Abstract
Amyloid β (Aβ) oligomers may play a decisive role in Alzheimer’s disease related neurodegeneration, but their structural properties are poorly understood. In this report, sedimentation velocity centrifugation, small angle neutron scattering (SANS) and molecular modelling were used to identify the small oligomeric species formed by the 42 amino acid residue long isoform of Aβ (Aβ42) in solution, characterized by a sedimentation coefficient of 2.56 S, and a radius of gyration between 2 and 4 nm. The measured sedimentation coefficient is in close agreement with the sedimentation coefficient calculated for Aβ42 hexamers using MD simulations at µM concentration. To the best of our knowledge this is the first report detailing the Aβ42 oligomeric species by SANS measurements. Our results demonstrate that the smallest detectable species in solution are penta- to hexamers. No evidences for the presence of dimers, trimers or tetramers were found, although the existence of those Aβ42 oligomers at measurable quantities had been reported frequently.
Introduction
An increasing number of human diseases are characterized by the accumulation of specific protein aggregates found in proteinaceous depositions with a common structural motif, which is characterized by a cross-β-sheet architecture called the amyloid fold1. While these amyloid fibril folds are already clarified for a number of proteins (reviewed in ref. 2), little is known about the first stages of the amyloid formation process, where dynamic, heterogeneous and often toxic intermediates are most likely populated. Such oligomeric species appear to be currently elusive to high-resolution structural methods such as solution based NMR or X-ray crystallography. Aside from the size limit of solution based NMR, the rather low fraction of the quested oligomer, its transient nature and heterogeneity renders NMR measurements at least extremely difficult3. Only with stabilized oligomeric preparations such studies have been feasible thus far4. Remarkably, Kotler et al. recently succeeded in the structural characterization of an Aβ40 oligomer of 5 to 15 nm size contributing to only 7% of the sample by magic angle spinning recoupling 1H-1H NMR experiments5. The above mentioned solution heterogeneity also interferes with the formation of diffracting crystals as required for X-ray crystallography.
In the case of Alzheimer’s disease (AD), which is the most common form of dementia worldwide, the accumulation of aggregated amyloid β protein (Aβ) in brain tissue is one of the disease hallmarks6. Aβ is generated as a proteolytic fragment from the transmembrane amyloid β-protein precursor (APP). Among the various isoforms that arise from variation of the location of the termini or by post-translational modification, the 42 amino acid long isoform (Aβ42) is the main constituent of extracellular senile plaques found post mortem in AD brains7.
Soluble Aβ oligomers, rather than the deposited fibrillar forms, are considered by many to be responsible for neurodegeneration in AD patients8, 9. The smallest Aβ assemblies reported in the literature are dimers, which exist in body fluids and brain tissues of AD patients10, 11. It should be noted that the finding of brain derived neurotoxic dimers by Shankar et al.10 had been refuted by later work12. Several studies of in vitro and in vivo experiments indicate the existence of a monomer-dimer equilibrium for Aβ13, 14. In photo-induced cross-linking of unmodified proteins (PICUP) experiments using Aβ, covalently linked dimers and other larger oligomeric species12 were detected for Aβ42 and for Aβ40, the shorter isoform missing the last two c-terminal residues15, 16. While for Aβ40 trimers and tetramers were the abundant species, for Aβ42 penta-/hexamers and also dodecamers and octadecamers could be detected16. Based on the PICUP technique the involvement of a penta- to hexameric oligomers in the Aβ42 assembly process had been reported in several studies17–19. Additionally, non-cross-linked Aβ oligomers have been identified in polyacrylamide gel electrophoresis as sodium dodecyl sulphate (SDS)-resistant bands and interpreted as dimers to tetramers20. Cultured neuronal cells were found to release small Aβ oligomers, among which the dimer represents a major species, whose size was determined by SDS-PAGE combined with western blotting21–23. Furthermore, dimers were found along with monomers and trimers as stable components, which could be purified by size exclusion chromatography under denaturing conditions, in neuritic plaques obtained from AD brain material24. Other studies have identified Aβ paranuclei as penta- or hexamers, which were detected by mass spectrometry based techniques25, 26. Recently, characterization of an enriched Aβ42 oligomer by NMR combined with AFM was achieved27. Further information on Aβ oligomers can be found in recently published reviews28–34.
Considering the plethora of different experimental approaches, even those techniques which produced the most consistent results on the early assembly states of Aβ42, i.e., PICUP and ISM-MS, possess some shortcomings which should be addressed for the sake of gain in knowledge. Crosslinked oligomers are only well reported by PICUP if all subunits become stably connected to withstand SDS-denaturation prior to SDS-PAGE. Detergence below or above their critical micellar concentration might stabilize assemblies by shielding hydrophobic regions, which would not be stable in hydrous solutions. Ionization coupled with transfer to the gas phase as required for mass spectrometry, on the other hand, might be a selective as well as destructive process. Recently, a critical evaluation of PICUP followed by SDS-PAGE analysis performed by IMS-MS challenged the penta- to hexameric building block in Aβ42 aggregation35. A clear understanding of the early steps in the self-association of Aβ is critical in order to understand the formation of toxic oligomeric species and the initiation of fibrillisation in Alzheimer’s disease. We therefore chose an approach with high detection sensitivity, which is applicable to proteins in solution, for the characterization of the smallest oligomers of Aβ.
Recently, sedimentation velocity (SV) analysis, which is an application of analytical ultracentrifugation (AUC), was shown to be a highly sensitive detection method for trace amounts of aggregates in protein samples. Sedimentation velocity centrifugation allows distinguishing multiple sedimenting species in free solution while maintaining reversibly formed complexes in a bath of their components at all times. This permits the study of self-association as well as heterogeneous protein interactions36. Using SV, <1% aggregates in a sample can be reliably detected, provided instrument and equipment are handled with sufficient care37. SV analysis can determine the size- and shape distributions for self-assembling proteins38–41. Due to the fractionating property of the sedimentation process, with large particles sedimenting before small particles, the method is resistant to the presence of large particles such as unresolved protein aggregates or dust particles. The method is characterized by an excellent signal-to-noise ratio. The evolution of the shape of the formed sedimentation boundary over time reveals not only information about the sedimentation velocity but also about the diffusion properties of the analytes42, 43. Importantly, the method requires no interactions of Aβ with surfaces that might induce aggregation. SANS is a well-established, non-destructive method to examine structure on length scales of 1 to 1000 nm. While only one report on a SANS study of full length Aβ was found in PubMed44, which is about the characterization of Aβ40 at acidic solvent conditions, some more reports exist on small angle X-ray scattering (SAXS) studies of Aβ4245, 46. X-ray scattering has the disadvantage of being destructive to proteins and the generation of radicals might impair the aggregation process. In a previous study, we demonstrated the applicability of SANS using monomeric Aβ47. While in SV the hydrodynamic properties of macromolecules are evaluated, yielding the hydrodynamic radius, in SANS the properties of macromolecules as neutron scatterers are evaluated yielding also the radius of gyration, aside from information about size, shape and interactions. Because the two solution-based methods are exploiting different physical properties they are ideally suited to complement each other.
By SV analysis, we previously demonstrated the existence of discrete oligomeric species in the s-value range from 4 to 15 S during the lag phase of fibril formation48. Analysis reproducibly yielded two Aβ42 assembly species at low salt conditions and physiological pH, independent of the total Aβ42 concentration; one corresponding to a 12-mer and the other to an 18-mer of Aβ42. These sizes suggest the involvement of a trimeric or hexameric building block. A pentameric to hexameric building block could be evaluated by PICUP for Aβ42 and has been defined by Bitan et al. as paranucleus15, 16, 44. We interpreted the recently observed gap in the determined s-value distributions between 0.69 S, corresponding to the Aβ42 monomer and 4 S as the range for the nucleus size48. In this s-value region corresponding to molecular weights between 5 and 55 kDa, we hypothesize further on-pathway to fibril reaction intermediates. Here, we aim to close this gap by combining two techniques for the structural characterization of macromolecules in solution, namely SV analysis and small angle neutron scattering (SANS). Because the two solution-based methods are exploiting different physical properties they are ideally suited to corroborate each other in order to provide important information about the first steps of the nucleation process. In addition, we further complement our experimental results by including results from molecular dynamics (MD) simulations of Aβ42 assemblies, which bridge our SV and SANS results. This combined approach reveals clear experimental evidence for a penta- or hexameric assembly of Aβ42 in solution.
Results
The goal of this study is to identify metastable intermediates during the aggregation of Aβ42 in solution and thus to elucidate the initial steps of Aβ42 assembly.
Sedimentation velocity centrifugation allows sensitive detection of Aβ oligomers in solution
The first step in searching for small Aβ assemblies was to identify the smallest species, i.e., the dimers, and prove that SV analysis can discriminate them from monomers. As a model, a stable synthetic Aβ40 dimer was generated from a monomer with a cysteine residue at position 0, which formed an intermolecular disulphide bridge under oxidizing conditions. SV analysis revealed that the Aβ monomer and the covalently linked dimer are sufficiently different, with s-values of 0.65 and 0.9 S, respectively (Fig. S2). None of the SV experiments performed for non-linked Aβ42 provided evidence of an Aβ42 dimer in solution. If Aβ42 dimers or trimers were in rapid equilibrium with the monomers, one would expect that at higher Aβ42 concentrations the equilibrium were shifted from monomers towards the dimers and trimers. For such a sample, the peak in c(s) would not represent a single s-value species, but a so-called reaction boundary. A diagnostic for the identification of such a reaction boundary would be a shift towards higher s-values at higher Aβ42 concentrations or at least peak broadening. The superposition of the c(s) peaks for the monomer from different data sets revealed no Aβ42 concentration dependent shift in s-value nor peak broadening of the c(s) signal at 0.62 S. We thus conclude that the dimer fraction is below the detection limit.
The smallest detectable assembly state in the SV experiments is a penta- to hexamer
Experimental evidence for oligomeric species between trimers and 18-mers were found mainly from SDS-PAGE analysis after PICUP15, 16 and mass spectrometry studies. For example, a trimer is postulated as the building block for the Aβ*56 aggregate found in in-vivo studies by Lesné and colleagues49, 50, and trimers along with tetramers were also detected by Bernstein et al.17, 51. We decreased the total Aβ42 concentrations to the low micro- to nanomolar concentration range in our SV experiments, expecting that at Aβ42 concentrations below the critical concentration for the formation of higher-order structures, the assembly of larger assemblies from small oligomers is suppressed. The necessary increase in detection sensitivity was gained by using AUC coupled with a fluorescence detection system. Thus, a lower limit of about 100 nM fluorophore labelled Aβ42 was achieved. For the position of the fluorescence label we chose the N-terminus of Aβ, because the N-terminus is not primarily involved in intermolecular contacts required for aggregate formation52 and similar conjugates had been successfully used before53. Additionally, those regions of Aβ42, which had been identified as critical for paranucleus formation, i.e. the central hydrophobic core as well as the C-terminal residues15, are distantly located from the fluorophore. A linker further separates the fluorophore from the Aβ42 peptide and introduces flexibility. Moreover, charged fluorophores also possess a smaller propensity to stack on each other than uncharged fluorophores. From our AFM measurements (Fig. S3) we concluded that the fluorescence label neither prohibits fibril formation nor leads to extensive amorphous aggregate formation. Nevertheless, it cannot be fully excluded that the presence of a fluorescence label at the N-terminus favours certain paranuclei conformations, which would be less or not at all populated by the wild-type Aβ42 peptide. Thus, we conclude that the application of the fluorophore-labelled conjugate allow us to expand our measurements into the submicromolar Aβ42 concentration range and to draw conclusions that can be transferred with the above mentioned restrictions to wild-type Aβ42.
SV analysis is a technique, which recently gained special relevance in the determination of aggregate concentration in samples of pharmaceutically relevant proteins54–57. According to theoretical considerations at high signal-to-noise ratios of absorbance and fluorescence detection, a superior sensitivity for traces well below 1% of total signal is achievable in sedimentation velocity experiments57. For example; this would correspond to 0.01 absorbance units of an oligomer in the presence of 0.99 absorbance units of the monomer at a suitable wavelength. The limit of detection (0.2% of total signal) and the limit of quantification (about 2.8% of total signal) have been determined from practical studies on a dimeric antibody37. For confirmation of the detected high molecular weight species, the calculation of the c(s) distribution was extended by applying a Bayesian approach followed by F statistics, as reported recently by Wafer et al.54. After repeating the c(s) calculation by alternating between the simplex and Marquardt-Levenberg algorithms until no further change in rmsd was observed, the c(s) model is exchanged by a model of non-interacting species. This model can then be tested by F-statistics for the significance of the high molecular weight species. This was achieved by calculating a critical relative root mean square deviation (rmsd) value based on the model including the oligomeric species. A species is considered as a significant part of a sample if the rmsd of a fit excluding this species exceeds the critical relative rmsd. Thus the significance of the oligomeric species was shown for four data sets by observing an increase of the rmsd above the critical rmsd upon removal of the larger species (Fig. 1a,b).
Figure 1.
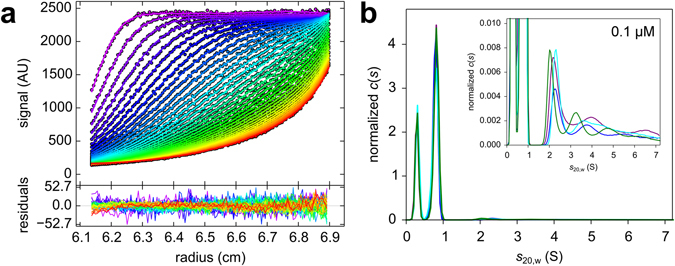
SV analysis of 0.1 µM AF488-Aβ42 with fluorescence detection reveals the existence of small oligomers in addition to the monomer. (a) Raw data of SV experiment with overlaid lines for the fitted data, showing a signal intensity of about 2500 RFU. Only every third scan, as well as every third data point is shown for clarity. In the box below the corresponding residuals within the fitted data range are shown. Rmsd for this sample was 11.86. (b) Area normalized c(s) results for four independent samples of 0.1 µM AF488-Aβ42 have been superimposed in the graph. The 0.8 S species is assigned to the monomer, the peak at 0.3 S is the result of either incomplete removal of unincorporated dye or the dark count rate or a combination of both. The inset shows the magnification of c(s) distributions. Sedimentation was performed at 60,000 rpm, 20 °C.
The total Aβ42 concentration was varied from 0.1 μM to 160 μM. All SV measurements were performed with Aβ42 incubation times of ~2 h before SV centrifugation. The s-value for the monomeric AF488-Aβ42 was determined as 0.80 ± 0.01 S, which is slightly higher than the s-value 0.6 ± 0.02 S found for the unlabelled monomer (Table 1). The monomer accounts for 61% of the total fluorescence signal. The s-values of the smallest detectable oligomers in this experiment were determined as 2.32 ± 0.14 S for AF488-Aβ42 and 2.56 ± 0.32 S for Aβ42. The smaller s-value for AF488-Aβ42 oligomer in comparison to the unlabelled Aβ42 oligomer could be explained by an increase of the frictional coefficient f or shape factor f/f 0 for AF488-Aβ42 due to the fluorophores protruding away from the oligomer. For the monomer, which is unstructured in aqueous solution46, the increase in mass seems to be the dominating effect leading to an increase of the s-value for AF488-Aβ42. In Fig. 1b the c(s) distributions for four independent samples of 0.1 μM AF488-Aβ42 dissolved in 10 mM phosphate buffer are shown. At this low concentration aside from the monomer and the residual free dye only a small fraction of small oligomeric species were detected. These oligomers are dominated by a species between 2 S and 3 S, which represents the smallest s-value of the oligomer distribution. The fraction of this species accounts for about 1.4% of the total signal. After subtracting the contribution of the free dye the value increases to 2% of total AF488-Aβ42. In order to evaluate whether by changing the temperature the equilibrium could be shifted from monomers to penta-/hexamers, we performed SV experiments at different temperatures between 10 and 30 °C. However, variation of the experimental temperature in this range did not lead to measurable changes in the oligomeric fractions. To exclude the possibility of contamination or that we observe a covalently bonded aggregate species, we treated the sample with 6 M guanidine hydrochloride. Under these denaturing conditions this oligomer species disappeared (data not shown).
Table 1.
Experimentally determined sedimentation coefficients for Aβ42 monomer and small oligomer.
| Monomer (S)* | Small Oligomer (S)* | |
|---|---|---|
| Aβ42 | 0.62 ± 0.02 | 2.56 ± 0.32 |
| AF488-Aβ42 | 0.80 ± 0.01 | 2.32 ± 0.14 |
*Sedimentation coefficients for the monomeric and small oligomeric species of Aβ42 were determined from c(s) distributions. Weight averaged s 20,w-values and standard deviations were calculated from at least 4 independent sample preparations.
Figure 2 shows the c(s) distributions obtained for 5 (Fig. 2a,c) and 10 μM Aβ42 (Fig. 2b,c). At these medium Aβ42 concentrations, larger aggregates become detectable in addition to the small oligomers. Figure 3 shows three c(s) distributions obtained for high Aβ42 concentrations, which demonstrate that the small oligomer is still detectable under these conditions, where larger aggregates have already formed. In Table 1, the mean s 20,w-values for the monomer and for the small oligomers are summarized for Aβ42 and AF488-Aβ42.
Figure 2.
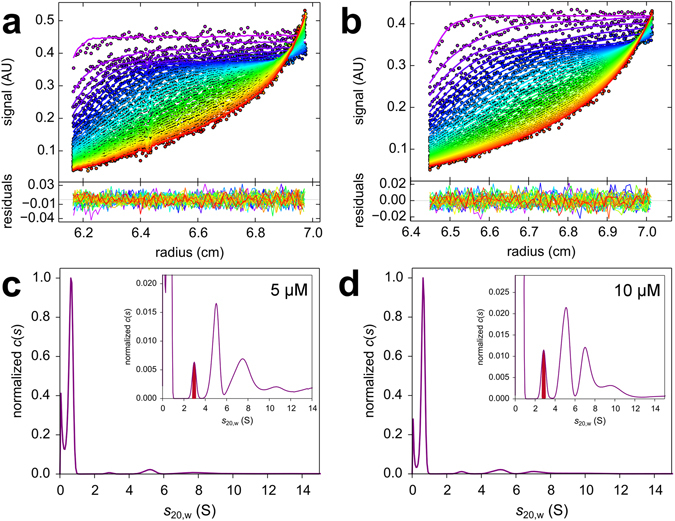
The SV analysis at low micromolar Aβ42 concentrations yields the existence of the small oligomer between 2 S and 3 S and a fraction of larger assemblies with s-values between 3 and 12 S. The raw data at 5 µM (a) and 10 µM Aβ42 (b) together with overlaid fit results and residuals are shown. The corresponding c(s) results for 5 µM (c) and 10 µM Aβ42 (d) are shown below. Sedimentation was performed at 50,000 rpm, 20 °C and monitored with absorbance detection. Detection wavelength for 5 µM was 208 and 220 nm for 10 µM Aβ42. The c(s) distribution was normalized to a maximum c(s) value. The insets show the distributions with an expanded y scale.
Figure 3.
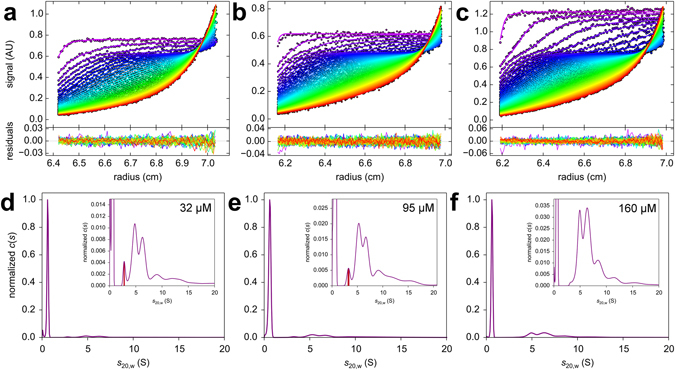
SV analysis at high Aβ42 concentrations. Measured data overlaid with fit data and corresponding residuals are shown for 32 μM (a), 95 μM (b) and 160 μM (c) Aβ42. Below, c(s) analysis reveals the existence of a high fraction of larger oligomeric species aside from the monomer and a comparably low fraction of the 2 to 3 S species for 32 µM (d), 95 µM (e) and 160 µM (f) Aβ42. Sedimentation was performed at 60,000 rpm, 10 °C and monitored with absorbance detection at 280 nm. Normalization was performed according to peak height of the monomeric Aβ42. The insets show the distributions with an expanded y scale.
In summary, SV measurements covering a concentration range between 0.1 μM and 160 μM consistently reveal the presence of a small oligomer at 2.56 S or 2.32 S for Aβ42 and AF488-Aβ42, respectively. According to MD simulations described below, these values correspond to Aβ42 pentamers or hexamers.
Small angle neutron scattering studies of Aβ42 in solution corroborate the presence of penta- to hexamer species
In contrast to SV with fluorescence detection, SANS measurements do not require dye labelling; although, an equivalent D2O buffer is used to reduce buffer scattering. For the relative low concentrations used in the aggregation studies, a long counting time was required to obtain reasonable statistical accuracy because the detected signal at larger wave vectors is two orders of magnitude below the buffer scattering. To slow down the aggregation process and reduce measurement times to ~8 h for several detector distances without significant change in the sample, the measurements have been performed at 7 °C. SANS measurements have been performed for Aβ42 with incubation times between 0.5 and 316 h (Fig. 4) for concentrations of 221 μM (1 mg/ml), 55 μM 0.25 mg/ml) and 22 μM (0.1 mg/ml).
Figure 4.
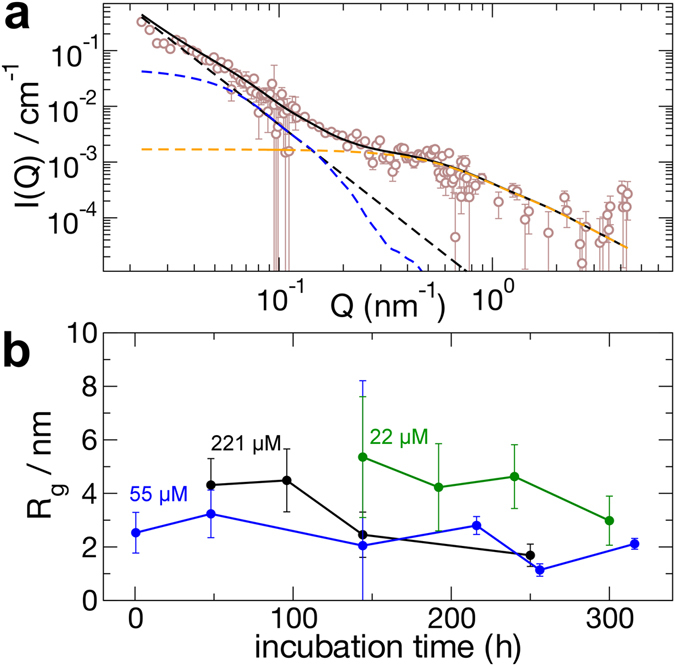
Results from SANS scattering experiments after incubation of Aβ42 at pH 7.4. Measurement and incubation was in 50 mM Napi plus 150 mM NaCl in 100% D2O at 7 °C. (a) Exemplary SANS scattering intensity after 0.5 h. Contributions to the scattered intensity arise from aggregates larger than 50 nm resulting in a power law (black broken line), mid-size aggregates modelled as ellipsoid of revolution (blue broken line) and small oligomers (orange broken line), which are modelled by a Beaucage function. A vanishing background of monomers is modelled as a Beaucage function with fixed R g = 1 nm. The combined fit result is shown as a black solid line. Ellipsoids have radii between 50 and 10 nm. (b) The radius of gyration of small oligomers from SANS analysis after incubation at concentrations of 221 μM (black), 55 µM (blue) and 22 µM (green).
SANS measurements show that even at low temperature and relative low concentrations aggregates of all sizes are formed after the addition of buffer to the dry Aβ. This in in contrast to SV experiments, where at the speed applied to study the small oligomers larger oligomers, protofibrils or fibrils are not detectable due to their fast sedimentation. The scattering intensity of the samples did not change measurably within 10 h as monitored by dynamic light scattering measurements (data not shown). The measured intensities after buffer subtraction were analysed by a model adding contributions of larger aggregates and complexes, midsize aggregates, small oligomers and monomers as shown in Fig. 4a. Except for the monomer, the populations of the different assembly states were determined in accompanying measurements by dynamic light scattering (Fig. S4). Large aggregates or complexes, such as assemblies of fibrils with radii of gyration (R g) larger than 50 nm contribute at low Q by a power law with d between 5/3 and 3 as expected for networks/gels58. Midsize aggregates, such as protofibrils or fibrils were modelled by an ellipsoid of revolution with R g between 50 and 10 nm1. Small oligomers were modelled by a Beaucage model providing the R g of the small aggregates with fixed dimensionality59. Monomers contribute with a fixed R g = 1 nm as a background contribution and cannot be resolved. Small oligomers with R g between 1 and 5 nm were observed at different concentrations throughout the experiments (Fig. 4b).
It should be emphasized that we observe oligomers and aggregates of different sizes, which are well separated in the SANS signal. If objects of all sizes were present in the sample, we would observe an undefined, decaying scattering intensity without a shoulder. Importantly, the small oligomers (identified as hexa-/pentamers, see below) can only be observed if species between the small oligomers and monomers are significantly missing and do not contribute intensity above the shoulder at ~0.6 nm−1. If smaller oligomers than pentamers were present, the scattered intensity could not be described by a Beaucauge function separated from the monomer background. Nevertheless, the presence of smaller species cannot be completely ruled out, but is limited to amounts which contribute to the scattering signal at the scale of the noise of the measurements.
MD simulations provide size and shape information about early oligomeric species
From the R g values determined from the SANS data it is not possible to determine the size of the oligomers. Therefore, in order to compare the oligomer sizes observed in the SV experiments with the R gvalues obtained from the SANS experiments, we performed five independent all-atom MD simulations, yielding a total simulation time was 1 μs, of twenty Aβ42 monomers inserted in a cubic box at a solute concentration of ~80 µM60. While this concentration is considerably larger than that used in the SV experiments, it should be noted that, to the best of our knowledge, it is lower compared to all other atomistic simulations studying Aβ aggregation. Moreover, as the current simulation results are used to provide R g and s-values for the different oligomer sizes, their relative population as sampled in the simulations is not important. Instead, of importance is that the oligomer structures are sampled realistically. In order to check that the oligomer structures are not affected by the high peptide concentration or the use of an implicit solvent, we performed additional MD simulations of isolated dimers, trimers and tetramers using an explicit water model. The resulting oligomer structures have similar s- and R g values and also exhibit similar structures as those obtained from the simulations involving twenty Aβ peptides. Thus, it can be concluded that our MD results are not affected by the rather high peptide concentration used in the current simulations. First, we estimated the sedimentation coefficients of the modelled Aβ42 oligomers. Normalized distributions of the sedimentation coefficient for different oligomer orders are shown in Fig. 5a. The resulting weight averaged s-values for the calculated distribution are listed in Table 2. Tetramers displayed the lowest sedimentation coefficients with an average value of 2.09 ± 0.06 S, followed by pentamers with 2.38 ± 0.06 S and hexamers with an average value of 2.61 ± 0.07 S. For the current analysis we used 576 tetramers, 123 pentamers and 299 hexamers. The s-values obtained for the small oligomeric species collected in five MD simulations show a distribution, e.g., hexamers are found with s-values between 2.42 S and 2.90 S, which seems to approach a Gaussian like shape with higher sampling. The distributions reflect the heterogeneity of the simulated oligomers with regard to their surface area. Nevertheless the overlap between two adjacent oligomer sizes does not exceed 0.3 S units, which allows us to conclude that we identified pentamers and hexamers as smallest oligomeric species in our SV experiments.
Figure 5.
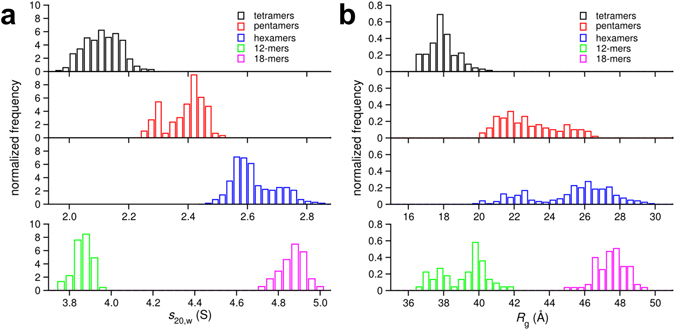
Calculated sedimentation coefficients and radii of gyration of Aβ42 oligomers obtained from MD simulations. (a) Normalized distributions of sedimentation coefficients for Aβ42 tetramers (black), pentamers (red), hexamers (blue), 12-mers (green), and 18-mers (magenta). (b) Normalized radii of gyration for the ensembles of Aβ42 tetramers (black), pentamers (red), hexamers (blue), 12-mers (green), and 18-mers (magenta). All values for tetra- to hexameric Aβ42 oligomers were calculated with the program HydroPro._ENREF_69 Corresponding weight averaged s-values are summarized in Table 2.
Table 2.
Average sedimentation coefficients for Aβ42 tetramers, pentamers and hexamers based on confirmations obtained from MD simulations.
| Oligomer | Tetramer | Pentamer | Hexamer |
|---|---|---|---|
| s-value (S) | 2.09 ± 0.06 | 2.38 ± 0.06 | 2.61 ± 0.07 |
| No. of conformations | 576 | 123 | 299 |
The number of conformations is the total number before the clustering algorithm was applied. All values were calculated with the program HydroPro._ENREF_75.
To correlate the R g values determined from the SANS experiments, we calculated the for the ensemble of structures generated by our MD simulations (Fig. 5b). For the lowest Aβ concentration of 55 µM used in the SANS experiments, the constant presence of oligomers with R g ~ 2.3 nm corresponds to pentamer and/or hexamer species. According to Fig. 5b, it is difficult to discriminate between pentamers and hexamers based on the R g value only. At 221 μM the small oligomers have R g values of around 4 nm at the beginning and after 120 h incubation we find smaller aggregates again with R g ~ 2 nm, which probably correspond to a pentamer/hexamer (Fig. 4b). Interestingly, the largest radius of gyration calculated from SANS measurements of ~5 nm corresponds to assemblies of 18 Aβ42 peptides, according to MD simulations. At the earliest measurement time it was not possible to discriminate between different size classes because of the strong smearing between the various species. Importantly, the SANS studies revealed no species in size between the monomer and the putative pentamer/hexamer.
Discussion
Our study shows evidence for the occurrence of small oligomeric assemblies built of five to six Aβ42 monomers in solution. This conclusion emerged from the application of two independent experimental approaches, which are based on different physical principles, SV analysis and SANS. The two approaches were bridged by MD simulations. The experimentally determined s-values of the smallest oligomers observed in SV experiments were compared to s-values calculated for a set of different oligomers generated by MD simulations. According to this comparison, a combination of pentamers and hexamers explains the best the experimentally observed s-values. From SANS measurements we calculated radii of gyration between 2 and 5 nm, which are in agreement with radii of gyration of assemblies ranging between pentamers and 18-mers calculated for the same set of MD simulated oligomers. Theoretically a sedimentation coefficient is informative on mass as well as shape of a molecular species, but since the detected oligomeric species represents only a very small fraction of the mixture the weight-average of f/f 0 does not provide reliable information on mass or shape of the oligomer. Therefore, based on the experimentally determined s-values we calculated hydrodynamic radii assuming a tetra-, penta-, and hexamer (Supporting Eq. 1 and Supporting Table 2). Assuming that hydrodynamic radius and radius of gyration are sufficiently similar for the oligomer we concluded that the measured s-value can be best explained by a pentamer or hexamer.
These comparisons led us to a model of a penta- to hexamer for the small oligomeric Aβ42 species that would explain the best the SV and SANS results. Data interpretation was further complemented by MD simulations of Aβ42 assembly, which were performed as close as possible to the experimental conditions. Deviating buffer conditions in SANS experiments were shown to give comparable results in SV experiments (Fig. S5) with regard to oligomer formation.
Remarkably, in both the SV and SANS experiments only a small fraction of about 1 to 10% of total Aβ42 could be detected in the pentamer/hexamer state. The dominant Aβ42 species is the monomeric state and larger oligomers present only at higher concentrations. The consistently low fraction of the small oligomeric species over a broad range of Aβ42 concentrations, which is reported by both methods, is a strong indicator of its role as an important reaction intermediate with low population.
Within the accessible Aβ42 concentration range used it was impossible to shift the equilibrium towards higher fractions of the pentamer/hexamer assembly. Higher fractions are most probably prevented by a loss of the pentamer/hexamer to larger oligomers, once a threshold Aβ concentration has been reached. At concentrations below 10 µM, the pentamer/hexamer is the sole oligomeric species. At higher concentrations, the pentamer/hexamer appears to be in a steady-state equilibrium with the monomer and larger oligomeric species, representing a lowly populated reaction intermediate. This classifies the pentamer/hexamer to be an on-pathway intermediate of fibril formation.
Ahmed et al. reported the characterization of a penta- or hexameric Aβ42 with structural information from solution and solid-state NMR27. They also used a low salt phosphate buffer and a similar pre-treatment of Aβ42. In contrast to our study, the oligomeric, disc-like particle they described has a size (diameter 10–15 nm, height ~2 nm) that would lead to a sedimentation coefficient of ~6 S, which is much larger than our species of ~2.6 S. Therefore our penta- to hexamer has to be a different oligomeric species. These differences might be caused by a deviating peptide conformation and/or packing. Furthermore we always observe a considerable amount of free monomer in the presence of our oligomeric species, which appears to be absent in the aforementioned study. A study on oligomeric species of Aβ based on high resolution atomic force microscopy also confirms the hexamer and 12 mer as dominant species in addition to monomers and dimers61. In earlier studies, small Aβ assemblies with 2 to 6 monomeric units have been identified mostly on the basis of SDS-PAGE analysis of either cross-linked or non-cross linked samples20, 62. In addition, a number of computational studies of Aβ oligomerization revealed the existence of several small assemblies, which considerably vary with regard to secondary structure content and overall appearance60, 63. In a study of Aβ42 fused to GroES and ubiquitin and performed at high pH in the presence of urea, Aβ42 assembled into SDS-resistant hexamers and tetramers64. This study suggests that only the most stable complexes will occur under the chosen reaction conditions.
Our SANS as well as SV results demonstrate that solutions of Aβ42 do not contain dimers, trimers or tetramers at detectable levels. Under the assumption of a rapid equilibrium between these small oligomers, the lower limit of detection would be increased due to broadening of the peak or missing resolution of single species to ~5% according to SV data simulations. A possible explanation for the appearance of these small species in a number of other studies would be that these species represent fragmentation products of larger species. The smallest detectable species is a penta- to hexamer. The fraction of this species is independent of the total Aβ42 concentration and close to 1% of the sedimentation boundary. Aβ42 penta-/hexamers are a recurring motif in literature and have been reported frequently over the last years (reviewed in refs 32 and 65). They appear not only to exist in solution but might also represent the Aβ42 conformation interacting with lipids or lipid bilayers66, 67. Nevertheless, we want to emphasize that our study, for the first time, provides evidence for the Aβ42 penta-/hexamer by two solution based methods without the addition of stabilizers. Other groups that showed the presence of similar small oligomers applied the IMS-MS technique17, 68, which required the transfer of ionized molecules and molecular complexes into the gas phase, leaving space for additional events to take place. Nevertheless, it remains reasonable to assume, that, because of the concentration dependency of the aggregation, there is a low concentration at which only dimers and trimers but no larger oligomers are in equilibrium with the monomers. Lowering the total Aβ42 concentration in our experiments has been one of the strategies we followed in order to suppress the conversion of monomers into large aggregates and to increase the fraction of the small oligomeric species. However, we could not detect dimers and trimers under our experimental conditions. These species seem to be less populated than the penta-/hexameric oligomer. Additionally, it might be possible to speculate that their higher abundance in other studies is a consequence of fragmentation of larger oligomers.
Our previous studies by sedimentation velocity centrifugation revealed the power of the method to characterize Aβ oligomeric species in equilibrium in solution. The formerly observed gap in the size distribution could be further restricted. We interpreted this gap as indicative for the size, i.e., s-value, range of a so called nucleus of aggregation according to the assumption of a nucleated aggregation mechanism. A nucleus would be the smallest assembly, which favours the addition of further monomers or oligomers to facilitate fibril formation versus disassembly. Therefore a nucleus is a kinetically unstable intermediate, which makes it elusive to experimental detection. The hydrodynamic radius of the nucleus has been estimated to be between 5 and 50 nm according to fluorescence correlation spectroscopy measurements performed by Garai and colleagues69. This boundary is similar to our estimated size region between 1 and 4 S, although the size of our determined penta-to hexameric species with an R H between 2.3 and 2.8 nm is clearly smaller. Nevertheless, our characterized oligomeric species resembles more likely a structure formerly defined as paranucleus as a building block for larger oligomeric species, like the 12- and 18 mers15, 16.
In the current study, two independent, solution based methods, which do not necessitate the addition of surfactants, crosslinking or surface interactions, have been combined for the first time to investigate the smallest detectable oligomers occurring during Aβ42 aggregation. It would be desirable to verify, whether these penta-/hexameric species exhibit the primary toxicity agent that leads to synapse failure and finally to memory loss. To address this question larger quantities of this species have to be obtained, which is our goal for future studies.
Methods
Experimental details
Amyloid β-Proteins
Synthetic Aβ42 was purchased from Bachem as a trifluoroacetate salt (Bachem H-1368). Aβ42 was first dissolved in 100% 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP) for at least 12 h in order to remove any preexisting aggregates. Before usage HFIP was removed by lyophilisation. Aliquots were stored at −80 °C until use. Repeated freeze thaw cycles were generally avoided.
For fluorescence detection measurements we used recombinantly produced Aβ42 with an additional cysteine residue at position 0 (Cys0). Cloning was based on a plasmid encoding wild-type Aβ(1–42) as a fusion protein with an N-terminal His6-tag, a solubilizing fusion partner (NANP)19 and a modified tobacco etch virus protease (TEV) recognition site, which was kindly provided by Finder, Glockshuber and co-workers70. Cys0-Aβ42 was labelled by maleimide chemistry with AlexaFluor488 (Molecular Probes, Thermo Fisher Scientific) including a short linker (CH2)5. The dye conjugate was further purified by SEC from unincorporated dye and is named AF488-Aβ42 (see Fig. S1 for structural information). The partial specific volume of the dye-protein conjugate was calculated according to Durchschlag71, 72 as cm3/g; see also values summarized (Table S1).
A synthetic, covalently linked Aβ40 dimer was obtained by using Aβ40 with an additional Cys residue at the N-terminus, Cys-Aβ40 (Bachem H-7368) under oxidizing conditions. The disulphide bridge linked dimer was purified by high performance liquid chromatography (HPLC). The partial specific volume calculated for the Cys-Aβ40-dimer with the help of sednterp (vs. 20120828 BETA)73 is = 0.7316 cm3/g at 20 °C and = 0.7295 cm3/g at 15 °C.
Sedimentation Velocity Centrifugation
AUC was performed with an Optima XL-A or ProteomLab XL-A (Beckman Coulter). The ProteomLab XL-A is equipped with standard absorbance optics and an additional fluorescence detection system (Aviv Biomedical Inc.). Samples were filled either in 12 mm titanium double sector cells with 400 µl filling volume (Nanolytics) for absorbance detection or in 3 mm titanium double sector cells with 100 µl filling volume for fluorescence measurements. An integrated spacer places the sample volume to the upper third of a cell assembly guaranteeing an optimal focus position (Nanolytics) for the fluorescence detection. For both cell assemblies quartz windows were used. Labelled or unlabelled Aβ42, pre-treated with HFIP, was dissolved in 10 mM sodium phosphate buffer (NaPi), pH 7.4 at concentrations between 0.1 to 160 µM directly prior to the SV experiments. All samples were thermally equilibrated within the centrifuge for ~2 h before starting the run. Sedimentation velocity runs with absorbance detection were performed either at 50,000 rpm or at 60,000 rpm, equivalent to 201,600 g or 289,000 g, respectively, at the maximum radius of 7.2 cm. At concentrations above 20 µM the runs were performed at 10 or 15 °C to suppress aggregation during centrifugation. For absorbance data collection a radial resolution of 30 µm was chosen together with the shortest possible scan interval, which was ~1.5 min. The detection wavelength was chosen such that sample absorbance was between 0.5 and 1.2 OD. In general, up to 25% loss of Aβ42 concentrations had been determined by comparing the signal of the first scan with the spectroscopically determined concentrations of the samples prior to filling the samples into the cells for SV measurements. This loss during the acceleration phase might be either due to incomplete dissolution, rapid aggregate formation, or attachment of Aβ42 to surfaces, i.e. glass windows. Higher recovery rates were not achievable with Aβ at neutral pH. It is a special feature of AUC that these losses can be easily quantified.
For the fluorescence detection system, based on laser excitation at 488 nm, the radial resolution was fixed at 20 µm and data acquisition was not done sequentially but simultaneously for all sample sectors, leading to scan intervals shorter than 1 min. Each sample sector was measured with an individually adjusted signal amplification factor resulting in 2000−3400 relative fluorescence units (RFU). The addition of carrier protein BSA, as recommended for fluorescence detection, was not appropriate for our system because Aβ directly interacts with BSA74. Low percentages of Tween 20 did not alter the signals and were therefore not applied either. All SV data was initially analysed using a continuous distribution c(s) with maximum entropy. Thereafter the c(s)-distribution analysis with the size-distribution option “with prior probabilities” was applied as implemented in the software package sedfit (vs. 14.7 g; May 2015) (http://www.analyticalultracentrifugation.com/)75. For data analysis a resolution of 0.05 S with a confidence level (F-ratio) of 0.95 was chosen for the appropriate s-value range. A more detailed description of the fitting process has been added to the supplement. All presented c(s) distributions have root mean square deviations of <1% of the total signal. Sedimentation coefficients are reported as s 20,w values in Svedberg units with 1 S = 10−13 s, which represent the apparent sedimentation coefficient normalized to the standard conditions of 20 °C and pure water solvent.
Small Angle Neutron Scattering
Proteins were dissolved in HFIP for three weeks and dried before being resolved in deuterated buffer (50 mM Napi, pH 7.4 + 150 mM NaCl in 100% D2O, filtered with Anopore Whatman, pore size 20 nm). Samples were incubated at 7 °C at rest. SANS experiments were performed at the small angle diffractometer, KWS-2 of Heinz Maier-Leibnitz Zentrum (MLZ Garching, Germany) with wavelengths between 0.47 nm and 0.81 nm and detector distances from 1.1 m to 19.7 m to cover a wave vector range of 0.001 Å−1 to 0.5 Å−1 76. The wavelength spread was and measurement times from 1 to 4 h dependent on detector distance and sample scattering were used. Samples and buffer measurements for background correction were performed in quartz cells of 2 mm thickness together with appropriate measurements for detector sensitivity and dark current. Appropriate standard methods for evaluation and background correction were used from the software QtiKWS77.
Computational details
Molecular Dynamics calculations
Five independent all-atom MD simulations of twenty Aβ42 monomers inserted in a cubic box with a side length of 350 Å and periodic boundary conditions were performed. We applied different initial velocity distributions for each simulation and simulated in total 2.5 μs, with 500 ns per simulation. We used the parallel processing MD software Gromacs 4.5.5 for performing the simulations with a leap-frog stochastic dynamics integrator, the OPLS/AA force field78–80, and implicit solvent using a Generalized Born model with a hydrophobic solvent accessible surface area term (GBSA)81. The temperature coupling was done via velocity rescaling with a stochastic term algorithm82 using a time constant for coupling of 2 ps and keeping the system at 300 K. Electrostatic interactions were treated with the cut-off method with a value of 1.2 nm and van der Waals interactions were also cut at 1.2 nm. Hydrogen atoms were treated as virtual interaction sites, permitting an integration time step of 4 fs while maintaining energy conservation83. From the 5 × 500 ns simulations we extracted all tetramer, pentamer and hexamer conformations and clustered all structures for each oligomer size using the method of Daura and colleagues84 with a cut-off of 0.2 nm. For the resulting cluster centres we calculated sedimentation coefficients using the program HydroPro85. To this end, we first estimated the partial specific volume as the total protein volume divided by the molecular weight of the protein. To estimate the total protein volume we used the program 3 V Volume Calculator86, and for determining the molecular weight of the protein we used VMD87.
Electronic supplementary material
Acknowledgements
The authors gratefully acknowledge the computing time granted on the supercomputers JUROPA and JURECA at the Jülich Supercomputing Centre (grant numbers JICS61 and JICS64). Further, the authors thank the Helmholtz Association for funding part of this work through the Helmholtz-Portfolio Topics “Technology and Medicine” (C.D., L.N.-S., D.W.) and “Drug Research” (D.W.). We would like to express our great appreciation to Dieter Richter for his valuable and constructive suggestions regarding SANS measurements. We thank Stephanie Schiefer for her skilful assistance in atomic force microscopy. This work is based on experiments performed at the Heinz Maier-Leibnitz Zentrum (MLZ), Garching, Germany.
Author Contributions
M.W., C.D. performed and evaluated SV experiments; A.R., B.Z.-H., R.B. performed and evaluated SANS measurements; M.S. contributed fluorescently labelled Aβ; B.B., B.S. contributed M.D. simulations. L.N.-S., R.B., B.S., D.W. are responsible for conception, design and data interpretation. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
M. Wolff and B. Zhang-Haagen contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-02370-3
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Meier BH, Bockmann A. The structure of fibrils from ‘misfolded’ proteins. Curr Opin Struct Biol. 2014;30C:43–49. doi: 10.1016/j.sbi.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Comellas G, Rienstra CM. Protein structure determination by magic-angle spinning solid-state NMR, and insights into the formation, structure, and stability of amyloid fibrils. Annu Rev Biophys. 2013;42:515–536. doi: 10.1146/annurev-biophys-083012-130356. [DOI] [PubMed] [Google Scholar]
- 3.Kotler SA, et al. High-resolution NMR characterization of low abundance oligomers of amyloid-beta without purification. Sci Rep. 2015;5:11811. doi: 10.1038/srep11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu L, et al. Structural characterization of a soluble amyloid beta-peptide oligomer. Biochemistry. 2009;48:1870–1877. doi: 10.1021/bi802046n. [DOI] [PubMed] [Google Scholar]
- 5.Lendel C, et al. A hexameric peptide barrel as building block of amyloid-beta protofibrils. Angew Chem Int Ed Engl. 2014;53:12756–12760. doi: 10.1002/anie.201406357. [DOI] [PubMed] [Google Scholar]
- 6.Alzheimer A. Über eine eigenartige Erkrankung der Hirnrinde. Allgemeine Zeitschrift für Psychiatrie und psychisch-gerichtliche Medizin (Berlin) 1907;64:146–148. [Google Scholar]
- 7.Joachim CL, Selkoe DJ. The seminal role of beta-amyloid in the pathogenesis of Alzheimer disease. Alzheimer Dis Assoc Disord. 1992;6:7–34. doi: 10.1097/00002093-199205000-00003. [DOI] [PubMed] [Google Scholar]
- 8.Klein WL, Krafft GA, Finch CE. Targeting small Abeta oligomers: the solution to an Alzheimer’s disease conundrum? Trends Neurosci. 2001;24:219–224. doi: 10.1016/S0166-2236(00)01749-5. [DOI] [PubMed] [Google Scholar]
- 9.Klein WL. Abeta toxicity in Alzheimer’s disease: globular oligomers (ADDLs) as new vaccine and drug targets. Neurochem Int. 2002;41:345–352. doi: 10.1016/S0197-0186(02)00050-5. [DOI] [PubMed] [Google Scholar]
- 10.Shankar GM, et al. Amyloid-[beta] protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nat Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mc Donald JM, et al. The aqueous phase of Alzheimer’s disease brain contains assemblies built from approximately 4 and approximately 7 kDa Abeta species. Alzheimers Dement. 2015;11:1286–1305. doi: 10.1016/j.jalz.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Nuallain B, et al. Amyloid beta-protein dimers rapidly form stable synaptotoxic protofibrils. J Neurosci. 2010;30:14411–14419. doi: 10.1523/JNEUROSCI.3537-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garai K, Frieden C. Quantitative analysis of the time course of Abeta oligomerization and subsequent growth steps using tetramethylrhodamine-labeled Abeta. Proc Natl Acad Sci USA. 2013;110:3321–3326. doi: 10.1073/pnas.1222478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garzon-Rodriguez W, Sepulveda-Becerra M, Milton S, Glabe CG. Soluble amyloid Abeta-(1–40) exists as a stable dimer at low concentrations. J Biol Chem. 1997;272:21037–21044. doi: 10.1074/jbc.272.34.21037. [DOI] [PubMed] [Google Scholar]
- 15.Bitan G, Vollers SS, Teplow DB. Elucidation of primary structure elements controlling early amyloid beta-protein oligomerization. J Biol Chem. 2003;278:34882–34889. doi: 10.1074/jbc.M300825200. [DOI] [PubMed] [Google Scholar]
- 16.Bitan G, et al. Amyloid beta -protein (Abeta) assembly: Abeta 40 and Abeta 42 oligomerize through distinct pathways. Proc Natl Acad Sci USA. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein SL, et al. Amyloid-beta protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roychaudhuri R, Yang M, Hoshi MM, Teplow DB. Amyloid beta-protein assembly and Alzheimer disease. J Biol Chem. 2009;284:4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ono K, Condron MM, Teplow DB. Structure-neurotoxicity relationships of amyloid beta-protein oligomers. Proc Natl Acad Sci USA. 2009;106:14745–14750. doi: 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burdick D, et al. Assembly and aggregation properties of synthetic Alzheimer’s A4/beta amyloid peptide analogs. J Biol Chem. 1992;267:546–554. [PubMed] [Google Scholar]
- 21.Podlisny MB, et al. Aggregation of secreted amyloid beta-protein into sodium dodecyl sulfate-stable oligomers in cell culture. J Biol Chem. 1995;270:9564–9570. doi: 10.1074/jbc.270.16.9564. [DOI] [PubMed] [Google Scholar]
- 22.Walsh DM, Tseng BP, Rydel RE, Podlisny MB, Selkoe DJ. The oligomerization of amyloid beta-protein begins intracellularly in cells derived from human brain. Biochemistry. 2000;39:10831–10839. doi: 10.1021/bi001048s. [DOI] [PubMed] [Google Scholar]
- 23.Muller-Schiffmann A, et al. Molecular engineering of a secreted, highly homogeneous, and neurotoxic abeta dimer. ACS Chem Neurosci. 2011;2:242–248. doi: 10.1021/cn200011h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roher AE, et al. Morphology and toxicity of Abeta-(1-42) dimer derived from neuritic and vascular amyloid deposits of Alzheimer’s disease. J Biol Chem. 1996;271:20631–20635. doi: 10.1074/jbc.271.34.20631. [DOI] [PubMed] [Google Scholar]
- 25.Roychaudhuri R, et al. C-Terminal turn stability determines assembly differences between Aβ40 and Aβ42. Journal of Molecular Biology. 2013;425:292–308. doi: 10.1016/j.jmb.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cernescu M, et al. Laser-induced liquid bead ion desorption mass spectrometry: an approach to precisely monitor the oligomerization of the beta-amyloid peptide. Anal Chem. 2012;84:5276–5284. doi: 10.1021/ac300258m. [DOI] [PubMed] [Google Scholar]
- 27.Ahmed M, et al. Structural conversion of neurotoxic amyloid-[beta]1-42 oligomers to fibrils. Nat Struct Mol Biol. 2010;17:561–567. doi: 10.1038/nsmb.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viola KL, Klein WL. Amyloid beta oligomers in Alzheimer’s disease pathogenesis, treatment, and diagnosis. Acta Neuropathol. 2015;129:183–206. doi: 10.1007/s00401-015-1386-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lesné SE. Toxic oligomer species of amyloid-beta in Alzheimer’s disease, a timing issue. Swiss Med Wkly. 2014;144:w14021. doi: 10.4414/smw.2014.14021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayden EY, Teplow DB. Amyloid beta-protein oligomers and Alzheimer’s disease. Alzheimers Res Ther. 2013;5:60. doi: 10.1186/alzrt226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahimi F, Shanmugam A, Bitan G. Structure-function relationships of pre-fibrillar protein assemblies in Alzheimer’s disease and related disorders. Curr Alzheimer Res. 2008;5:319–341. doi: 10.2174/156720508784533358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagel-Steger, L., Owen, M. C. & Strodel, B. An account of amyloid oligomers: facts and figures obtained from experiments and simulations. Chembiochem, doi:10.1002/cbic.201500623 (2016). [DOI] [PubMed]
- 33.Fändrich M. Oligomeric intermediates in amyloid formation: structure determination and mechanisms of toxicity. Journal of Molecular Biology. 2012;421:427–440. doi: 10.1016/j.jmb.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 34.Benilova I, Karran E, De Strooper B. The toxic A[beta] oligomer and Alzheimer’s disease: an emperor in need of clothes. Nat Neurosci. 2012;15:349–357. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- 35.Pujol-Pina R, et al. SDS-PAGE analysis of Abeta oligomers is disserving research into Alzheimer s disease: appealing for ESI-IM-MS. Sci Rep. 2015;5:14809. doi: 10.1038/srep14809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dam J, Velikovsky CA, Mariuzza RA, Urbanke C, Schuck P. Sedimentation velocity analysis of heterogeneous protein-protein interactions: Lamm equation modeling and sedimentation coefficient distributions c(s) Biophys J. 2005;89:619–634. doi: 10.1529/biophysj.105.059568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pekar A, Sukumar M. Quantitation of aggregates in therapeutic proteins using sedimentation velocity analytical ultracentrifugation: practical considerations that affect precision and accuracy. Anal Biochem. 2007;367:225–237. doi: 10.1016/j.ab.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 38.Brookes, E., Cao, W., Demeler, B. A two-dimensional spectrum analysis for sedimentation velocity experiments of mixtures with heterogeneity in molecular weight and shape. Eur Biophys J. 39, 405–414, doi:10.1007/s00249-009-0413-5 (2010). [DOI] [PubMed]
- 39.Demeler, B., Brookes, E., Nagel-Steger, L. Analysis of heterogeneity in molecular weight and shape by analytical ultracentrifugation using parallel distributed computing. Methods Enzymol. 454, 87–113, doi:10.1016/S0076-6879(08)03804-4 (2009). [DOI] [PubMed]
- 40.Nagel-Steger, L. et al. Modulation of aggregate size- and shape-distributions of the amyloid-beta peptide by a designed beta-sheet breaker.Eur Biophys J. 39, 415–422, doi:10.1007/s00249-009-0416-2 (2010). [DOI] [PubMed]
- 41.Schuck P, Perugini MA, Gonzales NR, Howlett GJ, Schubert D. Size-distribution analysis of proteins by analytical ultracentrifugation: strategies and application to model systems. Biophys J. 2002;82:1096–1111. doi: 10.1016/S0006-3495(02)75469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuck P. Sedimentation patterns of rapidly reversible protein interactions. Biophys J. 2010;98:2005–2013. doi: 10.1016/j.bpj.2009.12.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuck, P. Diffusion of the Reaction Boundary of Rapidly Interacting Macromolecules in Sedimentation Velocity. Biophys J98, 2741-2751, doi:10.1016/j.bpj.2010.03.004 (2010). [DOI] [PMC free article] [PubMed]
- 44.Yong W, et al. Structure determination of micelle-like intermediates in amyloid beta -protein fibril assembly by using small angle neutron scattering. Proc Natl Acad Sci USA. 2002;99:150–154. doi: 10.1073/pnas.012584899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ryan TM, et al. Small angle X-ray scattering analysis of Cu(2+)-induced oligomers of the Alzheimer’s amyloid beta peptide. Metallomics. 2015;7:536–543. doi: 10.1039/C4MT00323C. [DOI] [PubMed] [Google Scholar]
- 46.Ryan TM, et al. Stabilization of nontoxic Abeta-oligomers: insights into the mechanism of action of hydroxyquinolines in Alzheimer’s disease. J Neurosci. 2015;35:2871–2884. doi: 10.1523/JNEUROSCI.2912-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang-Haagen B, et al. Monomeric amyloid beta peptide in hexafluoroisopropanol detected by small angle neutron scattering. PLoS One. 2016;11:e0150267. doi: 10.1371/journal.pone.0150267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolff M, et al. Amyloid beta oligomeric species present in the lag phase of amyloid formation. PLoS One. 2015;10:e0127865. doi: 10.1371/journal.pone.0127865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Larson ME, Lesné SE. Soluble Abeta oligomer production and toxicity. J Neurochem. 2012;120(Suppl 1):125–139. doi: 10.1111/j.1471-4159.2011.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lesné S, et al. A specific amyloid-beta protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 51.Bernstein SL, et al. Amyloid beta-protein: monomer structure and early aggregation states of Abeta42 and its Pro19 alloform. J Am Chem Soc. 2005;127:2075–2084. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- 52.Kheterpal I, Williams A, Murphy C, Bledsoe B, Wetzel R. Structural features of the Abeta amyloid fibril elucidated by limited proteolysis. Biochemistry. 2001;40:11757–11767. doi: 10.1021/bi010805z. [DOI] [PubMed] [Google Scholar]
- 53.Gouwens LK, Makoni NJ, Rogers VA, Nichols MR. Amyloid-beta42 protofibrils are internalized by microglia more extensively than monomers. Brain Res. 2016;1648:485–495. doi: 10.1016/j.brainres.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wafer L, Kloczewiak M, Luo Y. Quantifying Trace Amounts of Aggregates in Biopharmaceuticals Using Analytical Ultracentrifugation Sedimentation Velocity: Bayesian Analyses and F Statistics. Aaps J. 2016;18:849–860. doi: 10.1208/s12248-016-9925-y. [DOI] [PubMed] [Google Scholar]
- 55.Arthur KK, Kendrick BS, Gabrielson JP. Guidance to Achieve Accurate Aggregate Quantitation in Biopharmaceuticals by SV-AUC. Methods Enzymol. 2015;562:477–500. doi: 10.1016/bs.mie.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 56.Krayukhina E, et al. Aggregation analysis of pharmaceutical human immunoglobulin preparations using size-exclusion chromatography and analytical ultracentrifugation sedimentation velocity. J Biosci Bioeng. 2013;115:104–110. doi: 10.1016/j.jbiosc.2012.07.021. [DOI] [PubMed] [Google Scholar]
- 57.Brown PH, Balbo A, Schuck P. A Bayesian approach for quantifying trace amounts of antibody aggregates by sedimentation velocity analytical ultracentrifugation. Aaps J. 2008;10:481–493. doi: 10.1208/s12248-008-9058-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shibayama M, Tanaka T, Han CC. Small angle neutron scattering study on poly(N‐isopropyl acrylamide) gels near their volume‐phase transition temperature. The Journal of Chemical Physics. 1992;97:6829–6841. doi: 10.1063/1.463636. [DOI] [Google Scholar]
- 59.Xiao Y, et al. Abeta(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015;22:499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barz B, Olubiyi OO, Strodel B. Early amyloid beta-protein aggregation precedes conformational change. Chem Commun (Camb) 2014;50:5373–5375. doi: 10.1039/C3CC48704K. [DOI] [PubMed] [Google Scholar]
- 61.Economou NJ, et al. Amyloid beta-Protein Assembly and Alzheimer’s Disease: Dodecamers of Abeta42, but Not of Abeta40, Seed Fibril Formation. J Am Chem Soc. 2016;138:1772–1775. doi: 10.1021/jacs.5b11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bitan G, Lomakin A, Teplow DB. Amyloid beta-protein oligomerization: prenucleation interactions revealed by photo-induced cross-linking of unmodified proteins. J Biol Chem. 2001;276:35176–35184. doi: 10.1074/jbc.M102223200. [DOI] [PubMed] [Google Scholar]
- 63.Nasica-Labouze J, et al. Amyloid beta protein and Alzheimer’s disease: when computer simulations complement experimental studies. Chem Rev. 2015;115:3518–3563. doi: 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ngo S, Guo Z. Key residues for the oligomerization of Abeta42 protein in Alzheimer’s disease. Biochem Biophys Res Commun. 2011;414:512–516. doi: 10.1016/j.bbrc.2011.09.097. [DOI] [PubMed] [Google Scholar]
- 65.Sengupta U, Nilson AN, Kayed R. The Role of Amyloid-beta Oligomers in Toxicity, Propagation, and Immunotherapy. EBioMedicine. 2016;6:42–49. doi: 10.1016/j.ebiom.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andreasen M, Lorenzen N, Otzen D. Interactions between misfolded protein oligomers and membranes: A central topic in neurodegenerative diseases? Biochim Biophys Acta. 2015;1848:1897–1907. doi: 10.1016/j.bbamem.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 67.Johnson RD, Steel DG, Gafni A. Structural evolution and membrane interactions of Alzheimer’s amyloid-beta peptide oligomers: new knowledge from single-molecule fluorescence studies. Protein Sci. 2014;23:869–883. doi: 10.1002/pro.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng X, Liu D, Roychaudhuri R, Teplow DB, Bowers MT. Amyloid beta-Protein Assembly: Differential Effects of the Protective A2T Mutation and Recessive A2V Familial Alzheimer’s Disease Mutation. ACS Chem Neurosci. 2015;6:1732–1740. doi: 10.1021/acschemneuro.5b00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garai K, Sahoo B, Sengupta P, Maiti S. Quasihomogeneous nucleation of amyloid beta yields numerical bounds for the critical radius, the surface tension, and the free energy barrier for nucleus formation. The Journal of Chemical Physics. 2008;128:045102. doi: 10.1063/1.2822322. [DOI] [PubMed] [Google Scholar]
- 70.Finder VH, Vodopivec I, Nitsch RM, Glockshuber R. The recombinant amyloid-beta peptide Abeta1-42 aggregates faster and is more neurotoxic than synthetic Abeta1-42. Journal of Molecular Biology. 2010;396:9–18. doi: 10.1016/j.jmb.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 71.Durchschlag, H. & Zipper, P. In Progress in Colloid & Polymer Science: Ultracentrifugation Vol. 94 (ed. G. Lagaly M.D. Lechner (guest editor) F. Kremer) Ch. Calculation of the partial volume of organic compounds and polymers, 20–39 (Springer Verlag Berlin Heidelberg GmbH, 1994).
- 72.Durchschlag, H. In Thermodynamic Data for Biochemistry and Biotechnology (ed. Hans-Jürgen, Hinz) Ch. Specific Volumes of Biological Macromolecules and Some Other Molecules of Biological Interest, 45–128 (Springer, 1986).
- 73.Hurton, T. et al. SEDNTERP, http://bitcwiki.sr.unh.edu/index.php/Main_Page (Date of access: 04/01/2017) (2016).
- 74.Reyes Barcelo AA, Gonzalez-Velasquez FJ, Moss MA. Soluble aggregates of he amyloid-beta peptide are trapped by serum albumin to enhance amyloid-beta activation of endothelial cells. J Biol Eng. 2009;3:5. doi: 10.1186/1754-1611-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Radulescu, A., V. Pipich, H. Frielinghaus and M.-S. Appavou. KWS-2, the high intensity/wide Q -range small-angle neutron diffractometer for soft-matter and biology at FRM II. Journal of Physics: Conference Series351 (2012).
- 77.Pipich, V. QtiKWS:: SA(N)S Data Reduction and Analysis Program, http://iffwww.iff.kfa-juelich.de/~pipich/dokuwiki/doku.php/qtikws (Date of access: 04/01/2017) (2015).
- 78.Jorgensen WL, Maxwell DS, TiradoRives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. doi: 10.1021/ja9621760. [DOI] [Google Scholar]
- 79.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. Journal of Physical Chemistry B. 2001;105:6474–6487. doi: 10.1021/jp003919d. [DOI] [Google Scholar]
- 80.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 81.Qiu, D., Shenkin, P. S., Hollinger, F. P. & Still, W. C. The GB/SA continuum model for solvation. A fast analytical method for the calculation of approximate Born radii. J Phys Chem A101, 3005–3014, doi:10.1021/Jp961992r (1997).
- 82.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J Chem Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 83.Feenstra KA, Hess B, Berendsen HJC. Improving efficiency of large time-scale molecular dynamics simulations of hydrogen-rich systems. J Comput Chem. 1999;20:786–798. doi: 10.1002/(SICI)1096-987X(199906)20:8<786::AID-JCC5>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 84.Daura, X. et al. Peptide folding: When simulation meets experiment. Angew Chem Int Edit38, 236–240, doi:10.1002/(Sici)1521-3773(19990115)38:1/2<236::Aid-Anie236>3.0.Co;2-M (1999).
- 85.Ortega A, Amoros D, de la Torre JG. Prediction of Hydrodynamic and Other Solution Properties of Rigid Proteins from Atomic- and Residue-Level Models. Biophys J. 2011;101:892–898. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Voss NR, Gerstein M, Steitz TA, Moore PB. The geometry of the ribosomal polypeptide exit tunnel. Journal of Molecular Biology. 2006;360:893–906. doi: 10.1016/j.jmb.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 87.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


