitivity to Endocrine Therapy for Breast Cancer. Journal of Clinical Oncology. 2010;28:4111–4119. doi: 10.1200/JCO.2010.28.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andre F, Hatzis C, Anderson K, Sotiriou C, Mazouni C, Mejia J, et al. Microtubule-Associated Protein-tau is a Bifunctional Predictor of Endocrine Sensitivity and Chemotherapy Resistance in Estrogen Receptor-Positive Breast Cancer. Clinical Cancer Research. 2007;13:2061–2067. doi: 10.1158/1078-0432.CCR-06-2078. [DOI] [PubMed] [Google Scholar]
- 40.Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A Multigene Assay to Predict Recurrence of Tamoxifen-Treated, Node-Negative Breast Cancer. New England Journal of Medicine. 2004;351:2817–2826. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 41.Paik S, Tang G, Shak S, Kim C, Baker J, Kim W, et al. Gene Expression and Benefit of Chemotherapy in Women With Node-Negative, Estrogen Receptor-Positive Breast Cancer. Journal of Clinical Oncology. 2006;24:3726–3734. doi: 10.1200/JCO.2005.04.7985. [DOI] [PubMed] [Google Scholar]
- 42.Harris LN, Ismaila N, McShane LM, Andre F, Collyar DE, Gonzalez-Angulo AM, et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. Journal of Clinical Oncology. 2016 doi: 10.1200/JOP.2016.010868. [DOI] [PubMed] [Google Scholar]
- 43.van ‘t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 44.Cortazar P, Zhang L, Untch M, Mehta K, Costantino JP, Wolmark N, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. The Lancet. 384:164–172. doi: 10.1016/S0140-6736(13)62422-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.