Abstract
Crohn's disease (CD) and ulcerative colitis (UC) are types of chronic inflammatory bowel disease (IBD) of which the actual causes remain unknown. Emerging data indicate that alterations in cytokine synthesis may be involved in IBD pathogenesis. The aim of the present study was to determine whether the tumor necrosis factor (TNF)-α mRNA expression level and rs1799964 polymorphism are the genetic susceptibility component of IBD development. The TNF-α mRNA expression level of peripheral blood mononuclear cells (PBMCs) was measured using comparative reverse-transcription quantitative polymerase chain reaction (PCR). Genomic DNA from 201 individuals (CD: n=15; UC: n=86; control subjects: n=100) was analyzed for the presence of the TNF-α-1031 polymorphism by PCR-restriction fragment length polymorphism. An increased TNF-α mRNA expression level was additionally observed in the CC genotype of the −1031 TNF-α gene polymorphism compared with the TC and TT genotypes (P<0.05). Furthermore, the present results revealed that there was no significant difference in the genotype/allele frequencies of the −1031 TNF-α gene polymorphism in Iranian IBD patients. By comparison, the TNF-α mRNA expression level was evaluated in patients with a history of taking medications and demonstrated a significant association in the group that received the 5-ASA + Pred + AZA,5. 5-ASA + Pred + AZA + IFX when compared with the other groups (P<0.05). Thus, these results support the hypothesis that overexpression of the TNF-α gene, which correlated with the CC genotype, may represent a genetic risk factor for Iranian IBD.
Keywords: inflammatory bowel disease, TNFα-1031 T>C polymorphism, Crohn's disease, ulcerative colitis, quantitative polymerase chain reaction
Introduction
Human inflammatory bowel diseases (IBD), ulcerative colitis (UC) and Crohn's disease (CD) are a group of chronic inflammatory conditions of the gastrointestinal tract (1). UC and CD are differentiated by the location of inflammation in the digestive tract (2).
Incidence and prevalence of IBD in the Nordic countries, North America and developing countries is steadily increasing (3,4). New Zealand has the highest rates of IBD in the world (5), and the exact rates of IBD in Iran are not clear, but it appears that the frequency of IBD is increasing (6,7). The exact causes of this disease remains unknown, but studies have proposed that it may be the result of complex interactions between multiple factors, such as genetic, environmental and immunological factors (8,9). There is direct evidence for the role of genetics in IBD, associated with the family history of patients, as well as increased rates of IBD in monozygotic twins (10,11). The predominant genetic association in IBD is divided into genes that contribute to innate and adaptive immune responses (12). Tumor necrosis factor (TNF)-α is a multifunctional cytokine involved in the advancement of inflammatory responses and is critical in the pathogenesis of inflammatory, autoimmune and malignant diseases (13). TNF-α is key in cell-mediated immunity, as demonstrated by high serum levels of TNF-α in IBD blood and tissue samples (14,15). TNF-α has numerous polymorphisms, the majority of which are located in its promoter region and certain polymorphisms affect the expression level of the gene (16,17). The present study hypothesized that differences in cytokine levels are due to gene polymorphisms, which may have an important role in the inflammatory response and influence the pathophysiology of IBD.
No association between the TNF-α promoter polymorphisms at position-1031 and susceptibility to IBD in Iranian patients was demonstrated in our previous findings (18). Subsequently, studies have proposed a direct correlation between the single nucleotide polymorphism (SNP) genotype with disease; an important reason for regulatory SNPs to cause human genetic disease is that SNPs are located in regulatory sequences, for example promoter regions or UTRs, that could effected the expression of the gene. Tt is therefore important to investigate the association between the TNF-α gene expression pattern and its association with the −1031 polymorphism. The present study investigated the association between the TNF-α gene expression pattern and its association with the −1031 polymorphism in an Iranian population of patients with IBD.
Materials and methods
Patients
Between 2011 and 2014, 101 IBD patients (15 CD and 86 UC) and 100 healthy control subjects, who had been referred to the Research Institute for Gastroenterology and Liver Diseases, Shahid Beheshti University of Medical Sciences (Tehran, Iran), were included in the present study. The control group individuals did not have a family history of gastrointestinal disorders. Furthermore, control group subjects were selected based on an absence of any evidence, personal or family history of IBD or inflammatory diseases, such as gastritis, ulcerative colitis and CD. The patients and healthy individuals were all Iranian. Colonoscopy was performed by a gastroenterologists and the diagnosis of IBD was confirmed by a pathologist. Patients with a history of using anti-inflammatory drugs, such as 5-aminosalicylic acid (5ASA), prednisone (Pred), azathioprine (AZA) and infliximab (IFX) were evaluated. Written informed consent was obtained from each patient and healthy volunteer, and peripheral blood samples (6 ml) were collected in EDTA tubes and stored at 4°C until DNA/RNA extraction. The Ethics Committee of the Research Institute for Gastroenterology and Liver diseases, Shahid Beheshti University of Medical Sciences provided approval for the present study.
Genotyping
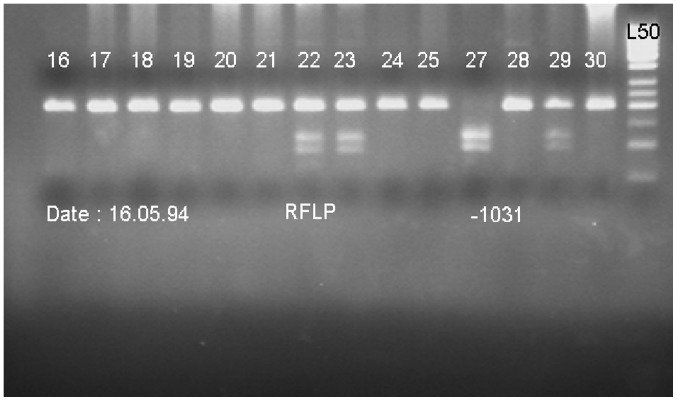
Genomic DNA was isolated from 5 ml peripheral blood mononuclear cells (PBMCs) using the standard salting out method (19). The quality and quantity of DNA was determined using a spectrophotometer and electrophoresis on agarose gel. Genotype determination was performed by polymerase chain reaction (PCR); the promoter −1031 TNF-α gene polymorphism was PCR amplified in a 186-bp fragment using the following primers: Forward, 5-CTT CAG GGA TAT GTG ATG GAC TC-3 and reverse, 5-GGA GAC CTC TGG GGA GAT GT-3. Followed by restriction fragment length polymorphism (RFLP). The PCR reaction was performed on a Mastercycler (Eppendorf; Hamburg, Germany) as follows: Denaturation for 5 min at 94°C, followed by 36 cycles of amplification (40 sec at 94°C, 35 sec at 62°C and 40 sec at 72°C). A final elongation step (5 min at 72°C) was applied at the end of the 36 cycles. The PCR product was 186 bp. Following amplification, PCR products were digested (at 37°C) by restriction endonuclease BbsI (Thermo Fisher Scientific, Inc.; C allele, 106 and 80 bp; T allele, 186 bp) overnight. The digested PCR products were visualized by electrophoresis in a 2.5% agarose gel (120 V for 40 min; Fig. 1).
Figure 1.
Polymorphism fragments on agarose gel following digestion with BbsI restriction enzyme. Ladder 16–21, 24, 25, 30: TT; ladder 22, 23, 29: TC; ladder 27: CC; L50, ladder 50 bp. RFLP, restriction fragment length polymorphism.
Sequencing
To confirm the results of genotyping, 10% of the PCR products were sequenced using the ABI PRISM 3130xL Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA) and the results were fully concordant.
RNA isolation with reverse transcription-PCR
Total RNA was extracted from PBMCs using a YTA RNA Extraction kit (Yekta Tajhiz Azma, Tehran, Iran) according to the manufacturer's instructions. The RNA quantity and quality was determined from spectrophotometric optical density measurement (wavelength, 260 and 280 nm). The cDNA was synthesized using a Revert Aid RT Reverse Transcription kit (cat. no. K1691; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The expression rate of the TNF-α gene was analyzed using an ABI 7500 real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) and SYBR Master Mix (Takara Bio, Inc., Otsu, Japan) according to the manufacturer's instructions. Primers were designed using Genscript and Primer3 online programs (http://primer3.ut.ee/). The following primer pairs were used: Forward, 5-CTGAACTTCGGGGTGATCG-3 and reverse, 5-GCT TGG TGG TTT GCT ACG AC-3 for TNF-α; forward, 5-ACA ACC TTC TTG CAG CTC CTC-3 and reverse, 5-TGACCCATACCCACCATCAC-3 for β-actin (ACTB). The expression levels of all target genes were normalized against the expression of ACTB, which served as the endogenous control (20). The first stage, initial denaturation at 95°C for 30 sec, was followed by a second 40-replication cycle (two-step cycling at 95°C for 5 sec and 60°C for 34 sec) and the third step (performed following creation of the PCR melting curve) was as follows: 95°C for 15 sec, 60°C for 60 sec and 95°C for 15 sec. The melting curve differentiates between the different products, and demonstrates contamination and the absorption peak. Melting curve analysis detects non-specific and primer dimer products. The ΔΔCq method was used according to Livak and Schmittgen (21).
Statistical analysis
SPSS statistical software version 21 (IBM Corp., Armonk, NY, USA) was used to perform statistical analysis of the genotyping. The χ2 test was used to evaluate the distribution of the allele and genotype frequencies. Furthermore, the Hardy-Weinberg equilibrium was performed along with the χ2 test to compare the observed genotype frequencies among the investigated cases and control subjects with the expected genotype frequencies. Logistic regression was applied to calculate odds ratio (OR) and 95% confidence intervals, and to adjust the data for confounding factors, such as age and gender. P<0.05 was considered to indicate a statistically significant difference. GraphPad prism 5 software (https://www.graphpad.com/scientific-software/prism/) was used to perform statistical analysis of the TNF-α mRNA expression levels. One-way ANOVA was used to examine the TNF-α mRNA expression level between groups.
Results
Demographics
A total 101 patients with the diagnosis of IBD were investigated, including 59 males (58.4%) and 42 females (41.6%). The control group consisted of 100 non-IBD subjects, 58 males (58%) and 42 females (42%). The percentage of male and female subjects in the two groups was not significantly different (P>0.05). The mean age of the IBD group was significantly higher than that in the healthy control subjects group (P<0.05). Furthermore, no significant differences were identified between the IBD group and healthy control subjects with regard to the body mass index and smoking behavior (P>0.05; Table I).
Table I.
Demographic characteristics of the study population, including inflammatory bowel disease patients and control subjects.
| Variable | Patient (n=101) | Control (n=100) | P-value |
|---|---|---|---|
| Age (mean ± standard deviation), years | 33.99±10.86 | 42.00±13.52 | <0.001 |
| Body mass index (kg/m2)b | 25.19±4.99 | 25.49±5.85 | 0.698 |
| Gender, n (%)b | 0.138 | ||
| Female | 42 (41.6) | 42 (42) | |
| Male | 59 (58.4) | 58 (58) | |
| Smoking, n (%)b | 0.462 | ||
| Smoker | 16 (15.8) | 15 (15) | |
| Non-smoker | 85 (84.2) | 85 (85) |
Student's t-test
χ2 test.
Lack of associations of the TNF-α-1031T>C polymorphism with IBD in the Iranian population
According to our previous study (18), the frequency of TT, TC and CC genotypes in patients were 64.4, 28.7 and 6.9%, respectively and 63, 29 and 8%, respectively in the control groups. No statistically significant differences were identified between genotypes during the observation and study groups (18) (Table II).
Table II.
Genotype distribution of −1031 polymorphism among inflammatory bowel disease patients and healthy control subjects.
| Genotype | Patients, n (%) | Controls, n (%) | Odds ratio (95% confidence interval) | P-value |
|---|---|---|---|---|
| TT | 65 (64.4) | 63 (63.0) | 1.00 (Ref.) | |
| TC | 29 (28.7) | 29 (29.0) | 0.986 (0.509–1.912) | 0.968 |
| CC | 7 (6.9) | 8 (8.0) | 1.308 (0.419–4.088) | 0.644 |
TNF-α gene expression patterns were associated with pathological IBD subtypes
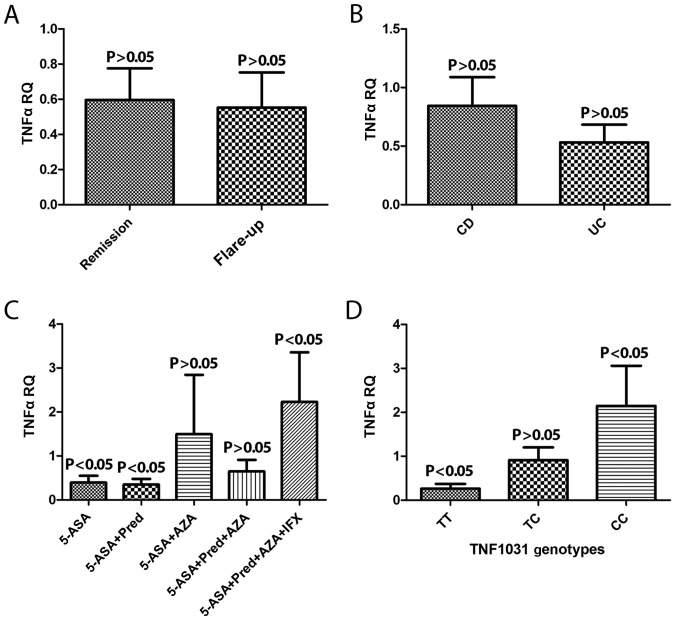
The extent of TNF-α gene expression in two disease phases, remission and flare up, were compared. The expression level of the TNF-α gene remained the same (Fig. 2A). In addition, the expression of TNFα mRNA levels between the CD and UC type of IBD patient were analyzed and were not statistically different (Fig. 2B).
Figure 2.
TNF-α mRNA expression level (A) at different IBD pathological phases, (B) according to the type of IBD (UC or CD) and (C) according to drug history. (D) Interaction between the −1031 genotype and TNF-α expression level. TNF-α, tumor necrosis factor-α; IBD, inflammatory bowel disease; UC, ulcerative colitis; CD, Crohn's disease; RQ, relative quantification; 5-ASA, 5-aminosalicylic acid; Pred, prednisone; AZA, azathioprine; IFX, infliximab.
An assessment of patients who had a history of taking medication was also performed and patients were classified into five different groups as follows: i) 5-ASA; ii) 5-ASA + Pred; iii) 5-ASA + AZA; iv) 5-ASA + Pred + AZA; v) 5-ASA + Pred + AZA + IFX according to medication usage. TNF-α gene upregulation of mRNA exhibited a significant association in the group that received all four types of drug when compared with the other groups (P<0.05; Fig. 2C).
Interaction between the −1031 T>C polymorphism genotype, phenotype and TNF-a expression levels
TNF-α gene expression was screened for the polymorphism −1031 T>C genotypes TT, TC and CC. Overexpression of TNF-α mRNA was observed in genotype CC when compared with CT and TT (P<0.05; Fig. 2D).
Discussion
IBD is complex multifactorial disease, the pathogenesis of which is not well understood (22). Allelic variants in cytokine genes have been shown to influence gene expression and subsequently the susceptibility to and severity of inflammation diseases (23). There is increasing evidence that TNF-α is key in the pathogenesis of IBD and may represent novel therapeutic targets (12). Studies indicate the TNF-α gene as an appropriate and functional candidate for IBD treatment (24,25). In the present study, the allele frequency of SNP-1031 T>C was greater in patients when compared with the control group, although the difference between the patients and control group was not significant. Similar studies to the current study, performed in Canada (26) and Japan (27) and Iran (28), demonstrated that the allele frequency between the control group and CD group in polymorphism −1031 was significant, but no significant association was observed between the UC and the control groups. This mutant allele frequency in certain studies, for example in the UK (24,29) and a Canadian study (30) was not significantly different. In another study, the T allele was shown to confer a significant risk for developing UC in Iranian Azeri Turkish patients (31). Previous studies have reported conflicting results in the TNF-α gene polymorphism susceptibility to IBD. The reasons for these contradictory findings remain unknown. Different ethnicities, distribution of genotype, environment-genetic interactions and sample size, may be the cause of the inconsistent results in the frequency of the −1031 TNF-α gene polymorphisms in the present study.
In the current study, a correlation between polymorphism −1031 T> C and the TNF-α mRNA expression level was significant. The results demonstrated that the level of TNF-α gene mRNA expression was reduced in genotype TT when compared with the other genotypes. A recent independent study by Rad et al (32) in 2004 demonstrated similar results with regard to the polymorphisms of cytokines and their effect on gene expression, which was increased in patients with the Helicobacter pylori infection. The results showed that variations in SNPs may change gene expression levels, leading to a significant association with IBD. Another study by Chen et al (33) in 1995, examined the effect of TNF-α gene polymorphisms with mRNA expression in patients with pre-eclampsia, and indicated that TNF-α gene mRNA expression may be associated with its gene polymorphism.
The findings of the current study provide evidence that patients who had taken the four drugs (5-ASA + Pred + AZA and 5. 5-ASA + Pred + AZA + IFX) for the treatment of IBD exhibit overexpression of TNF-α mRNA. To the best of our knowledge, no study has determined the correlation between drug use and TNF-α gene expression in IBD patients. However, it may be that the TNF-α gene expression may serve as a biomarker for the drug-resistant inflammation in IBD initiation. Therefore, it is proposed that, in future studies, an improved understanding of the association with other TNF-α gene promoter polymorphisms and the underlying mechanism of functional polymorphisms may provide novel therapeutic target gene expression for the treatment of IBD patients.
In conclusion, the present study examined the association between the TNF-α gene expression pattern and its association with the −1031 polymorphism, which demonstrated an increased expression of the CC genotype and a decreased expression of the TT genotype. The overexpression of the CC genotype may represent a genetic risk factor for IBD in an Iranian population. In addition, overexpression of TNF-α was observed in patients who had a history of taking the four types of drug (5-ASA, Pred, AZA or IFX). Thus, these analyses may be used in the prognosis, diagnosis and treatment of IBD.
References
- 1.Liu TC, Stappenbeck TS. Genetics and pathogenesis of inflammatory bowel disease. Annu Rev Pathol. 2016;11:127–148. doi: 10.1146/annurev-pathol-012615-044152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Okamoto R, Watanabe M. Role of epithelial cells in the pathogenesis and treatment of inflammatory bowel disease. J Gastroenterol. 2016;51:11–21. doi: 10.1007/s00535-015-1098-4. [DOI] [PubMed] [Google Scholar]
- 3.Portela F, Dias CC, Caldeira P, Cravo M, Deus J, Gonçalves R, Lago P, Morna H, Peixe P, Ramos J, et al. The who-when-why triangle of CAM use among portuguese IBD patients. Dig Liver Dis. 2017;49:388–396. doi: 10.1016/j.dld.2016.12.031. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein CN, Fried M, Krabshuis JH, Cohen H, Eliakim R, Fedail S, Gearry R, Goh KL, Hamid S, Khan AG, et al. World gastroenterology organization practice guidelines for the diagnosis and management of IBD in 2010. Inflamm Bowel Dis. 2010;16:112–124. doi: 10.1002/ibd.21048. [DOI] [PubMed] [Google Scholar]
- 5.Lakatos PL. Recent trends in the epidemiology of inflammatory bowel diseases: Up or down? World J Gastroenterol. 2006;12:6102–6108. doi: 10.3748/wjg.v12.i38.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology. 2011;140:1785–1794. doi: 10.1053/j.gastro.2011.01.055. [DOI] [PubMed] [Google Scholar]
- 7.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol. 2015;12:205–217. doi: 10.1038/nrgastro.2015.34. [DOI] [PubMed] [Google Scholar]
- 8.Leppkes M, Neurath MF, Herrmann M, Becker C. Immune deficiency vs. immune excess in inflammatory bowel diseases-STAT3 as a rheo-STAT of intestinal homeostasis. J Leukoc Biol. 2016;99:57–66. doi: 10.1189/jlb.5MR0515-221R. [DOI] [PubMed] [Google Scholar]
- 9.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sadler T, Bhasin JM, Xu Y, Barnholz-Sloan J, Chen Y, Ting AH, Stylianou E. Genome-wide analysis of DNA methylation and gene expression defines molecular characteristics of Crohn's disease-associated fibrosis. Clin Epigenetics. 2016;8:30. doi: 10.1186/s13148-016-0193-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGovern DP, Kugathasan S, Cho JH. Genetics of inflammatory bowel diseases. Gastroenterology. 2015;149:1163–1176.e2. doi: 10.1053/j.gastro.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francescone R, Hou V, Grivennikov SI. Cytokines, IBD, and colitis-associated cancer. Inflamm Bowel Dis. 2015;21:409–418. doi: 10.1097/MIB.0000000000000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang M, Viennois E, Prasad M, Zhang Y, Wang L, Zhang Z, Han MK, Xiao B, Xu C, Srinivasan S, Merlin D. Edible ginger-derived nanoparticles: A novel therapeutic approach for the prevention and treatment of inflammatory bowel disease and colitis-associated cancer. Biomaterials. 2016;101:321–340. doi: 10.1016/j.biomaterials.2016.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lykouras D, Sampsonas F, Kaparianos A, Karkoulias K, Tsoukalas G, Spiropoulos K. Human genes in TB infection: Their role in immune response. Monaldi Arch Chest Dis. 2008;69:24–31. doi: 10.4081/monaldi.2008.408. [DOI] [PubMed] [Google Scholar]
- 15.Mocellin S, Verdi D, Pooley KA, Nitti D. Genetic variation and gastric cancer risk: A field synopsis and meta-analysis. Gut. 2015;64:1209–1219. doi: 10.1136/gutjnl-2015-309168. [DOI] [PubMed] [Google Scholar]
- 16.Hajeer AH, Hutchinson IV. TNF-alpha gene polymorphism: Clinical and biological implications. Microsc Res Tech. 2000;50:216–228. doi: 10.1002/1097-0029(20000801)50:3<216::AID-JEMT5>3.3.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 17.Li CI, Li TC, Liao LN, Liu CS, Yang CW, Lin CH, Hsiao JH, Meng NH, Lin WY, Wu FY, Lin CC. Joint effect of gene-physical activity and the interactions among CRP TNF-α, and LTA polymorphisms on serum CRP, TNF-α levels and handgrip strength in community-dwelling elders in Taiwan-TCHS-E. Age (Dordr) 2016;38:46. doi: 10.1007/s11357-016-9909-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nourian M, Asgharian AM, Aghdaei H Asadzadeh. Lack of association between tumor necrosis factor alpha (TNFα) gene-1031C/T polymorphisms and susceptibility to inflammatory bowel disease (IBD) Arak Univ Med Sci J. 2016;19:71–79. [Google Scholar]
- 19.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moura AC, Lazzari VM, Agnes G, Almeida S, Giovenardi M, Veiga AB. Transcriptional expression study in the central nervous system of rats: What gene should be used as internal control? Einstein (Sao Paulo) 2014;12:336–341. doi: 10.1590/S1679-45082014AO3042. (In English, Portuguese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Chatzikonstantinou M, Konstantopoulos P, Stergiopoulos S, Kontzoglou K, Verikokos C, Perrea D, Dimitroulis D. Calprotectin as a diagnostic tool for inflammatory bowel diseases. Biomed Rep. 2016;5:403–407. doi: 10.3892/br.2016.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takeuchi O. Chronic Inflammation. Springer; 2016. Posttranscriptional regulation of cytokine mRNA controls the initiation and resolution of inflammation; pp. 319–332. [DOI] [PubMed] [Google Scholar]
- 24.van Heel DA, Udalova IA, de Silva AP, McGovern DP, Kinouchi Y, Hull J, Lench NJ, Cardon LR, Carey AH, Jewell DP, Kwiatkowski D. Inflammatory bowel disease is associated with a TNF polymorphism that affects an interaction between the OCT1 and NF(−kappa)B transcription factors. Hum Mol Genet. 2002;11:1281–1289. doi: 10.1093/hmg/11.11.1281. [DOI] [PubMed] [Google Scholar]
- 25.Chaparro M, Guerra I, Muñoz-Linares P, Gisbert JP. Systematic review: Antibodies and anti-TNF-α levels in inflammatory bowel disease. Aliment Pharmacol Ther. 2012;35:971–986. doi: 10.1111/j.1365-2036.2012.05057.x. [DOI] [PubMed] [Google Scholar]
- 26.Sanchez R, Levy E, Costea F, Sinnett D. IL-10 and TNF-alpha promoter haplotypes are associated with childhood Crohn's disease location. World J Gastroenterol. 2009;15:3776–3782. doi: 10.3748/wjg.15.3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Negoro K, Kinouchi Y, Hiwatashi N, Takahashi S, Takagi S, Satoh J, Shimosegawa T, Toyota T. Crohn's disease is associated with novel polymorphisms in the 5′-flanking region of the tumor necrosis factor gene. Gastroenterology. 1999;117:1062–1068. doi: 10.1016/S0016-5085(99)70390-2. [DOI] [PubMed] [Google Scholar]
- 28.Naderi N, Farnood A, Dadaei T, Habibi M, Balaii H, Firouzi F, Mahban A, Soltani M, Zali M. Association of tumor necrosis factor alpha gene polymorphisms with inflammatory bowel disease in Iran. Iran J Public Health. 2014;43:630–636. [PMC free article] [PubMed] [Google Scholar]
- 29.Tremelling M, Waller S, Bredin F, Greenfield S, Parkes M. Genetic variants in TNF-alpha but not DLG5 are associated with inflammatory bowel disease in a large United Kingdom cohort. Inflamm Bowel Dis. 2006;12:178–184. doi: 10.1097/01.MIB.0000217766.90766.37. [DOI] [PubMed] [Google Scholar]
- 30.Zipperlen K, Peddle L, Melay B, Hefferton D, Rahman P. Association of TNF-alpha polymorphisms in Crohn disease. Hum Immunol. 2005;66:56–59. doi: 10.1016/j.humimm.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Bonyadi M, Abdolmohammadi R, Jahanafrooz Z, Somy MH, Khoshbaten M. TNF-alpha gene polymorphisms in Iranian Azari Turkish patients with inflammatory bowel diseases. Saudi J Gastroenterol. 2014;20:108–112. doi: 10.4103/1319-3767.126322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rad R, Dossumbekova A, Neu B, Lang R, Bauer S, Saur D, Gerhard M, Prinz C. Cytokine gene polymorphisms influence mucosal cytokine expression, gastric inflammation, and host specific colonisation during Helicobacter pylori infection. Gut. 2004;53:1082–1089. doi: 10.1136/gut.2003.029736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen G, Wilson R, Wang SH, Zheng HZ, Walker JJ, McKillop JH. Tumour necrosis factor-alpha (TNF-alpha) gene polymorphism and expression in pre-eclampsia. Clin Exp Immunol. 1996;104:154–159. doi: 10.1046/j.1365-2249.1996.d01-647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]