Abstract
Extracellular vesicles are particles in mammalian body fluids that have attracted considerable attention as biomarkers for various diseases. In the present study, the authors isolated RNA aptamers with an affinity for extracellular vesicles from two library pools that encoded randomized sequences of different lengths. After the several rounds of selection, two conserved motifs are identified in the sequences that are obtained by next-generation sequencing. Most of the sequences were predicted to adopt a secondary structure that consisted of a non-conserved stem structure and a conserved loop sequence. Two minimal similar sequences are synthesized and confirmed the ability of these sequences to bind to extracellular vesicles. Circular dichroism spectroscopy and melting temperature analysis demonstrated that the aptamers were able to form a G-quadruplex structure in their loop regions and these structures were stabilized by potassium ions. Consistent with these structural data, the affinity of each aptamer for extracellular vesicles was dependent on potassium ions. The aptamers that were identified may be useful molecular tools for the development of diagnostic methods that utilize body fluids, such as blood, saliva and urine.
Keywords: RNA aptamer, extracellular vesicle, next-generation sequencing, non-conserved stem structure, G-quadruplex sequence
Introduction
Extracellular vesicles, which consist of lipid bilayer membranes, are secreted from various cells into mammalian body fluid, such as blood, saliva and urine (1). Such vesicles are generally classified into two groups: Exosomes and microvesicles, according to their size, physical properties and biogenesis (2). Exosomes are small particles with lipid bilayer membranes, ranging from 40 to 100 nm in diameter. They are associated with characteristic markers such as CD9, CD63, CD81 and HSP70 and are derived from the endocytic recycling pathway. Microvesicles are extracellular particles of 100 to 1,000 nm in diameter that are produced directly from the plasma membrane as a consequence of outward budding. Thus, the markers associated with microvesicles are considered to depend predominantly on the receptors that are present on the surface of the cells from which they are derived.
Extracellular vesicles contain proteins, microRNAs, mRNAs and DNA from their parent cells (3,4), and it appears that indicated these vesicles are important mediators of the transfer of information among cells (5). These features have suggested the potential utility of extracellular vesicles as diagnostic biomarkers of disease, as already proposed for ovarian cancer, lung cancer, pancreatic cancer and renal deficiency (6–10). Several methods have been developed for isolation of extracellular vesicles from biological samples, for example, ultracentrifugation, gel dialysis, immunoprecipitation, filtration and density-gradient centrifugation (11–15), but there is as yet no consensus as to optimal methods with respect to convenience and purity.
Aptamers are artificial DNA or RNA oligonucleotides that bind to targets with high affinity and specificity, and were initially investigated by Ellington and Szostak (16) and Tuerk and Gold (17) and their colleagues in the early 1990s. Aptamers can be selected using molecular-evolution technology, via in vitro selection or the systematic evolution of ligands by exponential enrichment (SELEX). Aptamers have been isolated that are specific for a large variety of targets, such as small molecules, nucleotides, peptides and proteins (18). In addition, live cells have been used as targets for selection of aptamers specific for receptors on cell surfaces (19–23).
Given that extracellular vesicles are more stable than cells and contain lipids, sugars and proteins within and/or on their membranes, as do cells themselves, the authors designed a procedure for isolation of RNA aptamers that bind to targets on extracellular vesicles, with the goal of developing new diagnostic systems. The current study reports the isolation of two aptamers specific for extracellular vesicles and the description of the target-specific affinity and structural features of these aptamers.
Materials and methods
Cell lines and preparation of extracellular vesicles
293T cells were a generous gift from Professor A. Fukamizu (Tsukuba University, Tsukuba, Japan). HeLa S3 cells were obtained from the RIKEN BioResource Center (Tsukuba, Japan). Both cell lines were maintained in Dulbecco's Modified Eagle's medium (DMEM; Thermo Fisher Scientific, Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific, Inc.). For experiments, cells were transferred to DMEM without serum and cultured for 2 or 3 days. The resultant conditioned medium was collected and passed through a 0.22 µm filter (EMD Millipore, Billerica, MA, USA) to remove cell debris. Then each sample was loaded into a 10 kDa ultrafiltration cartridge (Amicon Ultra; EMD Millipore), subjected to these exchanges of buffer with 40 ml PBS and concentrated to a final volume of 1 ml. The concentrated sample was placed in a 100 kDa ultrafiltration cartridge (VIVACON 500; Sartorius AG, Göttingen, Germany) and subjected to these exchanges of 500 µl PBS. The protein concentration of each preparation of vesicles was determined in terms of absorbance at 260 nm using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington DE, USA).
Selection of aptamers
Selection in vitro from two different library pools was performed as described previously (24–26). The libraries used are as follows: An N55 library that contained a region of 55 randomized nucleotides was used [5′-GGG AGG fUGG AAfC fUGA AGG AGA-(N55)-ACfU fUCG fCAA fUfCG fCfUfC fUAfC GfCA-3′]; an N30 library that contained a region of 30 randomized nucleotides [5′-GGfU AGA fUAC GAfU GGA-(N30)-fCAfU G AfC GfCG fCAG fCfCA-3′], where f represents a 2′-fluoro modification. Selections were performed in binding buffer [20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM KCl, 0.5 mM MgCl2 and 1.5 mM CaCl2].
Each pool of RNA was denatured at 98°C for 2 min and, following cooling, was incubated with extracellular vesicles in the presence of transfer (t)RNA (100 µg/ml) at room temperature. The mixture was passed through a 0.45 µm HAWP nitrocellulose filter (EMD Millipore) and the filter was washed three times with binding buffer. The RNA that had bound to extracellular vesicles and was retained on the filter was recovered by incubation with 7 M urea/10 mM EDTA at 98°C for 5 min, extraction with phenol/chloroform and precipitation in ethanol.
Reverse transcription was performed with a PrimeScript II first-strand cDNA synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China). Specifically, preparations of recovered RNA were supplemented with 1.25 µmol dNTPs and 25 pmol each reverse primer [reverse primer N55, 5′-TGC GTA GAG CGA TTG CGA AGT-3′; reverse primer N30, 5′-TGG CTG CGC GTC ATG-3′] then incubated at 65°C for 5 min. The RNase inhibitor (Takara Biotechnology Co., Ltd.) and 100 U reverse transcriptase (Takara Biotechnology Co., Ltd.) were added to the solution, which was incubated at 30°C for 10 min, 37°C for 10 min, 42°C for 40 min, 52°C for 30 min and 98°C for 5 min.
The cDNA was amplified by polymerase chain reaction (PCR) (98°C for 10 sec, 55°C for 30 sec and 72°C for 60 sec), with 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTPs, 0.5 U Ex Taq (Takara Biotechnology Co., Ltd.) and 0.125 µM each forward primer (N55 forward primer, 5′-TGTA ATA CGA CTC ACT ATA GGG AGG TGG AAC TGA AGG AGA-3′; N30 forward primer, 5′-TGT AAT ACG ACT CAC TAT AGG TAG ATA CGA TGG A-3′) and reverse primer (N55 reverse primer, 5′-TGC GTA GAG CGA TTG CGA AGT-3′; N30 reverse primer, 5′-TGG CTG CGC GTC ATG-3′), and products were precipitated with ethanol. RNAs, whose pyrimidine residues were 2-fluoro modified, were transcribed from template DNAs with a DuraScribe® kit (Epicentre; Illumina, Inc., San Diego, CA, USA) at 37°C for >6 h or overnight. Transcribed RNAs were treated with DNase I and purified on micro Bio-Spin columns (P-30; Bio-Rad Laboratories, Inc. Hercules, CA, USA) or by denaturing on 8% SDS-PAGE gels that contained 7 M urea in Tris/borate/EDTA buffer (Wako Pure Chemical Industries, Ltd., Osaka, Japan). Then, they were subjected to the next round of selection.
Sequencing
Sequencing was performed with a MiSeq sequencing system (Illumina, Inc.) next-generation sequencer and the reagent kit v2 (Illumina, Inc.) exactly according to the manufacturer's protocol. The sequences obtained were ranked in order of decreasing frequency.
Predictions of secondary structure and search for consensus sequences
Secondary structures were predicted using the CentroidFold prediction program (National Institute of Advanced Industrial Science and Technology, Tokyo, Japan) for RNA secondary folding (27). Consensus sequence were identified using the motif-discovering algorithm and the MEME Suite 4.10.0 online server (http://meme.nbcr.net/meme/tools/meme), applying the criterion of a minimum motif length of 8 nucleotides (28).
Minimized oligonucleotides
MO-1 [5′-GGC Cafc gafc gfcg gag gfug fugg ggg afufc gfuG GCC-3′] and MO-2 [5′-ggfu aga fuafc gafu gga aga ggg aaa ggg agg gfufu fcfua fcfc-3′] were purchased from Hokkaido System Science Co., Ltd. (Sapporo, Japan). Uppercase letters indicate deoxyribonucleic acids and lowercase letters indicate ribonucleic acids. The abbreviations fu and fc refer to 2-fluoro modified pyrimidines.
Filter-binding assay
For each assay, a 5′-biotinylated aptamer was incubated with extracellular vesicles in binding buffer that contained 100 µg/ml tRNA at room temperature for 2 h. The solution was passed through a nitrocellulose filter (EMD Millipore) and washed with 5 ml binding buffer. The filter was exposed to UV irradiation (254 nm) and was blocked in binding buffer that contained 3% BSA and 0.1% Tween-20 (Wako Pure Chemical Industries, Ltd.) for 60 min. Following washing, the filter was incubated with streptavidin-horseradish peroxidase conjugate (Thermo Fisher Scientific, Inc.; cat. no. N100; dilution, 1:1,000) for 60 min at room temperature and washed. Then, the Amersham ECL Prime Western Blotting Detection reagent (GE Healthcare Life Sciences, Chalfont, UK) was used to visualize the chemiluminescence of the 5′-biotinylated aptamer on the filter.
Surface plasmon resonance (SPR) assay
SPR assays were performed with Biacore X™ (GE Healthcare Life Sciences). Biotinylated oligonucleotides were immobilized on SA sensor chips (GE Healthcare Life Sciences). Then, extracellular vesicles were injected at the indicated concentrations (150, 75, 50, 43 and 38 µg/ml), and running buffer [binding buffer supplemented with 0.005% Tween-20 (Wako Pure Chemical Industries, Ltd)] was loaded onto flow cells 1 (blank) and 2 (aptamer) of the sensor chip. Data were analyzed with BiacoreTM evaluation software (GE Healthcare Life Sciences; version, 2.0). following subtracting the background (flow cell 1) from values for flow cell 2.
Prediction of G-quadruplex structure and measurements of circular dichroism (CD) and melting temperature (Tm)
The presence of a G-quadruplex structure in individual aptamers was predicted by the QGRS Mapper (http://bioinformatics.ramapo.edu/QGRS/index.php) (29).
Circular dichroism (CD) spectra were recorded with a spectropolarimeter (J-820; Jasco International Co., Ltd., Tokyo, Japan) as described previously (30). A cell with a 1 mm path length was used for analyses of titrations with KCl (at 0 mM, 0.1, 0.3, 1, 5, 10, 50 and 100 mM) in 20 mM Tris-HCl buffer (pH 7.5). Spectra were recorded four times, from 220 to 320 nm at 25°C. Then, CD intensities were expressed in terms of [θ] per residue.
Melting temperatures of aptamers were determined at 270 nm in buffer that included 100 mM KCl as the temperature was raised from 25 to 95°C.
Fractionation of extracellular vesicles and dot-blotting assay
Preparations of extracellular vesicles were fractionated on a column of sepharose 2B (GE Healthcare Life Sciences). Protein concentrations were determined with a DC protein assay kit (Bio-Rad Laboratories, Inc.) according to the manufacturer's instructions. Then, 2 µl aliquots of fractionated and concentrated extracellular vesicles were spotted on a nitrocellulose membrane, which was dried for 30 min at room temperature.
For detection of binding of aptamers, the membrane was blocked with binding buffer that contained 1% BSA and 100 µg/ml tRNA for 60 min at room temperature. The membrane was then incubated with 100 µM 5′-biotinylated aptamer in binding buffer supplemented with 0.1% Tween-20, 1% BSA and 100 µg/ml tRNA for 60 min at room temperature and washed three times with binding buffer supplemented with 0.1% Tween-20. The membrane was exposed to UV irradiation (254 nm) and then to streptavidin-horseradish peroxidase conjugate (Thermo Fisher Scientific, Inc.; cat. no. N100; dilution, 1:4,000) for 60 min at room temperature. It was washed three times and, finally, the Amersham ECL Prime Western Blotting Detection reagent (GE Healthcare Life Sciences; cat. no. RPN2232) was used to visualize the chemiluminescence of the aptamer on the membrane.
For detection of antibodies, the membrane was blocked in PBS supplemented with 0.1% Tween-20 (PBS-T) and containing 3% BSA for 60 min at room temperature. The membrane was incubated with human CD63-specific mouse monoclonal antibody (Santa Cruz Biotechnology, Inc., Dallas, TX, USA; cat. no. Mx-49.129.5; dilution, 1:1,000) for 60 min at room temperature and washed three times with PBS-T. The membrane was exposed to second antibody-HRP conjugate (GE Healthcare Life Sciences; cat. no. NA931; dilution, 1:2,000) for 60 min at room temperature and washed three times. Finally, the Amersham ECL Prime Western Blotting Detection reagent (GE Healthcare Life Sciences; RPN2232) was used to visualize chemiluminescence.
Results
Isolation of RNA aptamers with affinity for extracellular vesicles
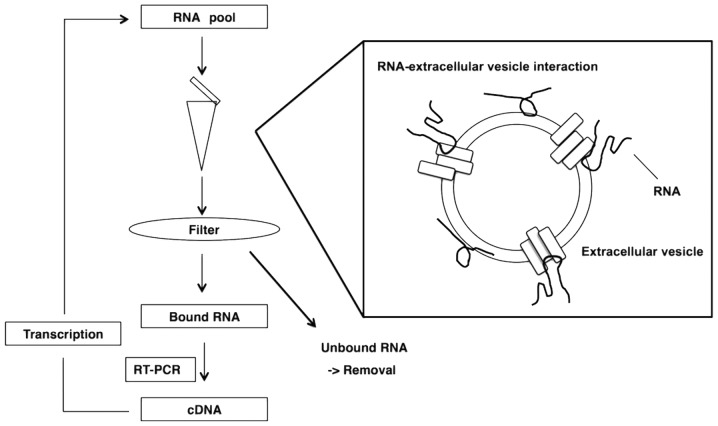
To isolate RNA aptamers with an affinity for extracellular vesicles, the authors used a selection method in vitro that was based on a previously reported method (Fig. 1) (24–26). A 97-nt RNA library pool (N55 pool) and a 60-nt RNA library pool (K pool) was prepared, with respective central randomized 55 and 30-nt sequences, by in vitro transcription with 2′-fluoro-modified pyrimidines from synthesized DNA libraries, as templates. Selection was performed as follows. A transcribed RNA library pool was incubated with extracellular vesicles in binding buffer for 30 min. The RNA-extracellular vesicle complexes were added to a nitrocellulose filter and washed. Bound RNAs were eluted, purified and subjected to reverse-transcription and amplification. An enriched RNA pool was transcribed from the amplified products of PCR and used for the next round of selection.
Figure 1.
Schematic representation of enrichment by SELEX for aptamers that bind to extracellular vesicles. The two RNA libraries were incubated, separately, with extracellular vesicles. RNA-extracellular vesicle complexes were added to nitrocellulose filters, which were then washed repeatedly. The bound RNAs were eluted, purified and subjected to reverse transcription and amplification (RT-PCR). Pools of enriched RNA were generated by transcription from the products of PCR and used for the next round of selection. SELEX, systematic evolution of ligands by exponential enrichment; RT-PCR, reverse transcription-polymerase chain reaction.
After 10 rounds of selection, the enriched pools were sequenced from the eighth, ninth and tenth rounds using a next-generation sequencer. The 50 most abundant unique sequences were ranked according to frequency and the results are presented in Tables I and II.
Table I.
The sequences obtained from S pool.
| Clone | Number | Sequence (5′-3′) |
|---|---|---|
| S1 | 935 | ACGCTAGCTGTGGAAAGACGCTAAATCGGGAGGTGGGTTGGGGTGCTAGCGATCA |
| S2 | 375 | CTAGGGTGGGGAAATATGGAGGGCGCTCAGGGCTACACTGACTACGTGATTAGAG |
| S3 | 365 | GTCCGCTTCCCGCCACCCGGTGGAAGGAGCCGCGAGGTCACGGAGGTGAGGGGGT |
| S4 | 362 | TGCTCATTCGTCATAGGTGGAGGGTAGTAGGGGGCGAACAGTTCCACGCTAGCGT |
| S5 | 357 | GGTGGGTTATAGAGGTTGGGTCCCCGACGTTGCTTCCCTATATAGAGTGGCGAGG |
| S6 | 344 | TCTCTCGCAGCGGTCTGCATGGTAGCGGGTCAACCACGGAGGAGTGGGGGTGTTG |
| S7 | 310 | CACGGAGGTGTGGGGGTTCTCAAAACGTTCTATGCTTGGCCAATCCCGAGCCGTC |
| S8 | 223 | CAGCATTGCGCACGACGCGGAGGTGTGGGGGATCGTGTGGAGAAATGGCTGGCTG |
| S9 | 180 | ATGGGATGGGAATAGTTCACCACAATTTTTGGGCTCCTTAGCTACACGCGATGGA |
| S10 | 169 | AGCAACGAAAATTACTGCCGCGGAGGAGAGGGGGAGCAGTATTCGTTGGGTCATA |
| S11 | 137 | TAGACCCACGCGCATGGCACGGAGGAGAGGGGGACACGCGCAAGGTCTTCACATG |
| S12 | 134 | AAATTCGCGTTGTAGGTGCACGGTGGTGTGGGGGTTATCCGGCAACGCGTTTGGT |
| S13 | 124 | GGTGCACCATCTCCGGTCTCGCGCGGTGGAGTGGGGGTTGAGACCGGGGTTGGTT |
| S14 | 107 | CTTACTCGCAGACACGGAGGTGTGGGGGTTCTGTGGGAGCTTGCACGAGACCTTC |
| S15 | 98 | TGAGTTAGTGGTGCTGCGCGCGGAGGAGTGGGGGTTGCAGCGTTACGTACGAGTC |
| S16 | 80 | TGCCGTGTCCGGAATTGCACGGAGGTGAGGGGGATATATTCTGGATAGCGCGCGG |
| S17 | 78 | TTTGGGGCGTTTCATCAAGATTCGGGTTGGGGACTCTAATAGTAGCCGGTTGTGG |
| S18 | 78 | ATTTTCGAATCCGCGTCCCGGCGGATAGCACGCGAGGTTACGGCGGAGTGGGGAA |
| S19 | 77 | CACGGAGGTGTGGGGGTTCTCAAAACGTTCTATGCTTGGCCAATCCCGAGCGGTC |
| S20 | 77 | ACTGTTGAACGCTCACGTGTACGGAGGTGTGGGGAATACGTGGGGGGTAACATCT |
| S21 | 75 | CACGGCGGAGTGGGGAGACTCTTGAAATGTTCCGGGTTGCGAACGTCGGTGCTGG |
| S22 | 71 | CACCGGGGGCAACATGGCGCACGGAGGAGAGGGGGTCGTCTAGTTAGTCCCGGTG |
| S23 | 63 | TGGCGCGGGTTCGCTGACACGGCGGTGTGGGGGTTCAGGAAACAATGCGCACGAA |
| S24 | 63 | GGTATTGGGCGGGGGTTCCCAAAAGTAGCGCGACGTCATACGAAGCCGTACGTCG |
| S25 | 62 | GGCGGAGCACGTATTGACACGGTGGAGAGGGGGTTCATACGGCTCAGGCACCTAA |
| S26 | 56 | TCGGGACGGCCTGAGGCGCACGGTGGTGTGGGGGTTGCCTACTGCCGTTTGGTCA |
| S27 | 54 | TCTCTCGCAGCGTCTGCATGGTAGCGGGTCAACCACGGAGGAGTGGGGGTGTTG |
| S28 | 43 | GAGCGCAATCTAACACGGAGGTGTGGGGGTTTAGCGTGCGCAAACTCAGATGCGT |
| S29 | 38 | CTCAGTTCATCGTCCCTAGACGCACGGCGGAATTGGGAGTACGGAGGTGGGGGGA |
| S30 | 38 | ACGAAGTTTAGGATCGCTCCTTCCACGGCGGTGAGGGGGTGAAGCTGCGACCCAG |
| S31 | 38 | AAGCCGCATCCGGCGGGCGACACGGAGGAGAGGGGGATCGCCACAGCCTGCGGTG |
| S32 | 36 | TTGGAGGGGGTGTGCCTATTTGGGGTTCAGCAGGGGCACATACGCGGTTGACGAG |
| S33 | 35 | CGTCAATCACGGCGGTGAGGGGGCATTGATGTCACGCAAAGTAGGCCTAATACCC |
| S34 | 29 | CACGGAGGAGTGGGGGTTCTCTGGGAAGTTTCGTCTTGTTGGTACGTAACCCGGC |
| S35 | 28 | AGGCCCGTACGGAGGTGTGGGGATTGGGTCTTTTGACTTGCGAGGCCGTTGTGCG |
| S36 | 27 | ACGCTAGCTGTGGAAAGACGCTAAATCGGGAGGTGGGTTGGGGTGCTAGCGATCG |
| S37 | 26 | TGAGGGATGGCCACGGAGGAGTGGGGGTGCCATCGCAACGTTGACACGGGTTGCA |
| S38 | 26 | GTGTGCCGAGCGATAGGGACACGGTGGAGTGGGGGGACCTATGCCCGGCAAGACA |
| S39 | 26 | GGGGTCATTAGGAGGGGCCTTTAGAAAAATAGTAGCCGCTGCGGGTCCTTTCGGG |
| S40 | 24 | ACGCTAGCTGTGGAAAGACGCTAAATCGGGAGGTGGGTTGGGGTGCTAGTGATCA |
| S41 | 21 | GATTTGGGGGCCACGGAGGTGAGGGGGCGCTCCTATTCTCGTGTTTTGCGTGCGA |
| S42 | 21 | CGGGCGCCGGGTTAGATCACGGAGGTGTGGGGGTATCTACCCCGTGAGGCGCCAC |
| S43 | 20 | GTGTTGCACCCGCCTGGGGCACGGAGGTGTGGGGGATCCCAGGGTGAGCGACTAA |
| S44 | 20 | CTAGGGTGGGGAAATATGGAGGGCGCTCAGGGCTACACTGACTATGTGATTAGAG |
| S45 | 20 | ATTTTTTTAATGCATCATTTTTACACTCCTTTTGGACCAACCCAACGGGCGCTGC |
| S46 | 19 | CACTCGGGTTGGAGGCGCACGGCGGTGTGGGGGTCGCTTCAAGTGACGGGTGTCA |
| S47 | 18 | TGCACAGGTGACACGCCGATCACGGAGGTGTGGGGAGATCGGCAATGGCACGGTG |
| S48 | 18 | GTCGAAGTGTAACTTATTTGTGTGATTTTTTTGTTTTATGCTTACACGCGGCTCA |
| S49 | 18 | GGTGGGTTATAGAGGTTGGGTCCCCGACGTTGCTTCCCTATATAGAGTGGCGCGG |
| S50 | 18 | ATTTTGTTTTGTTGTTTTTGTGTACTCCTTGAGATGCTGGCTACTGTCCGAGCCG |
Table II.
The sequences obtained from K pool.
| Clone | Number | Sequence (5′-3′) |
|---|---|---|
| K1 | 918 | ATAGCGGGAGGGAGGGTTCTACCTGGTGGG |
| K2 | 692 | GGGTGGAGGGAAGGAGTGGGGTTCTACCGG |
| K3 | 645 | AGAGGGAAAGGGAGGGTTCTACCGGGTGCA |
| K4 | 552 | ACGTGGGAGGGATTGGGGTATCTCCGGTTG |
| K5 | 475 | AGAGGGTGGGAAAGGGTTCTACCACAGTGC |
| K6 | 471 | AAGTGGGAGGGGAGGGTTCTACCGGGCCGC |
| K7 | 436 | GTAGGGAGGTATGTATCTCCTGGTTGGGGG |
| K8 | 403 | GAGGGAAGGGATATGGGGTATCTAGGGCCG |
| K9 | 367 | AAGAGGGAGGGTAGGGTTCTACCAGCTGGG |
| K10 | 361 | GTACGGAGGTGAGGGGAACTCCACGGTCGG |
| K11 | 356 | GCCCGGCACGGTGGAGAGGGGGTCCGGGGC |
| K12 | 355 | AGTGGGACTGGGATGGGGTGTATCGCCCGG |
| K13 | 349 | ATGAGGGAGGGTTGGGGTATCTCCCCGGTG |
| K14 | 338 | TGAGGGAATAGGGAAAGGGGTATCGTTGGG |
| K15 | 287 | ACGAGGGAGGGAGGGGTATCACCGGGCCGG |
| K16 | 276 | GAGGGATGAGGGTGGGCTCTACCTGGCCGG |
| K17 | 271 | GCCTACGCACGGTGGAGAGGGGGTTGTGGG |
| K18 | 236 | AGTGGGAAGGGTTTGGGTGGTCTACCGTGG |
| K19 | 230 | GAGGGAGGGAGGGCTCTACCTTTGTGGCCC |
| K20 | 225 | CACGGTGGAGTGGGGAGTTCATTGGGCGGG |
| K21 | 222 | AGTGGGATGGGTAGGGTTCTACATATGCTG |
| K22 | 207 | GCGGGATTGGGTTTTGGGGTATCTGGGCGG |
| K23 | 205 | CGGTCAATGCCCACGGTGGAGAGGGGGTGG |
| K24 | 188 | CCACGGAGGTGAGGGGGTGTCCACGGTGGC |
| K25 | 185 | TAGAGGGAGGGAGGGATCTACCAGGTGGGG |
| K26 | 174 | TAGTGGGAGGGAATGGGATTTCTACCGGGG |
| K27 | 170 | CACTGGAGGAGGGTGGGGAGTTCATCCGGG |
| K28 | 158 | TTGAGGGAAAGGGTGGGGCATCTACCGTGG |
| K29 | 151 | CCGTAACACGGAGGAGAGGGGGGAACGGTG |
| K30 | 146 | CGCGGAGGTGTGGGGGATCCGTCGTGGTGG |
| K31 | 149 | GAGGGAGGGACTGGGGTATCTTCAGGCGGC |
| K32 | 143 | ATGAGGGTAGGGAAAGGGGTATCTCGGCGG |
| K33 | 136 | GGGCACGGTGGAGTGGGGGTCCTTCCTGGG |
| K34 | 132 | GAGGGTCAGGGATTTGGGGTATCTGGGTGG |
| K35 | 127 | TTGGAGTGGTGGGTGGGGATCGTGAGGCGG |
| K36 | 124 | ACACGGTGGTGTGGGGGTTTCCAGGGCGGG |
| K37 | 121 | CTGTCGGTGCCCACGGTGGAGAGGGGGTGG |
| K38 | 117 | TGAGGGAGGGACAGGGGTATCTTGGTGCGG |
| K39 | 116 | CCCTGTCACGGTGGTGTGGGGGTATAGGGC |
| K40 | 114 | AGAGGGAGGGCAAGGGTTCTACCAGGTCG |
| K41 | 113 | CCAGCACGCGGTGGAGAGGGGGATGTTGGC |
| K42 | 111 | TGGCCGATCACGGAGGAGAGGGGGTATCGG |
| K43 | 106 | GAGGGTTAAGGGACGGGGTATCTGAGGCGG |
| K44 | 97 | GTGGGATAGGGTTTACGGGGTATCGGTGGG |
| K45 | 94 | CACGGAGGAGAGGGGGTTCCATCGTTGTCG |
| K46 | 92 | TTGAGGTGGGATAGGGTAGGGGTCGTGTGC |
| K47 | 92 | GTGGGAGGGTGGGCTCTGCCAGAACCGGC |
| K48 | 92 | AGTGGGTTGGGTAATGGGGTATCTACGGGG |
| K49 | 88 | CACGGTGGAGAGGGGGATCCTACTACTCGG |
| K50 | 88 | ACAGTGGGAGGGAGGGTTATCACCGGGCCG |
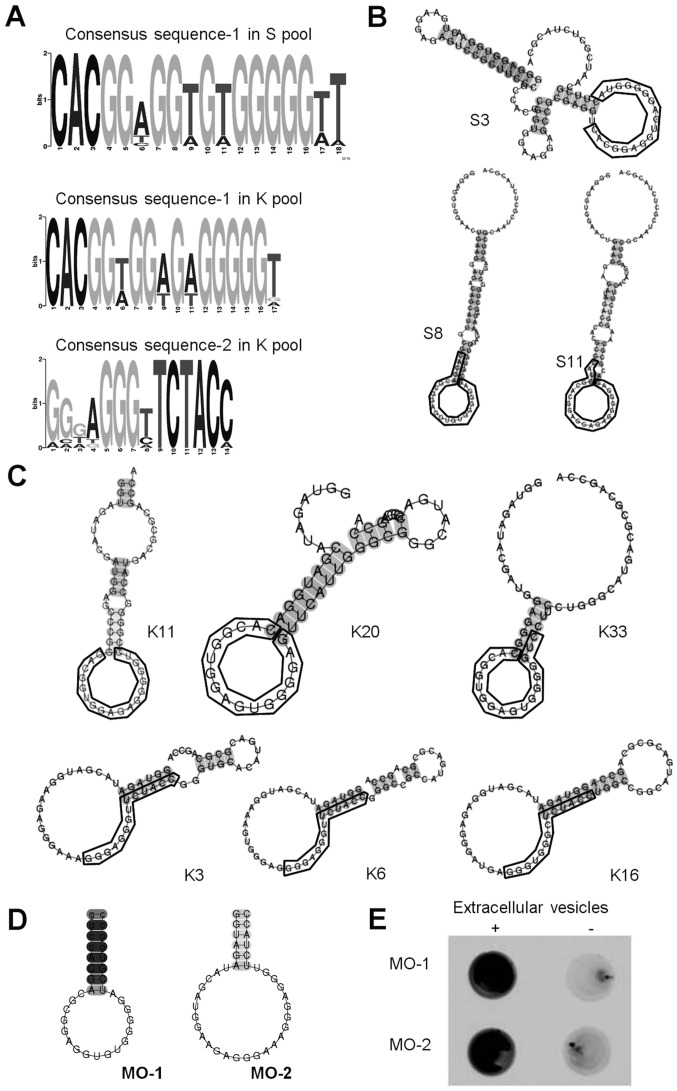
The MEME suite (28) was used to search for sequence similarities among all 50 sequences from the respective tenth rounds of enrichment (Fig. 2A). The analysis revealed two common sequences, the consensus sequence-1 ‘cac gg(t/a) gg(t/a) g(t/a) g ggg gt’ in the S pool (31 out of 50 clones), the K pool (5 out of 50 clones) and the consensus sequence-2 ‘ggg agg gtt cta cc’ in the K pool (17 out of 50 clones), respectively. Consensus sequence-1 was identified from both library pools, even through the design of the two libraries (lengths of randomized region and the fixed primer regions) were different. The independent isolation of consensus sequence-1 from the two very different libraries suggested that the sequence may have strong affinity for extracellular vesicles.
Figure 2.
Characterization of aptamers that bound to extracellular vesicles. (A) The consensus sequences shared among the top 50 selected sequences, as identified with the MEME suite. (B and C) Typical secondary structures (clones S3, S8 and S11 in the S pool and clones K3, K20, K33, K3, K6 and K16 in the K pool), as predicted by the CentroidFold program. Consensus sequences are enclosed in boxes. (D) Secondary structures of minimized aptamers MO-1 and MO-2. (E) Binding of selected aptamers to extracellular vesicles. Labeled aptamers (MO-1 and MO2) were incubated with extracellular vesicles and mixtures were filtered through a membrane. Aptamer-extracellular vesicle complexes were trapped on the membrane (+). The minus sign (−) represents negative controls without aptamers. MO, minimized oligonucleotides.
Next, the secondary RNA structures of the complete sequences of 53 clones were analyzed (36 clones that included consensus sequence-1 and 17 clones that included consensus sequence-2) with primer regions, using CentroidFold software, to identify common secondary structures (Fig. 2B) (27). All of the sequences were predicted to have a non-conserved stem with a loop. Some examples of this structure are presented in Fig. 2C.
Next, the authors attempted to shorten a full-length sequence to a minimized sequence with the step-loop structure (Fig. 2D). The sequences of clone S8 and clone K3 were selected as representatives of aptamers with each respective consensus sequence because both were in the upper ranks of respective rankings and were likely to be relatively stable as a result of their longer stems. The authors synthesized minimized oligonucleotide-1 (MO-1), including consensus sequence-1 referenced to clone S8, and minimized oligonucleotide-2 (MO-2), including consensus sequence-2 referenced to clone K3, respectively.
To examine the binding affinities of MO-1 and MO-2 for extracellular vesicles, filter-binding assays were performed with the respective 5′-biotinylated minimized aptamers. Both aptamers bound to the extracellular vesicles (Fig. 2E) and not to BSA or mouse IgG (data not shown), indicating that the minimized sequences had retained binding affinity and specificity with respect to extracellular vesicles.
Analysis of interactions of aptamers with extracellular vesicles
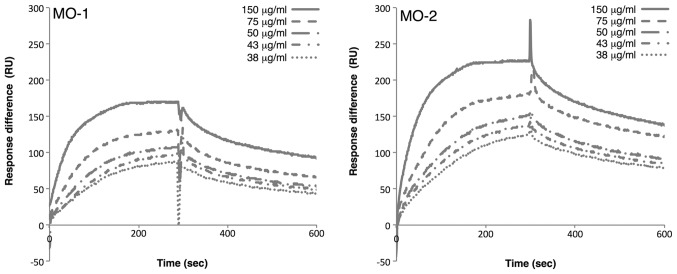
To examine the interaction of the each minimized aptamer with extracellular vesicles, affinities were measured by SPR. The 5′-biotinylated minimized oligonucleotides were immobilized on streptavidin-coated sensor chips. Extracellular vesicles at various concentrations were then passed over the immobilized oligonucleotides. SPR analysis revealed that MO-1 bound to extracellular vesicles with a KD of 1.17×10–15 mg/ml, while MO-2 bound to extracellular vesicles with a KD of 6.08×10–16 mg/ml (Fig. 3). These rather similar values suggested that two aptamers may bind to the same target on extracellular vesicles.
Figure 3.
Analysis by SPR spectroscopy of binding of aptamers to extracellular vesicles. The binding affinity of aptamers was examined by SPR spectroscopy with the Biacore X system. Extracellular vesicles at various concentrations (150, 75, 50, 43 and 38 µg/ml) were passed over aptamers, which had been immobilized on streptavidin-coated sensor chips. The sensorgrams demonstrate the significant binding of the aptamers to extracellular vesicles. RU, resonance units; SPR, surface plasmon resonance.
Since both aptamers bound to extracellular vesicles derived from 293T cells, the authors examined whether the aptamers could bind to extracellular vesicles derived from another cell line. Extracellular vesicles were prepared from HeLa S3 cells and the SPR analysis was repeated. The results demonstrated that the MO-1 aptamer bound to HeLa S3 extracellular vesicles with a KD of 6.83×10−15 mg/ml, and the MO-2 aptamer bound to the HeLa S3 extracellular vesicles with a KD of 2.82×10−16 mg/ml. These results were similar to those obtained with 293T-derived extracellular vesicles. Thus, these observations suggested that the two aptamers may recognize a common target on extracellular vesicles from a variety of sources.
Spectroscopic analysis of the structures of minimized aptamers
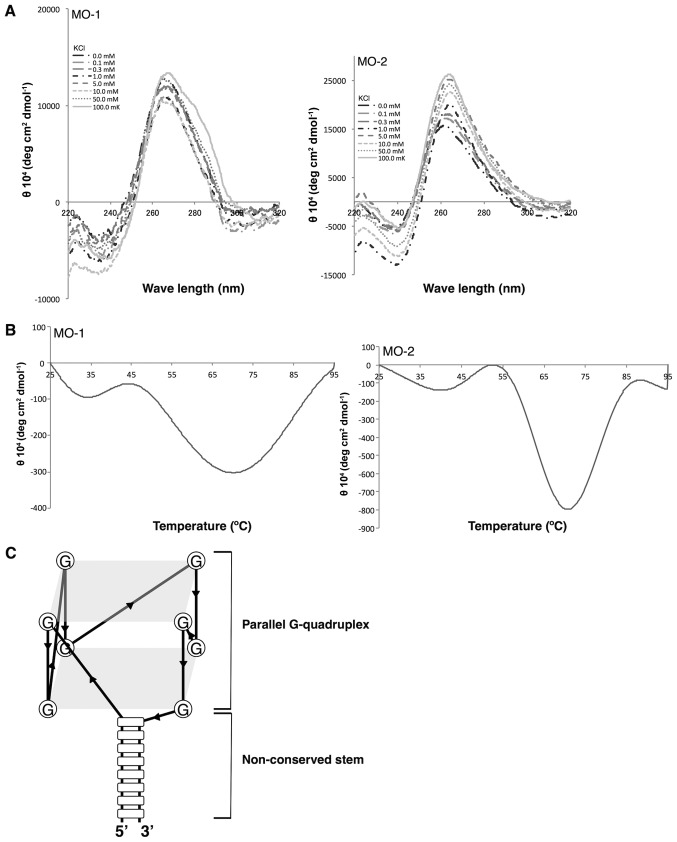
There are many examples of guanine-rich sequences folding into G-quadruplex structures in the presence of potassium ions (31,32). In previous studies of the authors, it was demonstrated that an RNA aptamer directed against a bovine prion forms a G-quadruplex structure (25,26,33–35). Since guanine-rich sequences were present in the MO-1 and MO-2 aptamers, the authors examined whether the guanine-rich sequences may resemble the G-quadruplex consensus motif, using the QGRS Mapper, namely, G-quadruplex analysis software (29). There was a high probability of the presence of a G-quadruplex structure in both the MO-1 aptamer (G-score=19) and the MO-2 aptamer (G-score=20).
To confirm these predictions experimentally, structural and thermodynamic analyses were performed of each aptamer by circular dichroism (CD) spectroscopy and thermal melting point (Tm) analysis (Fig. 4A). Both aptamers yielded a negative band at 240 nm and a positive band at 260 nm in the CD spectrum. The minimum band at 240 nm and the maximum band at 260 nm were induced by potassium ions in a concentration-dependent manner (K+ from 0 mM to 100 mM; Fig. 4A). The Tm values of the aptamers were relatively high (approximately 70°C; Fig. 4B), indicating that the aptamers were tightly folded, as expected from a G-quadruplex structure. All of the results supported the hypothesis that both aptamers were folded into a G-quadruplex.
Figure 4.
Analysis of quadruplex structures. (A) CD spectra of aptamers at various concentrations of KCl (0, 0.1, 0.3, 1, 5, 10, 50 and 100 mM). The aptamers yielded a negative peak at 240 nm and a positive peak at 260 nm, an indication of the formation of a parallel quadruplex structure. (B) CD melting-temperature curves of aptamers at 270 nm in 100 mM KCl. (C) Putative G-quadruplex structure of the aptamers. The loop region of the hairpin structure folds into the G-quadruplex structure. CD, circular dichroism; MO, minimized oligonucleotides; KCl, potassium chloride.
G-quadruplex structures have been classified into two characteristic spectral forms (36) on the basis of their CD spectra: The parallel G-quadruplex with a maximum at ~260 nm and a minimum at ~240 nm, and the anti-parallel G-quadruplex with a maximum at ~290 nm and a minimum at ~260 nm. Both the MO-1 and the MO-2 aptamers yielded a maximum at ~260 nm and a minimum at ~240 nm, strongly suggesting the presence of a parallel G-quadruplex in each one (25,26,33–35), presented schematically in Fig. 4C.
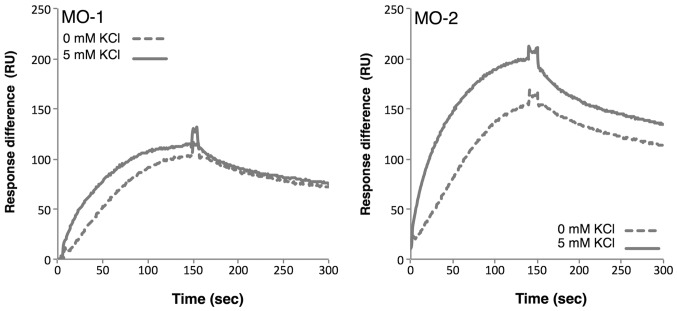
Next, the binding of aptamers to extracellular vesicles in the presence and absence of potassium ions was examined. SPR analysis indicated that the binding affinities of both aptamers increased in the presence of potassium ions (Fig. 5), reflecting the observation that the G-quadruplex structure is stabilized by potassium ions. The strong affinities of the aptamers for extracellular vesicles may be sustained by the tight structure of the G-quadruplex.
Figure 5.
Dependence of changes in the SPR spectra of aptamers on the concentration of potassium ions. The affinities of aptamers for extracellular vesicles (75 µg/ml) were analyzed by SPR spectroscopy in the presence or absence of potassium ions (5 mM). SPR, surface plasmon resonance; MO, minimized oligonucleotides; KCl, potassium chloride; RU, resonance units.
Properties of extracellular vesicles recognized by aptamer MO-1
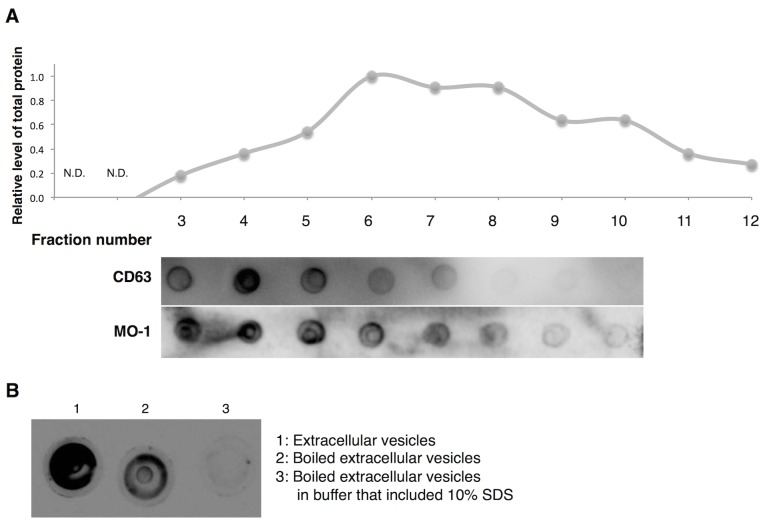
Extracellular vesicles, including exosomes and microvesicles, exist as heterogeneous populations of vesicles of different sizes, densities and constituent molecules because of difference in origin and mode of generation (37). Therefore, whether the MO-1 aptamer recognized all fractions of extracellular vesicles or only some members of populations of vesicles was investigated. The authors fractioned extracellular vesicles in terms of size on a sepharose 2B column and monitored the distribution of amounts of total protein (upper graph in Fig. 6A) and CD63 protein, a marker of exosomes (middle panel in Fig. 6A). The CD63-specific antibody bound strongly around fraction 4. By contrast, aptamer MO-1 exhibited strong affinity for fractions 3 through 6 (lower panel in Fig. 6A). Thus, the aptamer appeared to recognize the population of vesicles that included a majority of exosomes.
Figure 6.
Properties of extracellular vesicles that were recognized by aptamer MO-1. (A) Extracellular vesicles were fractionated by on a column of sepharose 2B. The distribution of proteins (by mass) in the fractionated extracellular vesicles (fraction numbers 1–12) is presented in the graph. Results of blotting analysis, indicating the affinities of CD63 and the aptamer for the fractions of extracellular vesicles, are presented in the middle panel and the lower panel, respectively. (B) The affinity of the MO-1 aptamer for denatured extracellular vesicles was evaluated by a dot-blotting assay. MO, minimized oligonucleotides.
Also examined was the binding of the MO-1 aptamer to denatured extracellular vesicles (Fig. 6B). The binding affinity of the aptamer (MO-1) for heat-denatured and for SDS-denatured extracellular vesicles was very much reduced. Thus, it seems likely that the aptamer may recognize a specific protein structure on the extracellular vesicles.
Discussion
This study is the first, to the best of the authors' knowledge, to identify RNA aptamers (33-nucleotide MO-1 and 38-nucleotide MO-2) that bind to extracellular vesicles. The sequences were identified with a next-generation sequencer and each consisted of a non-conserved stem with a conserved loop (Fig. 2D). SPR analysis revealed that the aptamers MO-1 and MO-2 had similar strong affinity for extracellular vesicles (Fig. 3). Structural analysis by CD spectroscopy demonstrated that both aptamers formed a G-quadruplex in the loop region (Fig. 4).
The two aptamers MO-1 and MO-2, with consensus sequence-1, were obtained from two independent libraries, in which the lengths of the randomized regions and the sequences of fixed primer regions were completely different (see Materials and methods). This result may reflect the fact that the aptamers were effectively and efficiently selected from a huge variety of randomized sequences. Alternatively, it is possible that the presence of consensus sequence-1 in both aptamers was due to its strong affinity and shorter length.
All the candidate aptamers with consensus sequence-1 and consensus sequence-2, respectively, had non-conserved stem regions. In other words, the regions surrounding the consensus sequences were different but were able to form stable double-strand structures. This method allowed the authors to concentrate related sequences from a massive diversity of libraries sequences effectively, comprehensively and individually.
Spectroscopic analysis suggested that the MO-1 and MO-2 aptamers formed a G-quadruplex structure in their loop regions, and the stable conformation of the G-quadruplex in the presence of potassium ions may contribute to the binding affinity of the aptamers for extracellular vesicles (Fig. 5). The stem-G-quadruplex structure of these aptamers may be one of essential structures for recognition of target molecules as a binding scaffold with strong affinity and compact size.
To confirm the interaction between MO-1 and extracellular vesicles, the authors designed ‘pull-down’ experiments using the 5′-biotinylated MO-1 aptamer. In spite of several attempts, these experiments failed, presumably because of difficulties in capturing vesicles of high molecular weight (data not shown). The binding of MO-1 to size-fractionated extracellular vesicles suggested that a majority of its targets may be exosomes but the targets may also be other entities of similar molecular weight. The SPR analyses demonstrated that extracellular vesicles derived from HeLa S3 cells also bound to the MO-1 aptamer, suggesting that the same molecule(s) derived from both 293T and HeLa S3 cells may be its target (Fig. 3). Moreover, binding affinity was severely depressed under denaturing conditions (Fig. 6B). Thus, it is very likely that the targets of the MO-1 and MO-2 aptamers are proteins on the surface of extracellular vesicles, for example, CD9, CD13, CD26, CD63, CD81, heat shock protein 70, integrins, intercellular adhesion molecule-1, major histocompatibility complex and tumor necrosis factor (38–41).
Previously, a patent was filed for a sequence very similar to that of MO-1 (42). The sequence was identified as an aptamer that bound to platelet-derived growth factor (PDGF). In the patent application, the claim was made that the sequence was able to bind to PDGF receptors, to vascular endothelial growth factor and its receptors. Therefore, the target of MO-1 may be such a ligand or receptor on extracellular vesicles.
For development of diagnostic technologies that exploit extracellular vesicles, aptamers have several advantages over antibodies. The most significant advantage is that extracellular vesicles can be purified intact, under mild conditions, from body fluids (11). Captured vesicles can be eluted from an aptamer-conjugated support by chelating agents, such as EDTA, which can unfold aptamers by sequestering magnesium ions. In addition, aptamers can target small molecules, lipids and sugars (18). Aptamers can be easily improved by inclusion of modified nucleic acids or artificial nucleic acids that enhance stability and/or the ability to bind to nucleic acids. Furthermore, a method using counter-SELEX (43) may allow isolation of aptamers against disease-specific extracellular vesicles via ‘subtractive’ use of disease-derived and normal vesicles. In such counter-SELEX, the aptamers identified in the present study, which recognize whole extracellular vesicles, may be useful in the future as blocking reagents.
It is hoped that the current work will lead to the development of extracellular vesicle-based diagnostic systems in which aptamers are used to target disease-specific molecules on extracellular vesicles.
Acknowledgements
The present study was supported by a Grant-in-Aid for Challenging Exploratory Research from the Japan Society for the Promotion of Science (grant no. 26640097). The authors would like to thank Dr. M. Ikemoto for his helpful comments and Dr. T. Yamasaki for her skilled technical assistance.
References
- 1.Simpson RJ, Jensen SS, Lim JW. Proteomic profiling of exosomes: Current perspectives. Proteomics. 2008;8:4083–4099. doi: 10.1002/pmic.200800109. [DOI] [PubMed] [Google Scholar]
- 2.Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: Extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012;21:R125–R134. doi: 10.1093/hmg/dds317. [DOI] [PubMed] [Google Scholar]
- 3.Valadi H, Ekström K, Bossios A, Sjostrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 4.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Jr, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simons M, Raposo G. Exosomes-vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 7.Zhou H, Pisitkun T, Aponte A, Yuen PS, Hoffert JD, Yasuda H, Hu X, Chawla L, Shen RF, Knepper MA, Star RA. Exosomal Fetuin-A identified by proteomics: A novel urinary biomarker for detecting acute kidney injury. Kidney Int. 2006;70:1847–1857. doi: 10.1038/sj.ki.5001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamashita T, Kamada H, Kanasaki S, Maeda Y, Nagano K, Abe Y, Inoue M, Yoshioka Y, Tsutsumi Y, Katayama S, et al. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie. 2013;68:969–973. [PubMed] [Google Scholar]
- 9.Zoller M. Pancreatic cancer diagnosis by free and exosomal miRNA. World J Gastrointest Pathophysiol. 2013;4:74–90. doi: 10.4291/wjgp.v4.i4.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, LeBleu VS, Mittendorf EA, Weitz J, Rahbari N, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006 doi: 10.1002/0471143030.cb0322s30. Chapter 3: Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 12.Rekker K, Saare M, Roost AM, Kubo AL, Zarovni N, Chiesi A, Salumets A, Peters M. Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem. 2014;47:135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 13.van der Pol E, Hoekstra AG, Sturk A, Otto C, van Leeuwen TG, Nieuwland R. Optical and non-optical methods for detection and characterization of microparticles and exosomes. J Thromb Haemost. 2010;8:2596–2607. doi: 10.1111/j.1538-7836.2010.04074.x. [DOI] [PubMed] [Google Scholar]
- 14.Grant R, Ansa-Addo E, Stratton D, Antwi-Baffour S, Jorfi S, Kholia S, Krige L, Lange S, Inal J. A filtration-based protocol to isolate human plasma membrane-derived vesicles and exosomes from blood plasma. J Immunol Methods. 2011;371:143–151. doi: 10.1016/j.jim.2011.06.024. [DOI] [PubMed] [Google Scholar]
- 15.Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 17.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 18.Stoltenburg R, Reinemann C, Strehlitz B. SELEX-a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Ohuchi SP, Ohtsu T, Nakamura Y. Selection of RNA aptamers against recombinant transforming growth factor-beta type III receptor displayed on cell surface. Biochimie. 2006;88:897–904. doi: 10.1016/j.biochi.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: Systematic evolution of ligands by exponential enrichment; Proc Natl Acad Sci USA; 2003; pp. 15416–15421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilner SE, Wengerter B, Maier K, de Lourdes Borba Magalhães M, Del Amo DS, Pai S, Opazo F, Rizzoli SO, Yan A, Levy M. An RNA alternative to human transferrin: A new tool for targeting human cells. Mol Ther Nucleic Acids. 2012;1:e21. doi: 10.1038/mtna.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li N, Nguyen HH, Byrom M, Ellington AD. Inhibition of cell proliferation by an anti-EGFR aptamer. PLoS One. 2011;6:e20299. doi: 10.1371/journal.pone.0020299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shigdar S, Lin J, Yu Y, Pastuovic M, Wei M, Duan W. RNA aptamer against a cancer stem cell marker epithelial cell adhesion molecule. Cancer Sci. 2011;102:991–998. doi: 10.1111/j.1349-7006.2011.01897.x. [DOI] [PubMed] [Google Scholar]
- 24.Sekiya S, Noda K, Nishikawa F, Yokoyama T, Kumar PK, Nishikawa S. Characterization and application of a novel RNA aptamer against the mouse prion protein. J Biochem. 2006;139:383–390. doi: 10.1093/jb/mvj046. [DOI] [PubMed] [Google Scholar]
- 25.Murakami K, Nishikawa F, Noda K, Yokoyama T, Nishikawa S. Anti-bovine prion protein RNA aptamer containing tandem GGA repeat interacts both with recombinant bovine prion protein and its beta isoform with high affinity. Prion. 2008;2:73–80. doi: 10.4161/pri.2.2.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishikawa F, Murakami K, Matsugami A, Katahira M, Nishikawa S. Structural studies of an RNA aptamer containing GGA repeats under ionic conditions using microchip electrophoresis, circular dichroism and 1D-NMR. Oligonucleotides. 2009;19:179–190. doi: 10.1089/oli.2008.0167. [DOI] [PubMed] [Google Scholar]
- 27.Hamada M, Sato K, Kiryu H, Mituyama T, Asai K. Predictions of RNA secondary structure by combining homologous sequence information. Bioinformatics. 2009;25:i330–i338. doi: 10.1093/bioinformatics/btp228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kikin O, D'Antonio L, Bagga PS. QGRS Mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsugami A, Ouhashi K, Kanagawa M, Liu H, Kanagawa S, Uesugi S, Katahira M. An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad and its dimeric interaction. J Mol Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]
- 31.Catasti P, Chen X, Moyzis RK, Bradbury EM, Gupta G. Structure-function correlations of the insulin-linked polymorphic region. J Mol Biol. 1996;264:534–545. doi: 10.1006/jmbi.1996.0659. [DOI] [PubMed] [Google Scholar]
- 32.Xia T, SantaLucia J, Jr, Burkard ME, Kierzek R, Schroeder SJ, Jiao X, Cox C, Turner DH. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry. 1998;37:14719–14735. doi: 10.1021/bi9809425. [DOI] [PubMed] [Google Scholar]
- 33.Mashima T, Matsugami A, Nishikawa F, Nishikawa S, Katahira M. Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein. Nucleic Acids Res. 2009;37:6249–6258. doi: 10.1093/nar/gkp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mashima T, Nishikawa F, Kamatari YO, Fujiwara H, Saimura M, Nagata T, Kodaki T, Nishikawa S, Kuwata K, Katahira M. Anti-prion activity of an RNA aptamer and its structural basis. Nucleic Acids Res. 2013;41:1355–1362. doi: 10.1093/nar/gks1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsugami A, Mashima T, Nishikawa F, Murakami K, Nishikawa S, Noda K, Yokoyama T, Katahira M. Structural analysis of r(GGA)4 found in RNA aptamer for bovine prion protein. Nucleic Acids Symp Ser (Oxf) 2008:79–180. doi: 10.1093/nass/nrn091. [DOI] [PubMed] [Google Scholar]
- 36.Zhang AY, Bugaut A, Balasubramanian S. A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology. Biochemistry. 2011;50:7251–7258. doi: 10.1021/bi200805j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kowal J, Arras G, Colombo M, Jouve M, Morath JP, Primdal-Bengtson B, Dingli F, Loew D, Tkach M, Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes; Proc Natl Acad Sci USA; 2016; pp. E968–E977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christianson HC, Svensson KJ, van Kuppevelt TH, Li JP, Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity; Proc Natl Acad Sci USA; 2013; pp. 17380–17385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 40.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Mark M Tesic, Molina H, Kohsaka S, Di Giannatale A, Ceder S, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clayton A, Court J, Navabi H, Adams M, Mason MD, Hobot JA, Newman GR, Jasani B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods. 2001;247:163–174. doi: 10.1016/S0022-1759(00)00321-5. [DOI] [PubMed] [Google Scholar]
- 42.Epstein D, Grate D, Stanton M, et al., inventors. US Patent No US20040253679 A1. Stabilized aptamers to platelet derived growth factor and their use as oncology therapeutics. 2004 Apr 21; issued December 16, 2004.
- 43.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]