Abstract
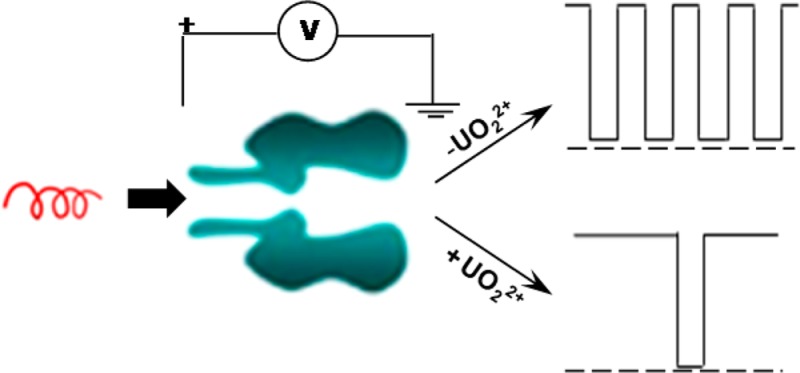
Uranium is one of the most common radioactive contaminants in the environment. As a major nuclear material in production, environmental samples (like soil and groundwater) can provide signatures on uranium production activity inside the facility. Thus, developing a new and portable analytical technology for uranium in aqueous media is significant not only for environmental monitoring, but also for nonproliferation. In this work, a label-free method for the detection of uranyl (UO22+) ions is developed by monitoring the translocation of a peptide probe in a nanopore. Based on the difference in the number of peptide events in the absence and presence of uranyl ions, nanomolar concentration of UO22+ ions could be detected in minutes. The method is highly selective; micromolar concentrations of Cd2+, Cu2+, Zn2+, Ni2+, Pb2+, Hg2+, Th4+, Mg2+, and Ca2+ would not interfere with the detection of UO22+ ions. In addition, simulated water samples were successfully analyzed.
Keywords: nanopore sensing, uranyl ion, peptide, biosensor, chelation
Because of uranium mining, nuclear power production, nuclear weapon development, as well as other industrial and medical application, uranium has been one of the most common radioactive contaminants in the environment, which raises concerns about its environmental impact and risk for human health.1,2 For example, UO22+ can disturb organ function by accumulating in the skeleton, kidneys, lungs, and liver.3−7 In aqueous solutions, uranium can exist in four different oxidation states such as +3, +4, +5, and +6, but uranyl (UO22+) and its complexing ions are the most stable and common species. Thus, far, various analytical techniques have been utilized to detect uranyl ions, including radiospectrometry,8 inductively coupled plasma mass spectrometry,9 fluorescence,10 and colorimetric11,12 and complexometric titration.13 However, most of these techniques are laborious, time-consuming, and require the use of sophisticated instruments or fluorescent labels/dyes. In addition, environmental samples like soil and groundwater near a uranium processing facility can provide significant signatures about its production activities inside the facility, and to detect such early signatures at declared or undeclared areas is significant for preventing nuclear materials from the wrong people.14 Therefore, development of other fundamentally different techniques, which are especially label-free, easy to operate, and potentially field-deployable, for uranyl detection is highly desirable not only for environmental monitoring but also for nonproliferation of nuclear materials or weapons.
Nanopore stochastic sensing has attracted substantial interest as an emerging label-free and amplification-free technique for measuring single molecules.15−18 Under an applied voltage bias, the movement of an analyte in a nanopore produces a measurable ionic current blockage. The identity and the concentration of the analyte could be revealed from its characteristic current signatures such as the event residence time, amplitude, frequency, and even the shape of the blockage.19 In addition to its biosensing application,20−29 nanopore sensing technology has been successfully applied to study a variety of other research areas, for example, covalent and noncovalent bonding interactions,30,31 biomolecular folding and unfolding,32−35 and enzyme kinetics.36−38 It should be noted that nanopore biosensing is generally achieved by modifying the nanopore interior to introduce binding sites for molecular recognition of target analytes.39−43 Recently, Long et al. utilized the effect from electrochemical confined space to efficiently convert the single DNA/peptide characteristics into measurable electrochemical signatures with high temporal and current resolution, which has been successfully achieved in the study of the heterogeneous structure–function relationship of biomolecules.44−46 More recently, Wang and co-workers reported a coordination chemistry-based stochastic nanopore sensing method for the detection of Cu2+ ions by using a peptide as a ligand probe.47 The detection was based on the effect of Cu2+ on peptide translocation in a nanopore. Briefly, in the absence of Cu2+, the translocation of the copper-chelating agent in the nanopore produced only one major type of event. In contrast, in the presence of Cu2+ ions, they interacted with the copper-chelating agent to form copper chelates, which produced a new type of events in the nanopore. By taking advantage of these new events, quantitative detection of Cu2+ ions could be achieved.
In this work, based on the similar metal ion-chelating agent interaction principle but with a different detection mechanism, we developed a new nanopore method for the detection of UO22+ in aqueous media. We also investigated the effect of metal ions on the detection of UO22+. The results demonstrated that this nanopore detection method is highly sensitive and selective to UO22+ in aqueous media, and the presence of Ni2+, Cu2+, Zn2+, Cd2+, Pb2+, Hg2+, Th4+, Ca2+, and Mg2+ showed little impact on its detection and quantification.
Experimental Section
Chemicals and Reagents
Peptide HH14, a 14-amino-acid peptide with a sequence of HHHHHHKHHHYHHH, was obtained from WatsonBio sciences (Houston, TX). Other chemicals such as UO2(NO3)2 (99.999%), Ca(NO3)2 (99.999%), Mg(NO3)2 (99.999%), Ni(NO3)2 (99.999%), Zn(NO3)2 (99.999%), Cu(NO3)2 (99.999%), Cd(NO3)2 (99.999%), Pb(NO3)2 (99.999%), Hg(NO3)2 (99.999%), Th(NO3)4 (99.999%), NaCl (99.999%), HCl (ACS reagent, ≤1 ppm heavy metals), NaH2PO4 (BioXtra grade, ≥99.5%), H3PO4 (ACS reagent, ≤0.002% heavy metals), C2H3NaO2 (BioXtra grade, ≥99.0%), CH3CO2H (ACS reagent, ≥99.7%), NaC6H7O7 (≥99%), C6H8O7 (FG, ≥99.5%), and Trizma base (BioXtra grade, ≥99.9%) were bought from Sigma (St. Louis, MO). All the chemicals, including the HH14 peptide, were dissolved in HPLC-grade water (ChromAR, Mallinckrodt Baker). The stock solutions of the peptide and metal salts were prepared at concentrations of 10 mM each, and were kept at −20 °C before and after use. The buffer solutions used in this study included: (1) 1.0 M NaCl and 10 mM tris with pH values adjusted to 6.5 and 7.5 using HCl; (2) 1.0 M NaCl and 10 mM NaH2PO4 with pH values adjusted to 4.5 and 5.5 using H3PO4; (3) 1.0 M NaCl and 10 mM CH3COONa with pH values adjusted to 4.5 and 5.5 using CH3COOH; and (4) 1.0 M NaCl and 10 mM NaC6H7O7 “sodium citrate” with pH values adjusted to 4.5 and 5.5 using C6H8O7 “citric acid”. Lipid 1,2-diphytanoylphosphatidylcholine was purchased from Avanti Polar Lipids (Alabaster, AL). Teflon film was obtained from Goodfellow (Malvern, PA). The α-hemolysin (αHL) (M113F)7 protein pores was made according to our previous work.48
Electrical Recording and Data Analysis
Single channel recordings were carried out at 24 ± 1 °C in a two-compartment chamber, which is separated by a Teflon septum having a 150 μm diameter aperture (refer to Supporting Information, Figure S1, for a schematic illustration of the nanopore sensor system). Briefly, the planar bilayer was formed on the aperture of the Teflon film using 1,2-diphytanoylphosphatidylcholine. Unless otherwise noted, the experiments were performed under symmetrical buffer conditions, with the αHL proteins added to the grounded cis compartment, while metal ion salts and the peptide probe were introduced to the trans side of the chamber device. Currents were recorded with an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA, USA), filtered with a built-in four-pole low-pass Bessel filter at 10 kHz, and then sampled at 50 kHz with a Digidata 1440 A/D converter (Molecular Devices).
The signatures of current blockage events were obtained using Clampfit 10.5 software (Molecular Device). Specifically, the conductance values and the mean residence time (τoff) for the HH14 peptide were derived from the amplitude and the residence time histograms by fitting the distributions to Gaussian and single exponential functions, respectively.31 The change (Δn) in the number of peptide HH14 events after addition of metal ions, including UO22+, to the solution was calculated by using the equation Δn = n0 – n1, where n0 represented the number of HH14 events in the absence of metal ions, while n1 depicted the number of peptide HH14 events in the presence of metal ions. Therefore, a positive value of Δn indicated a reduction in the number of peptide events after addition of metal ions to the solution. Each single-channel current trace was recorded for 10 min. At least three separate experiments, in each of which a new protein nanopore was used, were performed for each sample.
Results and Discussion
Detection of UO22+ Ions Using Peptide HH14
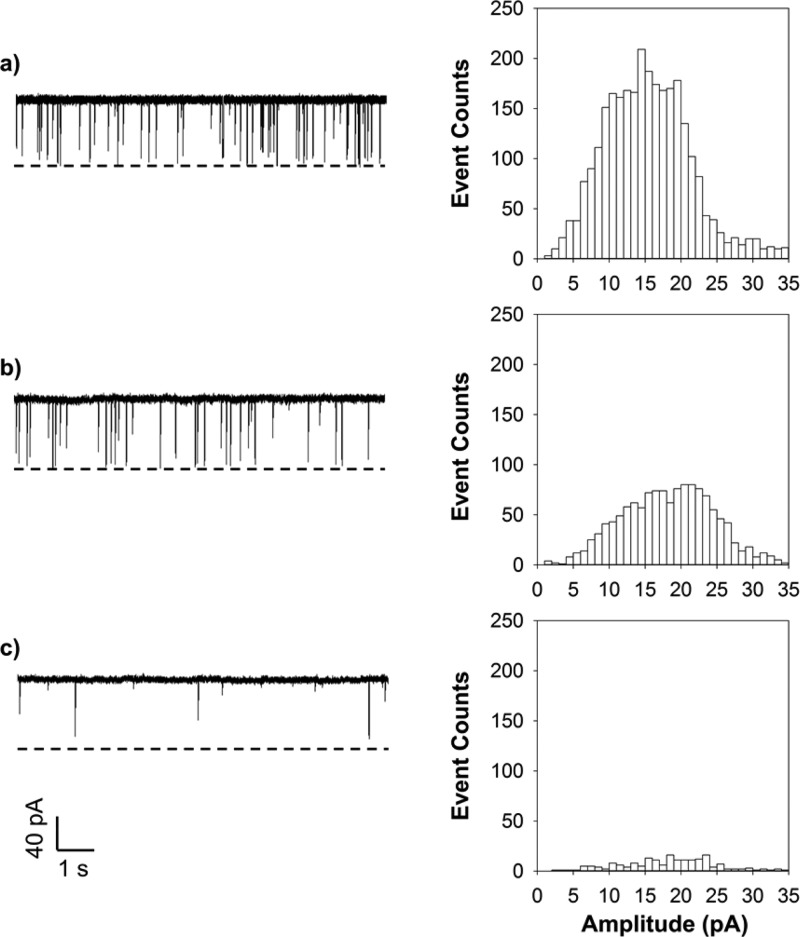
Peptides possess a range of potential donor atoms, and are very effective ligands for a variety of metal ions with high specificities. As a noted example, the amyloid beta (Aβ) peptides, which is crucially involved in Alzheimer’s disease, binds Cu2+ ions in vitro, and binding results in aggregation of the Aβ peptide. In particular, it is well documented that the histidine and cystine residues in the peptide display strong affinity for divalent or trivalent metal ions due to the chelation/coordination interaction.49 Since uranyl ions themselves could not produce any current modulation events in the nanopore (Supporting Information, Figure S2), in order to detect UO22+, we utilized a 14-amino-acid peptide (i.e., HH14) as the ligand probe. The three histidines in the 6-, 13-, and 14-positions of the peptide HH14 sequences were designed based on the finding that, in the Cu(II)-Aβ complex, the Cu2+ ions were coordinated by three histidine amino acids (i.e., His-6, His-13, and His-14) in the Aβ peptide.49 The other nine histidines were introduced to increase the coordination possibility between the peptide ligand and the target metal ion. Similar to the Cu2+ sensor reported previously, the peptide probe HH14 produced only one major type of event (Figure 1a). However, unlike the Cu2+ sensor, no new events were observed after addition of UO22+ to the peptide solution. Instead, the number of peptide events decreased. Furthermore, we noticed that, with an increase in the concentration of added UO22+, the peptide events become fewer and fewer. Specifically, when 0.5 μM UO22+ ions were added to the peptide HH14 (40 μM) solution, the number of peptide events decreased by 56.5 ± 2.4% (Figure 1b). As the concentration of uranyl ions increased to 10 μM, 92.8 ± 2.2% of the HH14 peptide events disappeared (Figure 1c). Since the I–V curves of HH14, UO22+, and their mixtures showed that the existence of uranyl in the nanopore did not rectify ionic current (Supporting Information, Figure S3), one possible reason for our observation that addition of UO22+ to the peptide HH14 solution did not produce new types of events, but only decreased the peptide event count is because the interactions between peptide HH14 and uranyl ions led to formation of UO22+–HH14 complexes, which passed through the nanopore too rapidly to be captured by the nanopore sensor (∼200 μs resolution). Note that the isoelectric point of histidine is around 7.5, while that of lysine is ∼9.7. Therefore, under our experimental conditions, peptide HH14 was positively charged. After chelation with UO22+, the net positive charge of the peptide-uranyl ion complex increased, and hence, the complex would be electrophoretically driven through the nanopore more rapidly than the uncomplexed peptide. Alternatively, the metal ion–peptide complexes might have larger molecular sizes than the nanopore opening so that they could not enter and pass through the pore. However, stoichiometric consideration of the UO22+–HH14 interaction could not explain such a large (56.5%) reduction in the peptide HH14 events after addition of 0.5 μM to 40 μM HH14. Furthermore, dynamic light scattering experiment (data not shown) demonstrated that uranyl would not induce HH14 aggregation. Therefore, the most likely mechanism behind our finding was that the binding of uranyl to the peptide HH14 enabled other uncomplexed peptide molecules to undergo conformational change. It is worth mentioning that disappearance of the biomolecule events or change in the event signatures due to conformational change has been reported previously.35,44,50
Figure 1.
Nanopore detection of UO22+ ions using peptide HH14. (a) 40 μM HH14; (b) 40 μM HH14 + 0.5 μM UO22+; and (c) 40 μM HH14 + 10 μM UO22+. (Left) Typical single-channel current recording trace segments; and (Right) the corresponding event amplitude histograms. The experiments were performed at +100 mV with the (M113F)7 αHL pore in an electrolyte solution containing 1.0 M NaCl and 10 mM Tris·HCl (pH 6.5). Both the peptide and uranyl ions were added to the trans compartment of the nanopore sensing chamber. Dashed lines represent the levels of zero current.
pH Effect on the Sensitivity of the Nanopore Sensor
Nanopore sensor usually has an extremely low background, and hence increasing the peptide HH14 event frequency in the nanopore offers the potential to greatly improve the detection limit for UO22+ quantitation. It has been well documented that the electrolyte pH affects the properties such as conductance and ion selectivity of the protein pores.51−53 Therefore, a change in the pH of the electrolyte solution may influence peptide translocation in the nanopore, thus affecting the sensor sensitivity and resolution. Note that nanopore experiments are usually carried out at/near the physiology pH. In our previous studies, we demonstrated that, as the electrolyte pH decreased, not only did the event frequency and residence time of target analytes (e.g., biomolecules and terrorist agents) increase, but the contrast in the event signatures between the target analyte and other matrix components also improved.54,55 On the other hand, it is well-known that the coordination properties of a histidine residue within a peptide sequence depend enormously on its position in a peptide chain; further, the metal-peptide complexes formed can exist in a variety of conformations that are dependent not only on the concentrations of both the peptide and metal ion but also on the pH of the reaction medium.56−58 To achieve highly sensitive detection of UO22+, translocation of peptide HH14 in the αHL nanopore was carried out in a series of electrolyte solutions with different pH values (from pH 4.5 to 7.5) and different buffer components. Our experimental results (Figure 2 and Supporting Information, Table S1) showed that, as the pH value of the electrolyte solution decreased from pH 7.5 to pH 4.5, the frequency and the mean residence time of the peptide events decreased by ∼10-fold and ∼60-fold, respectively. The results were not unreasonable considering the net charges of peptide HH14 at these various pH values. As discussed in the previous section, the isoelectric point of histidine is ∼7.5, while that of lysine is ∼9.7. Therefore, in our various investigated buffer solutions of different pH values (from pH 4.5 to pH 7.5), peptide HH14 had net positive charges. By systematic calculation of the charge state of HH14 (Supporting Information, Table S2), we found that peptide HH14 had a +1.05, +4.05, +10.31, and +12.69 charge at pH 7.5, pH 6.5, pH 5.5, and pH 4.5, respectively. Therefore, under a positively applied potential bias, in theory, a decrease in the pH of the electrolyte solution would lead to a decrease in the peptide event residence time and an increase in the peptide event frequency. However, due to the resolution of the single channel recording setup, most of the rapid peptide events (e.g., at pH 4.5 and pH 5.5) were missed under our experimental conditions. The electrolyte buffer solution of pH 6.5 rather than pH 7.5 was used in the remaining experiments because a larger percent reduction in the number of peptide events after addition of uranyl ion (1 μM) to the peptide solution (10 μM) was obtained at this pH. Specifically, after addition of uranyl, the number of peptide events was reduced by 83.0% in the pH 6.5 solution compared to 6.2% for the pH 7.5 solution, again suggesting that the net charge of the uranyl-peptide complex played an important role in the disappearance of the biomolecule events.
Figure 2.
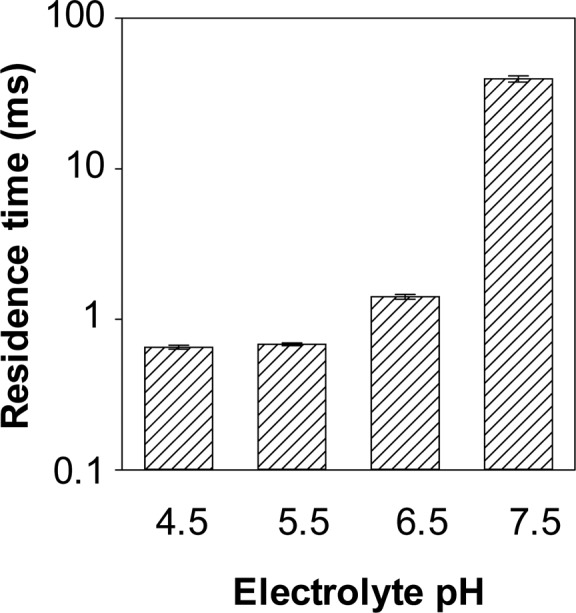
Effect of electrolyte pH on the mean residence time of the peptide HH14 events. The experiments were performed at +80 mV with the (M113F)7 αHL nanopore in a series of electrolyte solutions with different pH values and in the presence of 10 μM peptide HH14.
Effect of Voltage Bias and Peptide Concentration on UO22+ Detection
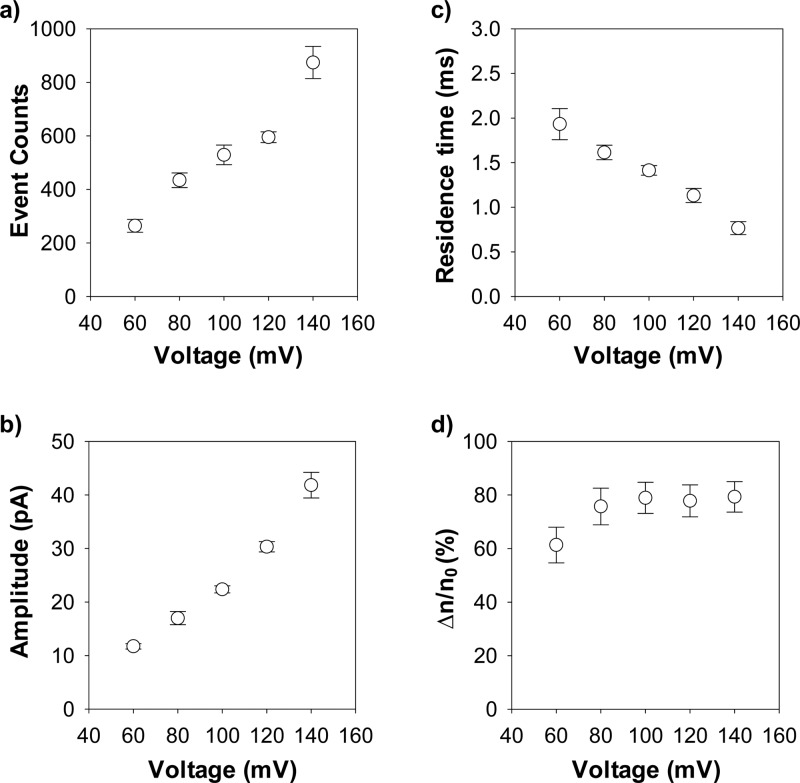
To identify the optimum conditions needed to achieve the maximum nanopore resolution for the detection of UO22+, we further investigated the translocation of peptide HH14 (without/with UO22+) in the nanopore at different voltages, ranging from +60 mV to +140 mV. Our experimental results (Figure 3) showed that, in the absence of UO22+, both the frequency and the blockage amplitude of the peptide events increased, while the peptide event residence time decreased as the applied potential bias increased. After addition of UO22+ to the peptide HH14 solution, the percent reduction in the number of peptide events first increased and then did not change significantly with an increase in the voltage. Although the percent event reduction at +100 mV was slightly smaller than that of +140 mV (78.9 ± 5.8% vs 79.4 ± 5.7%), +100 mV was chosen as the optimum applied potential, and this voltage was used in the remaining experiments because the bilayer at +140 mV was not as stable as that at +100 mV. Note that the principle for our nanopore sensor to detect UO22+ was based on the effect of uranyl on the frequency of the peptide HH14 events. In order to achieve highly sensitive detection of UO22+, two conditions need to be satisfied: one is a large number of peptide events in the absence of UO22+; and the other is a large percent peptide event reduction in the presence of UO22+.
Figure 3.
Effect of the applied potential bias on the (a) number of occurrences; (b) blockage amplitude; and (c) residence time of the peptide HH14 events; as well as (d) percent reduction in the number of HH14 events after addition of uranyl ions. The experiments were performed with the (M113F)7 αHL protein nanopore in an electrolyte solution containing 1 M NaCl and 10 mM Tris (pH 6.5) and in the presence of 10 μM peptide HH14 at various voltages ranging from +60 mV to +140 mV. The concentration of UO22+ used in part d was 0.5 μM.
In addition, the effect of the peptide HH14 concentration on the nanopore sensor resolution was examined. We found that the number of peptide events was linearly proportional to the peptide concentration, suggesting that the concentration of the peptide would not affect the sensitivity of the nanopore significantly (Supporting Information, Figure S4). A concentration of 40 μM HH14 was used in the remaining experiments since it produced enough events for statistical data analysis within a relatively short recording time.
Sensitivity and Selectivity of the UO22+ Nanopore Sensor
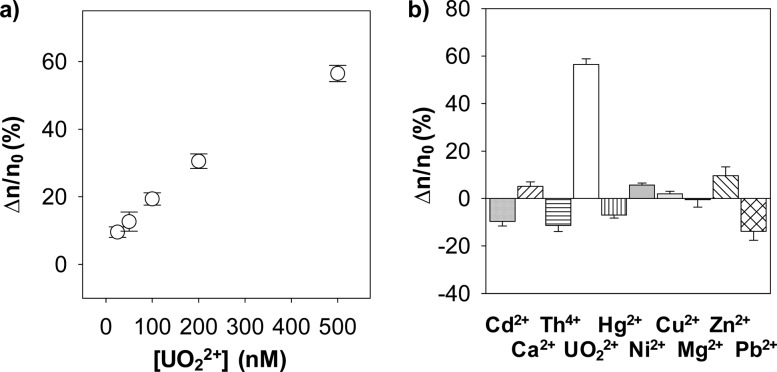
By utilizing the current physical condition (i.e., pH 6.5, + 100 mV applied potential bias, and 40 μM HH14 peptide), the dose response curve for UO22+ detection was constructed by monitoring the interaction between peptide HH14 and the nanopore in the presence of UO22+ ions at various concentrations, ranging from 25 nM to 500 nM. Our experimental results showed that the percent peptide event reduction was linearly correlated with the UO22+ concentration from 25 nM to ∼200 nM (Figure 4a). It was found that the detection limit (which is defined as the UO22+ concentration corresponding to three times the standard deviation of blank signal) in a 10 min electrical recording was 10 nM, which is more than good enough for analyzing uranyl ion in natural water (note that the maximum contamination level for UO22+ in drinking water defined by U.S. Environmental Protection Agency is 130 nM).
Figure 4.
Sensitivity and selectivity of the UO22+ nanopore sensor. (a) Dose–response curve. (b) Interference study. The experiments were performed at +100 mV with the (M113F)7 αHL protein nanopore in an electrolyte solution containing 1 M NaCl and 10 mM Tris (pH 6.5) and in the presence of 40 μM peptide HH14. With the exception of Th4+ (5 μM), Ca2+ (500 μM), and UO22+ (0.5 μM), the concentrations of all the other metal ions shown in part b were 20 μM each. In parts a and b, the change (Δn) in the number of peptide HH14 events after addition of UO22+ to the solution was calculated by using the equation: Δn = n0 – n1, where n0 represented the number of HH14 events in the absence of uranyl, while n1 depicted the number of peptide HH14 events in the presence of UO22+.
Nine metal ions, including Ni2+, Cu2+, Zn2+, Cd2+, Pb2+, Hg2+, Th4+, Ca2+, and Mg2+, were selected as potential interfering species to examine the selectivity of the nanopore UO22+ sensor because of their similar chemical properties and/or abundances in water. With the exception of Th4+ (5 μM) and Ca2+ (500 μM), the concentrations of all the other metal ions used in this investigation were 20 μM each. Our single-channel recording experimental results suggested that these nine metal ions did interact with peptide HH14 to form metal-peptide complexes. However, the existence of these cationic species would not affect uranyl ion detection significantly. As shown in Figure 4b, in the presence of Mg2+, Cd2+, Pb2+, Hg2+, and Th4+, the number of peptide events increased by 0.5 ± 3.1%, 9.7 ± 1.9%, 13.9 ± 3.8%, 7.0 ± 1.2%, and 11.3 ± 2.6%, respectively. Similar to UO22+, the existence of Ca2+, Ni2+, Cu2+, or Zn2+ ions in the solution led to a decrease in the peptide event count. However, considering that only small event count decreases (5.1 ± 1.9%, 5.6 ± 0.9%, 2.0 ± 1.1%, and 9.6 ± 3.7% for Ca2+, Ni2+, Cu2+, and Zn2+, respectively) were obtained in the presence of relatively large (20 to 500 μM) concentrations of interfering metal ions, the effect is negligible (note that, in comparison, 56.5 ± 2.4% decreases in the number of peptide events were observed after addition of 0.5 μM uranyl ions to the solution). Taking together, the combined results suggest that our nanopore sensor is highly selective to UO22+. It should be noted that histidines can bind to a variety of divalent and trivalent metal ions, and especially display high affinity to Ni2+, Co2+, Cu2+, and Zn2+.47,49 The high selectivity of the nanopore to UO22+ based on unselective histidine–ion coordination suggests that stoichiometry and the strength of the binding affinity between metal ions and peptide HH14 were less likely to be the sensing mechanism of the nanopore uranyl sensor, thus favoring our interpretation that addition of UO22+ to the peptide HH14 solution caused significant reduction in the peptide event count was because the binding of uranyl to the peptide HH14 enabled other uncomplexed peptide molecules to undergo conformational change.
Effect of the Salt Gradient on the Sensitivity of the UO22+ Nanopore Sensor
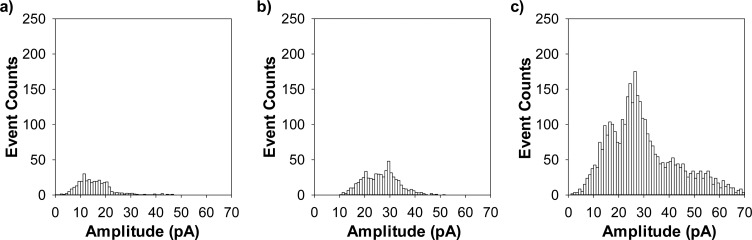
Use of an asymmetric electrolyte gradient instead of the conventional symmetric electrolyte solution is a well-established approach to significantly increase the event frequency for the translocation of DNA/RNA molecules through a nanopore, thus improving the sensor sensitivity for nucleic acid analysis.59 To examine whether this strategy can be employed to improve the sensitivity for UO22+ detection, two experiments were performed to investigate the translocation of peptide HH14 in the nanopore under asymmetric electrolyte conditions. In one experiment, the cis chamber compartment contained a solution comprising 1.5 M NaCl and 10 mM Tris (pH 6.5), while the trans compartment contained a solution comprising 0.5 NaCl and 10 mM Tris (pH 6.5). In the other experiment, we increased the salt concentration in the cis compartment to 3 M, while maintained the 0.5 M NaCl in the trans compartment. Our experimental results (Figure 5) showed that, with a salt gradient solution instead of the conventional symmetric electrolyte solution, there was indeed a significant increase in the number of peptide translocation events. Specifically, compared with the symmetric electrolyte solution of 1.0 M NaCl (cis)/1.0 M NaCl (trans), a salt gradient of 1.5 M NaCl (cis)/0.5 M NaCl (trans) and a salt gradient of 3 M NaCl (cis)/0.5 M NaCl (trans) resulted in a ∼1.75-fold and 10-fold increase in the number of peptide events, respectively. It is not unreasonable to expect tens- and even hundreds- fold increase in the peptide event count if a larger salt gradient than that of 3 M NaCl (cis)/0.5 M NaCl (trans) is used. As discussed in the voltage effect on HH14 translocation, with an increase in the applied potential bias, the number of peptide events increased. Under the symmetric electrolyte solution of 1.0 M NaCl (cis)/1.0 M NaCl (trans) and at +100 mV, the detection limit of our nanopore sensor for UO22+ was 10 nM. By taking advantage of the salt gradient effect and using a larger voltage bias (>100 mV), the detection limit for UO22+ would be greatly improved. As a noted example, with a salt gradient of 3 M NaCl (cis)/0.5 M NaCl (trans), the detection limit of the nanopore uranyl sensor was 2 nM at +100 mV (Supporting Information, Figure S5). Using the same salt gradient but with the applied potential increased to +140 mV, another 4-fold enhancement in the sensor sensitivity could be achieved (Supporting Information, Figure S6). This sensitivity is comparable with those of other reported highly sensitive electrochemical, optical, and radiospectrometry methods,8,10−12 with detection limits ranging from ∼100 pM to 50 nM.
Figure 5.
Effect of salt gradient on UO22+ detection. (a) Symmetric electrolyte solution of 1.0 M NaCl and 10 mM Tris·HCl (pH 6.5) (cis)/1.0 M NaCl and 10 mM Tris·HCl (pH 6.5) (trans), (b) salt gradient of 1.5 M NaCl and 10 mM Tris·HCl (pH 6.5) (cis)/0.5 M NaCl and 10 mM Tris·HCl (pH 6.5) (trans), and (c) salt gradient of 3 M NaCl and 10 mM Tris·HCl (pH 6.5) (cis)/0.5 M NaCl and 10 mM Tris·HCl (pH 6.5) (trans). The experiments were performed at +100 mV with the (M113F)7 αHL nanopore in the presence of 10 μM peptide HH14, which was added to the trans compartment of the nanopore sensing chamber. The event counts in parts a–c were calculated based on 5 min single channel recording trace segments.
Simulated Water Sample Analysis
To demonstrate the potential application of our nanopore sensor in real-world sample analysis, three simulated uranyl ion-contaminated water samples were created by spiking 100 nM uranyl ions into the tap water (obtained from our life science building), lake water (from Lake Michigan), and Ice Mountain brand bottled spring water. The simulated water samples were analyzed by our nanopore sensor under the symmetrical buffer conditions. Our experimental results (Figure 6) showed that the percent event reduction values (22.7 ± 2.3%, 19.9 ± 0.2%, and 19.7 ± 1.4%) of the three simulated water samples were similar to that (19.3 ± 1.8%) of the control sample (i.e., uranyl ion standard solution), suggesting the matrix component in the water would not affect uranyl ion detection significantly.
Figure 6.
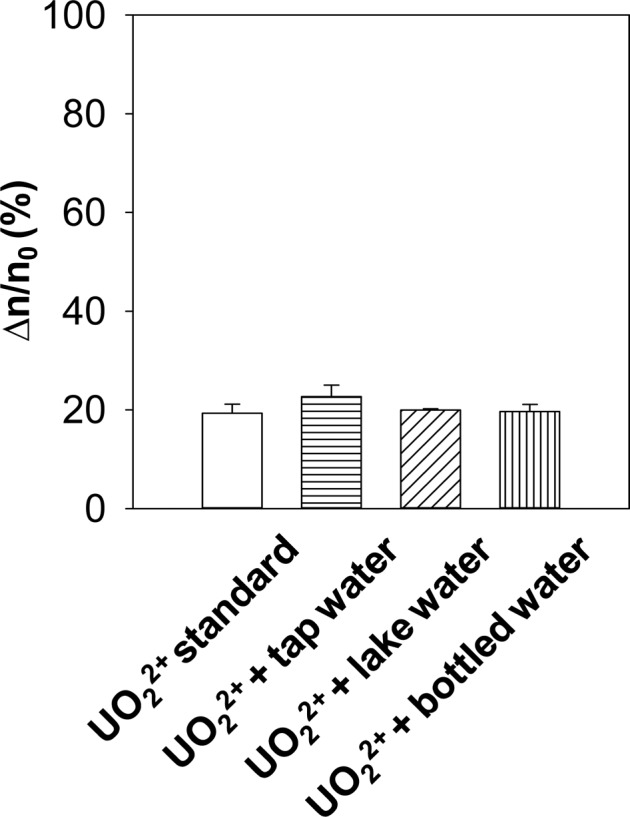
Simulated water sample analysis. The experiments were performed at +100 mV with the (M113F)7 αHL protein nanopore in an electrolyte solution containing 1 M NaCl and 10 mM Tris (pH 6.5) and in the presence of 40 μM peptide HH14.
Conclusions
In summary, a highly selective and sensitive nanopore sensor was successfully developed to detect UO22+ ions by using a peptide molecule as a chelating agent and taking advantage of peptide translocation in the nanopore. Although the formation of the UO22+-peptide complex did not produce new types of events in the nanopore, the percent reduction in the uncomplexed peptide translocation events could be utilized for UO22+ quantitation. The high selectivity for UO22+ of our nanopore sensor was supported by two experiments, i.e., the interference study and simulated water analysis. Our study showed that, in spite of their similar chemical properties and/or large concentration in the real-world samples, metal ions such as Cd2+, Th4+, Cu2+, Zn2+, Ni2+, Pb2+, Hg2+, Mg2+, and Ca2+ and other matrix components would not interfere with UO22+ detection significantly. Our developed nanopore sensor may find useful applications in detection of uranyl ions in natural water for environmental monitoring or for signatures on nuclear material production activity inside a processing facility.
Acknowledgments
This work was financially supported by the National Institutes of Health (1R15GM110632).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssensors.7b00210.
Additional tables and figures, including effect of buffer solution on peptide translocation in the nanopore; net charges of peptide HH14 at different pH values; schematic illustration of the uranyl nanopore sensor; typical single-channel recording trace segment of uranyl ions; I–V curves of peptide HH14, UO22+, and their mixtures in the nanopore; dose response curves for peptide HH14 and uranyl in asymmetric electrolyte conditions; and voltage effect on the sensitivity of the nanopore uranyl sensor (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Brugge D.; Buchner V. Health effects of uranium: new research findings. Rev. Environ. Health 2011, 26, 231–49. 10.1515/REVEH.2011.032. [DOI] [PubMed] [Google Scholar]
- The World Health Organization. Depleted Uranium: Sources, Exposure and Health Effects; WHO/SDE/PHE/01.1, Geneva, 2001.
- Gongalsky K. B. Impact of pollution caused by uranium production on soil macrofauna. Environ. Monit. Assess. 2003, 89, 197–219. 10.1023/A:1026031224658. [DOI] [PubMed] [Google Scholar]
- Cantaluppi C.; Degetto S. Civilian and military uses of depleted uranium: environmental and health problems. Ann. Chim. 2000, 90, 665–676. [PubMed] [Google Scholar]
- Craft E.; Abu-Qare A.; Flaherty M.; Garofolo M.; Rincavage H.; Abou-Donia M. Depleted and natural uranium: chemistry and toxicological effects. J. Toxicol. Environ. Health, Part B 2004, 7, 297–317. 10.1080/10937400490452714. [DOI] [PubMed] [Google Scholar]
- Fisenne I. M.; Perry P. M.; Harley N. H. Uranium in Humans. Radiat. Prot. Dosim. 1988, 24, 127–131. 10.1093/rpd/24.1-4.127. [DOI] [Google Scholar]
- Zamora M. L.; Tracy B. L.; Zielinski J. M.; Meyerhof D. P.; Moss M. A. Chronic ingestion of uranium in drinking water: a study of kidney bioeffects in humans. Toxicol. Sci. 1998, 43, 68–77. 10.1093/toxsci/43.1.68. [DOI] [PubMed] [Google Scholar]
- Holzbecher J.; Ryen D. E. Determination of uranium by thermal and epithermal neutron activation in natural waters and in human urine. Anal. Chim. Acta 1980, 119, 405–408. 10.1016/S0003-2670(01)93645-1. [DOI] [Google Scholar]
- Caddia M.; Iversen B. S. Determination of uranium in urine by inductively coupled plasma mass spectrometry with pneumatic nebulization. J. Anal. At. Spectrom. 1998, 13, 309–313. 10.1039/a704444e. [DOI] [Google Scholar]
- Romanovskaya G. I.; Pogonin V. I.; Chibisov A. K. Fluorescence determination of trace amounts of uranium(VI) in various materials by a repetitive laser technique. Talanta 1987, 34, 207–210. 10.1016/0039-9140(87)80028-0. [DOI] [PubMed] [Google Scholar]
- Lee J. H.; Wang Z.; Liu J.; Lu Y. Highly sensitive and selective colorimetric sensors for uranyl (UO22+): development and comparison of labeled and label-free DNAzyme-gold nanoparticle systems. J. Am. Chem. Soc. 2008, 130, 14217–14226. 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivens D. A.; Zhang Y.; Angel S. M. Detection of uranyl ion via fluorescence quenching and photochemical oxidation of calcein. J. Photochem. Photobiol., A 2002, 152, 167–173. 10.1016/S1010-6030(02)00229-0. [DOI] [Google Scholar]
- Mlakar M.; Branica M. Applicability of synergistic adsorption in electroanalysis of dissolved uranium in seawater. Mar. Chem. 1994, 46, 61–66. 10.1016/0304-4203(94)90045-0. [DOI] [Google Scholar]
- Axelsson A.; Fischer D. M.; Penkin M. V. Use of data from environmental sampling for IAEA safeguards. Case study: uranium with near-natural U-235 abundance. J. Radioanal. Nucl. Chem. 2009, 282, 725–729. 10.1007/s10967-009-0149-9. [DOI] [Google Scholar]
- Wang G.; Wang L.; Han Y.; Zhou S.; Guan X. Nanopore stochastic detection: diversity, sensitivity, and beyond. Acc. Chem. Res. 2013, 46, 2867–2877. 10.1021/ar400031x. [DOI] [PubMed] [Google Scholar]
- Howorka S.; Siwy Z. Nanopore analytics: sensing of single molecules. Chem. Soc. Rev. 2009, 38, 2360–2384. 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- Matile S. Transport across membranes. Acc. Chem. Res. 2013, 46, 2741–2742. 10.1021/ar400234d. [DOI] [PubMed] [Google Scholar]
- Majd S.; Yusko E. C.; Billeh Y. N.; Macrae M. X.; Yang J.; Mayer M. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Curr. Opin. Biotechnol. 2010, 21, 439–476. 10.1016/j.copbio.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller A.; Nivon L.; Brandin E.; Golovchenko J.; Branton D. Applications of biological pores in nanomedicine, sensing, and nanoelectronics. Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 1079–1084. 10.1073/pnas.97.3.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.; Han Y.; Zhou S.; Wang G.; Guan X. Real-time label-free measurement of HIV-1 protease activity by nanopore analysis. Biosens. Bioelectron. 2014, 62, 158–162. 10.1016/j.bios.2014.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. S. Nanopore analysis of the effect of metal ions on the folding of peptides and proteins. Protein Pept. Lett. 2014, 21, 247–255. 10.2174/09298665113209990075. [DOI] [PubMed] [Google Scholar]
- Apetrei A.; Ciuca A.; Lee J. K.; Seo C. H.; Park Y.; Luchian T. A Protein Nanopore-Based Approach for Bacteria Sensing. Nanoscale Res. Lett. 2016, 11, 501. 10.1186/s11671-016-1715-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asandei A.; Schiopu I.; Chinappi M.; Seo C. H.; Park Y.; Luchian T. Electroosmotic Trap Against the Electrophoretic Force Near a Protein Nanopore Reveals Peptide Dynamics During Capture and Translocation. ACS Appl. Mater. Interfaces 2016, 8, 13166–13179. 10.1021/acsami.6b03697. [DOI] [PubMed] [Google Scholar]
- Kasianowicz J. J.; Brandin E.; Branton D.; Deamer D. W. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 13770–13773. 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.; Zhao Q.; Kang X.; Guan X. Probing mercury(II)-DNA interactions by nanopore stochastic sensing. J. Phys. Chem. B 2013, 117, 4763–4769. 10.1021/jp309541h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movileanu L.; Schmittschmitt J. P.; Scholtz J. M.; Bayley H. Interactions of peptides with a protein pore. Biophys. J. 2005, 89, 1030–1045. 10.1529/biophysj.104.057406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefureac R.; Long Y. T.; Kraatz H. B.; Howard P.; Lee J. S. Transport of alpha-helical peptides through alpha-hemolysin and aerolysin pores. Biochemistry 2006, 45, 9172–9179. 10.1021/bi0604835. [DOI] [PubMed] [Google Scholar]
- Zhao Q.; Wang D.; Jayawardhana D. A.; Guan X. Stochastic sensing of biomolecules in a nanopore sensor array. Nanotechnology 2008, 19, 505504. 10.1088/0957-4484/19/50/505504. [DOI] [PubMed] [Google Scholar]
- Apetrei A.; Asandei A.; Park Y.; Hahm K. S.; Winterhalter M.; Luchian T. J. Unimolecular study of the interaction between the outer membrane protein OmpF from E. coli and an analogue of the HP(2–20) antimicrobial peptide. J. Bioenerg. Biomembr. 2010, 42, 173–180. 10.1007/s10863-010-9273-z. [DOI] [PubMed] [Google Scholar]
- Luchian T.; Shin S. H.; Bayley H. Single-molecule covalent chemistry with spatially separated reactants. Angew. Chem., Int. Ed. 2003, 42, 3766–3771. 10.1002/anie.200351313. [DOI] [PubMed] [Google Scholar]
- Zhao Q.; Jayawardhana D. A.; Guan X. Stochastic study of the effect of ionic strength on noncovalent interactions in protein pores. Biophys. J. 2008, 94, 1267–1275. 10.1529/biophysj.107.117598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oukhaled A.; Bacri L.; Pastoriza-Gallego M.; Betton J. M.; Pelta J. Sensing proteins through nanopores: fundamental to applications. ACS Chem. Biol. 2012, 7, 1935–1949. 10.1021/cb300449t. [DOI] [PubMed] [Google Scholar]
- Pastoriza-Gallego M.; Rabah L.; Gibrat G.; Thiebot B.; van der Goot F. G.; Auvray L.; Betton J. M.; Pelta J. Dynamics of unfolded protein transport through an aerolysin pore. J. Am. Chem. Soc. 2011, 133, 2923–2931. 10.1021/ja1073245. [DOI] [PubMed] [Google Scholar]
- Pastoriza-Gallego M.; Breton M. F.; Discala F.; Auvray L.; Betton J. M.; Pelta J. Evidence of unfolded protein translocation through a protein nanopore. ACS Nano 2014, 8, 11350–11360. 10.1021/nn5042398. [DOI] [PubMed] [Google Scholar]
- Stefureac R. I.; Madampage C. A.; Andrievskaia O.; Lee J. S. Nanopore analysis of the interaction of metal ions with prion proteins and peptides. Biochem. Cell Biol. 2010, 88, 347–358. 10.1139/O09-176. [DOI] [PubMed] [Google Scholar]
- Safarian S.; Moosavi-Movahedi A. Binding patterns and kinetics of RNase a interaction with RNA. J. Protein Chem. 2000, 19, 335–344. 10.1023/A:1026414928279. [DOI] [PubMed] [Google Scholar]
- Zhao Q.; Wang D.; Jayawardhana D. A.; de Zoysa R. S.; Guan X. Real-time monitoring of peptide cleavage using a nanopore probe. J. Am. Chem. Soc. 2009, 131, 6324–6325. 10.1021/ja9004893. [DOI] [PubMed] [Google Scholar]
- Fennouri A.; Daniel R.; Pastoriza-Gallego M.; Auvray L.; Pelta J.; Bacri L. Kinetics of enzymatic degradation of high molecular weight polysaccharides through a nanopore: experiments and data-modeling. Anal. Chem. 2013, 85, 8488–8492. 10.1021/ac4020929. [DOI] [PubMed] [Google Scholar]
- Braha O.; Walker B.; Cheley S.; Kasianowicz J. J.; Song L.; Gouaux J. E.; Bayley H. Designed protein pores as components for biosensors. Chem. Biol. 1997, 4, 497–505. 10.1016/S1074-5521(97)90321-5. [DOI] [PubMed] [Google Scholar]
- Braha O.; Gu L. Q.; Zhou L.; Lu X.; Cheley S.; Bayley H. Simultaneous stochastic sensing of divalent metal ions. Nat. Biotechnol. 2000, 18, 1005–1007. 10.1038/79275. [DOI] [PubMed] [Google Scholar]
- Movileanu L.; Howorka S.; Braha O.; Bayley H. Detecting protein analytes that modulate transmembrane movement of a polymer chain within a single protein pore. Nat. Biotechnol. 2000, 18, 1091–1095. 10.1038/80295. [DOI] [PubMed] [Google Scholar]
- Cheley S.; Gu L. Q.; Bayley H. Stochastic sensing of nanomolar inositol 1,4,5-trisphosphate with an engineered pore. Chem. Biol. 2002, 9, 829–838. 10.1016/S1074-5521(02)00172-2. [DOI] [PubMed] [Google Scholar]
- Guan X.; Gu L. Q.; Cheley S.; Braha O.; Bayley H. Stochastic sensing of TNT with a genetically engineered pore. ChemBioChem 2005, 6, 1875–1881. 10.1002/cbic.200500064. [DOI] [PubMed] [Google Scholar]
- Ying Y. L.; Wang H. Y.; Sutherland T. C.; Long Y. T. Monitoring of an ATP-binding aptamer and its conformational changes using an α-hemolysin nanopore. Small 2011, 7, 87–94. 10.1002/smll.201001428. [DOI] [PubMed] [Google Scholar]
- Wang H. Y.; Ying Y. L.; Li Y.; Kraatz H. B.; Long Y. T. Nanopore analysis of β-amyloid peptide aggregation transition induced by small molecules. Anal. Chem. 2011, 83, 1746–1752. 10.1021/ac1029874. [DOI] [PubMed] [Google Scholar]
- Cao C.; Ying Y. L.; Hu Z. L.; Liao D. F.; Tian H.; Long Y. T. Discrimination of oligonucleotides of different lengths with a wild-type aerolysin nanopore. Nat. Nanotechnol. 2016, 11, 713–716. 10.1038/nnano.2016.66. [DOI] [PubMed] [Google Scholar]
- Wang G.; Wang L.; Han Y.; Zhou S.; Guan X. Nanopore detection of copper ions using a polyhistidine probe. Biosens. Bioelectron. 2014, 53, 453–458. 10.1016/j.bios.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D.; Zhao Q.; de Zoysa R. S. S.; Guan X. Detection of nerve agent hydrolytes in an engineered nanopore. Sens. Actuators, B 2009, 139, 440–446. 10.1016/j.snb.2009.02.069. [DOI] [Google Scholar]
- Minicozzi V.; Stellato F.; Comai M.; Serra M. D.; Potrich C.; Meyer-Klaucke W.; Morante S. Identifying the minimal copper- and zinc-binding site sequence in amyloid-beta peptides. J. Biol. Chem. 2008, 283, 10784–10792. 10.1074/jbc.M707109200. [DOI] [PubMed] [Google Scholar]
- Kakish J.; Allen K. J.; Harkness T. A.; Krol E. S.; Lee J. S. Novel dimer compounds that bind α-synuclein can rescue cell growth in a yeast model overexpressing α-Synuclein. A possible prevention strategy for Parkinson’s disease. ACS Chem. Neurosci. 2016, 7, 1671–1680. 10.1021/acschemneuro.6b00209. [DOI] [PubMed] [Google Scholar]
- Gu L.; Bayley H. Interaction of the noncovalent molecular adapter, beta-cyclodextrin, with the staphylococcal alpha-hemolysin pore. Biophys. J. 2000, 79, 1967–1975. 10.1016/S0006-3495(00)76445-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L.; Cheley S.; Bayley H. Prolonged residence time of a noncovalent molecular adapter, beta-cyclodextrin, within the lumen of mutant alpha-hemolysin pores. J. Gen. Physiol. 2001, 118, 481–493. 10.1085/jgp.118.5.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merzlyak P. G.; Capistrano M. F.; Valeva A.; Kasianowicz J. J.; Krasilnikov O. W. Conductance and ion selectivity of a mesoscopic protein nanopore probed with cysteine scanning mutagenesis. Biophys. J. 2005, 89, 3059–3070. 10.1529/biophysj.105.066472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zoysa R. S.; Krishantha D. M.; Zhao Q.; Gupta J.; Guan X. Translocation of single-stranded DNA through the α-hemolysin protein nanopore in acidic solutions. Electrophoresis 2011, 32, 3034–3041. 10.1002/elps.201100216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta J.; Zhao Q.; Wang G.; Kang X.; Guan X. Simultaneous detection of CMPA and PMPA, hydrolytes of soman and cyclosarin nerve agents, by nanopore analysis. Sens. Actuators, B 2013, 176, 625–631. 10.1016/j.snb.2012.10.058. [DOI] [Google Scholar]
- Daniele P. G.; Zerbinati O.; Zelano V.; Ostacoli G. Thermodynamic and spectroscopic study of copper(II)–glycyl-L-histidylglycine complexes in aqueous solution. J. Chem. Soc., Dalton Trans. 1991, 10, 2711–2715. 10.1039/DT9910002711. [DOI] [Google Scholar]
- Sjöberg S. Critical evaluation of stability constants of metal-imidazole and metal-histamine systems. Pure Appl. Chem. 1997, 69, 1549–1570. 10.1351/pac199769071549. [DOI] [Google Scholar]
- Gaggelli E.; D’Amelio N.; Valensin D.; Valensin G. 1H NMR studies of copper binding by histidine-containing peptides. Magn. Reson. Chem. 2003, 41, 877–883. 10.1002/mrc.1250. [DOI] [Google Scholar]
- Wanunu M.; Morrison W.; Grosberg A. Y.; Meller A. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat. Nanotechnol. 2010, 5, 160–165. 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.