Abstract
Repair of uracils in DNA is initiated by uracil DNA glycosylases (UDGs). Family 1 UDGs (Ung) are the most efficient and ubiquitous proteins having an exquisite specificity for uracils in DNA. Ung are characterized by motifs A (GQDPY) and B (HPSPLS) sequences. We report a novel dimeric UDG, Blr0248 (BdiUng) from Bradyrhizobium diazoefficiens. Although BdiUng contains the motif A (GQDPA), it has low sequence identity to known UDGs. BdiUng prefers single stranded DNA and excises uracil, 5-hydroxymethyl-uracil or xanthine from it. BdiUng is impervious to inhibition by AP DNA, and Ugi protein that specifically inhibits family 1 UDGs. Crystal structure of BdiUng shows similarity with the family 4 UDGs in its overall fold but with family 1 UDGs in key active site residues. However, instead of a classical motif B, BdiUng has a uniquely extended protrusion explaining the lack of Ugi inhibition. Structural and mutational analyses of BdiUng have revealed the basis for the accommodation of diverse substrates into its substrate binding pocket. Phylogenetically, BdiUng belongs to a new UDG family. Bradyrhizobium diazoefficiens presents a unique scenario where the presence of at least four families of UDGs may compensate for the absence of an efficient family 1 homologue.
INTRODUCTION
Uracil (as dUMP) can be incorporated into DNA by DNA polymerases (from dUTP, forming an A:U pair) or arise by deamination of cytosines (resulting in G:U pairs) (1,2). If unrepaired, A:U base pairs can hinder recognition by DNA binding proteins (3,4) while G:U base pairs can cause G:C to A:T mutations in the subsequent rounds of DNA replication, posing a threat to genomic integrity. Enzymes of the uracil DNA glycosylase (UDG) superfamily initiate base excision repair to remove uracil residues (5–7). Five distinct families of UDGs have so far been established which differ in their primary amino acid sequences and substrate specificities. However, UDGs possess a conserved α/β structural fold and seemingly a common evolutionary origin (8,9). Ubiquitously found family 1 UDGs (Ung), exemplified by the well-studied ‘master catalyst’ EcoUng, are the most efficient of UDGs and show an exquisite specificity for uracil in DNA (10). Conserved orthologues of family 1 are found in nearly all organisms studied, including many viruses (5,10). This UDG family is characterized by motif A (GQDPY) responsible for catalysis and motif B (HPSPLS) implicated in the stabilization of the enzyme substrate complex (11–14). Interestingly, phage PBS-1 or 2 that infects Bacillus subtilis encodes a protein inhibitor, Ugi that forms a physiologically irreversible non-covalent complex in 1:1 stoichiometry specifically with family 1 UDGs to allow occurrence of uracil in their genomes (15,16).
While EcoUng was the first UDG to be identified and extensively studied, several UDGs have been reported since then (5,17–21). Family 2 UDGs referred to as Mug/TDG have GINPG as motif A and MPSSSAR as motif B. While Mug/TDG excise uracil from double stranded (ds) DNA (G:U and A:U pairs) with a low efficiency, they also excise thymine from T:G pairs (18,22–24). GMNPG and HPSPRN define motif A and motif B for family 3 UDGs (SMUG). SMUGs excise uracil from single stranded (ss) DNA with higher efficiency than from dsDNA (19,25). Family 4 and family 5 UDG members contain a 4Fe–4S cluster. Motif A and Motif B are represented by GE(A/G)PG and HPAAVL, respectively, for family 4, and by GLAPA and HPSPLN for family 5 (20,21,26,27). While family 4 UDGs act on ssDNA and dsDNA substrates, UDG activity of family 5 is enhanced on dsDNA substrates.
Members of Rhizobiaceae are an important group of soil bacteria that are recognized for their ability to fix atmospheric nitrogen through a symbiotic association with root nodules of leguminous plants. Formation and maintenance of root nodules in leguminous plants represents an interesting scenario of oxidative and nitrosative stress to invading rhizobial cells. To establish symbiotic associations with leguminous plants, rhizobia should efficiently overcome the initial defense response elicited by the plant. Similar to the response to pathogens, plants also respond to rhizobial infection through an oxidative burst, with enhanced production of superoxide and H2O2 (28–30). Further, high rates of respiration to support the process of nitrogen fixation along with autoxidation of leghemoglobins in root nodules can produce high levels of reactive oxygen species (ROS) (31–41). In addition to ROS, nodules also generate reactive nitrogen species (RNS) like nitric oxide (NO) and peroxynitrite (ONOO-) (42,43).
Oxidative and nitrosative stress result in various kinds of lesions in DNA including deamination of cytosines, posing a threat to the genomic integrity of rhizobia (44,45). Interestingly, their genome contains high G+C content making them inherently more susceptible to cytosine deamination events that produce uracil in the genome which can lead to transition mutations. Hence, one could expect these organisms to have evolved sophisticated strategies of DNA repair to cope with their hostile environment. However, aspects of DNA repair in rhizobium are currently under explored. In the present study, we used a member of α-proteobacteria, Bradyrhizobium diazoefficens (previously B. japonicum) (46) as our model organism, which is well known for its ability to form nitrogen fixing root nodules through a symbiotic association with Glycine max (47,48) to explore aspects of uracil excision repair and report the presence of at least four UDGs in this organism with a distinct absence of a classical family 1 UDG. Furthermore, we demonstrate that in Bradyrhizobium diazoefficens there exists a unique base excision repair system catalyzed by a member of a new UDG family.
MATERIALS AND METHODS
Bacterial strains, plasmids, DNA oligomers, media and growth conditions
Bacterial strains and plasmids are listed in Supplementary Table S1. DNA oligomers are listed in Supplementary Table S2. Escherichia coli CC102 (49) was obtained from Coli Genetic Stock Centre (CGSC). E. coli strains were grown in Luria-Bertani broth (LB) or on LB containing 2% (w/v) agar (Difco, USA) at 37°C. Bradyrhizobium diazoefficiens was grown in TY medium (0.5% tryptone, 0.3% yeast extract, 6 mM CaCl2) or on TY with 2% agar at 30°C. Ampicillin (Amp), kanamycin (Kan) and chloramphenicol (Cm) were added to media at a concentration of 100, 25 and 30 μg ml−1 respectively.
Cell lysate preparation
Saturated cultures were obtained in 2 ml media by incubation at 37°C overnight for E. coli strains and at 30°C for 5–6 days for B. diazoefficiens and then subcultured (1% for E. coli and 10% for B. diazoefficiens) into 2 ml media and incubated until the OD600 reached ∼0.6. Cells were pelleted, re-suspended in 200 μl TME (25 mM Tris–HCl pH 8.0, 2 mM β-mercaptoethanol, 1 mM Na2EDTA), sonicated with 2 s alternate pulses for 20 s, six to seven times with 50–60 s intervals and centrifuged at 13 000 rpm for 1.5 h on table top centrifuge at 4°C. The supernatant was collected, mixed with an equal volume of 2× storage buffer (80% glycerol, 1 mM β-mercaptoethanol, 20 mM Tris–HCl pH 8.0 and 200 mM NaCl) and stored at –20°C. Protein concentration in cell extracts was estimated by the Bradford assay using BSA as standard (50).
Cloning of putative UDGs
Bdi-ung ORF was PCR amplified from genomic DNA with forward (BdiUng Fp, 5΄-GCATTTCATATGCTCACAGAATTCG-3΄) and reverse (BdiUng Rp, 5΄-GCCCAAAGCTTCGGGTTTGCGGCCG-3΄) primers containing NdeI and HindIII sites, respectively and Pfu DNA polymerase. The reaction was heated at 94°C for 4 min followed by 35 cycles of incubations at 94°C for 1 min, 50°C for 45 s and 70°C for 50 s. The PCR product (856 bp) was ligated into pTrcNdeIHis vector through NdeI and HindIII sites, such that a hexa-His tag is attached to the N-terminus of the protein. The clones were confirmed by DNA sequencing. Other putative UDG genes, including bll3023, blr5068 and blr6661, were also similarly cloned into pTrcNdeIHis vector (with hexa-His tag, through NdeI and HindIII sites).
Purification of proteins
For protein overexpression and purification, plasmid pTrcBdiUng was introduced into E. coli Δung strain by transformation. The E. coli TG1Δung::cm harbouring pTrcBdiUng was inoculated into 10 ml LB containing Amp and Cm and grown overnight. The inoculum was diluted 1:100 (v/v) (1%) into 600 ml fresh medium and grown at 37°C to an OD600 of ∼0.6. The culture was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and grown for another 4–5 h. Cells were harvested by centrifugation (7500 rpm, 4°C, 5 min), resuspended in buffer A (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 10% glycerol v/v, 2 mM β-mercaptoethanol) containing 30 mM imidazole, sonicated and centrifuged at 29 000 rpm (Avanti™ J-30I, JA30.50 Ti) for 2 h 30 min at 4°C. The supernatant was loaded onto a 5 ml Ni-NTA column pre-equilibrated with buffer A, washed with 30 ml of buffer A and eluted with a linear gradient of 30 mM to 1 M imidazole in buffer A. Aliquots from the fractions were analyzed on 15% SDS-PAGE, fractions containing pure protein were pooled, dialyzed against buffer A, then against storage buffer (20 mM Tris–HCl pH 8.0, 300 mM NaCl, 50% glycerol v/v, 2 mM β-mercaptoethanol), and stored at –20°C. Protein concentration was estimated by the Bradford assay using BSA as standard (50). Other UDGs were also purified using the same procedure, except that Bll3023 was purified from E. coli TG1, cultured in the presence of 0.01% FeCl3.
DNA oligomers and their radiolabelling
The 24 nucleotide long ssU9 (ctcaagtgUaggcatgcaagagct) and the 37 nucleotide long tetra-loop hairpin GU9 (ctcaagtgUaggcatgcttttgcatgcctgcacttga) containing uracil in the stem region, were used as ssDNA and dsDNA substrates, respectively. Other DNA oligomers are listed in Supplementary Table S2. DNA oligomers (10 pmol) were 5΄ 32P-end labelled using 10 μCi of [γ-32P] ATP (6000 Ci/mmol) and T4 polynucleotide kinase and purified on Sephadex G-50 minicolumns (1 ml). To generate duplexes (HmU and ssX9 with ssU9-Compl-G, Compl-T with G-oligo, and I with Compl-G), 5΄ 32P- end labelled DNA oligomers (with modified bases) were annealed with complementary oligomers in approximately equimolar ratios in 40 mM Tris–HCl pH 7.5, 20 mM MgCl2 and 50 mM NaCl, heated at 90°C for 5 min and then slowly cooled to room temperature.
Base excision and inhibition assays
Appropriate amounts of cell extract/pure protein were incubated with 5΄ 32P-end labelled DNA oligomers (∼10000 cpm; ∼10 nM concentration) in 10 μl reaction volumes in UDG buffer (50 mM Tris–HCl pH 8.0, 1 mM Na2EDTA, 1 mM DTT and 25 μg ml−1 of BSA) at 37°C for the indicated times. The reactions were stopped by the addition of 8 μl each of 0.2 N NaOH and formamide dye (80% formamide, 0.05% each of bromophenol blue and xylene cyanol FF, 10 mM NaOH and 2 mM Na2EDTA) and boiled at 90°C for 15 min. Aliquots (20 μl) were analyzed on 15% polyacrylamide (19:1) 8 M urea gels, exposed to phosporimaging screens and imaged using Fujifilm analyzer. To see if BdiUng is inhibited by Ugi, BdiUng (1 μg, ∼32 pmol of monomer) was pre-incubated with excess Ugi (1 μg; ∼100 pmol) in 4 μl volume for 30 min before the reaction.
Gel filtration chromatography
Proteins were subjected to gel filtration chromatography using Superdex 75 (GE Healthcare) column equilibrated with a buffer containing 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 2 mM β-mercaptoethanol, 10% glycerol. Elution was performed with the same buffer at a flow rate of 1 ml/min and the absorbance at 280 nm was recorded to obtain the elution profile.
EMSA for checking Ugi binding
Ugi (∼8 μg, ∼850 pmol) was incubated with BdiUng (5 μg, ∼160 pmol of monomer) or EcoUng (2 μg, ∼80 pmol) in 25 μl volumes in UDG buffer at room temperature for 20 min and then stored on ice (4°C) for 15 min. Loading dye (5 μl) containing 30% (v/v) glycerol, 0.25% (w/v) bromophenol blue and 0.25% (w/v) xylene cyanol FF was added, and the samples were analyzed on 15% native PAGE (29:1 crosslinking, pH 6.8). After electrophoresis, the gel was stained using Coomassie brilliant blue.
Impact of uracil and AP DNA on activity of BdiUng
To check the effect of free uracil and AP DNA on uracil excision enzymes, varying concentrations of free uracil or unlabelled ssF9 (ctcaagtgFaggcatgcaagagct, where F is tetrahydrofuran, a stable mimic of AP site provided by Midland Certified Reagent Co.) were pre-incubated with 100 pg (∼4 fmol) EcoUng or 10 ng (∼0.32 pmol monomer) BdiUng at room temperature followed by incubation on ice for 15 min. The reaction mixtures were followed by addition of 5΄ end 32P-labelled ssU9 (∼0.1 pmol) and incubated at 37°C for 30 min for uracil excision.
Generation of site directed mutation in BdiUng, and its characterization
PCR based method (see supplementary material) was used to generate A59Y mutant of BdiUng. Mutant protein was purified from E. coli TG1Δung::cm strain using Ni-NTA column chromatography as described for the wild type BdiUng.
Generation of ung knockout in CC102 strain
A P1 phage lysate generated using E. coli MG1655Δung::kan was used to transduce the Δung::kan allele into E. coli strain CC102. Transfer of the Δung::kan allele into the transductants was verified by diagnostic PCR (with EcoUng-up-Fp and EcoUng-dn-Rp primers which flank the ung locus) and uracil excision assays (Supplementary Figure S1).
Assay for reversion of Lac− to Lac+ in E. coli
E. coli CC102 with a mutation in lacZ was used to assay for reversion to Lac+. Isolated colonies of this strain and its Δung::kan derivative were transformed with pTrc99c or EcoUng/BdiUng expression constructs. Cells were inoculated in replicates of ten in 2 ml LB supplemented with Amp and grown for 24 h with shaking at 37°C. Serial dilutions of saturated cultures were made and 50 μl of 10−5 dilution was plated onto M9 minimal media plates containing 0.2% glucose for viable counts. The saturated culture (1.5 ml) was spun down and plated onto M9 minimal media containing 0.2% lactose for counts of Lac+ revertants. The plates were incubated at 37°C until colonies appeared (24 h for glucose plates and 72 h for lactose plates). The reversion frequency was obtained by dividing the number of revertants (per ml culture) by the number of bacteria plated (viable count).
Crystallization and refinement
The purified BdiUng was dialyzed against 20 mM Tris–HCl, 50 mM NaCl, 5 mM β-mercaptoethanol, pH 8.5 and concentrated to 10 mg/ml before freezing in liquid nitrogen and storage at -70°C. Crystals of BdiUng were generated using sitting-drop vapor-diffusion in 20% PEG3350, 200 mM sodium citrate, 100 mM sodium citrate/citric acid pH 4.0 with a protein:precipitant ratio of 1:1. BdiUng was co-crystallized with ligands (uracil, and xanthine) using 9:1 ratio (BdiUng:saturated solution of uracil or xanthine) with an incubation period of 15 min before crystallization setup. Before data collection, the crystals were cryocooled to 93 K using a cryoprotectant consisting of mother liquor with 30% glycerol. The native BdiUng crystal diffracted at 2 Å, selenomethionine-labelled BdiUng crystal at 2.3 Å, BdiUng-uracil crystal at 2.9 Å and BdiUng-xanthine crystal at 2.8 Å. The diffraction data were processed and scaled using HKL2000 (53). Structure refinement and model building were performed using the programs COOT (54) and PHENIX (55). The structure was visualized using Pymol software (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC).
RESULTS
Identification of UDG homologues and Blr0248 (BdiUng) as putative UDGs
Analysis of B. diazoefficiens USDA110 genome through blast searches indicated the absence of a conserved family 1 homologue in this organism. Further analysis based on motif based BLAST (NCBI) searches revealed many putative UDGs (Table 1), among which most could be classified into established families of UDGs: blr6661 (family 2 MUG), bll2523 (family 4 UdgA), blr5068 (family 5 UdgB) and bll3023 (family 4 UdgX). Of particular interest among the identified ORFs was blr0248 which codes for a 272 amino acid long protein Q89XRO_BRAJA containing a putative motif A (GQDPA) similar to family 1 UDGs (referred to as BdiUng). However, BdiUng lacked the motif B of family 1 UDGs and showed low sequence similarity to established UDG families (Supplementary Figure S2). Homology searches through BLAST revealed several homologues of BdiUng from many organisms (Figure 1), which form the putative members of a new family of UDGs. Homologues of BdiUng were found in various lineages of bacteria including actinobacteria (Streptacidiphilus rugosus, Kitasatospora azatica, Intrasporangium calvum, Streptomyces xylophagus, Frankia sp. EuI1c and Janibacter sp. HTCC2649) and β-proteobacteria (Burkholderia ubonensis, Chromobacterium haemolyticum, Nitrosospira sp. NpAV). Evolutionary analysis based on neighbor joining method (Supplementary Figure S3) showed that these bacteria possess a new family of UDGs related to family 1.
Table 1. Putative UDGs identified from Bradyrhizobium diazoefficiens through genome analysis.
| Gene | Protein | Motif |
|---|---|---|
| blr0248 | Q89XRO_BRAJA | GQDPA |
| bll2523 | Q89S82_BRAJA | UDG F4 TTUDGA like |
| bll3023 | Q89QV1_BRAJA | UDG F4 TTUDGA like |
| blr5068 | Q89K49_BRAJA | UDG F5 TTUDGB like |
| blr6661 | Q89FNB_BRAJA | UDG F2 MUG |
Figure 1.
Multiple sequence alignment of BdiUng with putative homologues. The protein sequence of BdiUng was aligned against the protein sequences of FUDG (from Frankia sp. EuI1c), SrUDG (from Streptacidiphilus rugosus), KaUDG (from Kitasatospora azatica) and NsUng (from Nitrosospira sp. NpAV) using clustal omega. The schematic representation was obtained by using BOXSHADE (www.ch.embnet.org). Identical residues are shaded in black while similar residues in grey. Putative motif A and B are additionally shaded in red and blue, respectively.
Cloning and purification of putative UDGs
We cloned ORFs of blr0248, blr6661, blr5068 and bll3023 from B. diazoefficiens USDA110 into the pTrcNdeIHis expression vector along with a 20 amino acid long pre-sequence containing a hexa-His tag. All UDGs other than Bll3023 were purified from an E. coli TG1Δung::cm strain to ensure that the UDG preparations were free from EcoUng contamination (Supplementary Figure S4). Since UdgX proteins are toxic in ung− strains (56), Bll3023 was purified from E. coli TG1. As shown in Supplementary Figure S5, Bll3023, Blr6661, Blr5068 possessed uracil excision activities, which corresponded to their known classes of proteins, and were not pursued any further.
BdiUng: a novel Ugi insensitive UDG with broad substrate specificity
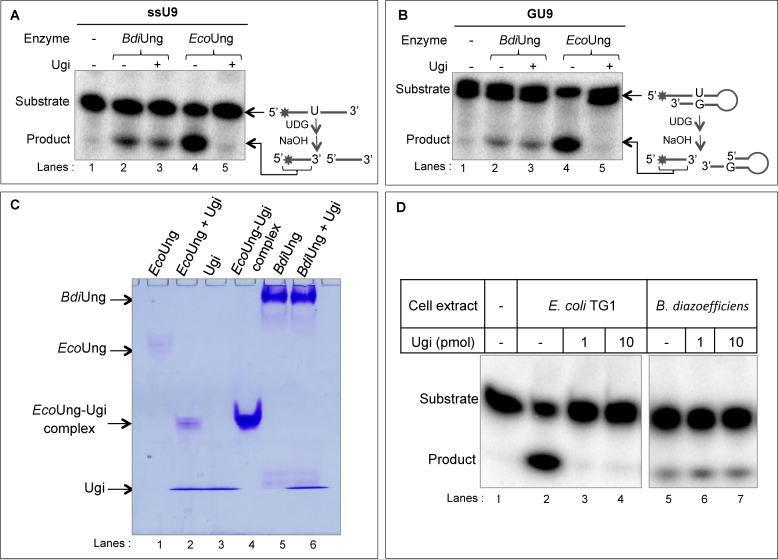
The activity of purified BdiUng was analyzed on uracil containing ssDNA and dsDNA oligomers (ssU9 and GU9) and was found to be active on both the substrates (Figure 2A and B, compare lanes 2 with 1). BdiUng reactions with/without preincubation with Ugi showed product bands of similar intensity (compare lanes 2 and 3). However, under the same conditions, preincubation of EcoUng with Ugi completely inhibited uracil excision activity (compare lanes 5 with 4 and 1). These observations suggest that Ugi does not inhibit BdiUng to any detectable levels and reveals a fundamental difference of BdiUng to family 1 UDG members.
Figure 2.
Uracil excision assays with single stranded and double stranded substrates. Assays were carried out at 37°C for 30 min with 1 μg of BdiUng (∼32 pmol monomer) or EcoUng (∼4 pmol) with (+) or without (–) pre-incubation with 1 μg of Ugi (∼100 pmol) as described in Materials and Methods using ∼0.1 pmol of ssU9 (A) or GU9 (B) DNAs, and resolved on 8 M urea PAGE (15%). (C) Electrophoretic mobility shift assay to analyze complex formation of BdiUng with Ugi. After incubation of enzyme with/without Ugi, reaction products were resolved on 15% native PAGE (29:1 crosslinking, pH 6.8). While complex EcoUng-Ugi complex formed is found to migrate adjacent to pure complex control, no band indicative of BdiUng-Ugi complex could be seen. (D) Uracil excision assay on ssU9 with 2 μg of B. diazoefficiens cell free extracts with/without pre-incubation with varying amounts of Ugi. E. coli TG1 cell free extract (2 μg) was used as a control. Reaction products were resolved on 8 M urea PAGE (15%).
Although the activity of BdiUng was not inhibited by Ugi, it was still possible that it formed a complex with Ugi (like family 1 UDGs). Therefore, electrophoretic mobility shift assays were carried out using native PAGE after pre-incubation of BdiUng or EcoUng with Ugi (Figure 2C). As a control, EcoUng–Ugi complex formed in vivo was also loaded (lane 4). While pre-incubation of EcoUng with Ugi resulted in a shift in the migration of EcoUng in native PAGE (compare lane 2 with lanes 1 and 3), BdiUng did not show any shift in its migration (nor a new band) upon pre-incubation with Ugi (compare lanes 5 and 6) suggesting the absence of complex formation. The EcoUng-Ugi (lane 2) complex migrated to the same position for purified EcoUng–Ugi complex (lane 4) formed in vivo (57).
Even though the bioinformatics and biochemical analyses with BdiUng suggested that B. diazoefficiens lacks a classical family 1 UDG, to rule out other unidentified proteins as family 1 UDG (Ung), we decided to carryout UDG assays with cell free extracts prepared from B. diazoefficiens (Figure 2D). As expected, the assays revealed UDG activity (compare lane 5 with 1). However, the activity was not diminished by Ugi (compare lane 5 with lanes 6 and 7). As a control, UDG activity of E. coli cell free extracts was inhibited by Ugi (compare lane 2 with lanes 3 and 4). These observations suggest that B. diazoefficiens lacks Ugi sensitive UDG activity.
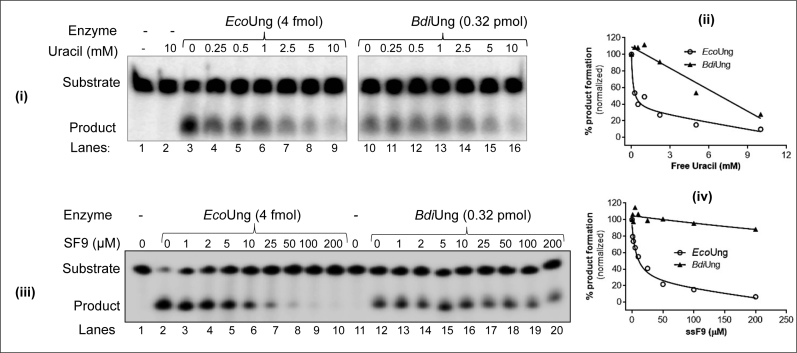
To check if BdiUng is inhibited by either of the reaction products (uracil, or AP DNA) we employed a uracil excision assay with 5΄ end 32P-labelled ssU9, in the presence of varying concentrations of uracil or a tetrahydrofuran-containing DNA (ssF9), a stable mimic of AP DNA (Figure 3). While free uracil caused a 5-fold reduction in BdiUng activity at 10 mM concentration (panels i and ii), ssF9, in spite of its high concentrations (of up to 200 μM) did not significantly inhibit BdiUng (panels iii and iv). As a control, both the products inhibited uracil excision activity of EcoUng (panels i–iv).
Figure 3.
Uracil excision assays for product inhibition. Uracil excision assay using ∼0.1 pmol 5΄ end 32P-labelled ssU9 substrate with the indicated amounts of EcoUng or BdiUng in the presence of varying concentrations of uracil (i and ii), and ssF9 (a stable AP DNA mimic harbouring tetrahydrofuran at position 9) (iii and iv) were carried out as described in Materials and Methods. Reactions were resolved on 8M urea PAGE (15%) and imaged. The estimated values (%) of substrate (S) to product (P) conversion [P/(S + P) × 100] are plotted against the concentrations of the inhibitor in the reaction. The data were fitted with the one site-total of saturation binding equations (Y = Bmax * X/(Kd + X) + NS*X + background) (where Y is specific binding/activity, Bmax is the maximum specific binding/activity in the same units as Y, Kd is the equilibrium binding constant in the same units as X, NS is the slope of nonspecific binding in Y unit divided by X unit and background is the amount of nonspecific binding) using GraphPad Prism software. Amounts of product formed in control (without free uracil/AP DNA) reaction in panels i and iii were scaled to 100%, and the remaining activities shown relative to this reference to generate plots in panels ii and iv.
Substrate specificity of BdiUng
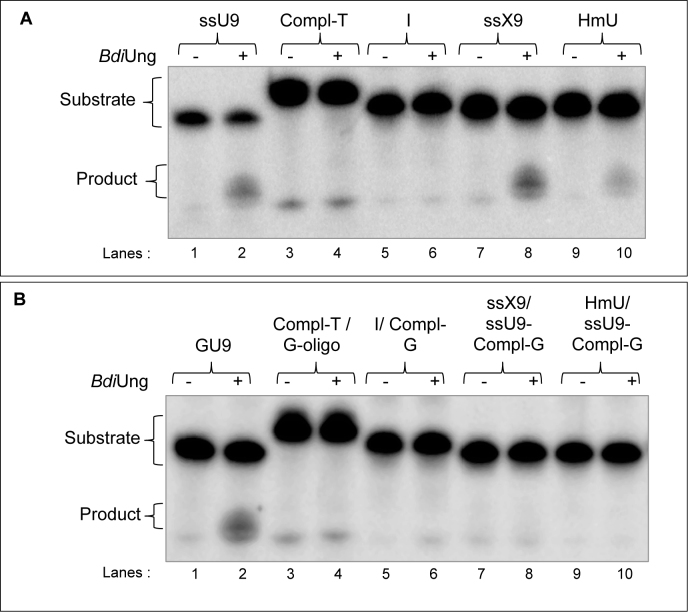
To investigate if BdiUng is highly specific towards uracil in DNA (like family 1 UDGs, Ung) or relaxed in substrate choice like UDGs belonging to some other families, we used 5΄ end 32P-labelled DNA oligomers containing different base modifications. BdiUng utilized DNA oligomers containing xanthine (X) (Figure 4A, compare lane 8 with 7) and a uracil analogue 5-hydroxymethyluracil (HmU) (Figure 4A, compare lane 10 with 9) in ssDNA as substrates. However, it did not utilize DNA oligomer containing inosine (I) in ssDNA or dsDNA contexts to any detectable levels (Figure 4A and B). Thymine DNA glycosylase activity was also not detected (over the control reaction) using a G:T pair containing dsDNA (Figure 4B, lanes 3 and 4).
Figure 4.
BdiUng shows broad substrate specificity. Excision assays using BdiUng (1 μg, ∼32 pmol monomer) with (A) ssDNA, and (B) dsDNA containing modified bases (∼0.1 pmol each); inosine (I), xanthine (X) and 5-hydroxymethyluracil (HmU) were carried out at 37°C for 30 min as described in Materials and Methods. The reactions were resolved on 8 M urea PAGE (15%) and imaged.
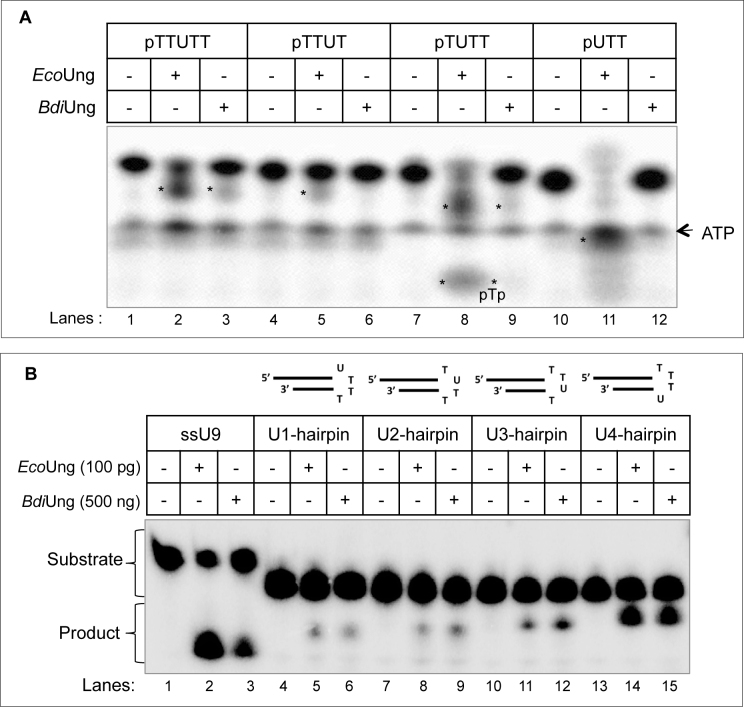
To determine the minimum substrate from which BdiUng could excise uracil, we used 5΄ end 32P-labelled DNA oligomers having a different number of bases on the 5΄ and 3΄ sides of the uracil (Figure 5A). EcoUng whose activity on these substrates has been well characterized (51) was used as control. With reference to EcoUng, BdiUng activity was detectable on oligomers having one phosphorylated nucleotide 5΄ of the uracil (compare lane 9 with 8) and the two nucleotides 3΄ of it (compare lane 3 with 2). However, excision of uracil from the 5΄ terminal or the penultimate position (from the 3΄ end) was undetectable. Thus, the minimum size substrate required for detectable excision of uracil by BdiUng is pNUNN (Figure 5A).
Figure 5.
Excision of uracil from different structural contexts. Uracil excision assays with DNA oligomers (∼0.1 pmol each) containing a different number of nucleotides on either side of uracil (A) or with the hairpin DNA oligomers containing uracil in different positions in tetraloop (B) were carried out at 37°C as described in Materials and Methods. (A) Reactions were carried out with 1 μg of either protein (∼40 pmol EcoUng or ∼32 pmol BdiUng monomer) for 30 min. More than one product band is observed in some reactions because incomplete alkaline cleavage of AP site (first reaction leaves a semialdehyde residue on 3΄ end of 5΄ product and the second reaction removes it, producing a 3΄ phosphate on the 5΄ product; products are marked with asterisks). (B) Reactions were performed for 30 min with 100 pg (4 fmol) and 500 ng (∼16 pmol monomer) of EcoUng and BdiUng, respectively. Reaction products were resolved on 8 M urea PAGE (15%).
Genomes with a higher G+C content are inherently prone to forming stable intra-molecular structures such as the hairpin loops due to various topological changes during physiological processes like transcription and replication. The hairpin like structures formed on single stranded regions may lead to the appearance of unpaired C residues highly prone to deamination. In addition, in the loop structures such as the tetraloops, uracils may be fixed into extrahelical (flipped out) positions (52,58). Earlier studies have shown inefficient excision of uracils from the tetraloops by EcoUng (59). However, the uracil excision activity of Ung from Mycobacterium smegmatis, a G+C rich organism (like B. diazoefficiens) was found to be more efficient than that of EcoUng (60). Thus, we checked the uracil excision activity of BdiUng on four hairpin DNA structures that contained uracil at different positions of a tetraloop hairpin (Figure 5B). BdiUng excised uracil from all the loop positions. However, unlike EcoUng, which showed a significant decrease in excision of uracil from the U1-hairpin, U2-hairpin and U3-hairpin substrates when compared with the excision of uracil from ssDNA (compare lanes 5, 8 and 11 with lane 2), BdiUng was not as severely compromised on the same hairpin substrates when compared with its activity on ssDNA (compare lanes 6, 9, 12 with lane 3). Excision of uracil by BdiUng from U4-hairpin was as efficient as its excision of ssDNA (compare lane 15 with 3).
Overall structure of BdiUng
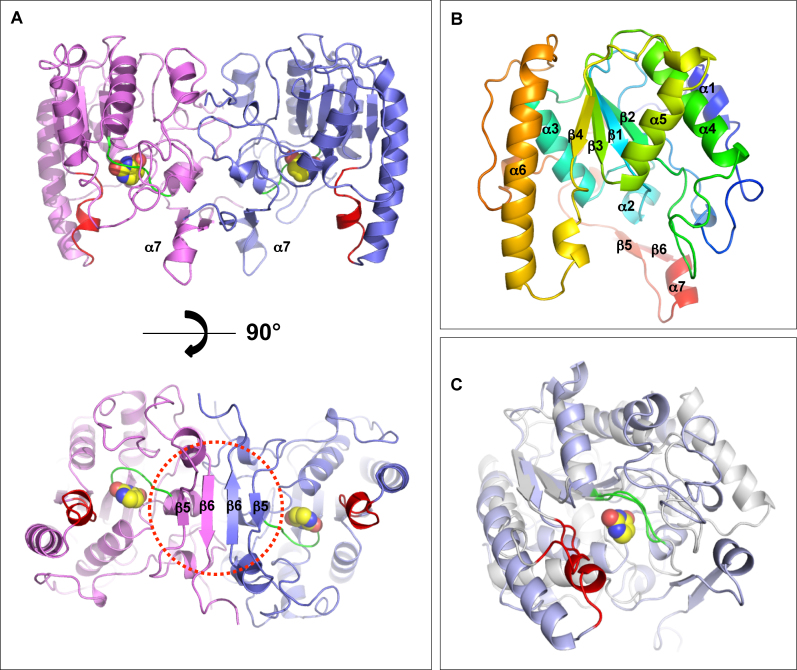
The structure of BdiUng was determined using the single anomalous dispersion (SAD) method. The detailed statistics for the structure are described in Table 2. In agreement with the gel filtration chromatography results (Supplementary Figure S6), the overall structure is a dimer with a typical α/β/α sandwich fold of UDG proteins for each monomer such that a central β-sheet is layered by α-helixes on both sides (Figure 6A and B). Using the Dali server, the nearest structural homologue occurred with a family 4 UDG from Sulfolobus tokadaii (sequence identity, 12%, Supplementary Figure S7) with a Z-score of 14.4 and a 1.1 Å rmsd over 31 Cα atoms (61). However, unlike family 4 UDGs, BdiUng lacks the conserved cysteine residues for metal chelation and hence does not contain an iron-sulfur cluster [4Fe–S] in the structure, which is a characteristic feature of family 4 UDGs. The superposition with family 1 Ung shows the conservation of the scaffold core domain with four β-strands in the center and α-helices on both sides, with 7.6 Å rmsd over 70 Cα atoms (Figure 6C). The C-terminal region extends to constitute a unique hairpin like β-strands β5β6 (residues A248–R250, T261–T265) in each monomer that associate to form a β-sheet in the dimer (Figure 6A). The dimer association of BdiUng constituted by both the N-terminal and the C-terminal of each monomer yields 1781 Å2 of buried interface, 13.4% of the total surface area. The N terminal is mostly involved in hydrophobic interactions with residues (M1, L2, T3, F5, E11, R15, A19, N20, Y21) whereas the C terminal interface (255–272) is stabilized by hydrogen bonds of the antiparallel β-sheet (R260, I262, I264, I266, D268) with additional charged interaction between D268 and R260 of the adjacent monomers (Supplementary Figure S8). The surface analysis of BdiUng shows a narrow groove with predominately positively charged residues (69-75, 165–168 and 190–194), a probable site for the binding of negatively charged phosphate backbone of ssDNA while the additional positively charged region (146–151) could be utilized for binding of separated strand after flipping of uracil towards the ligand binding pocket (Supplementary Figure S9).
Table 2. Data collection and structure refinement statistics.
| Ligand | BdiUng native | Selenomethionine-labelled BdiUng | Uracil bound BdiUng | Xanthine bound BdiUng |
|---|---|---|---|---|
| X-ray source | 7A, PAL | 7A, PAL | Home source | 5C, PAL |
| Space group | P212121 | P212121 | C121 | P212121 |
| Unit cell dimension | ||||
| a, b, c (Å) | 70.65, 90.03, 255.67 | 70.95, 90.06, 256.26 | 209.53, 89.63,143.78 | 70.02, 89.90, 255.34 |
| α, β, γ (°) | 90.00, 90.00, 90.00 | 90.00, 90.00, 90.00 | 90.00, 96.20, 90.00 | 90.00, 90.00, 90.00 |
| Resolution (Å) | 1.95 | 2.28 | 2.86 | 2.80 |
| R sym (%)a,b | 13.2 (56.7) | 6.5 (60.8) | 14.0 (33.3) | 14.0 (52.8) |
| I/σ(I) | 13.34 (1.78) | 21.39 (9.41) | 09.67 (3.43) | 15.13 (4.59) |
| Completeness (%) | 98.71 | 91.73 | 83.20 | 99.91 |
| Redundancy | 4.0 (3.2) | 7.0 (6.5) | 2.7 (3) | 7.0 (6.3) |
| Refinement | ||||
| Resolution (Å) | 1.95 | 2.28 | 2.86 | 2.80 |
| Unique reflection | 117958 | 70019 | 50734 | 40422 |
| R work/Rfree (%)c | 24.98/28.93 | 18.20/21.08 | 25.25/30.21 | 22.47/24.67 |
| rmsd | ||||
| Bond lengths (Å)/angles (°) | 0.009/1.30 | 0.010/1.22 | 0.009/1.00 | 0.015/1.00 |
| Average B values (Å2) | 47.40 | 33.20 | 24.80 | 41.10 |
| Ramachandran plot (%) | ||||
| Favoured region | 96.60 | 96.50 | 95.00 | 96.50 |
| Allowed region | 3.40 | 3.50 | 5.00 | 3.50 |
| PDB code | 5GN2 | 5GN3 | 5GNW | 5GRK |
aNumbers in parentheses are statistics from the highest-resolution shell.
bRsym=∑|Iobs−Iavg|/Iobs, where Iobs is the observed intensity of individual reflection and Iavg is the average over symmetry equivalents.
cRwork=∑||Fo|−|Fc||/∑|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated using 5% of the data.
Figure 6.
Overall structure of BdiUng. (A) A dimer of BdiUng. The molecule (top) is rotated perpendicular to highlight the formation of β-sheet in the dimeric arrangement. The corresponding regions of motifs A and B in family 1 UDG are colored in green and red, respectively, with the bound uracil molecule depicted in spheres. (B) A monomer of BdiUng with the typical α/β/α sandwich fold of UDG proteins. Rainbow color scheme from blue (N terminus) to red (C terminus) is shown. (C) Overlap of BdiUng monomer (blue) with the EcoUng (PDB: IEUI) (white). Motif A (green) is well superposed whereas the corresponding region of motif B (red) is shaped significantly different in BdiUng. The bound uracil of BdiUng is depicted in spheres.
Motif analysis
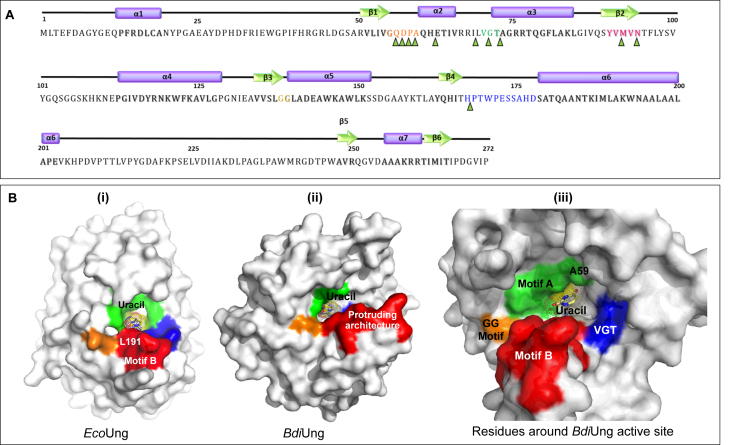
We analyzed BdiUng structure for motifs common in UDG family proteins (Figure 7A, B). The motif ‘GQDPA’ in BdiUng occupies a position analogous to ‘GQDPY’ motif in family 1 UDG with respect to the active site and hence was identified as the ‘water-activating’ loop or motif A. The motif of ‘PPS’ known to be involved in ‘pinching’ target DNA is substituted by VGT residues in BdiUng and positioned in a groove that appears to bind DNA. Residues 90–94, YVMVN, constitute the fourth strand of the core β-sheet corresponding to VLLLN motif in family 1 whereas GS motif involved in ‘holding’ DNA substrate is substituted by GG residues in BdiUng. Interestingly, the motif B of HPSPLS residues, commonly called the Leu-intercalation loop, shows a significantly different shape and composition in BdiUng. This motif is substituted by residues 168–178 in BdiUng and forms an irregularly extended helical geometry that connects further to a long helix α6 at the C-terminal region. Due to the significantly extended protruding architecture of this region (Figure 7B(ii)), the uracil binding pocket is located distant from the tip of this region and no residues previously known for intercalation function such as leucine, arginine or tyrosine were identified in this motif. The higher B-factor of this region, 33.1 compared to 25.3, suggests the flexible characteristic of this region. This unique architecture is not observed in any other UDG family known to date and could be a signature motif in this family of BdiUng. The leucine intercalation loop in family 1 UDGs provides the major binding surface for the inhibitor protein Ugi, which enables docking to the minor-groove in DNA. The altered geometry and surface potential of this region in BdiUng also explains the absence of the binding affinity to the Ugi molecule (Supplementary Figure S10).
Figure 7.
Primary sequence and motifs of BdiUng. (A) Secondary structure elements and motifs are represented along with the primary sequence. Alpha helices are represented as purple cylinders while beta strands are shown as green arrows and corresponding regions in the primary sequence are shaded. Sequences are colored as follows: motif A: orange, PPS motif: green, VLLLN motif: pink, GS motif: yellowish green, sequence forming protruding structure (motif B): blue. Residues constituting ligand binding cavity are marked by green triangles. The nomenclatures of the motifs are done with respect to family 1 UDGs. (B) Surface diagrams of EcoUng (PDB: IEUI) and BdiUng generated using Pymol. (i) The conserved motifs in Ung are colored (motif A; green, motif PPS; blue, motif VLLLN; yellow, motif GS; orange and motif B; red). (ii) The corresponding regions of BdiUng to Ung have the same color. (iii) The residues surrounding the uracil binding pocket are shown in the surface diagram.
Active site
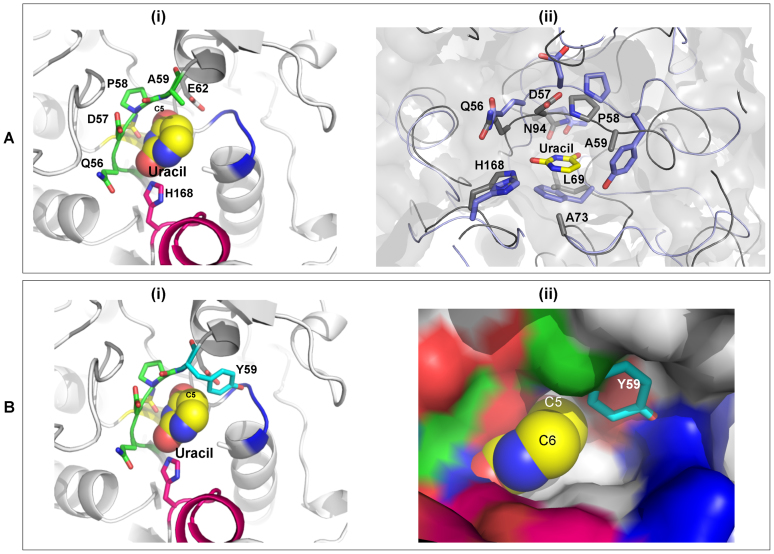
The uracil binding pocket in the active site is lined with residues such as Q56, P58, A59, E62, L69, G71, A73, M92 and N94 (Figure 8A, Supplementary Figure S12). Residues surrounding the O2 and O4 of uracil moieties are conserved similarly to family 1 UDG members. Essential residues for catalysis in family 1 UDGs such as H187 and D64 are also conserved in BdiUng (represented by H168 and D57), suggesting a catalytic mechanism for uracil cleavage similar to family 1 UDGs. The invariant aromatic residue that provides stable π-π stacking interaction with uracil ring in most UDGs, such as F62 in Ung, is absent and replaced by M92. Interestingly, the region surrounding uracil position 5 is differently shaped with a significantly larger cavity. The presence of A59 instead of a bulky residue (Y66 in EcoUng) in motif A provides sufficient cavity space in the active site. Analysis of the uracil binding pocket between the apo and the uracil bound form reveals no significant conformational changes in the cavity region indicating that a structural change may not be required for the recognition of the uracil base by BdiUng. Given that the L191 of EcoUng forms stable interactions with residues in the hydrophobic pocket of Ugi, the protruding loop of BdiUng, mainly constituted of polar residues such as E173, S174, S175, H177, D178, and S179, would disrupt any stable interaction with hydrophobic pocket of Ugi (Supplementary Figure S10). Similarly, a dimeric Ung protein from vaccinia virus, that has been reported to be resistant to Ugi inhibition, contains polar Arg at the position corresponding to L191 of EcoUng affecting the stable hydrophobic interactions with Ugi (Supplementary Figure S11) (62).
Figure 8.
Active site of BdiUng. (A) (i) The active site of BdiUng is shown along with uracil depicted as spheres. Motif A and H168 are highlighted. (ii) The ligand binding cavity of BdiUng and E. coli Ung (PDB Id: 1EUI) show high similarity. Overlap of BdiUng (grey) and E. coli Ung (blue) active site. The residues are labelled for BdiUng. (B) Model structures for mutation A59Y (i) BdiUng active site showing the A59Y mutation. (ii) Surface diagram of BdiUng showing the steric hindrance to substituents at C5 of uracil molecule caused by the A59Y mutation.
Mutational analysis of BdiUng
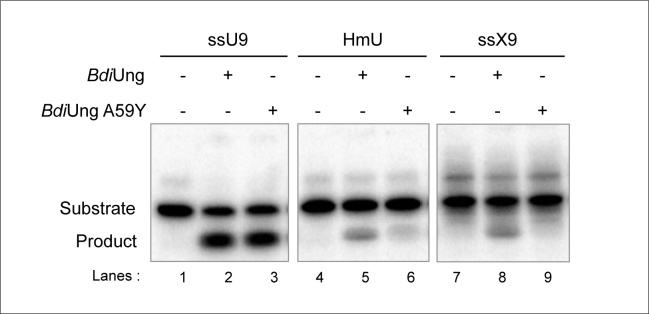
In spite of the very low homology with family 1 UDGs, the crystal structure of UDGs revealed a conservation of most of the key residues implicated in catalysis by family 1 UDGs (Figure 8A ii). The presence of Y66 in EcoUng allows uracil (with no bulky substitutions at position 5 of the pyrimidine ring) but not the 5-hydroxymethyluracil (with a bulky substitution at position 5 of the pyrimidine ring), or the xanthine (a purine) in the active site pocket. Thus, to test the importance of A59 in BdiUng in conferring broad substrate specificity of excising uracil, xanthine, and 5-hydroxymethyluracil, we generated the A59Y mutant and purified the mutant protein from the TG1Δung::cm strain (Supplementary Figure S4B). Interestingly, the A59Y mutation led to a decrease in its activity on xanthine, and 5-hydroxymethyluracil containing ssDNAs but not on uracil containing ssDNA (Figure 9, compare lane 9 with 8; 6 with 5; and 3 with 2).
Figure 9.
Activities of BdiUng A59Y mutant. The excision assays were carried out with oligomers (∼0.1 pmol each) containing different lesions (uracil in ssU9, xanthine in ssX9 and 5-hydroxymethyluracil in HmU) using BdiUng and BdiUng A59Y mutant (each 1 μg, ∼32 pmol monomer) at 37°C for 30 min as described in Materials and Methods. Products were resolved on 8 M urea PAGE (15%).
BdiUng does not rescue ung deficiency in E. coli
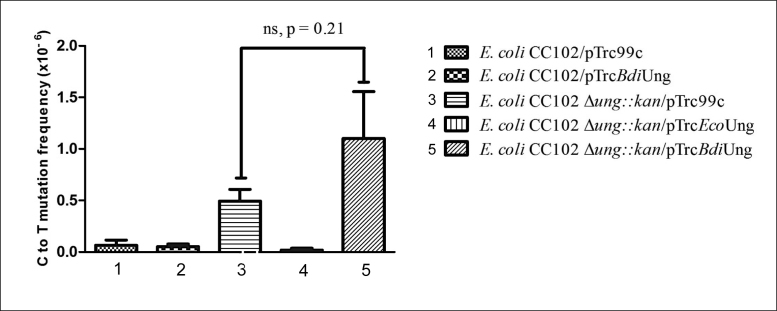
A LacZ reversion assay can be used to detect UDG deficiency using an E. coli CC102 Lac− background. This strain possesses a mutation at the 461st codon in its lacZ gene converting the codon GAG to GGG (E461G) which is crucial for LacZ activity. The absence of UDG will enable C to T mutations to occur, enabling a reversion of the central G in codon 461 to A. As expected, a deficiency of EcoUng (in CC102Δung::kan strain) resulted in an increased Lac− to Lac+ phenotype and introduction of EcoUng (pTrcEcoUng) decreased the reversion frequency efficiently (Figure 10). However, under the same conditions, BdiUng did not significantly change Lac− to Lac+ reversion. This observation suggests that BdiUng does not substitute for the deficiency of EcoUng in E. coli in reducing C to T mutations.
Figure 10.
Mutation frequency (Lac+ reversion) of E. coli CC102 strain and its Δung::kan derivatives harboring plasmid pTrc99c or its derivatives containing EcoUng or BdiUng. Mutation frequency values are represented as mean ± SD from 10 independent colonies (replicates), calculated as the ratio of a number of colonies that appeared on minimal lactose plates compared to minimal glucose plates (per 1 ml culture). In Δung::kan strain, provision of BdiUng did not significantly change the C to T mutation frequency (comparing 3 and 5, P value = 0.21).
DISCUSSION
Uracil DNA glycosylases are essential to prevent incorporation of uracil in DNA. The high G+C content in the root nodule bacterium B. diazoefficiens could predispose this organism to the accumulation of uracils in DNA, especially in cells exposed to reactive oxygen and nitrogen species collectively referred to as ROS/RNS. Both ROS (31–41) and RNS species (such as NO and ONOO-) (42,43) are produced in nitrogen fixing nodule environments. B. diazoefficiens contains five different UDGs (four belonging to families 2, 4 and 5) with a distinct absence of the classical family 1 UDG, which were thought to be ubiquitously present in all organisms since they provide one of the most efficient DNA repair mechanisms.
Interestingly, in the case of Drosophila also, while the earlier studies have shown the presence of UDG activity (63), the genome sequencing has now revealed lack of the classical family 1 UDG (UNG) protein (64). Thus, the presence of weak UDG activity in these organisms may be attributed to the other members of the UDG family (64). In fact, recent findings have shown that the lack of the highly efficient family 1 UDG (UNG) in Drosophila and the Holometabola in general, could be for an important physiological purpose (65). Developmental patterns in these organisms show strong correlation between the presence of uracil in their genomes and the lowered level of expression of dUTPase, an enzyme that hydrolyses dUTP to dUMP to avoid its incorporation in DNA. Thus, the lack of family 1 UDG (UNG) activity in these organisms is physiologically relevant in allowing occurrence of uracil in DNA (66). Whether the lack of highly efficient family 1 UDG in B. diazoefficiens contributes to its specialized physiology during nodulation or otherwise is not known.
To study the role of UDGs in B. diazoefficiens, we have identified and characterized a novel uracil DNA glycosylase from B. diazoefficiens (Q89XRO_BRAJA, referred to as BdiUng, encoded by blr0248) containing a motif (GQDPA) similar to the active site motif A (GQDPY) of family 1 UDGs. BdiUng was found to be active on both ssDNA and dsDNA containing uracil with pNUNN as the minimum substrate required for detectable activity. BdiUng did not interact with Ugi and was therefore resistant to Ugi mediated inhibition in uracil excision. Further, unlike family 1 UDGs, the BdiUng showed broader substrate specificity and excised 5-hydroxymethyl uracil and xanthine present in ssDNA. The co-crystal structures of BdiUng with uracil, and xanthine provide a rationale for the relaxed substrate specificity (Supplementary Figure S12). While BdiUng was inhibited by free uracil, AP DNA (ssF9) did not inhibit its activity to any significant level even at 200 μM concentration (Figure 3).
Substrate specificity of EcoUng is due to the well-tailored active site pocket made to specifically fit uracil (13). The efficient recognition of uracil is proposed to occur through a ‘pinch, push, plug, and pull’ mechanism (68–70). Initial backbone phosphate contacts pinch the DNA backbone. Then a conserved Leu (L191 in EcoUng) acts as a wedge/plug to retain the flipped out uracil into the active site of the enzyme (67,68). Many specific hydrogen bonds and stacking interactions stabilize the uracil in extra-helical conformation within the active site (pull) (70). The side chain of Tyr in the GQDPY motif is in van der Waals’ interaction with C5 of uracil, thus blocking thymine (with a methyl group on C5) and bulky purine rings (11,13,71).
While BdiUng exhibited no significant sequence similarity with any of the established families of UDGs, the crystal structure of the protein revealed that the overall fold is similar to family 4 UDGs in the absence of the characteristic 4Fe–S cluster and that most of the key amino-acid residues implicated in EcoUng catalysis of family 1 UDG are present in the active site. Structural alignment of BdiUng with EcoUng showed that the GQDPA motif of BdiUng was located in a position analogous to motif A of EcoUng which is involved in activation of a water molecule that performs a nucleophilic attack on C1’ of ribose during catalysis. BdiUng also contains Asn and His residues similar to those in EcoUng, which form specific contacts with a uracil residue in the active site. In spite of the drastically different architecture of the active site periphery compared to EcoUng, the similarity observed with respect to the key active site residues implies BdiUng catalysis to be similar to that of EcoUng. But the substitution of the classical Leu intercalation loop by the extended protruding architecture with no apparent functional residue similar to L191 of EcoUng (implicated in pushing the uracil/plugging the duplex void) could be the reason for its reduced activity on dsDNA as well as the absence of its complex formation with Ugi (68,69).
A conserved Tyr in the motif A of Ung proteins prevents entry of bulky bases and position 5- substituted uracil into the active site of the family 1, Ung proteins. In BdiUng the Tyr residue of motif A is substituted by Ala explaining its broad substrate specificity. Consistent with this observation, BdiUng A59Y mutant revealed a much decreased activity on 5-hydroxymethyl uracil and xanthine containing ssDNAs (Figure 9). Still, the active site seems carefully designed to exclude any of the normal bases. Most interestingly, while all the UDGs studied so far show a conserved aromatic residue (Phe) in the active site that provides stability to the flipped out uracil in the active site, no such supporting aromatic residue was seen in the active site of BdiUng.
As revealed by the crystal structure and biochemical analyses, BdiUng is a dimeric protein. UNG from vaccinia virus was also characterized as a dimer (72). However, the dimeric interfaces of the two enzymes are significantly different, in that the BdiUng dimerizes primarily by the C-terminal extended hairpin strand with each active site of the monomer, facing the same side of the dimer, whereas the vaccinia virus UNG dimerizes through the main α/β/α sandwich fold with two active sites positioned opposite (Supplementary Figure S13). The functional significance of the dimerization is not clear at this stage and may need further investigation.
From the analysis of the impact of BdiUng in the mutation frequency, using ung− strains of E. coli CC102 (monitoring Lac+ reversion frequency), we found that BdiUng cannot carry out a physiological function in rescuing C to T mutations similar to that of EcoUng in E. coli. We attribute this to the poor efficiency of uracil excision from dsDNA and also to the fact that these studies were carried out in a heterologous system with a relatively much higher growth rate. Besides, the much higher efficiency with which BdiUng acts on ssDNA compared to dsDNA and the higher relative efficiencies of uracil excision from loop regions of DNA tells us that it is a single strand specific UDG. This could be further supported by the surface analysis of BdiUng. The unique protruding loop yields a narrow DNA binding groove at the uracil binding pocket, a suitable architecture for ssDNA substrate interaction (Supplementary Figure S9).
Interestingly, most organisms with homologues of BdiUng possessed genomes with high G+C contents (Supplementary Table S3). Organisms having G+C rich genomes are intrinsically prone to accumulating hairpin-loops in their genome. The relatively less compromising efficiency of BdiUng on uracil found in loop regions of DNA (as compared to ssU9), along with the presence of its homologues in high G+C containing organisms might be pointing to a physiological function of this family of UDGs.
In addition to BdiUng, we have identified and characterized three other UDGs from B. diazoefficiens. Besides a family 2 homologue (Blr6661) and a family 5 homologue (Blr5068), the organism also contains a homologue of the UdgX protein, which we recently identified as a novel uracil–DNA binding protein in M. smegmatis (56). Blr5068 was found to efficiently excise uracil from ssDNA and dsDNA. Further, bll2523 was identified as a gene coding for a putative family 4 homologue. The unusually high number of UDGs and the presence of novel UDGs like BdiUng might compensate for the lack of a family 1 UDG homologue in this slow growing organism.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the DBT-IISc partnership programme, University Grants Commission, New Delhi for the Centre of Advanced Studies, the DST-FIST level II infrastructure supports and the KRIBB Research Initiative Program to carry out this work. UV is a J.C. Bose Fellow of the Department of Science and Technology (DST), New Delhi. S. Sah is a senior research associate supported by the Council of Scientific and Industrial Research, New Delhi.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology (DBT), New Delhi, and facilitated by the Distinguished Collaborator Award of the Murdoch University, Australia (to U.V. and R.P.T.); Murdoch University small grant scheme; National Research Fund [NRF-2015R1A2A2A03006970 to E.J.W.], Korea. Funding for open access charge: Research grants (to U.V.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Lindahl T. DNA repair enzymes. Annu. Rev. Biochem. 1982; 51:61–87. [DOI] [PubMed] [Google Scholar]
- 2. Friedberg E.C., Walker G.C., Siede W.. DNA Repair and Mutagenesis. 1995; ASM Press. [Google Scholar]
- 3. Jancso A., Botfield M.C., Sowers L.C., Weiss M.A.. An altered-specificity mutation in a human POU domain demonstrates functional analogy between the POU-specific subdomain and phage lambda repressor. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:3887–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rogstad D.K., Liu P., Burdzy A., Lin S.S., Sowers L.C.. Endogenous DNA lesions can inhibit the binding of the AP-1 (c-Jun) transcription factor. Biochemistry. 2002; 41:8093–8102. [DOI] [PubMed] [Google Scholar]
- 5. Lindahl T. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc. Natl. Acad. Sci. U.S.A. 1974; 71:3649–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krokan H.E., Drablos F., Slupphaug G.. Uracil in DNA–occurrence, consequences and repair. Oncogene. 2002; 21:8935–8948. [DOI] [PubMed] [Google Scholar]
- 7. Pearl L.H. Structure and function in the uracil-DNA glycosylase superfamily. Mutat. Res. 2000; 460:165–181. [DOI] [PubMed] [Google Scholar]
- 8. Aravind L., Koonin E.V.. The alpha/beta fold uracil DNA glycosylases: a common origin with diverse fates. Genome Biol. 2000; 1, RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lucas-Lledo J.I., Maddamsetti R., Lynch M.. Phylogenomic analysis of the uracil-DNA glycosylase superfamily. Mol. Biol. Evol. 2011; 28:1307–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lindahl T., Ljungquist S., Siegert W., Nyberg B., Sperens B.. DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J. Biol. Chem. 1977; 252:3286–3294. [PubMed] [Google Scholar]
- 11. Savva R., McAuley-Hecht K., Brown T., Pearl L.. The structural basis of specific base-excision repair by uracil-DNA glycosylase. Nature. 1995; 373:487–493. [DOI] [PubMed] [Google Scholar]
- 12. Handa P., Acharya N., Varshney U.. Effects of mutations at tyrosine 66 and asparagine 123 in the active site pocket of Escherichia coli uracil DNA glycosylase on uracil excision from synthetic DNA oligomers: evidence for the occurrence of long-range interactions between the enzyme and substrate. Nucleic Acids Res. 2002; 30:3086–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mol C.D., Arvai A.S., Slupphaug G., Kavli B., Alseth I., Krokan H.E., Tainer J.A.. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell. 1995; 80:869–878. [DOI] [PubMed] [Google Scholar]
- 14. Parikh S.S., Putnam C.D., Tainer J.A.. Lessons learned from structural results on uracil-DNA glycosylase. Mutat. Res. 2000; 460:183–199. [DOI] [PubMed] [Google Scholar]
- 15. Cone R., Bonura T., Friedberg E.C.. Inhibitor of uracil-DNA glycosylase induced by bacteriophage PBS2. Purification and preliminary characterization. J. Biol. Chem. 1980; 255:10354–10358. [PubMed] [Google Scholar]
- 16. Ravishankar R., Bidya Sagar M., Roy S., Purnapatre K., Handa P., Varshney U., Vijayan M.. X-ray analysis of a complex of Escherichia coli uracil DNA glycosylase (EcUDG) with a proteinaceous inhibitor. The structure elucidation of a prokaryotic UDG. Nucleic Acids Res. 1998; 26:4880–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee H.W., Dominy B.N., Cao W.. New family of deamination repair enzymes in uracil-DNA glycosylase superfamily. J. Biol. Chem. 2011; 286:31282–31287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neddermann P., Jiricny J.. The purification of a mismatch-specific thymine-DNA glycosylase from HeLa cells. J. Biol. Chem. 1993; 268:21218–21224. [PubMed] [Google Scholar]
- 19. Haushalter K.A., Todd Stukenberg M.W., Kirschner M.W., Verdine G.L.. Identification of a new uracil-DNA glycosylase family by expression cloning using synthetic inhibitors. Curr. Biol.: CB. 1999; 9:174–185. [DOI] [PubMed] [Google Scholar]
- 20. Sandigursky M., Franklin W.A.. Thermostable uracil-DNA glycosylase from Thermotoga maritima a member of a novel class of DNA repair enzymes. Curr. Biol.: CB. 1999; 9:531–534. [DOI] [PubMed] [Google Scholar]
- 21. Sartori A.A., Fitz-Gibbon S., Yang H., Miller J.H., Jiricny J.. A novel uracil-DNA glycosylase with broad substrate specificity and an unusual active site. EMBO J. 2002; 21:3182–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gallinari P., Jiricny J.. A new class of uracil-DNA glycosylases related to human thymine-DNA glycosylase. Nature. 1996; 383:735–738. [DOI] [PubMed] [Google Scholar]
- 23. Neddermann P., Jiricny J.. Efficient removal of uracil from G.U mispairs by the mismatch-specific thymine DNA glycosylase from HeLa cells. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:1642–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moe E., Leiros I., Smalas A.O., McSweeney S.. The crystal structure of mismatch-specific uracil-DNA glycosylase (MUG) from Deinococcus radiodurans reveals a novel catalytic residue and broad substrate specificity. J. Biol. Chem. 2006; 281:569–577. [DOI] [PubMed] [Google Scholar]
- 25. Wibley J.E., Waters T.R., Haushalter K., Verdine G.L., Pearl L.H.. Structure and specificity of the vertebrate anti-mutator uracil-DNA glycosylase SMUG1. Mol. Cell. 2003; 11:1647–1659. [DOI] [PubMed] [Google Scholar]
- 26. Hinks J.A., Evans M.C., De Miguel Y., Sartori A.A., Jiricny J., Pearl L.H.. An iron-sulfur cluster in the family 4 uracil-DNA glycosylases. J. Biol. Chem. 2002; 277:16936–16940. [DOI] [PubMed] [Google Scholar]
- 27. Sartori A.A., Schar P., Fitz-Gibbon S., Miller J.H., Jiricny J.. Biochemical characterization of uracil processing activities in the hyperthermophilic archaeon Pyrobaculum aerophilum. J. Biol. Chem. 2001; 276:29979–29986. [DOI] [PubMed] [Google Scholar]
- 28. Santos R., Herouart D., Sigaud S., Touati D., Puppo A.. Oxidative burst in alfalfa-Sinorhizobium meliloti symbiotic interaction. Mol. Plant-Microbe Interact.: MPMI. 2001; 14:86–89. [DOI] [PubMed] [Google Scholar]
- 29. Ramu S.K., Peng H.M., Cook D.R.. Nod factor induction of reactive oxygen species production is correlated with expression of the early nodulin gene rip1 in Medicago truncatula. Mol. Plant-Microbe Interact.: MPMI. 2002; 15:522–528. [DOI] [PubMed] [Google Scholar]
- 30. D’Haeze W., Holsters M.. Nod factor structures, responses, and perception during initiation of nodule development. Glycobiology. 2002; 12:79R–105R. [DOI] [PubMed] [Google Scholar]
- 31. Becana M., Klucas R.V.. Transition metals in legume root nodules: iron-dependent free radical production increases during nodule senescence. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:8958–8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Becana M., Klucas R.V.. Oxidation and reduction of leghemoglobin in root nodules of leguminous plants. Plant Physiol. 1992; 98:1217–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Becana M., Rodríguez-Barrueco C.. Protective mechanisms of nitrogenase against oxygen excess and partially-reduced oxygen intermediates. Physiol. Plantarum. 1989; 75:429–438. [Google Scholar]
- 34. Becana M., Dalton D.A., Moran J.F., Iturbe‐Ormaetxe I., Matamoros M.A., Rubio M.C.. Reactive oxygen species and antioxidants in legume nodules. Physiol. Plantarum. 2000; 109:372–381. [Google Scholar]
- 35. Puppo A., Halliwell B.. Generation of hydroxyl radicals by soybean nodule leghaemoglobin. Planta. 1988; 173:405–410. [DOI] [PubMed] [Google Scholar]
- 36. Becana M., Moran J.F., Iturbe-Ormaetxe I.. Iron-dependent oxygen free radical generation in plants subjected to environmental stress: toxicity and antioxidant protection. Plant Soil. 1998; 201:137–147. [Google Scholar]
- 37. Fridovich I. Superoxide dismutases: studies of structure and mechanism. Adv. Exp. Med. Biol. 1976; 74:530–539. [DOI] [PubMed] [Google Scholar]
- 38. Moreau S., Davies M.J., Mathieu C., Herouart D., Puppo A.. Leghemoglobin-derived radicals. Evidence for multiple protein-derived radicals and the initiation of peribacteroid membrane damage. J. Biol. Chem. 1996; 271:32557–32562. [DOI] [PubMed] [Google Scholar]
- 39. Puppo A., Halliwell B.. Formation of hydroxyl radicals from hydrogen peroxide in the presence of iron. Is haemoglobin a biological Fenton reagent. Biochem. J. 1988; 249:185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Puppo A., Rigaud J., Job D.. Role of superoxide anion in leghemoglobin autoxidation. Plant Sci. Lett. 1981; 22:353–360. [Google Scholar]
- 41. Schneider K., Schlegel H.G.. Production of superoxide radicals by soluble hydrogenase from Alcaligenes eutrophus H16. Biochem. J. 1981; 193:99–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Becana M., Matamoros M.A., Udvardi M., Dalton D.A.. Recent insights into antioxidant defenses of legume root nodules. New Phytologist. 2010; 188:960–976. [DOI] [PubMed] [Google Scholar]
- 43. Baudouin E., Pieuchot L., Engler G., Pauly N., Puppo A.. Nitric Oxide Is Formed in Medicago truncatula-Sinorhizobium meliloti Functional Nodules. Mol. Plant-Microbe Interact. 2006; 19:970–975. [DOI] [PubMed] [Google Scholar]
- 44. Halliwell B., Gutteridge J.M.C.. Free Radicals in Biology and Medicine. 1999; Oxford University Press. [Google Scholar]
- 45. Kreutzer D.A., Essigmann J.M.. Oxidized, deaminated cytosines are a source of C –>T transitions in vivo. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:3578–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Delamuta J.R., Ribeiro R.A., Ormeno-Orrillo E., Melo I.S., Martinez-Romero E., Hungria M.. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int. J. Syst. Evol. Microbiol. 2013; 63:3342–3351. [DOI] [PubMed] [Google Scholar]
- 47. Mathis J.N., McMillin D.E., Champion R.A., Hunt P.G.. Genetic variation in two cultures of Bradyrhizobium japonicum 110 differing in their ability to impart drought tolerance to soybean. Curr. Microbiol. 1997; 35:363–366. [DOI] [PubMed] [Google Scholar]
- 48. Kaneko T., Nakamura Y., Sato S., Minamisawa K., Uchiumi T., Sasamoto S., Watanabe A., Idesawa K., Iriguchi M., Kawashima K. et al. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110 (supplement). DNA Res. 2002; 9:225–256. [DOI] [PubMed] [Google Scholar]
- 49. Cupples C.G., Miller J.H.. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:5345–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sedmak J.J., Grossberg S.E.. A rapid, sensitive, and versatile assay for protein using Coomassie brilliant blue G250. Anal. Biochem. 1977; 79:544–552. [DOI] [PubMed] [Google Scholar]
- 51. Varshney U., van de Sande J.H.. Specificities and kinetics of uracil excision from uracil-containing DNA oligomers by Escherichia coli uracil DNA glycosylase. Biochemistry. 1991; 30:4055–4061. [DOI] [PubMed] [Google Scholar]
- 52. Kumar N.V., Varshney U.. Contrasting effects of single stranded DNA binding protein on the activity of uracil DNA glycosylase from Escherichia coli towards different DNA substrates. Nucleic Acids Res. 1997; 25:2336–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Otwinowski Z., Minor W.. Methods in Enzymology. 1997; 276:Academic Press; 307–326. [DOI] [PubMed] [Google Scholar]
- 54. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 55. Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D. 2010; 66:213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sang P.B., Srinath T., Patil A.G., Woo E.J., Varshney U.. A unique uracil-DNA binding protein of the uracil DNA glycosylase superfamily. Nucleic Acids Res. 2015; 43:8452–8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Roy S., Purnapatre K., Handa P., Boyanapalli M., Varshney U.. Use of a coupled transcriptional system for consistent overexpression and purification of UDG-Ugi complex and Ugi from Escherichia coli. Protein Express. Purif. 1998; 13:155–162. [DOI] [PubMed] [Google Scholar]
- 58. Ghosh M., Kumar N.V., Varshney U., Chary K.V.. Structural characterisation of a uracil containing hairpin DNA by NMR and molecular dynamics. Nucleic Acids Res. 1999; 27:3938–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kumar N.V., Varshney U.. Inefficient excision of uracil from loop regions of DNA oligomers by E. coli uracil DNA glycosylase. Nucleic Acids Res. 1994; 22:3737–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Purnapatre K., Varshney U.. Uracil DNA glycosylase from Mycobacterium smegmatis and its distinct biochemical properties. Eur. J. Biochem./FEBS. 1998; 256:580–588. [DOI] [PubMed] [Google Scholar]
- 61. Holm L., Rosenström P.. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010; 38:W545–W549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schormann N., Grigorian A., Samal A., Krishnan R., DeLucas L., Chattopadhyay D.. Crystal structure of vaccinia virus uracil-DNA glycosylase reveals dimeric assembly. BMC Struct. Biol. 2007; 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Morgan A.R., Chlebek J.. Uracil-DNA glycosylase in insects. Drosophila and the locust. J. Biol. Chem. 1989; 264:9911–9914. [PubMed] [Google Scholar]
- 64. Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F. et al. The genome sequence of Drosophila melanogaster. Science. 2000; 287:2185–2195. [DOI] [PubMed] [Google Scholar]
- 65. Muha V., Horvath A., Bekesi A., Pukancsik M., Hodoscsek B., Merenyi G., Rona G., Batki J., Kiss I., Jankovics F. et al. Uracil-containing DNA in Drosophila: stability, stage-specific accumulation, and developmental involvement. PLoS Genet. 2012; 8:e1002738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Horvath A., Bekesi A., Muha V., Erdelyi M., Vertessy B.G.. Expanding the DNA alphabet in the fruit fly: uracil enrichment in genomic DNA. Fly. 2013; 7:23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Handa P., Roy S., Varshney U.. The role of leucine 191 of Escherichia coli uracil DNA glycosylase in the formation of a highly stable complex with the substrate mimic, ugi, and in uracil excision from the synthetic substrates. J. Biol. Chem. 2001; 276:17324–17331. [DOI] [PubMed] [Google Scholar]
- 68. Jiang Y.L., Kwon K., Stivers J.T.. Turning on uracil-DNA glycosylase using a pyrene nucleotide switch. J. Biol. Chem. 2001; 276:42347–42354. [DOI] [PubMed] [Google Scholar]
- 69. Parikh S.S., Mol C.D., Slupphaug G., Bharati S., Krokan H.E., Tainer J.A.. Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA. EMBO J. 1998; 17:5214–5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Slupphaug G., Mol C.D., Kavli B., Arvai A.S., Krokan H.E., Tainer J.A.. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature. 1996; 384:87–92. [DOI] [PubMed] [Google Scholar]
- 71. Kavli B., Slupphaug G., Mol C.D., Arvai A.S., Peterson S.B., Tainer J.A., Krokan H.E.. Excision of cytosine and thymine from DNA by mutants of human uracil-DNA glycosylase. EMBO J. 1996; 15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 72. Burmeister W.P., Tarbouriech N., Fender P., Contesto-Richefeu C., Peyrefitte C.N., Iseni F.. Crystal structure of the vaccinia virus uracil-DNA glycosylase in complex with DNA. J. Biol. Chem. 2015; 290:17923–17934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.