Abstract
Previously we reported the production and characterization of monoclonal antibody 1H6 raised against (T4G4)2 intermolecular guanine quadruplex (G4) DNA structures (Henderson A. et al. (2014) Nucleic Acids Res., 42, 860–869; Hoffmann R.F. et al. (2016) Nucleic Acids Res., 44, 152–163). It was shown that 1H6 strongly stains nuclei and has exquisite specificity for heterochromatin by immuno-electron microscopy. Here we extend our studies of 1H6 reactivity using enzyme-linked immunosorbent assay (ELISA) and microscale thermophoresis (MST). As previously reported, 1H6 was found to strongly bind intermolecular G4 structures with a (T4G4)2 sequence motif. However, using both methods we did not detect significant binding to G4 structures without thymidines in their sequence motif or to G4 structures made with (T2G4)2 oligonucleotides. In addition, we observed strong, sequence-specific binding of 1H6 by ELISA to immobilized single stranded poly(T) DNA but not to immobilized poly(C) or poly(A) homo-polymers. Cross-reactivity of 1H6 to poly(T) was not measured in solution using MST. 1H6 was furthermore found to bind to selected areas on DNA fibers but only after DNA denaturation. Based on these observations we propose that 1H6 binds with high affinity to adjacent T's that are restricted in their movement in selected G4 structures and denatured DNA. Cross-reactivity of 1H6 to immobilized single stranded T-rich DNA next to its previously reported specificity for bona fide G4 structures needs to be taken into account in the interpretation of 1H6 binding to (sub-) cellular structures.
INTRODUCTION
In vitro, single stranded guanine-rich RNA or DNA readily adopts higher order structures known as guanine quadruplex (G4) structures (1). Despite accumulating evidence supporting a role for G4 RNA and DNA in diverse biological processes (2,3) detection of G4 structures in situ has been problematic in part because suitable reagents to detect G4 structures have been lacking. We recently described 1H6, a mouse monoclonal antibody that strongly binds to synthetic inter- as well as intra-molecular G4 DNA structures (4,5). The specificity of 1H6 for various G4 DNA structures was validated in vitro and in vivo and epitopes recognized by 1H6 in cells were found to be sensitive to DNAse treatment but resistant to RNAse. Most nuclei of cultured cells were labeled by 1H6 and this staining intensity increased following incubation with G4 stabilizing ligands (4). Human and murine metaphase chromosomes were strongly labeled by 1H6 as were the nuclei of cells in most human tissues (4). In subsequent studies the binding to metaphase chromosomes was confirmed by immuno-electron microscopy and it was shown that 1H6 selectively binds to heterochromatic areas in the nucleus as well as heterochromatic bands in salivary gland polytene chromosomes from Drosophila (5). In this and other species nuclei of somatic cells stained much stronger with 1H6 than mitotic cells of the germline (5). In the current study, we extended our studies on the specificity of the 1H6 antibody following the observation that binding of 1H6 to DNA fibers is greatly increased upon DNA denaturation.
MATERIALS AND METHODS
Immunofluorescence on DNA fibers
DNA fibers from mouse embryonic stem cells and HEK293 cells were obtained as described (6). Briefly, cells were harvested, 104–105 cells were spotted on a microscope slide and after 30–60 min cells were lysed (1% sodium dodecyl sulphate, 200 mM Tris and 50 mM ethylenediaminetetraacetic acid (EDTA), pH7.4). After 2 min the slide was tilted to a 15–30 degree angle to obtain DNA fibers. Slides with DNA fibers were typically air-dried and fixed with methanol and acetic acid (3:1). For experiments slides were rehydrated with phosphate buffered saline (PBS) and treated with RNAse A (Invitrogen, 0.4 mg/ml in PBS) for 1 h at 37°C under a coverslip. DNA fibers were denatured by incubation for 15 min in 3 M HCl or 3 M NaOH or the slides were treated with 50% formamide in Na-citrate pH7.0 for 15 min at 75°C on a slide heater. Slides were washed three times and blocked (5% bovine serum albumin (BSA) in PBS containing 300 mM glycine) for 30 min at room temperature followed by incubation with purified 1H6 or an isotype control antibody (mouse IgG2b, clone MOPC-141, Sigma) at 1 μg/ml in blocking solution for 2 h at room temperature in a humidified slide incubator. Slides were incubated for 2–4 h with Alexa-Fluor-488 anti-mouse IgG at 1:2000 (Invitrogen) and DNA was counterstained using DAPI, NucRed™ Dead 647 or YOYO-1. Fluorescence microscopy was done using a Zeiss-LSM780 NLO confocal microscope.
Oligonucleotides used
All 5΄-biotinylated, 5΄-Cy5 labeled or unmodified oligonucleotides were from IDT (Leuven, Belgium). Only 5΄-Cy5 labeled oligonucleotides were High-performance liquid chromatography (HPLC) purified, others were standard desalted. Lyophilized oligonucleotides were reconstituted to 100 μM in water and were stored at −20°C. See Supplementary Table S1 for sequences of oligonucleotides used in this study.
In vitro formation of G4 structures
G4 structures were folded as described previously (4). Briefly, oligonucleotides listed in Supplementary Table S1 were diluted to 10 μM in TE buffer + 100 mM KCl (10 mM Tris–HCl pH = 7.5, 1 mM EDTA, 100 mM KCl). Following denaturation for 10 min at 95°C, G4 structures could form overnight by slowly cooling to room temperature in the heating block. In Supplementary Table S1 oligonucleotides folded into G4 structures are marked in the seventh column.
Circular dichroism
The formation of G4 was confirmed using circular dichroïsm. Samples prepared for circular dichroism (CD) analysis were diluted in TE buffer + 100 mM KCl to a final concentration of 5 μM. CD spectra were measured using a Jasco J-815 spectropolarimeter. Readings were recorded over a wavelength range of 215–350 nm in a quartz cuvette with a 1 cm path length. Measurements were averaged between three accumulations with an instrument scanning speed of 200 nm/min, a response time of 0.5 s, 1 nm data pitch and 2 nm bandwidth.
Enzyme-linked immunosorbent assay (ELISA)
Enzyme-linked immunosorbent assay (ELISA) experiments were essentially performed as described previously (4). Briefly, 3.3 pmol biotinylated oligonucleotides were bound per well to streptavidin-coated microtiter plates (Pierce Cat# 15125 Thermo Scientific) and incubated for 1 h in 100 μl PBS at room temperature (prepared from 20× solution Pierce, no potassium). Upon washing of the plates three times with PBS + 0.05%Tween-20 (PBS-T) plates were incubated for 2 h with 1H6 antibody at half maximal binding concentration (50 ng/ml) in 400 mM sodium PBS + 0.5% BSA at room temperature. Plates were washed three times with PBS-T, followed by incubation with goat-anti-mouse horseradish peroxidase (HRP) antibody (Sigma) at 50 ng/ml in PBS + 0.5% BSA. After washing five times with PBS-T, 100 μl of the HRP substrate TMB (3.3΄,5.5΄-tetramethylbenzidine, Merck Millipore) in was added to each well. Reactions were stopped with 100 μl 0.3 M Sulphuric Acid and signal intensity was detected at λ = 450 nm using a Multiskan absorption meter (Thermo Scientific). Absorbance was calculated after correcting for background from negative controls. All ELISA experiments were performed at least three times and each reaction was performed in triplicate.
Microscale thermophoresis (MST)
Binding reactions were prepared in 400 mM Na PBS + 0.5% BSA and 0.05% Tween-20 to a total volume of 40 μl. For binding reaction 25 nM 5΄Cy5 labeled oligonucleotides (folded G4s and controls) were used (see Supplementary Table S1 for details). Different concentrations of 1H6 antibody, ranging from 1 μM to 0.03 nM and a constant concentration of DNA (25 nM) were used. Microscale thermophoresis (MST) analysis was performed using standard Capillaries from Nanotemper. MST analysis was performed with LED 80%, 40% MST power, on the Monollith NT.115 instrument temperature 24°C. Kd was calculated with Kd fit using the MO Affinity analysis software. MST analysis was performed in triplicates. To determine the stoichiometry of the interaction between 1H6 and (T4G4)2 a saturation curve with a constant DNA concentration of 200 nM (1:1 unlabeled and 5΄Cy5 labeled) was used. In an initial experiment a broad range of 1H6 concentrations, ranging from 1 μM to 0.03 nM, was used to determine the point of saturation. Here, MST power was 40% and LED 40%. In a second experiment a narrower range of 1H6 concentration, ranging from 330 to 50 nM, was used to determine the saturation point more precisely. Linear regression lines of the saturated and non-saturated data points were set manually.
RESULTS
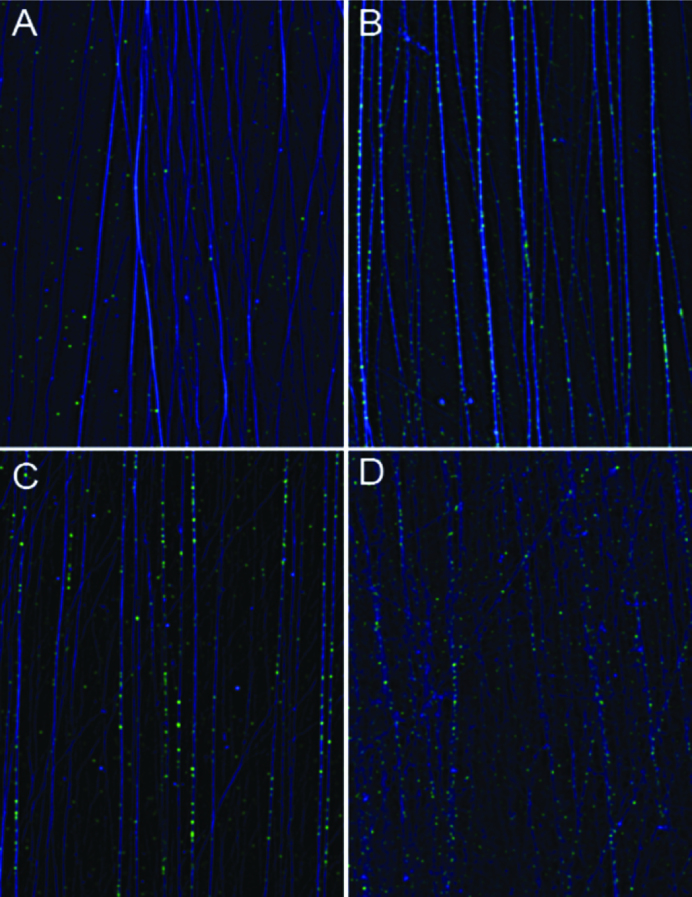
We initially observed variable, spotted, staining of 1H6 of DNA fibers when preparations of DNA fibers were air-dried following fixation with a 3:1 methanol acetic acid fixative (Supplementary Figure S1). Most likely the source of this variable staining was (partial) acid denaturation of DNA by the fixative since more uniform spotted staining was seen when DNA fibers were fixed with higher concentrations of acetic acid (Supplementary Figure S2). No staining was observed when DNA fibers were not allowed to dry following fixation (Figure 1A). However, when such DNA fibers were denatured using either 3M HCl (Figure 1B), heat and formamide (Figure 1C) or 3M NaOH (Figure 1D), the spotted staining of the fibers was again observed. None of the G4 structures used in this study survived these denaturing conditions as measured using CD (Supplementary Figure S3A).
Figure 1.
Denaturation of DNA fibers reveals novel 1H6 binding sites. Binding of 1H6 (light grey spots) to DNA fibers (dark grey lines) before (A) and after denaturation of DNA with 3 M HCl (B), heat and formamide (C) or 3 M NaOH (D).
We considered the possibility that single stranded DNA, following denaturation of DNA fibers, could fold back into intra-molecular G4 structures. However, for such structures to form the single stranded G-rich DNA would presumably have to detach from the slide in order to allow a fold-back G4 structure. This seems unlikely because DNA is stretched in fibers and detachment from the slide would result in loss of staining unless (i) the complementary C-rich strand would remain attached to the slide and (ii) the G-rich strand was nicked to allow the formation of a fold-back G4 structure. Both propositions are untested but do not seem likely in view of the observed density of staining on denatured DNA fibers. Indeed we calculated that 1H6 spots on denatured DNA fibers (∼1 per every 2 kb or more than 1 million spots per genome, see legend Supplementary Figure S1) were too numerous to fit even the most generous estimates of the number of ‘G4 motifs’ in the human genome (6). Instead, the spotted staining of denatured DNA fibers suggested that 1H6 cross-reacts with epitopes that are present on denatured DNA fibers.
Cross-reactivity of 1H6 to poly(T) oligonucleotides
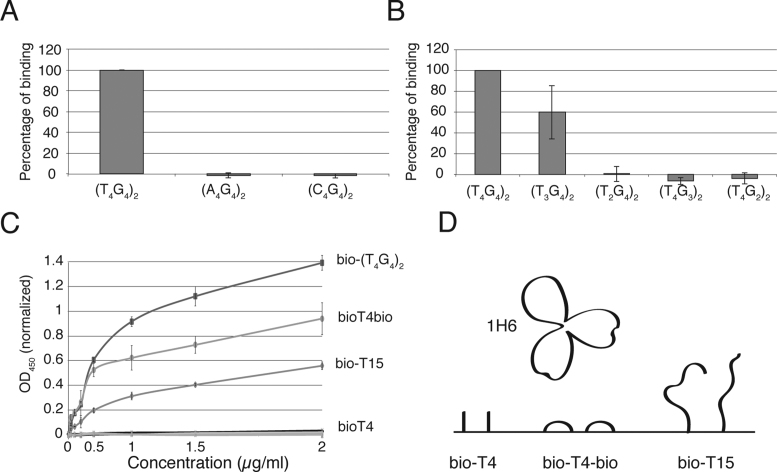
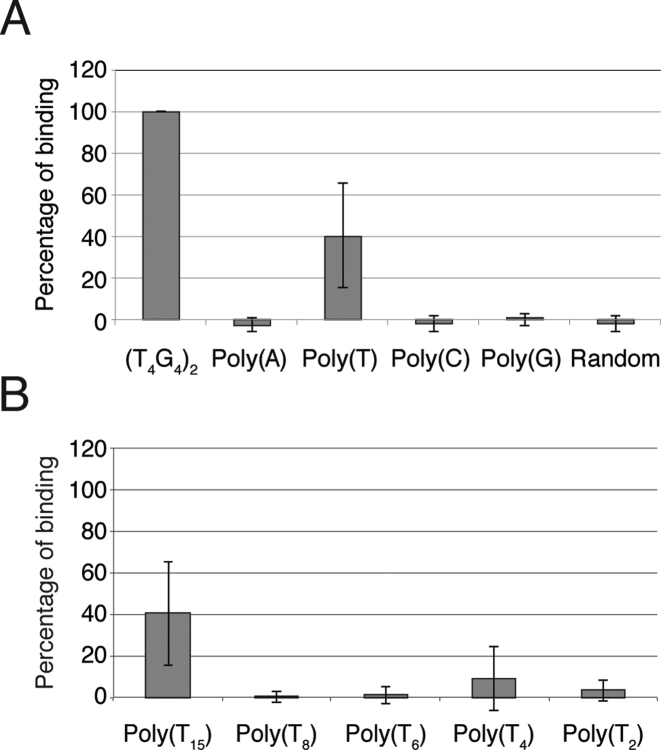
In previous studies the specificity of 1H6 was evaluated using ELISA assays (4). Therefore, we used ELISA to re-evaluate the reactivity of 1H6 to immobilized single stranded DNA's and various G4 structures. In a typical experiment, biotinylated oligonucleotides or preformed G4 structures made with biotinylated oligonucleotides were bound to streptavidin coated ELISA plates and incubated with 1H6 followed by wash steps and detection of the bound antibody. For all experiments (except Figure 3C) 1H6 was used at a half-maximal binding in ELISA at a concentration of 50 ng/ml. Initially, we tested 1H6 binding to the (T4G4)2 G4 structures to which the 1H6 antibody was raised next to 15-mers of poly(T), poly(A), poly(C) as well as longer single stranded DNA with a random sequence (Figure 2A). We observed very robust binding of 1H6 to (T4G4)2 as expected and no binding to poly(A), poly(C) and a random single stranded DNA sequence. Surprisingly, 1H6 also bound to immobilized poly(T). Although this binding was not as strong as the binding to (T4G4)2, it was readily detectable. This ELISA result suggested that 1H6 binds, in addition to intermolecular (T4G4)2 G4 structures, also to poly(T) oligonucleotides. In this first experiment poly(T) oligonucleotides consisted of 15 thymidines. To gain a better understanding of this cross-reactivity we tested 1H6 binding to poly(T) oligo's having a variable number of T's by ELISA. When reactivity to poly(T) with 2, 4, 6, 8 and 15 thymidines was compared binding was only observed to T15 (Figure 2B).
Figure 3.
1H6 does not bind to G4 structures without thymidines, but binding of 1H6 to these G4 structures requires at least three thymidines in the oligonucleotides. (A–C) ELISA experiments of 1H6 binding to various G4 structures. All ELISA experiments were performed in triplicate and 1H6 binding to (T4G4)2 was set at 100%. Error bar represent SEM. (A) Comparison of 1H6 binding to (T4G4)2, versuss (A4G4)2 and (C4G4)2. (B) Comparison of 1H6 binding to (T4G4)2, versus (T3G4)2 (T2G4)2, (T4G3)2 (T4G2)2 by ELISA. (A and B) 1H6 does not bind to (A4G4)2, (C4G4)2, (T2G4)2, (T4G3)2 and (T4G2)2. Statistical significance compared to (T4G4)2 was determined by Student's t-test. **: P ≤ 0.0001, * P ≤ 0.001. (C) 1H6 binds to oligo's with 4 thymidines (poly(T4)) if these are bound to ELISA plate at both the 5΄ and the 3΄ end via biotin (bio-poly(T4)-bio) but not if the are bound only via the 5΄ end (bio-poly(T4)). Controls: (T4G4)2 G4 and T15 with just one biotin (bio-poly(T15)). (D) Cartoon showing how the presentation of short stretches of T's could determine whether 1H6 binds or not.
Figure 2.
1H6 cross reacts with single stranded poly-thymidine. (A) Binding of 1H6 to different oligonucleotides determined by ELISA. Results of three independent experiments performed in triplicate for each oligonucleotide or G4 structure. Error bars represent SEM. 1H6 binding to (T4G4)2 was set at 100% (A) Binding of 1H6 to (T4G4)2 G4 structures and poly(T) but not poly(A), poly(C) or random single stranded DNA. (B) Binding of 1H6 to poly(T) in ELISA requires more than eight thymidines. Statistical significance compared to T4G4 was determined by Student's t-test. All tested substrates showed a P ≤ 0.0001.
Further studies of 1H6 binding to G4 structures
1H6 antibody was raised against intermolecular G4 structures using (T4G4)2 oligonucleotides to mimic telomeric repeats in the ciliate, Stylonychia (4). In our previous ELISA analysis, we found strong reactivity of 1H6 to the intermolecular (T4G4)2 G4 structures. In view of the cross-reactivity of 1H6 with poly(T) we wanted to revisit 1H6 binding to various G4 structures. Strikingly, when G4 structures were made with A's (A4G4)2 or C's (C4G4)2 instead of T's oligonucleotides, no reactivity with 1H6 was observed (Figure 3A). This was not because A- or C-containing oligonucleotides could not fold into G4 structures as CD analysis showed a typical absorption peak at 260 nm characteristic of G4 structures (Supplementary Figure S3B). We next examined binding of 1H6 to G4 structures made with different numbers of guanines in the G-tract or different numbers of T's in the loop regions (Figure 3B): (T4G4)2, (T4G3)2, (T4G2)2, (T3G4)2 and (T2G4)2 oligonucleotides. CD analysis revealed that all tested G4 motifs folded into G4 structures except (T4G2)2 and (T4G3)2, (Supplementary Figure S3C). As expected, (T4G4)2 showed strong reactivity with 1H6, but, surprisingly, no reactivity of 1H6 with (T2G4)2 was observed and reactivity of 1H6 with (T3G4)2 appeared to be less pronounced (Figure 3B). These results support the conclusion that 1H6 has exquisite specificity for intermolecular G4 structures with four thymidines in the spacer region but fails to recognize G4 structures that have either C's or G's or less than three thymidines in the backbone oligonucleotides. To further study the binding of 1H6 to poly(T) oligo's we compared the binding of 1H6 to two different poly(T4) constructs in ELISA. We used one oligo with four thymidines poly(T4) and a biotin on the 5΄end (bio-poly(T4)) and a second with biotin at both the 5΄ and 3΄end (bio-poly(T4)-bio). Titrating of 1H6 binding to these substrates revealed that as before, 1H6 showed robust binding to immobilized (T4G4)2 and poly(T15) and not to poly(A15), double stranded AT-rich DNA or hairpin. Strikingly, 1H6 bound strongly to the immobilized poly(T4) construct (bio-poly(T4)-bio) and not to the bio-poly(T4) construct (Figure 3C). These data support that 1H6 binds readily to poly(T4) immobilized on two sides and poly(T15) immobilized on one side but not to poly(T4) immobilized on just one side.
No 1H6 binding to poly(T) oligonucleotides in solution
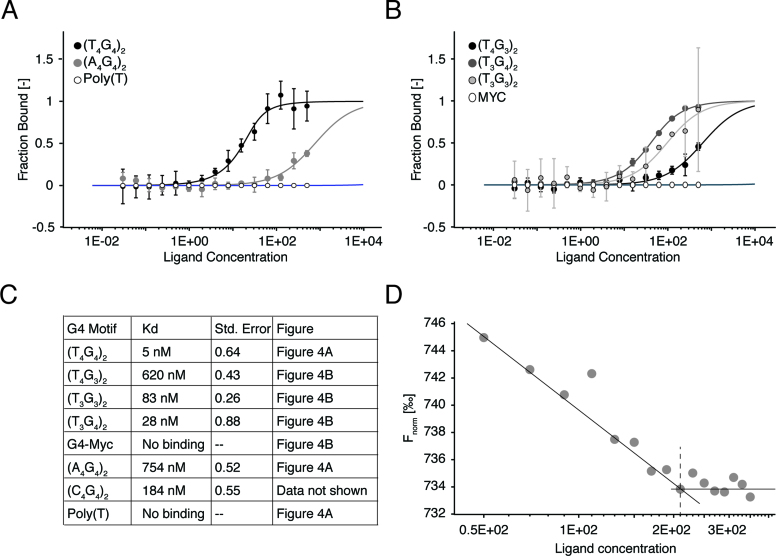
In our ELISA assays, biotinylated oligonucleotides and G4 structures were immobilized using streptavidin in the wells of ELISA plates. This assay configuration, with immobilized DNA substrates, results in a high local concentration of oligonucleotides most likely favoring bivalent binding of monoclonal antibodies. To analyze the binding of 1H6 to G4 structures and poly(T) in solution we turned to MST analysis, which is a powerful method to quantify protein–DNA interactions (7). The result of this analysis revealed strong reactivity (kd 5 nM) of 1H6 with (T4G4)2 but only non-specific binding of 1H6 to poly(T) (Figure 4A). The affinity of 1H6 for (T4G4)2 using MST (5 nM) was considerably less than previously measured in ELISA (0.3 nM (4)). Most likely, this difference reflects the immobilization of antigens in ELISA assays allowing for stronger binding by (bivalent) antibodies. To study 1H6 binding to other G4 structures using MST we analyzed G4 structures with different loop size and composition as well as G-tract length (T4G3)2, (T3G4)2, (T3G3)2, (A4G4)2, (Figure 4A and B). MST analysis clearly showed that 1H6 preferentially binds to (T4G4)2 compared to other G4 structures (see Figure 4C for Kd's of binding). In the past multiple studies on G4 structures were performed using the G4 motif located in the c-myc region (8,9). Interestingly, we did not observe any binding of 1H6 to the G4 structure formed with the c-myc sequence (Figure 4B and C).
Figure 4.
1H6 binds strongly to (T4G4)2 but not to poly(T) in solution. (A and B) MST measurements of 5΄Cy5-labeled oligonucleotides (A) (T4G4)2, (A4G4)2 and poly(T) and (B) (T4G3)2, (T3G4)2 (T3G3)2 and MYC (25 nM), binding with 1H6 at several concentrations (0.03 nM-1 μM). Error bars represent standard deviation with n = 3. (C) Table listing G4 motifs and their respective binding affinities. (D) Determination of the stoichiometry binding of 1H6, titrated in a narrow concentration range (50–330 nM) and 5΄Cy5-labeled (T4G4)2 (200 nM). Linear regression of the saturated and non-saturated data points reveal that 200 nM (T4G4)2 was bound by 215 nM 1H6, yielding a stoichiometry of 1:1. (n = 1).
These data agree with the binding of 1H6 observed in ELISA (Figure 3) and indicate that 1H6 cross-reacts to poly(T) when immobilized in ELISA plates and that 1H6 binding to different G4s is highly variable. MST analysis allowed us to address the binding status of 1H6 to its targets. In titration experiments of 1H6 to (T4G4)2 G4 structures we found that 215 nM of 1H6 binds 200 nM of (T4G4)2, supporting a 1:1 interaction between these molecules (Figure 4D).
DISCUSSION
We previously reported the generation of monoclonal antibody 1H6 with apparent specificity for a variety of intra-and intermolecular G4 structures (4). Using this antibody, we found remarkable differences in nuclear staining between cells of the germline and soma as well as specificity for heterochromatin in the nucleus (5). Here we report additional studies with 1H6 which indicate that this antibody, apart from its exquisite specificity for Stylonchia G4 structures, cross-reacts to single stranded thymidine's that are restricted in their movement. These novel observations complicate the interpretation of 1H6 binding to (sub-) cellular structures and need to be considered when this antibody is used.
We initially became concerned about possible cross-reactivity of 1H6 with non-G4 targets when we observed binding to denatured DNA fibers (Figure 1; Supplementary Figures S1 and 2) at densities that seemed higher than the frequencies reported for ‘G4 motifs’ in the human genome (6,10–12). In subsequent studies, we found that 1H6 also reacts with poly(T) in ELISA (Figure 1A) when the number of T-s exceeded eight nucleotides (Figure 2B). However, when poly(T4) oligo's were immobilized on both ends to ELISA plates strong binding was also observed. Most likely the interaction of poly(T) oligo's with 1H6 requires a certain position or distance of the poly(T)'s relative to the surface of the ELISA plate (Figure 3D). Reactivity between poly(T) and 1H6 was not observed in solution, suggesting that the reactivity in ELISA may reflect very high local densities of thymidines in ELISA in combination with bivalent binding to immobilized poly(T) by 1H6. Strikingly, G4 structures without T's were nor recognized by 1H6 and 1H6 also failed to react with G4 structures made from oligonucleotides with only two T's next to three or more G's (Figure 3). G4 structures that are bound by 1H6 revealed a mixed G4 conformation by CD whereas the ones with less T's showed a clear parallel conformation. This raises the possibility that 1H6 preferentially binds to mixed G4 structures.
Taken together our results suggest that 1H6 has exquisite specificity for adjacent T's, in (T4G4)2 G4 structures as well as for adjacent T's in denatured DNA fibers. This conclusion complicates the interpretation of 1H6 binding. The staining pattern of 1H6 to Stylonychia (5) was found to be essentially identical to what was previously reported with a phage display antibody (13). Most likely in this organism 1H6 staining accurately reflects the binding to actual G4 structures as the 1H6 antibody was raised against Stylonychia telomeric G4 structures. The observation that 1H6 was found to have high affinity for certain G4 structures in solution is encouraging and suggest that some of the previously reported staining with 1H6 could reflect the presence of bona fide G4 structures. This could be the case for a recent study describing the formation and localization of T-rich G4 structures within cells following infection with the human herpes simplex-1 virus (14).
Our confidence that G4 structures are also recognized by 1H6 in metaphase chromosomes (4,5) and heterochromatin (5) is considerably less in view of the novel observation reported here. The 1H6 antibody was found to cross-react in immunofluorescence with specific areas in denatured DNA fibers (Figure 1) and with short stretches (T4) of poly(T) single stranded oligo's in ELISA. Indeed, our observation that G4 structures are enriched in heterochromatin (5) seems difficult to reconcile with a recent antibody-based G4 chromatin immune-precipitation study suggesting that G4 structures are preferentially found in transcribed regions of the genome (6). Of note, in that study 1H6 was reported to co-localize with G4 structures at open chromatin. In Drosophila polytene chromosomes 1H6 staining was mutually exclusive with sites of transcription (5). The observation that cells of the germline in various species show less 1H6 staining than cells of the soma (5) remains puzzling. Could it be that cells of the germline have less condensed DNA and that, as a result, upon fixation, the chance that 1H6 binds to structured T-rich DNA is less? Also, the 1H6 banding pattern observed on salivary gland polytene chromosomes remains intriguing. We reported that the 1H6 banding pattern overlaps with the banding pattern observed with antibodies to the SUUR protein (5). SUUR binding sites are known to reflect late and under-replicated regions of the genome that contain DNA damage (15). Does 1H6 recognize single stranded T-rich DNA at SUUR binding sites instead of G4 structures as we originally proposed? The new findings regarding the specificity of 1H6 reported in this study complicate the interpretation of 1H6 staining results and point to the need to perform additional studies to confirm that 1H6 binding to DNA indeed reflects the presence of G4 structures.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Brad Johnson, Robert Brosh, Yuri Moshkin and Dipankar Sen and Stefan Juranek for comments on the manuscript. Roland Hoffmann, Wierd Kooistra, William Kennedy, Sarra Merzouk and Evert-Jan Uringa are thanked for discussions and experiments with DNA fibers. We thank Wesley Browne (University of Groningen) for his help with Circular Dichroic measurements and Arnold Driessen and Sabrina Koch for help with MST analysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Research Council Advanced Grant (ROOTS-Grant Agreement) [294740 to P.M.L.]; European Research Council Starting Grant (to K.P.). Funding for open access charge: European Research Council [638988-G4DSB to KP].
Conflict of interest statement. None declared.
REFERENCES
- 1. Sen D., Gilbert W.. Guanine quartet structures. Methods Enzymol. 1992; 211:191–199. [DOI] [PubMed] [Google Scholar]
- 2. Weldon C., Eperon I.C., Dominguez C.. Do we know whether potential G-quadruplexes actually form in long functional RNA molecules?. Biochem. Soc. Trans. 2016; 44:1761–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rhodes D., Lipps H.J.. G-quadruplexes and their regulatory roles in biology. Nucleic Acids Res. 2015; 43:8627–8637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M. Jr, Sen D., Lansdorp P.M.. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014; 42:860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hoffmann R.F., Moshkin Y.M., Mouton S., Grzeschik N.A., Kalicharan R.D., Kuipers J., Wolters A.H., Nishida K., Romashchenko A.V., Postberg J. et al. . Guanine quadruplex structures localize to heterochromatin. Nucleic Acids Res. 2016; 44:152–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. et al. . G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 7. Zhang W., Duhr S., Baaske P., Laue E.. Microscale thermophoresis for the assessment of nuclear protein-binding affinities. Methods Mol. Biol. 2014; 1094:269–276. [DOI] [PubMed] [Google Scholar]
- 8. Simonsson T., Sjoback R.. DNA tetraplex formation studied with fluorescence resonance energy transfer. J. Biol. Chem. 1999; 274:17379–17383. [DOI] [PubMed] [Google Scholar]
- 9. Rangan A., Fedoroff O.Y., Hurley L.H.. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001; 276:4640–4646. [DOI] [PubMed] [Google Scholar]
- 10. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S.. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 11. Eddy J., Maizels N.. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008; 36:1321–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bedrat A., Lacroix L., Mergny J.L.. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016; 44:1746–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schaffitzel C., Berger I., Postberg J., Hanes J., Lipps H.J., Pluckthun A.. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:8572–8577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Artusi S., Perrone R., Lago S., Raffa P., Di Iorio E., Palu G., Richter S.N.. Visualization of DNA G-quadruplexes in herpes simplex virus 1-infected cells. Nucleic Acids Res. 2016; 44:10343–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andreyeva E.N., Kolesnikova T.D., Belyaeva E.S., Glaser R.L., Zhimulev I.F.. Local DNA underreplication correlates with accumulation of phosphorylated H2Av in the Drosophila melanogaster polytene chromosomes. Chromosome Res. 2008; 16:851–862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.